Summary
The aciduricity of Streptococcus mutans is an important virulence factor of the organism, required to both out-compete commensal oral microorganisms and cause dental caries. In this study, we monitored transcriptional changes that occurred as a continuous culture of either an acid-tolerant strain (UA159) or an acid-sensitive strain (fabM::Erm) moved from steady-state growth at neutral pH, experienced glucose-shock and acidification of the culture, and transitioned to steady-state growth at low pH. Thus, the timing of elements of the acid tolerance response (ATR) could be observed and categorized as acute vs. adaptive ATR mechanisms. Modulation of BCAA biosynthesis, DNA/protein repair mechanisms, ROS metabolizers, and PTS occurred in the initial acute phase, immediately following glucose-shock, while up-regulation of F1F0-ATPase did not occur until the adaptive phase, after steady-state growth had been re-established. In addition to the archetypal ATR pathways mentioned above, glucose-shock led to differential expression of genes suggesting a re-routing of resources away from synthesis of fatty acids and proteins, and towards synthesis of purines, pyrimidines and amino acids. These adjustments were largely transient, as upon establishment of steady-state growth at acidic pH, transcripts returned to basal expression levels. During growth at steady-state pH 7, fabM::Erm had a transcriptional profile analogous to that of UA159 during glucose-shock, indicating that even during growth in rich media at neutral pH, the cells were stressed. These results, coupled with a recently established collection of deletion strains (Quivey et al., 2015), provide a starting point for elucidation of the acid tolerance response in S. mutans.
Keywords: Oral streptococci, acid adaptation, stress response, transcriptome, genomic characterization
Introduction
The oral pathogen Streptococcus mutans, an acidogenic and aciduric Gram-positive organism, is the chief causative agent of the disease dental caries (De Stoppelaar et al., 1969, Loesche, 1986, Nyvad et al., 2013). Dental caries has a high prevalence in nations where refined sugar consumption is a large part of the diet, with a strong trend for occurrence of the disease in segments of the population that can least afford treatment (Dye et al., 2007, Dye & Thornton-Evans, 2010). S. mutans is found exclusively on the surfaces of teeth in dental plaque, a matrix composed of bacteria, bacterially-produced polysaccharides, mainly water-insoluble glucans, salivary proteins, and food particles (Bowen & Koo, 2011). An important virulence trait of S. mutans is its ability to metabolize a wide array of human dietary carbohydrates from which it produces a large amount of organic acids through fermentation, which acidify the dental plaque (Carlsson et al., 1997). Demineralization of the tooth surface occurs from these transient drops in pH, but is typically repaired due to the immersion of the tooth in mineral-rich saliva. In the absence of scrupulous oral hygiene, plaque buildup can prevent salivary minerals from re-crystallizing the tooth surface at a rate that can keep pace with the demineralization, allowing progression to a disease state (Koo et al., 2002).
As S. mutans is more aciduric than many organisms with which it frequently competes, namely the mitis group of Streptococci and other early colonizers of the tooth surface, it was unsurprising that metagenomic studies identified S. mutans as a more dominant member of supragingival flora as plaque matures and becomes more acidic (Gross et al., 2010, Gross et al., 2012). The collective adjustments to low pH made by S. mutans have been termed the acid-adaptive response (ATR) and have been reviewed in depth (Lemos & Burne, 2008, Matsui & Cvitkovitch, 2010). The ATR of S. mutans is dependent on a number of traits and systems including the ability to carry out glycolysis at a lower pH than many other oral microbes (Belli & Marquis, 1991, Svensater et al., 1997). Importantly, S. mutans also maintains a cytosolic pH that is significantly higher than the extracellular pH (Quivey et al., 2000, Lemos et al., 2005, Lemos & Burne, 2008). This is accomplished by several mechanisms, most notably the up-regulation of the F1F0 ATPase, which, in this case, uses hydrolysis of ATP to pump protons out of the cytosol (Kuhnert & Quivey Jr, 2003, Sheng & Marquis, 2006). The S. mutans F1F0 ATPase can operate at low pH much more efficiently than the ATPase of several other competing oral bacterial species such as S. salivarius and S. sanguinis (Bender et al., 1986). Other important systems for cytosolic alkalization include up-regulation of branched-chain amino acid (BCAA) biosynthesis (Len et al., 2004a, Santiago et al., 2012) and the agmatine deiminase system (Griswold et al., 2006, Lemos & Burne, 2008) as well as induction of malolactic acid fermentation (Sheng & Marquis, 2007). It was also shown that during growth at low pH, S. mutans incorporates a larger percentage of unsaturated fatty acids (UFAs) into the plasma membrane (Fozo et al., 2004, Fozo & Quivey, 2004b). Production of de novo UFAs requires the fabM gene, encoding a trans-2-cis-3-decenoyl-ACP isomerase (Fozo & Quivey, 2004a). The loss of the ability to synthesize UFAs in a fabM mutant strain, fabM::Erm, resulted in a significantly smaller ΔpH across the organism’s membrane compared to the parental strain (Fozo & Quivey, 2004a), suggesting alterations in the ability of the mutant strain to maintain normal membrane permeability for protons, or in its ability to pump protons out of the cell. The fabM::Erm strain also exhibited a reduced ability to survive acid challenge, and was considerably less virulent in a rat model for dental caries (Fozo & Quivey, 2004a, Fozo et al., 2007). S. mutans encodes several DNA repair enzymes which are also induced in response to acid (Hanna et al., 2001, Faustoferri et al., 2005, Gonzalez et al., 2012b), supporting the concept that the response of S. mutans to acidification includes both defensive and repair capabilities.
While the ATR pathways discussed above are well known, the mechanisms by which these pathways are induced, and the timing of their induction on a large scale, are not fully understood. Many previous studies in S. mutans have used cDNA microarray analysis or RNA-seq to elucidate the regulon of a particular gene or condition on a global scale. These have included the studies of the oxidative-stress regulon and redox control through aeration (Ahn et al., 2007, Baker et al., 2014), the Nox regulon (Baker et al., 2014), the Rex regulon (Bitoun et al., 2011), and the SpxA1 and SpxA2 regulons (Kajfasz et al., 2010). Studies have also been conducted on the global response to nutrients through growth in various sugars (Zeng et al., 2013, Moye et al., 2014) and elucidation of the CodY regulon (Lemos et al., 2008), the CcpA regulon (Zeng et al., 2013), the ManL regulon (Abranches et al., 2006, Moye et al., 2014), and the CcpA-regulated FabT regulon (Faustoferri et al., 2014). Although the changes in the S. mutans proteome in response to growth at steady-state pH 5 versus steady-state pH 7 and the transcriptomic effects of growth in acidic buffered batch cultures have been described (Wilkins et al., 2002, Len et al., 2004a, Len et al., 2004b, Crowley et al., 2004, Gong et al., 2009, Krastel et al., 2010), global transcriptional changes in response to glucose-shock (which leads to a rapid acidification of the environment through the production of lactic acid) and steady-state growth at acidic pH have not been elucidated in S. mutans.
In this study, cDNA microarray analysis was used to examine the global transcriptional changes occurring when a continuous culture of S. mutans grown to steady-state at pH 7 was administered a glucose-shock, which resulted in rapid acidification of the culture, and then transitioned to steady-state growth at pH 5. The conditions described here represent an examination of glucose and/or acid-inducible genes in S. mutans using cells harvested from tightly controlled growth conditions. This is in contrast to previous transcriptomic studies describing acid-inducible genes in S. mutans that utilized buffered media in batch cultures (Gong et al., 2009, Krastel et al., 2010). To take a step further, the transcriptional profile from both an acid-tolerant strain, UA159, and an acid-sensitive strain, fabM::Erm, was obtained. Based on the timing of transcriptomic changes observed, we divided several of the major trends into three groups: the acute ATR, the adaptive ATR, and the transient ATR/glucose-shock response. This study demonstrates that during steady-state growth at pH 7, fabM::Erm had an elevated basal level of stress responses compared to UA159, suggesting that even during favorable growth conditions, the homeostasis of this mutant is under duress.
Materials and Methods
Bacterial strains and growth conditions
The S. mutans UA159 (parent strain) and UR117 (“fabM::Erm”, (Murchison et al., 1986, Fozo & Quivey, 2004a) strains were maintained on brain heart infusion agar (BHI; BD/Difco, Franklin Lakes, NJ) at 37°C in a 5% (vol/vol) CO2/95% air environment. Erythromycin was added to a final concentration of 5μg ml−1, where necessary. Organisms were cultured in TY medium (3% tryptone, 0.1% yeast extract, 0.5% KOH, 1 mM H3PO4) + 1% (w/v) glucose and were grown in liquid culture or in continuous culture in a Sixfors fermenter (Infors, Laurel, MD) as described previously (Quivey et al., 1995, Fozo & Quivey, 2004b). Continuous cultures of UA159 or fabM::Erm were grown at a dilution rate of 0.24 h−1 under glucose-limiting conditions (2.3 mM), with a continuous impeller speed of 200 rpm. Steady-state pH levels of 7.0 or 5.0 (5.5 for fabM::Erm) were maintained by the addition of 2 N KOH. The culture pH was continuously monitored throughout the experiment by using an in-dwelling pH probe (Mettler Toledo, Columbus, OH). After continuous culture at pH 7 had been maintained for a minimum of 10 generations, aliquots of the culture were removed and cells were collected by centrifugation (referred to as steady-state pH 7). Glucose was added to 200mM and the pH control was turned off to allow acidification of the culture. Upon reaching pH 5.5, aliquots were removed and cells harvested. These aliquots were referred to as “glucose-shock.” At this point, the pH control was set to 5.0 for UA159, or 5.5 for fabM::Erm and the culture was maintained for 10 generations, at which time the final aliquots were removed (referred to as “steady-state pH 5” or “steady-state pH 5.5”). All cell pellets were washed and stored frozen at −80°C. Five steady-state cultures were grown for each strain.
cDNA microarray analysis
S. mutans UA159 microarray slides were provided by The Institute for Genomic Research (TIGR) through a cooperative agreement with the National Institutes for Allergy and Infectious Disease (NIAID) and Dental and Craniofacial Research (NIDCR). Reference RNA was isolated from UA159 cells grown in BHI medium to an optical density at 600nm of 0.5 and purified as previously described (Abranches et al., 2006). Total RNA from the experimental and reference conditions was isolated and purified as previously described (Abranches et al., 2006, Kajfasz et al., 2010). cDNA was synthesized and labeled using Cy3-dUTP or Cy5-dUTP (GE Healthcare, Piscataway, NJ) as previously outlined (Abranches et al., 2006). Cy3-dUTP-labeled test cDNA and Cy5-dUTP-labeled reference cDNA were hybridized overnight at 42°C. Hybridization was carried out using a MAUI Hybridization system (BioMicro Systems, Inc., Salt Lake City, UT), washed according to protocols provided by J. Craig Venter Institute (JCVI), and scanned using a GenePix 4000b Microarray Scanner (Molecular Devices, Inc., Sunnyvale, CA). After scanning, the images were analyzed using TIGR Spotfinder, and normalized, as previously described (Abranches et al., 2006). Four replicate slides were used for each sample and statistical analysis was carried out using BRB array tools (http//linus.ncinih.gov/BRB-ArrayTools.html). Microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE64236.
Results
Experimental Design
Although the S. mutans adaptive acid tolerance response (ATR) has been thoroughly characterized, how the response itself is activated and controlled remains less clear. Two previous studies have explored acid-inducible genes in S. mutans via microarray analysis, however, these studies were done by suspending batch cultures in acidic media (Gong et al., 2009, Krastel et al., 2010). In contrast, this study used continuous cultures in which the pH was lowered by the acidic end products of carbon metabolism by S. mutans; no exogenous acid was added. The scheme described here is more analogous to what happens in the oral cavity, where the pH in dental plaque is lowered not by exogenous acid, but by bacterial fermentation products. The UA159 and fabM::Erm strains were grown in continuous culture at pH 7 for ten generations. At this point, samples were taken for downstream microarray analysis, and referred to as “steady-state pH 7,” (Figure 1A) which was used as a baseline for expression levels. Upon sampling of the pH 7 steady-state cells, glucose was added to the culture to a final concentration of 200mM and pH control via KOH was turned off, allowing for acidification of the culture, as previously described (Fozo & Quivey, 2004b). When culture pH values had fallen to pH 5.5, samples were harvested for microarray analysis. This second time point was referred to as “glucose-shock” (Figure 1A). After the glucose-shock samples had been harvested, the pH control was turned on, such that the culture was not permitted to acidify below pH 5.0 for UA159 or pH 5.5 for fabM::Erm, the lowest achievable steady-state pH value for this strain (Fozo & Quivey, 2004a). The final set of samples, referred to as “steady-state pH 5” (or “steady-state 5.5”, for fabM::Erm), were harvested after an additional ten generations of growth in the chemostat (Figure 1A). Microarray analysis was performed on all the samples and seven comparisons were made between the strains and time points, as shown in Figure 1B. Figure 1B also displays the number of genes that were up- or down-regulated in each comparison. In this manuscript, we cannot address all of the differences that were observed, but we present here a synopsis of the major trends. We implemented a p-value cutoff of 0.01 and a 2-fold change cutoff for the data detailed below. Table 1 displays notable genes of interest, many of which are specifically discussed in the text below, and shows the fold-change in the comparisons in which they appeared. The full list of genes appearing in the comparisons using our parameters are shown in Supplemental Tables S1–S7. The complete raw data from all the comparisons is available in the GEO Database under accession number GSE64236.
Figure 1. Overview of experimental design and comparisons used for data analysis.
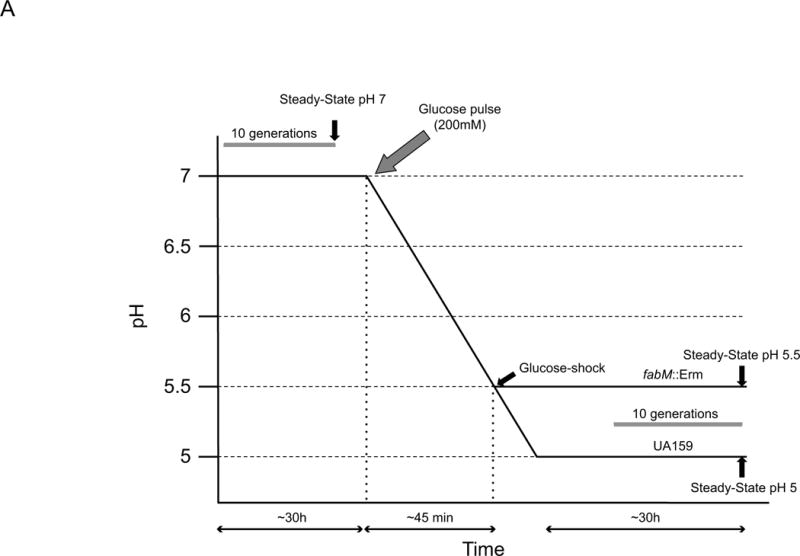
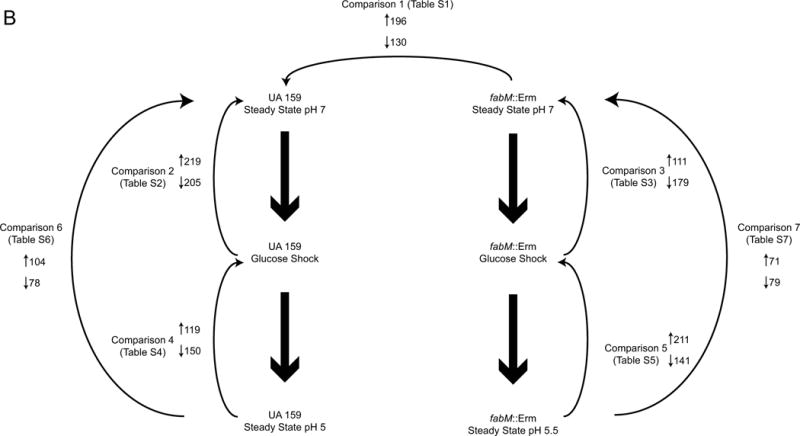
(Panel A) Overview of experimental design. Black arrows denote time points where samples (50ml) were taken from the culture vessel to prepare RNA for cDNA microarray analysis. “Steady-State pH 7” sample was harvested after 10 generations (~30 hours) of growth in the chemostat at pH 7. Glucose was added to the culture to a final concentration of 200mM (grey arrow) and the pH was allowed to fall. When the pH reached 5.5, the “Glucose-shock” sample was taken (typically occurred about 45 minutes after addition of glucose). pH control was re-established at pH 5 for UA159 and pH 5.5 for fabM::Erm. Cultures were allowed to continue growth for another 10 generations (~30 hours) and the final time point, “Steady-State pH 5/5.5,” was harvested. (Panel B) Overview of comparisons used for analysis of cDNA microarray data. Numbers next to upward pointing arrows denote number of genes up-regulated in that comparison and numbers next to downward pointing arrows denote number of genes down-regulated in that comparison. Table designations reflect supplemental tables where complete lists of data analysis for that particular comparison may be found, using cutoffs for a p-value of 0.01 and a 2-fold change in gene expression.
Table 1.
Genes up- and down-regulated in UA159 and fabM::Erm during glucose-shock and steady-state growth. This table includes genes commented on in the text. The complete dataset is available in Supplementary Tables S1–S7. Statistically significant changes in expression are given as a ratio of the first strain/condition listed versus the 2nd strain/condition listed. Data is grouped according to functional class. Number in parenthesis (where applicable) indicates value for comparison calculated from other available comparisons. ‘SS’ = steady-state; ‘GS’ = glucose-shock.
| Comparison # | 1 | 2 | 4 | 6 | 3 | 5 | 7 | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Gene | Locus | Description | fabM::Erm SS 7 vs UA159 SS 7 | UA159 GS vs. SS 7 | UA159 SS 5 vs. GS | UA159 SS 5 vs. SS 7 | fabM::Erm GS vs. SS 7 | fabM::Erm SS 5.5 vs. GS | fabM::Erm SS 5.5 vs. SS 7 |
| Oxidative Stress | |||||||||
| sod | SMU.629 | superoxide dismutase | 8.809 | 0.256 | (2.25) | 2.891 | |||
| ahpC | SMU.764 | alkyl hydroperoxide reductase | 2.41 | 13.46 | 0.211 | 2.84 (2.84) | |||
| ahpF | SMU.765 | H2O2-forming NADH oxidase | 12.397 | 0.242 | 3.00 (3.00) | 3.058 | |||
| tpx | SMU.924 | thiol peroxidase | 3.141 | 9.811 | 0.307 | (3.01) | 2.983 | ||
| gor | SMU.838 | glutathione oxidoreductase | 3.982 | 8.894 | 0.295 | (2.62) | 4.481 | ||
| dpr | SMU.540 | peroxide resistance protein | 2.429 | 0.375 | (0.91) | 3.442 | |||
| Amino Acid Biosynthesis | |||||||||
| glnA | SMU.364 | glutamine synthase | 2.857 | 0.345 | (0.99) | ||||
| proC | SMU.1974 | pyrroline carboxylate reductase | 2.2 | 0.382 | |||||
| hisA | SMU.1265 | ProFAR | 4.025 | 2.01 | |||||
| hisB | SMU.1268 | IGPD | 2.843 | 0.250 | |||||
| ilvE | SMU.1203 | BCAA aminotransferase | 2.391 | 4.571 | 3.521 | 2.19 | 0.417 | ||
| ilvB | SMU.231 | acetolactate synthase, large subunit | 2.718 | 0.480 | |||||
| ilvC | SMU.233 | ketol-acid reductoisomerase | |||||||
| nifS | SMU.249 | class-V aminotransferase | 3.677 | 0.381 | (1.40) | 2.255 | |||
| cysK | SMU.496 | cysteine synthetase | 2.848 | 3.035 | 0.313 | ||||
| cysD | SMU.1173 | O-acetylhomoserine sulfhydrylase | 1.894 | 1.877 | |||||
| hisF | SMU.1264 | imidazoleglycerol-phosphate synthase | 2.542 | 0.352 | (0.895) | ||||
| argJ | SMU.664 | ornithine acetyltransferase | 0.44 | ||||||
| Toxin Production and Resistance | |||||||||
| bip | SMU.1914c | 12.916 | 74.647 | 117 | 4.454 | 7.97 | |||
| SMU.1906c | 12.204 | 118.112 | 169.786 | 7.016 | 7.93 | ||||
| SMU.423 | 5.165 | 46.787 | 48.724 | 6.809 | |||||
| DNA Repair | |||||||||
| mutY | SMU.1865 | A/G-specific DNA glycosylase | 3.4 | 0.312 | (1.06) | 3.374 | |||
| smn (end3) | SMU.1650 | endonuclease III | 6.755 | 0.304 | (2.06) | 3.214 | |||
| Nucleotide Synthesis | |||||||||
| purB | SMU.59 | adenylosuccinate lyase | 0.220 | 0.36 | |||||
| purL | SMU.30 | FGAM synthase | 2.44 | 12.676 | 6.342 | ||||
| purD | SMU.48 | GARS | 2.672 | ||||||
| purF | SMU.32 | PPAT | 6.066 | 4.659 | |||||
| purM | SMU.34 | AIRS | 8.39 | 4.632 | 0.290 | ||||
| purN | SMU.35 | GART | 5.714 | 0.258 | |||||
| pyrB | SMU.858 | aspartate transcarbamoylase | 6.68 | 0.150 | (1.00) | ||||
| pyrA | SMU.859 | carbamoyl phosphate synthetase | 14.654 | 0.104 | (1.52) | ||||
| pyrAB | SMU.860 | carbamoyl phosphate synthetase | 9.405 | 0.115 | (1.09) | ||||
| pyrD | SMU.595 | dihydroorotate dehydrogenase | 3.778 | 5.206 | 3.691 | ||||
| pyrE | SMU.1221 | orotate phosphoribosyltransferase | 5.128 | 0.117 | (0.60) | ||||
| pyrF | SMU.1222 | orotidine-5-decarboxylase | 5.711 | 0.144 | (0.82) | ||||
| pyrDB | SMU.1223 | dihydroorotate dehydrogenase | 5.877 | 0.089 | (0.53) | 0.409 | |||
| pyrK | SMU.1224 | dihydroorotate dehydrogenase | 37.417 | 0.029 | (1.10) | ||||
| fhs | SMU.1073 | formyl-tetrahydrofolate synthetase | 4.277 | 1.963 | |||||
| Two-Component Systems | |||||||||
| comD | SMU.1916 | HK of competence regulon | 7.47 | 3.341 | |||||
| comE | SMU.1917 | RR of the competence regulon | 8.176 | ||||||
| ciaR | SMU.1129 | response regulator | 6.733 | ||||||
| PTS | |||||||||
| EI | SMU.675 | general PTS EI protein | 0.341 | ||||||
| ptsH | SMU.674 | general PTS histidine protein | 0.363 | ||||||
| bglP | SMU.980 | beta-glucoside-specific EII component | 3.037 | 0.11 | 10.841 | (1.193) | 0.356 | ||
| scrA | SMU.1841 | sucrose-specific IIABC component | 0.188 | 0.491 | |||||
| manL | SMU.1877 | mannose-specific component IIAB | 0.353 | 0.147 | 2.969 | ||||
| manM | SMU.1878 | mannose-specific component IIC | 0.387 | 0.162 | 2.863 | (0.464) | |||
| pttB | SMU.2038 | trehalose-specific IIABC component | 0.149 | 0.236 | |||||
| ptcC | SMU.1596 | cellobiose-specific IIC component | 6.058 | 0.435 | 0.265 | ||||
| ptsG | SMU.2047 | glucose-specific IIABC component | 0.353 | 0.214 | |||||
| pthP | SMU.754 | HPr kinase/phosphatase | 2.807 | ||||||
| Transport & Binding | |||||||||
| trk | SMU.1562 | potassium uptake protein | 4.655 | 3.075 | 3.63 | ||||
| trkA | SMU.1708 | potassium uptake protein | 2.585 | 2.661 | 3.212 | ||||
| trkB | SMU.1561 | potassium uptake protein | 3.447 | 2.486 | 3.678 | ||||
| trkH | SMU.1709 | potassium uptake protein | 2.914 | ||||||
| copA | SMU.426 | copper transporting ATPase | 8.104 | 3.125 | 5.023 | ||||
| copZ | SMU.427 | copper chaperone | 8.271 | 3.085 | 2.804 | ||||
| sloA | SMU.182 | ABC transporter | 0.157 | 0.192 | 0.190 | 0.223 | |||
| sloB | SMU.183 | Mn/Zn ABC transporter | 0.203 | 0.288 | 0.107 | 0.15 | |||
| sloC | SMU.184 | metal-binding ABC transporter | 0.345 | 2.202 | 0.147 | 0.324 (0.32) | 0.313 | 0.361 | |
| malF | SMU.1569 | maltose/maltodextrin ABC transporter | 0.421 | 0.385 | |||||
| malX | SMU.1568 | maltose/maltodextrin ABC transporters | 0.162 | 0.317 | |||||
| malG | SMU.1570 | maltose/maltodextrin ABC transporters | 0.389 | ||||||
| msmE | SMU.878 | MSM sugar-binding protein precursor | 6.351 | 6.615 | |||||
| msmF | SMU.879 | MSM permease protein | 4.270 | 3.714 | |||||
| msmG | SMU.880 | MSM permease preotein | 3.852 | 3.595 | 2.026 | ||||
| msmK | SMU.882 | MSM ATP-binding protein | 3.667 | 3.326 | |||||
| Replication and Cell Division | |||||||||
| ssb | SMU.1859 | single-stranded DNA-binding protein | 0.23 | 0.357 | 0.166 | 3.773 | (0.626) | ||
| dnaI | SMU.1921 | DNA replication protein primosome | 0.502 | 0.568 | |||||
| holB | SMU.1662 | DNA polymerase III, delta subunit | 0.348 | ||||||
| recJ | SMU.1472 | single-stranded DNA exonuclease | 0.268 | 2.942 | (0.788) | 0.175 | 3.354 | (0.587) | |
| rpoA | SMU.2001 | RNA polymerase, alpha subunit | 0.245 | 0.199 | 0.141 | 12.000 | (1.69) | ||
| ftsH | SMU.15 | cell division protein | 0.332 | 6.449 | |||||
| ftsX | SMU.1324 | cell division protein | 2.261 | 0.35 | |||||
| ftsQ | SMU.550 | cell division protein | |||||||
| ftsW | SMU.713 | cell division protein | 2.063 | 2.876 | 2.224 | ||||
| Protein Repair & Degradation | |||||||||
| clpE | SMU.562 | ATP-dependent protease | 2.821 | ||||||
| thdF | SMU.1235 | thioprene & furan degradation protein | 4.456 | 3.647 | |||||
| grpE | SMU.81 | heat shock protein (HSP-70) co-factor | 0.387 | 2.123 | 0.292 | (0.620) | 2.554 | ||
| clpB | SMU.1425 | Clp proteinase, ATP-binding subunit | 4.922 | ||||||
| Energy Metabolism | |||||||||
| pfk | SMU.1191 | phosphofructokinase | 2.977 | 1.806 | |||||
| gapN | SMU.676 | GAD-3-P dehydrogenase | 1.759 | 2.152 | |||||
| adhA | SMU.127 | acetoin dehydrogenase | 5.114 | 0.252 | (1.29) | 2.648 | |||
| adhB | SMU.128 | acetoin dehydrogenase | 6.335 | 0.246 | (1.56) | 7.386 | 4.328 | ||
| adhC | SMU.129 | dihydrolipoamide acetyltransferase | 7.334 | 0.230 | (1.69) | 8.811 | |||
| adhD | SMU.130 | dihydrolipoamide dehydrogenase | 11.891 | 0.222 | (2.64) | 9.068 | 4.05 | ||
| alsS | SMU.1452 | α-acetolactate synthase | 2.518 | 10.263 | 0.198 | (2.03) | 4.21 | 0.281 | (1.18) |
| aldB | SMU.1451 | α-acetolactate decarboxylase | 8.274 | 2.549 | |||||
| citB | SMU.670 | aconitate hydratase aconitase | 0.149 | 0.055 | 0.025 | 0.338 | 0.333 | ||
| citZ | SMU.671 | citrate synthase | 0.134 | 0.083 | 0.041 | ||||
| idh | SMU.672 | isocitrate dehydrogenase | 0.156 | 0.075 | 0.036 | 0.426 | |||
| nox | SMU.1117 | H2O2-forming NADH oxidase | 0.182 | 0.319 | 2.615 | (0.834) | |||
| glk | SMU.542 | glucose kinase | 0.411 | ||||||
| eno | SMU.1247 | enolase | 0.507 | 0.453 | 2.816 | (1.28) | |||
| pfkB | SMU.871 | fructose-1-P kinase | 0.167 | 4.048 | (0.676) | 2.269 | |||
| adhE | SMU.148 | alcohol/acetaldehyde dehydrogenase | 0.186 | 0.142 | |||||
| ackA | SMU.1978 | acetate kinase | 0.382 | ||||||
| scrB | SMU.1843 | sucrose-6-P hydrolase | 0.181 | 5.082 | (0.920) | ||||
| treA | SMU.2037 | trehalose-6-P hydrolase | 0.161 | 2.704 | (0.435) | 0.093 | 6.423 | (0.598) | |
| dex A | SMU.2042 | dextranase precursor | |||||||
| dex B | SMU.883 | dextran glucosidase | 3.689 | ||||||
| gtfA | SMU.881 | sucrose phosphorylase | 3.974 | ||||||
| pykF | SMU.1190 | pyruvate kinase | 2.507 | 2.182 | |||||
| pgk | SMU.361 | phosphoglycerate kinase | 3.318 | ||||||
| capP | SMU.712 | phosphoenolpyruvate carboxylase | 3.046 | ||||||
| gtfC | SMU.1005 | glucosyltransferase-SI | 0.386 | 6.047 | 2.205 | ||||
| pgm B | SMU.1747c | beta-phosphoglucomutase | 0.29 | ||||||
| citE | SMU.1020 | citrate lyase | 2.26 | ||||||
| ldh | SMU.1115 | lactate dehydrogenase | |||||||
| mleS | SMU.137 | malolactic enzyme | 12.874 | ||||||
| mleP | SMU.138 | malate permease | 5.778 | ||||||
| pfl | SMU.402 | pyruvate formate-lyase | |||||||
| glgD | SMU.1537 | glycogen biosynthesis protein | 0.194 | 4.567 | (0.886) | ||||
| glgC | SMU.1538 | glucose-1-P adenylyltransferase | 0.31 | 2.942 | (0.912) | ||||
| glgB | SMU.1539 | 1,4-alpha-glucan branching enzyme | 0.177 | 3.066 | (0.543) | ||||
| glgP | SMU.1564 | glycogen phosphorylase | 0.357 | 0.387 | |||||
| Cell Envelope | |||||||||
| spaP | SMU.610 | cell surface antigen | 6.012 | 2.704 | |||||
| mreC | SMU.20 | cell shape-determining protein | 0.104 | 7.711 | (0.802) | ||||
| mreD | SMU.21 | cell shape-determining protein | 0.287 | 3.438 | (0.987) | 0.34 | 3.459 | (1.18) | |
| dltC | SMU.1689 | putative D-alanyl carrier protein | 0.228 | 3.405 | (0.776) | 0.279 | |||
| dltD | SMU.1688 | putative extramembranal protein | 0.302 | 4.720 | (1.425) | ||||
| murC2 | SMU.1429 | UDP-N-acetylmuramyl tripeptide synthetase | 0.35 | ||||||
| pbp2a | SMU.1949 | carboxypeptidase | 0.366 | 5.880 | |||||
| pbp2x | SMU.455 | penicillin-binding protein 2X | 0.32 | 2.572 | (0.823) | 0.422 | 2.644 | (1.12) | |
| pgm A | SMU.1077 | alpha phosphoglucomutase | 0.247 | 4.449 | (1.099) | ||||
| mraY | SMU.456 | undecaprenyl-phosphate-UDP-MurNAc-pentapeptide transferase | 0.378 | 2.487 | (0.940) | ||||
| ffh | SMU.1060 | signal recognition particle protein | 0.355 | ||||||
| pbp1b | SMU.1991 | penicillin-binding protein 1 b | 0.417 | 2.929 | (1.22) | ||||
| thiD | SMU.85 | phosphomethylpyrimidine kinase | 4.511 | 2.362 | 0.288 | (0.68) | |||
| Fatty Acid Biosynthesis | |||||||||
| accA | SMU.1734 | acetyl-CoA carboxylase, alpha subunit | 0.324 | ||||||
| accD | SMU.1735 | acetyl-CoA carboxylase, beta subunit | 0.381 | 0.371 | 0.263 | 4.759 | (1.25) | ||
| fabZ | SMU.1737 | 3-hydroxymyristoyl-ACP dehydratase | 0.395 | 2.303 | |||||
| fabG | SMU.1740 | 3-oxoacyl-ACP reductase | 0.331 | ||||||
| fabD | SMU.1741 | malonyl-CoA (ACP) transacylase | 0.338 | 2.562 | |||||
| fabK | SMU.1742c | trans-2-enoyl-ACP reductase | 0.384 | 3.029 | |||||
| fabH | SMU.1744 | 3-oxoacyl-ACP synthase III | 0.314 | 0.351 | 3.400 | (1.19) | |||
| fabF | SMU.1739 | 3-oxylacyl-ACP-synthase | 0.481 | 2.097 | (1.01) | ||||
| acp | SMU.1743 | acyl carrier protein | 0.258 | 0.21 | |||||
| fabK2 | SMU.1335c | enoyl-ACP-reductase | 4.262 | ||||||
| cls | SMU.988 | cardiolipin synthase | 0.421 | 0.249 | 2.911 | (0.725) | |||
| Ribosome | |||||||||
| rs1 | SMU.1200 | putative ribosomal protein S1 | 0.275 | 0.409 | |||||
| rl1 | SMU.1626 | 50S ribosomal protein L1 | 0.341 | 2.831 | (0.97) | 0.363 | |||
| rl11 | SMU.1627 | 50S ribosomal L11 protein | 2.171 | 0.366 | |||||
| rs11 | SMU.2002 | 30S ribosomal protein S11 | 0.328 | 0.364 | 0.283 | 6.347 | (1.80) | ||
| rs13 | SMU.2003 | 30S ribosomal protein S13 | 0.23 | 0.265 | 11.605 | (3.08) | |||
| rs5 | SMU.2009 | 30S ribosomal protein S5 | 0.489 | 0.38 | 2.699 | (1.03) | 0.265 | 3.500 | (0.928) |
| rl18 | SMU.2010 | 50S ribosomal protein L18 | 0.39 | 2.189 | 0.366 | 3.994 | (1.46) | ||
| rl6 | SMU.2011 | 50S ribosomal protein L6 (BL10) | 0.415 | 0.38 | 0.418 | 5.677 | (2.37) | ||
| rs8 | SMU.2012 | 30S ribosomal protein S8 | 0.276 | 0.403 | 0.149 | 10.983 | (1.64) | ||
| rs14 | SMU.2014 | 30S ribosomal protein S14 | 0.471 | 0.428 | 5.719 | (2.45) | |||
| rl5 | SMU.2015 | 50S ribosomal protein L5 | 0.348 | 0.464 | 0.412 | 5.117 | (2.11) | ||
| rl24 | SMU.2016 | 50S ribosomal protein L24 | 0.331 | 0.303 | 3.662 | (1.11) | 0.252 | 10.765 | (2.71) |
| rl14 | SMU.2017 | 50S ribosomal protein L14 | 0.398 | 0.428 | 5.719 | (2.48) | |||
| rl29 | SMU.2019 | 50s ribosomal protein L29 | 0.335 | 0.274 | 0.242 | 8.009 | (1.94) | ||
| rl16 | SMU.2020 | 50S ribosomal protein L16 | 0.473 | 0.493 | 0.412 | 4.477 | (1.84) | ||
| rs2 | SMU.2032 | 30S ribosomal protein S2 | 0.512 | 0.45 | |||||
| F1F0 ATPase | |||||||||
| atpA | SMU.1527 | epsilon subunit | 2.468 | 2.538 | 2.609 | ||||
| atpB | SMU.1528 | beta subunit | 2.968 | 3.327 | 2.456 | ||||
| atpC | SMU.1529 | gamma subunit | 2.343 | 2.386 | 2.937 | ||||
| atpD | SMU.1530 | alpha subunit | 2.488 | 2.779 | 2.121 | 2.825 | |||
| atpE | SMU.1531 | delta subunit | 2.271 | ||||||
| atpF | SMU.1532 | b subunit | 3.050 | 2.626 | 3.20 | ||||
| atpG | SMU.1533 | a subunit | 3.109 | 2.452 | 3.555 | 2.942 | |||
| atpH | SMU.1534 | c subunit | 3.228 | 2.978 | 3.909 | 2.954 | |||
| Transcriptional Regulators | |||||||||
| sloR | SMU.186 | metal-dependent transcriptional regulator | 0.150 | 0.537 | |||||
| glnR | SMU.363 | glutamine synthase regulator | 2.879 | 0.218 | (0.628) | ||||
| copY | SMU.424 | regulator of the copper operon | 15.129 | 4.779 | 3.545 | ||||
| furR | SMU.593 | ferric uptake regulator protein | 2.853 | ||||||
| malR | SMU.1566 | maltose operon regulator | 2.841 | ||||||
| fabT | SMU.1745c | regulator of the fab operon | 0.365 | ||||||
| ccpA | SMU.1591 | carbon catabolite control protein | 0.319 | ||||||
| brpA | SMU.410 | transcriptional regulator | 0.412 | 0.445 | |||||
| treR | SMU.2040 | regulator of the trehalose operon | 0.19 | ||||||
| spxA1 | SMU.1142c | redox regulator | 0.527 | ||||||
| spxA2 | SMU.2084c | redox regulator | 5.546 | ||||||
| codY | SMU.1824 | GTP binding transcriptional regulator | 0.372 | ||||||
| fruR | SMU.870 | regulator of fructose metabolism | 0.244 | 3.085 | |||||
| pyrR | SMU.856 | regulator of the pyrimidine operon | 7.095 | 0.075 | (0.532) | ||||
| cysR | SMU.852 | regulator of cysteine biosynthesis | 6.849 | ||||||
| cpsY | SMU.1225 | transcriptional regulator | 2.249 | 0.343 | |||||
| yhcF | SMU.1193 | transcriptional regulator | 5.510 | ||||||
| hrcA | SMU.80 | transcriptional regulator | 0.165 | 0.254 | 2.841 | ||||
| dnaA | SMU.01 | chromosomal replication initiator protein | 5.492 | ||||||
| rggD | SMU.1509 | transcriptional regulator | 3.524 | ||||||
Differences in the fabM::Erm transcriptome compared to UA159 during growth at steady-state pH 7 (Comparison 1, Table S1)
We compared the transcriptomes of fabM::Erm and UA159 during growth of each strain to steady-state at pH 7. This comparison allowed us to estimate the effects of the loss of the ability to synthesize UFAs on gene expression in the fabM::Erm strain and, importantly, to establish a baseline for comparisons involving the fabM::Erm strain during glucose-shock and growth at acidic pH. Comparison 1 revealed 196 genes up-regulated and 130 genes down-regulated in fabM::Erm; of these, about one third of the genes (n=107) encoded hypothetical proteins or proteins of unknown function (Figure 1B).
Genes encoding proteins involved in detoxification, particularly those involved in redox/oxidative stress-sensing including alkyl hydroperoxidase (ahpC, SMU.764), thiol peroxidase (tpx, SMU.924), and glutathione oxidoreducase (gor, SMU.838), were up-regulated in fabM::Erm. Several well-established pathways of the ATR were also up-regulated in the fabM::Erm strain even though the strain was growing at neutral pH. The genes encoding the malolactic fermentation (MLF) enzymes mleS (SMU.137) and mleP (SMU.138) were up-regulated 5.8-fold and 12.9-fold, respectively. Additionally, ilvE (SMU.1203), encoding an aminotransferase required for branched-chain amino acid (BCAA) synthesis was up-regulated by 2.3-fold. Both MLF and BCAA synthesis are mechanisms employed to alkalinize the cytoplasm as part of the S. mutans ATR (Sheng & Marquis, 2006, Sheng & Marquis, 2007, Sheng et al., 2010, Lemme et al., 2010). The gene encoding acetolactate synthase, alsS (SMU.1452), was also up-regulated (2.5-fold). Not only is acetolactate a precursor for BCAA biosynthesis, production of acetolactate leads to formation of acetoin as a fermentation end-product, routing carbon away from production of the acidic end-products lactate and acetate. Moreover, the glycolytic enzymes pfk (SMU.113) and gapN (SMU.676) also exhibited elevated expression in fabM::Erm, possibly reflecting increased need of NADPH for the elevated BCAA biosynthesis (Len et al., 2004a). These observations are consistent with the concept that the fabM::Erm strain experiences elevated levels of acid stress, and with a need to protect stress-sensitive glycolytic enzymes (Baldeck & Marquis, 2008), carbon is routed away from the production of lactic acid. Reduced lactic acid production by the fabM::Erm strain is consistent with our previously reported data showing that the fabM::Erm strain cannot lower the environmental pH below 4.2 compared to a glycolytic minima of 3.3 for UA159 (Fozo & Quivey, 2004a).
Transcription of trkA (SMU.1708) and trkH (SMU.1709), involved in potassium uptake, was up-regulated, consistent with the role of potassium as the major counter-ion for outward-driven proton pumping via F-ATPases (Sato et al., 1989, Dashper & Reynolds, 1992, Iwami et al., 1997, Kashket & DePaola, 2002, Sheng & Marquis, 2006, Gong et al., 2009, Krastel et al., 2010). Regulation of PTS genes was complex, with the genes encoding the HPr kinase (SMU.754) and components of the β-glucoside (bglP, SMU.980) and cellobiose (ptcC, SMU.1596) PTS up-regulated, and manL (SMU.1877) and manM (SMU.1878) components of the glucose/mannose PTS down-regulated. Collectively, the data indicate an up-regulation of alternative carbon transport mechanisms, and a reduction in the reliance for glucose uptake via PTS, possibly indicating a shift in carbon metabolism in the fabM::Erm strain.
We also observed a down-regulation of genes encoding ribosomal subunits, suggesting a re-rerouting of resources away from protein production, or a reduction in the relative abundance of proteins. Metabolically, S. mutans does not code for a complete TCA cycle, only citB (SMU.670), citZ (SMU.671), and idh (SMU.672) are predicted in the genome (Ajdic et al., 2002). All three of these genes encoding TCA cycle enzymes were down-regulated, which may suggest a response to reduced need for amino acids in the glutamate family. This agrees with the observation that the argJ (SMU.664) gene, also involved in glutamate family biosynthesis was down-regulated in this comparison. Overall, there was a general up-regulation of genes known to be involved in stress responsiveness, suggesting that even during growth at neutral pH, the fabM::Erm strain experiences a higher degree of stress than the parent strain.
Transcriptomic changes in S. mutans UA159 following glucose-shock, compared to steady-state growth at pH 7 (Comparison 2, Table S2)
The removal of pH control and addition of a bolus of glucose to chemostat cultures resulted in rapid acidification of the cultures (Belli & Marquis, 1991, Fozo & Quivey, 2004b, Baker et al., 2014). Similarly, it has been known for many years that dental plaque samples from humans are also fully capable of rapid metabolism of glucose to organic acids (Kleinberg, 2002). Here, our goal was to establish the effects of such a “glucose-shock”, mimicking sugar consumption by the human host, on the transcriptome of the parent strain UA159, with the express purpose of identifying, if possible, genes that might be uniquely involved with a rapid acid-response. Following addition of glucose, we observed that 219 genes were up-regulated and 205 genes were down-regulated, compared to UA159 samples from steady-state pH 7 growth (Comparison 2, Figure 1B). Broadly, the changes in the transcriptome of cells subjected to glucose-shock acidification indicate several hallmarks of the acid stress response, as well as a shift away from protein synthesis and replication, and towards production of nucleotides and amino acids. Five genes encoding proteins involved in detoxification were up-regulated, including sod (SMU.629), ahpC, ahpF, tpx, gor, and the peroxide resistance protein (dpr (SMU.540)) which acts to sequester iron in order prevent damaging Fenton chemistry (Yamamoto et al., 2000). Eight amino acid biosynthesis genes were up-regulated, including those involved in glutamate, histidine, cysteine, and BCAA biosynthesis. Nine genes predicted to encode proteins involved in toxin production or resistance were up-regulated. In particular, SMU.1914c, SMU.1906c and SMU.423 were highly up-regulated, at 118-fold, 75-fold and 47-fold, respectively. Genes involved in nucleotide and nucleoside synthesis displayed increased expression, including the fifteen genes listed in Table 1, representing the majority of both the pur and pyr operons, as well as fhs, shown to be important in purine biosynthesis (Crowley et al., 1997). The genes encoding the DNA repair enzymes Smn (SMU.1650) and MutY (SMU.1865) (Gonzalez et al., 2012) were up regulated by 6.8-fold and 3.4-fold, respectively. Six genes encoding two-component systems were up-regulated, including the comDE system (SMU.1916 and SMU.1917) involved in regulation of competence. Thirty genes involved in transport and binding were up-regulated, with 11 of those genes encoding iron and cation carrying proteins. Among these were genes involved in potassium uptake (trk (SMU.1562), trkA, and trkB (SMU.1561)), copper transport (copA (SMU.426) and copZ (SMU.427)). Ten genes encoding known and putative transcription factors and nine genes encoding proteins involved in repair and degradation of proteins were up-regulated. Transcription was also elevated in 19 genes involved in energy and central intermediary metabolism. These included the genes encoding the glycolytic enzymes Pfk, AlsS and GapN, discussed in the previous paragraph, as well as the acetoin dehydrogenase complex (AdhABCD (SMU.127-SMU.130) and acetolactate decarboxylase (AldB (SMU.1451)). As mentioned previously, up-regulation of these enzymes serves to re-route carbon metabolism away from production of acidic end products and towards production of acetoin.
Twenty-five genes encoding proteins involved in cell envelope maintenance and eight genes involved in DNA replication, transcription and cell division were down-regulated. The fatty acid biosynthesis pathway was also broadly down-regulated. Twenty-three genes involved in protein synthesis were down-regulated, including several encoding tRNA synthetases and the majority of the genes encoding ribosomal subunits. Five genes encoding transcription factors were down-regulated, notably including the gene encoding the carbon catabolite control protein, CcpA (SMU.1591c). Genes encoding PTS systems were generally down-regulated, including the general Enzyme I (EI) and histidine protein HPr, as well as components of the ß-glucoside (bglP), sucrose (scrA, SMU.1841), glucose/mannose (manL and manM), trehalose (pttB, SMU.2038), cellobiose (ptcC), and glucose (ptsG, SMU.2047) PTS. This is likely due to the abundance of available glucose, and may suggest a switch from PTS transport to permease sugar transport systems, as external acidic pH values would create an environment that would greatly facilitate proton-motive force-driven uptake of glucose via permease activity, thus conserving ATP (Buckley & Hamilton, 1994). Ffh, the gene encoding the signal recognition particle protein (SMU.1060) (Gutierrez et al., 1999), saw a 0.3-fold decrease in expression, correlating with reduced membrane biosynthetic genes. Likewise, twenty-three genes encoding transport and binding proteins were also down-regulated, including the maltose permease system. Fifteen genes involved in energy metabolism were down-regulated, including the NADH oxidase (nox, SMU.1117), the glycolytic enzymes glk (SMU.542), eno (SMU.1247), and pfkB (SMU.871), alcohol/acetaldehyde dehydrogenase (adhE, SMU.148), acetate kinase (ackA, SMU.1978), sucrose and trehalose hydrolases (scrB, SMU.1843 and treA, SMU.2037), and the glycogen biosynthesis operon (glgBCD, SMU.1537-SMU.1539).
As a group, these results indicate that during glucose-shock, transcriptional changes are made to induce several well-characterized facets of the ATR including: up-regulation of ROS defense, DNA repair, BCAA biosynthesis, down-regulation of PTS, and re-routing of fermentation away from acidic end products. In addition, it appears that the cells are shifting metabolism away from production of new proteins and cell envelope components, and towards repair of existing macromolecules and synthesis of amino acids and nucleotides.
Changes in the transcriptome of the fabM::Erm strain following glucose-shock, as compared to steady-state pH 7 (Comparison 3, Table S3)
cDNA microarray analysis revealed 111 genes up-regulated and 179 genes down-regulated in the fabM::Erm strain following glucose-shock compared to the transcriptome derived from the strain at steady-state pH 7 (Figure 1B). Interestingly, when making the same comparison of growth conditions for UA159 (Comparison 2), we observed an additional 134 genes that were differentially regulated in the UA159 strain, indicating that the loss of fabM results in either a reduced response to glucose-shock, or that stress responses are already engaged to some extent during growth at steady-state pH 7. The data above comparing fabM::Erm to UA159 at steady-state pH 7 (Comparison 1) would suggest the latter, as many of the changes in expression seen in Comparison 2 are similar to those observed in Comparison 1. Similar to Comparison 2, genes involved in oxidative stress, amino acid biosynthesis, toxin production and resistance, acetolactate fermentation, copper transport, DNA repair, acetoin production and protein repair and degradation were up-regulated in Comparison 3. Additionally, similar to Comparison 2, genes involved in glycolysis, the cell envelope, fatty acid biosynthesis, cellobiose PTS, and maltose transport were down-regulated. Although these general trends were similar to those seen in Comparison 2, there were fewer genes differentially regulated in many functional classes and many of the genes that were differentially regulated had a less sizable fold-change in Comparison 3 than in Comparison 2. For example, in Comparison 3, genes encoding two-component systems, potassium uptake, purine and pyrimidine synthesis, Zn/Mn transport, PTS and ethanol fermentation were not differentially regulated to the same extent as they had been in Comparison 2. Reiteratively, this reduced response may be because fabM::Erm struggles to maintain cell homeostasis, even at neutral pH. Interestingly, the genes encoding ribosomal subunits were down-regulated to a greater extent (in both number of genes and fold-change of the genes) in Comparison 3 vs. Comparison 1. Taken together, these data show that following glucose-shock, the fabM::Erm strain exhibits many transcriptional changes; however, the changes are more modest compared to the changes made by UA159 during the same transition. This suggests that the acid-sensitive fabM::Erm strain, already in an elevated state of stress responsiveness, does not require as robust of a transcriptional response to fully adapt to a glucose-shock.
Genes differentially regulated in UA159 after transition from glucose-shock to steady-state pH 5 (Comparison 4, Table S4)
Next, we compared the transcriptome of cells grown to steady-state at pH 5 versus cells experiencing glucose-shock. In this category, there were 119 genes up-regulated and 150 genes down-regulated (Figure 1B). We observed the reversal of several trends in expression seen in Comparison 2. Seventeen genes encoding proteins involved in cell envelope maintenance were up-regulated, many of which were reduced in expression in Comparison 2, in accord with the well-established thickening of peptidoglycan in mutans streptococci during growth in acidic conditions (van Houte & Saxton, 1971). Several genes involved in energy metabolism that were reduced in expression levels in Comparison 2 also exhibited elevated expression levels after the transition from glucose-shock to steady-state pH 5. These included: glgBCD, scrB, treA, and pfkB, which suggested mobilization of the glycogen formation/utilization pathway and involvement of the trehalose pathway.
Several genes involved in energy metabolism were up-regulated in Comparison 4 that were not differentially regulated in Comparison 2. These included the genes encoding the subunits of the F1F0 ATPase (SMU.1527-SMU.1534), in order to preserve membrane ΔpH (Dashper & Reynolds, 1992, Sheng & Marquis, 2006). The reduced expression of the PTS enzymes and the ribosomal subunits observed during glucose-shock were partially reversed, with six genes encoding ribosomal proteins and the glucose/mannose (manLM) and ß-glucoside (bglP) PTS genes displaying an increase in expression. The genes encoding the multiple sugar-binding ABC transporter complex (msmEFGK, SMU.878-SMU.882) were up-regulated at steady-state pH 5, but had not been differentially regulated during glucose-shock conditions in the parent strain (Comparison 2), likely reflecting the conditions that excluded sugars other than glucose. Altogether, 22 genes involved in transport and binding were up-regulated in Comparison 4.
As with the genes up-regulated in this comparison, many of the genes down-regulated exhibited a reversal of expression patterns observed in Comparison 2, including decreased expression of many of the genes involved in detoxification, such as sod, ahpCF, tpx, dpr, and gor. Expression of the pyrimidine biosynthesis operon was decreased, while expression of the purine biosynthesis operon was not significantly altered, compared to glucose-shock. Eighteen genes involved in solute binding and transport were down-regulated in Comparison 4, including the sloABC Mn/Zn transporter operon (SMU.182-SMU.184) (Rolerson et al., 2006). Finally, many of the genes involved in energy metabolism that exhibited elevated expression during glucose-shock were reduced in expression levels in Comparison 4, including the adhABCD operon, aldB, and alsS. The genes encoding the incomplete TCA cycle (citB, citZ, and idh), were all significantly down-regulated in Comparison 4, and had not been differentially expressed following glucose-shock (Comparison 2), possibly indicating carbon flow away from the production of intermediates that result in the production of lactic acid.
The data obtained from Comparison 4 indicates that while some transcriptomic adjustments made following glucose-shock remain in effect during growth at steady-state pH 5, a large number of the transcriptional changes observed following glucose-shock were transient. The expression levels for many genes observed during growth at steady-state pH 5 were comparable to those seen during growth at steady-state pH 7, demonstrating that the cells were adapted to the acidic environment, and that steady-state metabolism has resumed (i.e. the excess glucose has been consumed). Thus, the genes which experienced transient differential regulation, were either required for metabolism of the excess glucose or were needed to adapt the cells to an acidic environment.
Changes in the fabM::Erm transcriptome at steady-state pH 5.5 compared to glucose-shock (Comparison 5, Table S5)
We compared the transcriptome of the fabM::Erm strain grown to steady-state at pH 5.5 to the transcriptome following glucose-shock (Comparison 5). In Comparison 5, 211 genes were up-regulated (92 more genes than Comparison 4) and 141 (9 fewer than the Comparison 4) genes were down-regulated. As with the parent strain UA159 grown to steady-state at low pH, the transcriptome of the fabM::Erm strain revealed a reversal of many of the trends seen immediately following glucose-shock. However, we observed a broader up-regulation of genes involved in central metabolic functions, including those of glycolysis, fatty acid biosynthesis, and protein synthesis, coupled with down-regulation of amino acid synthesis. Down-regulation of genes encoding the incomplete TCA cycle, acetoin production enzymes, pyrimidine synthesis enzymes, DNA repair enzymes, and oxidative stress enzymes did not appear in Comparison 5. With the exception of citB, citZ, and idh, we interpreted these results in the context of the inability of fabM::Erm to cope with acidic stress to the same extent as the parent strain (Fozo & Quivey, 2004a). In the case of citB, citZ, and idh, this may because they were already down-regulated in fabM::Erm at steady-state pH 7 (Comparison 1). The spxA2 gene (formerly designated spxB), encoding a redox-sensing transcriptional regulator (Kajfasz et al., 2010), was up-regulated 5.5-fold in this comparison, supporting previously published observations from our group indicating an overlap in the oxidative stress response with the acid stress response (Derr et al., 2012, Baker et al., 2014). Importantly, the atp operon, encoding the membrane-bound F1F0 ATPase was up-regulated in this comparison to a similar extent as seen in the same comparison in UA159 (Comparison 4), indicating that the acid sensitivity exhibited by fabM::Erm was not directly due to dysregulation of the atp operon.
Steady-state pH 5 compared to steady-state pH 7 in UA159 (Comparison 6, Table S6)
Here, we used cDNA microarray analysis to examine gene expression levels in cells grown at low pH in a tightly-controlled environment. We observed 104 genes up-regulated and 78 genes down-regulated in cells grown to steady-state pH 5 compared to steady-state pH 7. There were 36 genes appearing in Comparison 6 that did not appear in Comparison 2 or Comparison 4. This is most likely due to the fold-change or p-value of these 36 genes in Comparisons 2 and 4 being outside the cutoff applied here (2-fold and p ≤ 0.01). The F1F0 ATPase encoding genes were up-regulated in Comparison 6. While the atp operon had not changed expression in Comparison 2, it had been up-regulated in Comparison 4. Comparison 6 also displayed an up-regulation of genes encoding enzymes involved in BCAA synthesis, oxidative stress, purine and pyrimidine synthesis, copper transport, potassium uptake, and a down-regulation in genes involved with alcohol fermentation, Zn/Mn transport, maltose transport, TCA cycle and several PTS systems.
Comparison 6 also served as an internal control for Comparisons 2 and 4. The multiplicative product of the fold-changes from Comparisons 2 and 4 should be similar to the fold-change observed in the direct comparison (Comparison 6). For example, one would predict that by calculating the product of the fold-change in ahpC expression in Comparisons 2 and 4, that up-regulation of ahpC in Comparison 6 should be approximately 2.8-fold. Indeed, the direct comparison (Comparison 6) showed that ahpC was up-regulated 2.84-fold. Overall, in Comparison 6, transcriptional changes in UA159 indicate an elevated stress response compared to growth at neutral pH; however, the organism does not appear to be experiencing the same level of stress as it was immediately following glucose-shock due to acid-adaptation via the ATR.
The transcriptome of the fabM::Erm strain at steady-state pH 5.5 compared to steady-state pH 7 (Comparison 7, Table S7)
Our final comparison was that of fabM::Erm grown to steady-state pH 5.5 versus steady-state pH 7. As described above for Comparison 6, Comparison 7 served to validate both Comparisons 3 and 5. In Comparison 7, 71 genes were up-regulated and 79 genes were down-regulated during growth at low pH. The data from this analysis agreed with Comparisons 3 and 5, corroborating transcriptional shifts seen in genes from samples examined following glucose-shock.
Sixty-five genes appeared in Comparison 7 that did not appear in either Comparison 3 or Comparison 5. Again, the changes in expression that occurred in these genes in Comparisons 3 and 5 were outside of our pre-determined cutoff values. In Comparison 7, similar to Comparison 6 in UA159, genes involved in potassium uptake and the F1F0 ATPase were up-regulated, and the Zn/Mn transport slo operon was down-regulated. Unlike UA159 in Comparison 6, fabM::Erm in Comparison 7 did not show any differential regulation of BCAA biosynthesis, oxidative stress, copper transport, alcohol fermentation or maltose transport. Several genes involved in purine and pyrimidine biosynthesis, toxin production and resistance, the cell envelope, and PTS exhibited differential regulation in the same direction as Comparison 6, but to a lesser extent. Interestingly, of all the analyses, Comparison 7 had the fewest number of differentially regulated genes. As we hypothesized with the results from Comparison 3, we attribute the smaller number of changes in transcription in Comparison 7 to a higher basal level of gene expression in fabM::Erm involved in response to stress, even during growth at steady-state pH 7, the baseline for our comparisons.
Discussion
In this study, we show that as the environmental pH decreases due to the rapid accumulation of lactic acid, as a byproduct of glucose fermentation, many changes occur at the transcriptional level in S. mutans. In addition, upon adaptation to the low pH environment, many, but not all, of these changes are reversed. The transcriptional changes outlined above can be grouped into several categories, depicted in Figure 2. The differences in transcription observed in fabM::Erm in Comparison 1 are similar to those observed in UA159 in Comparison 2, when the homeostasis of steady-state growth is disrupted by the glucose-shock. Therefore, Comparison 1 defines the heightened basal stress level experienced by the fabM::Erm strain at pH 7. The genes that are differentially regulated promptly following glucose-shock, which remain differentially regulated even after the transition to steady-state acidic pH, represent the immediate or acute ATR. The late or adaptive ATR consists of the genes that are differentially regulated between glucose-shock and steady-state acidic pH, but were not differentially regulated between steady-state pH 7 and glucose-shock conditions. These mechanisms of the ATR appear to require the cells to become adapted to growth at low pH for some time. Finally, a large amount of genes were up- or down- regulated following glucose-shock, but transcripts returned to basal (steady-state pH 7) levels once the cells were acid-adapted (steady-state low pH). These transcriptional changes represent either a response to the glucose bolus, which is metabolized by the time steady-state low pH is reached, or may indicate portions of the ATR that have functioned to allow cells to acid-adapt and whose expression is not required after adaptation has occurred (transient ATR/glucose-shock response).
Figure 2. Summary of trends observed during glucose-shock and steady-state growth at pH 5 (or 5.5).
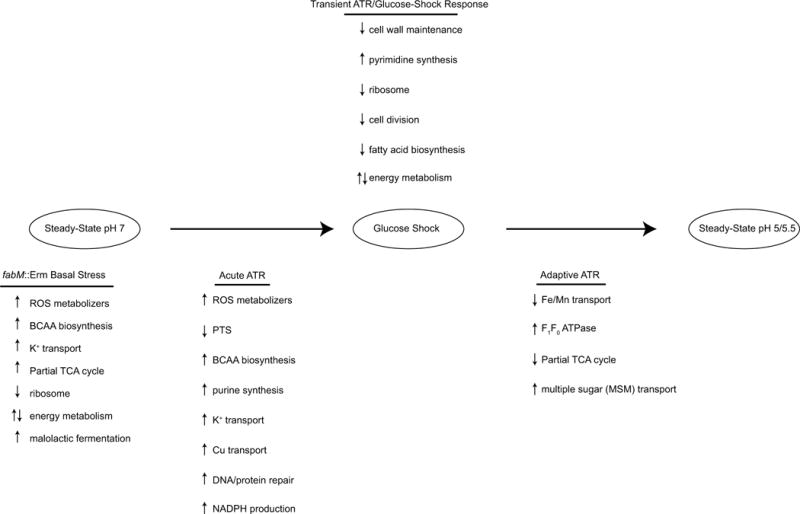
A diagram summarizing observed responses as UA159 and fabM::Erm move from steady-state pH 7 through glucose-shock to steady-state pH 5 (or 5.5). The responses were divided into categories to represent their respective contribution to the ATR of S. mutans. “fabM::Erm basal stress” indicates the differences discovered in gene expression in the fabM::Erm strain at steady-state pH 7 compared to those in UA159 at steady-state pH 7. “Acute ATR” was defined as genes/pathways that were differentially regulated at the glucose-shock time point and remained differentially regulated through steady-state growth at low pH. The “adaptive ATR” was defined as genes/pathways that were differentially regulated between glucose-shock and growth to steady-state at low pH that were not differentially regulated between steady-state pH 7 and glucose-shock. The “transient ATR/glucose-shock” response was defined as those genes/pathways that were differentially regulated at the glucose-shock time point, but were not differentially regulated between steady-state pH 7 and steady-state pH 5/5.5. ‘ROS’ = reactive oxygen species; ‘BCAA’ = branched-chain amino acid; ‘TCA’ = tricarboxylic acid; ‘PTS’ = phosphoenolpyruvate:phosphotransferase system.
Our results uphold several observations from three previous omics studies of the S. mutans ATR, although it should be noted that only our study allows temporal separation of the ATR (Crowley et al., 2004, Len et al., 2004a, Gong et al., 2009). Specifically, both the Gong et al. report and our report demonstrate up-regulation of copY, copA, comDE, the F1F0 ATPase, and the trk potassium transport system, and down-regulation of citB, citZ, idh, and ptsG during growth in an acidic environment. Proteomic and enzymatic data from the Crowley et al. study showed that growth at steady-state pH 5 compared to steady-state pH 7 caused an increase in ATPase activity and expression of the genes encoding AhpCF and FabK, and a decrease in PTS activity and expression of the genes encoding Eno, AckA (Crowley et al., 2004). These results were in agreement with the transcriptomic data provided in this study. Our comparisons also support results from the Len et al. proteomic study, that illustrated up-regulation of ATPase, GapN, Pgk, Pfk, GlnA, IlvE and down-regulation of ManL, AdhE, AckA, and Eno during growth at steady-state pH 5, compared to steady-state pH 7 (Len et al., 2004a). Len et al. hypothesized that the glycolytic adjustments indicated a decrease in the phosphoenolpyruvate (PEP) pool consistent with reduced need for PEP in PTS, specifically EIIman (encoded by manLMN), whose protein levels were also reduced (Len et al., 2004a). Our transcriptomic analysis showed that the adjustments to glycolysis and EIIman occurred immediately following glucose-shock (acute ATR), in accordance with the idea that as culture pH falls and glucose becomes readily available, energy, in the form of ATP, no longer needs to be expended to power the PTS, as glucose uptake can be powered by H+ symport via the permease system. Transcription of ilvE and glnA were also elevated following glucose-shock, suggesting that BCAA biosynthesis is also a portion of the acute ATR. BCAA biosynthesis serves to both re-route carbon towards less-acidic end products and alkalinize the cytoplasm through generation of NH3 (Len et al., 2004a). The prompt increase in gapN transcription is conceivably to generate the additional NADPH needed to support an increase in BCAA biosynthesis, as well as an increase in nucleotide synthesis suggested by the sweeping up-regulation of the pur and pyr operons, and fhs, following glucose-shock. Genes involved in potassium transport were also part of the acute ATR, likely in an effort to maintain the electrochemical gradient across the organism’s membrane.
Up-regulation of the atp operon and down-regulation of genes encoding metal transporters (slo) and citB, citZ, and idh did not occur until cells were acid-adapted (steady-state acidic pH, adaptive ATR), suggesting that these arms of the ATR are not activated until either low pH has been maintained for a considerable amount of time, or steady-state growth at acidic pH has been reached. The atp operon, encoding F1F0 ATPase, is utilized by S. mutans to extrude protons from the cell to maintain a more alkaline cytoplasm (Kuhnert & Quivey Jr, 2003, Sheng & Marquis, 2006). It is possible that following glucose-shock, the acid stress is not yet severe enough to warrant ATP usage, and more energy-efficient pathways, such as the early ATR pathways listed above (and in Figure 2), are used to maintain a more neutral intracellular space. Down-regulation of the slo operon suggests that either iron and/or manganese are not limiting at steady-state pH 5, or that iron uptake needs to be limited to prevent damaging Fenton chemistry from occurring. This is likely to be mediated in part by the metalloregulator of the slo operon, sloR, which has been implicated in control of certain aspects of the ATR, including the genes encoding the partial TCA cycle, also down-regulated at steady-state pH 5 (O’Rourke et al., 2010). Down-regulation of the genes encoding the partial TCA cycle may signify a reduced need for amino acids in the glutamate family, due to either lack of necessity, or because glutamate is highly acidic. An increase in transcription of the multiple sugar metabolism system (msm) operon also appeared to be induced with the adaptive ATR. This is presumably because under the glucose-limited conditions in a chemostat culture, the cells are searching for alternate carbohydrate sources to generate ATP for proton extrusion. Interestingly, the agmatine deiminase system, and the enzymes responsible for malolactic fermentation, both recognized components of the S. mutans ATR (Griswold et al., 2006, Lemos & Burne, 2008, Sheng et al., 2010, Lemme et al., 2010) did not appear to be differentially regulated as either strain went through transitions in pH (although fabM::Erm had a higher level of transcription of mleS and mleP), suggesting that the activity of these two pathways are regulated at the post-transcriptional level during the ATR.
Genes involved in ROS stress response were highly up-regulated following glucose-shock, then subsequently down-regulated following the transition from glucose-shock to steady-state at low pH. However transcription of these genes at steady-state low pH was still moderately up-regulated compared to levels observed at steady-state pH 7. These results indicate that the ROS defense genes are part of both the acute ATR and the transient ATR/glucose-shock response. This likely reflects the need to prevent and respond to higher levels of ROS formed following glucose-shock, due to increased metabolic rate and the concomitant drop in culture pH. After acid-adaptation has occurred, the expression levels presumably reflect induction of the acute ATR only, as excess glucose has been exhausted and metabolism has returned to steady-state rates. As stated above, following glucose-shock, both the pur and pyr operons were highly up-regulated. The pyr genes followed a similar pattern to the ROS stress response genes, and were down-regulated at steady-state pH 5 compared to glucose-shock, but their expression was not significantly altered between steady-state pH 5 versus steady-state pH 7, indicating that their induction was part of the transient ATR/glucose-shock response. The pur genes, on the other hand, remained up-regulated at steady-state pH 5, suggesting that they are, in fact, important for long-term acid tolerance. Purine biosynthesis has been shown previously to be critical for acid-tolerance (Crowley et al., 1997). These observations are consistent with a need to synthesize adenine to generate ATP for ATPase-mediated proton extrusion (Belli and Marquis, 1991).
Genes involved in cell wall maintenance, replication and division, fatty acid biosynthesis, and genes encoding the ribosomal proteins had an opposite pattern of expression to the ROS stress response and nucleotide synthesis genes, in that they were down-regulated following glucose-shock compared to steady-state pH 7, but not differentially regulated at steady-state low pH versus steady-state pH 7. Again, this suggests that these pathways were part of the transient ATR/glucose-shock response and that their repression was no longer needed once acid adaptation had occurred and glucose was once again limiting. While glucose is in excess, it appears that the priority is shifted away from proteins, DNA, and cell-wall macromolecules, and towards production of small molecules: amino acids and nucleotides.
Previous work has elucidated that an increase in the percentage of UFAs in the UA159 plasma membrane occurs immediately upon a decrease in culture pH (acute ATR) and does not reverse once the cells are acid-adapted (Fozo & Quivey, 2004a). It is noteworthy, however, that in the current study, no difference in expression of the fab gene cluster between steady-state pH 7 and steady-state pH 5 was observed. Therefore, this current study does not provide any novel clues as to how this shift in membrane composition occurs. Since the fabM::Erm strain cannot synthesize UFAs and is much less aciduric than UA159, we hypothesized that either due to, or in addition to, the absence of UFAs, there would be differences in the transcriptome between fabM::Erm and UA159 that would explain the apparent malfunction of the ATR in fabM::Erm. Expression levels in the fabM::Erm strain displayed similar patterns to UA159 throughout the transition from steady-state pH 7 to glucose-shock to steady-state low pH, but the transcriptional response was less pronounced, particularly following the transition from steady-state pH 7 to glucose-shock. In fact, expression levels of many genes in the fabM::Erm strain at steady-state pH 7 are more similar to those observed in UA159 following glucose-shock, rather than the transcript levels measured in UA159 at steady-state pH 7. These results indicate that even at neutral pH, the fabM::Erm strain experiences a heightened basal level of stress. Further, the magnitude of the response of fabM::Erm to the bolus of glucose and subsequent pH drop suggests that the fabM::Erm stress response, at the transcriptional level, either cannot be elevated beyond basal levels, or is already sufficiently heightened, so that the organism can cope with the changes in the growth environment. The results of a previous study indicated that ATPase activity in fabM::Erm is altered with respect to UA159 (Fozo & Quivey, 2004a). An attractive hypothesis to then explain the acid-sensitivity of the fabM::Erm strain is dysfunction of transmembrane proteins, notably the F1F0 ATPase, due to the absence of UFAs in the plasma membrane. The data presented here suggests that the cause of aberrant ATPase activity in fabM::Erm is post-transcriptional, as the levels of the F1F0 ATPase transcript were not disparate between UA159 and fabM::Erm at any of the three conditions in our experimental design. Further studies are, therefore, required to elucidate specifically why fabM::Erm is exceptionally acid-sensitive.
As the ability of S. mutans to cause disease is intimately tied to its ability to thrive and out-compete other oral microbes in an acidic feast-or-famine environment, understanding the interplay between metabolism and the ATR is crucial. We envision that the extensive data set presented here, coupled with information derived from a genome-wide collection of genetic deletion strains (Quivey et al., 2015), will provide a platform to analyze how the ATR in Streptococcus mutans is activated and controlled. Further endeavors coupling this data to promoter analysis and construction of the transcriptional regulatory response network of the S. mutans ATR and oxidative stress response are currently in progress.
Supplementary Material
Acknowledgments
This study was supported by Public Health Service grants from the National Institute for Dental and Craniofacial Research DE013683 (R.G.Q), DE017157 (R.G.Q.), DE019783 (J.A.L.) and T90 DE-021985 (J.L.B.).
References
- Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SJ, Wen ZT, Burne RA. Effects of oxygen on virulence traits of Streptococcus mutans. J Bacteriol. 2007;189:8519–8527. doi: 10.1128/JB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Derr AD, Karuppaiah K, et al. Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol. 2014;196:2166–2177. doi: 10.1128/JB.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeck JD, Marquis RE. Targets for hydrogen-peroxide-induced damage to suspension and biofilm cells of Streptococcus mutans. Can J Microbiol. 2008;54:868–875. doi: 10.1139/w08-078. [DOI] [PubMed] [Google Scholar]
- Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender GR, Sutton SV, Marquis RE. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun JP, Nguyen AH, Fan Y, Burne RA, Wen ZT. Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol Lett. 2011;320:110–117. doi: 10.1111/j.1574-6968.2011.02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley ND, Hamilton IR. Vesicles prepared from Streptococcus mutans demonstrate the presence of a second glucose transport system. Microbiology. 1994;140(Pt 10):2639–2648. doi: 10.1099/00221287-140-10-2639. [DOI] [PubMed] [Google Scholar]
- Carlsson A, Petersson M, Twetman S. 2-year clinical performance of a fluoride-containing fissure sealant in young schoolchildren at caries risk. Am J Dent. 1997;10:115–119. [PubMed] [Google Scholar]
- Crowley PJ, Gutierrez JA, Hillman JD, Bleiweis AS. Genetic and physiologic analysis of a formyl-tetrahydrofolate synthetase mutant of Streptococcus mutans. J Bacteriol. 1997;179:1563–1572. doi: 10.1128/jb.179.5.1563-1572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Svensäter G, Snoep JL, Bleiweis AS, Brady LJ. An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol Lett. 2004;234:315–324. doi: 10.1016/j.femsle.2004.03.043. [DOI] [PubMed] [Google Scholar]
- Dashper SG, Reynolds EC. pH regulation by Streptococcus mutans. J Dent Res. 1992;71:1159–1165. doi: 10.1177/00220345920710050601. [DOI] [PubMed] [Google Scholar]
- De Stoppelaar J, Van Houte J, Backer DO. The relationship between extracellular polysaccharide-producing streptococci and smooth surface caries in 13-year-old children. Caries Research. 1969;3:190–199. doi: 10.1159/000259582. [DOI] [PubMed] [Google Scholar]
- Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG., Jr Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol. 2012;78:1215–1227. doi: 10.1128/AEM.06890-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BA, Tan S, Smith V, et al. Vital and health statistics. Series 11, Data from the national health survey. 2007. Trends in oral health status: United States, 1988–1994 and 1999–2004; pp. 1–92. [PubMed] [Google Scholar]
- Dye BA, Thornton-Evans G. Trends in oral health by poverty status as measured by Healthy People 2010 objectives. Public Health Rep. 2010;125:817–830. doi: 10.1177/003335491012500609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustoferri RC, Hahn K, Weiss K, Quivey RG., Jr Smx nuclease is the major, low-pH-inducible apurinic/apyrimidinic endonuclease in Streptococcus mutans. J Bacteriol. 2005;187:2705–2714. doi: 10.1128/JB.187.8.2705-2714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustoferri RC, Hubbard CJ, Santiago B, Buckley AA, Seifert TB, Quivey RG., Jr Regulation of fatty acid biosynthesis by the global regulator CcpA and the local regulator FabT in Streptococcus mutans. Mol Oral Microbiol. 2014;30:128–146. doi: 10.1111/omi.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Kajfasz JK, Quivey RG., Jr Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol Lett. 2004;238:291–295. doi: 10.1016/j.femsle.2004.07.047. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J Bacteriol. 2004a;186:4152–4158. doi: 10.1128/JB.186.13.4152-4158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl Environ Microbiol. 2004b;70:929–936. doi: 10.1128/AEM.70.2.929-936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Scott-Anne K, Koo H, Quivey RG., Jr Role of unsaturated fatty acid biosynthesis in virulence of Streptococcus mutans. Infect Immun. 2007;75:1537–1539. doi: 10.1128/IAI.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Tian XL, Sutherland G, et al. Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology. 2009;155:3322–3332. doi: 10.1099/mic.0.031591-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez K, Faustoferri R, Quivey RG., Jr Role of DNA base excision repair in the mutability and virulence of Streptococcus mutans. Mol Microbiol. 2012;85:361–377. doi: 10.1111/j.1365-2958.2012.08116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AR, Jameson-Lee M, Burne RA. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J Bacteriol. 2006;188:834–841. doi: 10.1128/JB.188.3.834-841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: Dental Caries Onset Linked to Multiple Species by 16S rRNA Community Analysis. PLoS One. 2012;7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross EL, Leys EJ, Gasparovich SR, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JA, Crowley PJ, Cvitkovitch DG, et al. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiol. 1999;145(Pt 2):357–366. doi: 10.1099/13500872-145-2-357. [DOI] [PubMed] [Google Scholar]
- Hanna MN, Ferguson RJ, Li YH, Cvitkovitch DG. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J Bacteriol. 2001;183:5964–5973. doi: 10.1128/JB.183.20.5964-5973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwami Y, Guha-Chowdhury N, Yamada T. Effect of sodium and potassium ions on intracellular pH and proton excretion in glycolyzing cells of Streptococcus mutans NCTC 10449 under strictly anaerobic conditions. Oral Microbiol Immunol. 1997;12:77–81. doi: 10.1111/j.1399-302x.1997.tb00621.x. [DOI] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol. 2010;192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket S, DePaola DP. Cheese consumption and the development and progression of dental caries. Nutrition reviews. 2002;60:97–103. doi: 10.1301/00296640260085822. [DOI] [PubMed] [Google Scholar]
- Kleinberg I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med. 2002;13:108–125. doi: 10.1177/154411130201300202. [DOI] [PubMed] [Google Scholar]
- Koo H, Pearson SK, Scott-Anne K, et al. Effects of apigenin and tt-farnesol on glucosyltransferase activity, biofilm viability and caries development in rats. Oral Microbiol Immunol. 2002;17:337–343. doi: 10.1034/j.1399-302x.2002.170602.x. [DOI] [PubMed] [Google Scholar]
- Krastel K, Senadheera DB, Mair R, Downey JS, Goodman SD, Cvitkovitch DG. Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance. J Bacteriol. 2010;192:984–993. doi: 10.1128/JB.01169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert WL, Quivey RG., Jr Genetic and biochemical characterization of the F-ATPase operon from Streptococcus sanguis 10904. J Bacteriol. 2003;185:1525–1533. doi: 10.1128/JB.185.5.1525-1533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemme A, Sztajer H, Wagner-Dobler I. Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 2010;10:58. doi: 10.1186/1471-2180-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiol. 2008;154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J Bacteriol. 2008;190:5291–5299. doi: 10.1128/JB.00288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Len AC, Harty DW, Jacques NA. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiol. 2004a;150:1353–1366. doi: 10.1099/mic.0.26888-0. [DOI] [PubMed] [Google Scholar]
- Len AC, Harty DW, Jacques NA. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiol. 2004b;150:1339–1351. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui R, Cvitkovitch D. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 2010;5:403–417. doi: 10.2217/fmb.09.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye ZD, Zeng L, Burne RA. Modification of gene expression and virulence traits in Streptococcus mutans in response to carbohydrate availability. Appl Environ Microbiol. 2014;80:972–985. doi: 10.1128/AEM.03579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison HH, Barrett JF, Cardineau GA, Curtiss R., III Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986;54:273–282. doi: 10.1128/iai.54.2.273-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. Dental caries from a molecular microbiological perspective. Caries Res. 2013;47:89–102. doi: 10.1159/000345367. [DOI] [PubMed] [Google Scholar]
- O’Rourke KP, Shaw JD, Pesesky MW, et al. Genome-wide characterization of the SloR metalloregulome in Streptococcus mutans. J Bacteriol. 2010;192:1433–1443. doi: 10.1128/JB.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri RC, Clancy KA, Marquis RE. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett. 1995;126:257–261. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Kuhnert WL, Hahn K. Adaptation of oral streptococci to low pH. Adv Microb Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Grayhack EJ, Faustoferri RC, et al. Functional Profiling in Streptococcus mutans: construction and examination of a genomic collection of gene deletion mutants. Mol Oral Microbiol. 2015 doi: 10.1111/omi.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolerson E, Swick A, Newlon L, et al. The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J Bacteriol. 2006;188:5033–5044. doi: 10.1128/JB.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago B, MacGilvray M, Faustoferri RC, Quivey RG., Jr The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J Bacteriol. 2012;194:2010–2019. doi: 10.1128/JB.06737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Noji S, Suzuki R, Taniguchi S. Dual mechanism for stimulation of glutamate transport by potassium ions in Streptococcus mutans. J Bacteriol. 1989;171:4963–4966. doi: 10.1128/jb.171.9.4963-4966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Baldeck JD, Nguyen PT, Quivey RG, Jr, Marquis RE. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can J Microbiol. 2010;56:539–547. doi: 10.1139/w10-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Marquis RE. Enhanced acid resistance of oral streptococci at lethal pH values associated with acid-tolerant catabolism and with ATP synthase activity. FEMS Microbiol Lett. 2006;262:93–98. doi: 10.1111/j.1574-6968.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- Sheng J, Marquis RE. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol Lett. 2007;272:196–201. doi: 10.1111/j.1574-6968.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Svensäter G, Larsson UB, Greif EC, Cvitkovitch DG, Hamilton IR. Acid tolerance response and survival by oral bacteria. Oral Microbiol Immunol. 1997;12:266–273. doi: 10.1111/j.1399-302x.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- van Houte J, Saxton CA. Cell wall thickening and intracellular polysaccharide in microorganisms of the dental plaque. Caries Res. 1971;5:30–43. doi: 10.1159/000259730. [DOI] [PubMed] [Google Scholar]
- Wilkins JC, Homer KA, Beighton D. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl Environ Microbiol. 2002;68:2382–2390. doi: 10.1128/AEM.68.5.2382-2390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Higuchi M, Poole LB, Kamio Y. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J Bacteriol. 2000;182:3740–3747. doi: 10.1128/jb.182.13.3740-3747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Choi SC, Danko CG, Siepel A, Stanhope MJ, Burne RA. Gene regulation by CcpA and catabolite repression explored by RNA-Seq in Streptococcus mutans. PLoS One. 2013;8:e60465. doi: 10.1371/journal.pone.0060465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


