Abstract
A recent analysis of the genomes of Darwin's finches revealed extensive interspecies allele sharing throughout the history of the radiation and identified a key locus responsible for morphological evolution in this group. The radiation of Darwin's finches on the Galápagos archipelago has long been regarded as an iconic study system for field ecology and evolutionary biology. Coupled with an extensive history of field work, these latest findings affirm the increasing acceptance of introgressive hybridization, or gene flow between species, as a significant contributor to adaptive evolution. Here we review and discuss these findings in relation to both classical work on Darwin's finches and contemporary work showing similar evolutionary signatures in other biological systems. The continued unification of genomic data with field biology promises to further elucidate the molecular basis of adaptation in Darwin's finches and well beyond.
Introduction
The fields of ecology, evolutionary biology, and animal behavior are deeply rooted in organismal natural history. For centuries, curious naturalists have observed and catalogued the spectacular biology of diverse plants and animals in their natural environment, and no natural historian is more famous than Charles Darwin, the founder of modern evolutionary theory. In developing his theory of evolution by natural selection, Darwin wove together many detailed observations of organismal biology to produce a compelling argument that left little room for doubt regarding his basic tenets of descent with modification and the power of natural selection to produce remarkable phenotypic adaptation [1,2]. Today, as we continue to work out the ancestral relationships among taxa and explore the specific evolutionary processes responsible for adaptation, it is becoming increasingly clear that essential historical clues lie hidden in the genes that control organismal phenotypes [3-6]. However, the quest to find these genes is often an arduous one [7-10]. Genomics research, and the sub-fields of comparative and population genomics specifically, offers huge promise to unlock the molecular basis of biodiversity [11,12]. With the rapid advance of genome sequencing technology, we appear to be entering a ‘golden age’ for evolutionary genetics [11], one in which the hunt for genes underlying adaptation is progressing rapidly.
Recently, there have been a number of instances in which genomic approaches have produced major insights into classic evolutionary systems and questions, such as the diversification of cichlid [13] and stickleback [14] fishes, the evolution of mimicry in Heliconius butterflies [15], and the genetics of migration in the monarch butterfly [16]. A recent example of the power of population-level genomics to generate new understanding of an age-old question is a recent publication by Lamichhaney et al. [17] on Darwin's finches. Here, whole-genome sequencing of an entire adaptive radiation has been coupled with a study system that has an extensive record of field research focused on natural history and ecology. The results reveal the history of diversification and gene flow among species as well as identifying specific genes associated with an iconic morphological adaptation, beak shape. The findings also further unify observational evidence for interspecific hybridization with genetic evidence for adaptive interspecies allele sharing. This study, along with several other recent investigations [15,18-20], reinforces the increasing acknowledgement of adaptive introgression as a potentially important and widespread evolutionary phenomenon.
A tradition of field biology research on Darwin's finches
Darwin's finches are a group of about 14 species that evolved from a common ancestor on the Galápagos archipelago, a 15th species inhabiting Cocos Island (Figure 1). Since Charles Darwin's voyage on the HMS Beagle, this radiation has been the focal point of novel insights into evolutionary biology. Darwin's observations of the finch radiation led him to develop foundational ideas about evolution, including descent from a common ancestor, island colonization by mainland species, and adaptive radiation [1]. It was years after the Beagle voyage, however, that Darwin received input on the group's morphological variation and systematics from taxonomist John Gould and formulated the insights for which he is famous [21]. Darwin became particularly drawn to beak shape, and noting the incredible diversity in this trait among the closely related finches, he stated in the second edition of Journal of Researches (Voyage of the Beagle) [22] “Seeing this gradation and diversity of structure in one small, intimately related group of birds, one might really fancy that from an original paucity of birds in this archipelago, one species had been taken and modified for different ends.” Darwin's statement here about the Galápagos finches, 14 years before On the Origin of Species [1], reveals the early emergence of his evolutionary thinking.
Figure 1. Representatives of the Darwin's finch radiation.
Illustrations from Birds Part 3 No. 4 (1839) and Birds Part 3 No. 5 (1841) of The zoology of the voyage of H.M.S. Beagle by John Gould, edited by Charles Darwin. Reproduced with permission from John van Wyhe ed. 2002-. The Complete Work of Charles Darwin Online. (http://darwin-online.org.uk/)
More recently, the detailed work of Peter and Rosemary Grant has established the connection between climatic fluctuation, seed availability, and natural selection on beak morphology [23-25]. Furthermore, their work also documented detailed observations of immigration and hybridization producing viable offspring [25-27]. Additional analyses of song revealed the directionality of gene flow from hybrids into the parental populations [28-30]. The synthesis of 40 years of observations combined with analyses of beak morphology and body size, song, and microsatellite genetic data showed convergent evolution in Darwin's finches owing to introgressive hybridization and natural selection [31,32].
Advances by Podos, Huber, Hendry, DeLeón, and colleagues addressed the processes underlying adaptive radiation in Darwin's finches [33-36]. Podos [33] demonstrated that divergence in beak morphology drove evolution in vocal mating signals, likely promoting reproductive isolation and rapid speciation in the finch radiation. Huber et al. [34] showed beak size polymorphism in a Santa Cruz Island G. fortis population, presumably owing to ecological divergence, and subsequent assortative mating of the two morphs reminiscent of incipient speciation. Parallel studies related these findings to human impacts, showing how increased human population density can reduce the correlations between beak shape, size, bite force, and diet, thereby increasing the frequency of intermediate phenotypes and negatively impacting adaptive radiation [35, 36].
Genetic insight on beak development reveals the developmental modules underlying morphological change
Previous work by Abzhanov and colleagues [37-39] has investigated the developmental genetics of beak variation among Darwin's finches. Two anatomical components, the prenasal cartilage (pnc) and premaxillary bone (pmx), determine adult beak morphology. One study compared embryonic pnc development of the six Geospiza finch species and analyzed expression patterns of candidate growth factor genes involved in avian craniofacial development [38]. This study found a correlation between earlier and spatially broader expression of Bone morphogenetic protein 4 (Bmp4) in the developing upper beak and the deep, wide beak morphology of the ground finches [38]. Bmp4, however, was not implicated in the alternative elongated beak phenotype, suggesting the involvement of other genes [37]. To investigate the genetics of beak elongation, and go beyond candidate genes with a known role in craniofacial development, Abzhanov et al. [37] subsequently used a DNA microarray analysis. Using the sharp-beaked finch G. difficilis as a reference, transcripts up-regulated in the long-beaked cactus finches were distinguished from transcripts that were down-regulated or whose expression remained unchanged in the ground finches [37]. This approach revealed Calmodulin (CaM), a Ca2+ binding protein involved in Ca2+-dependent signal transduction, as a top candidate for beak morphogenesis. Further experiments misexpressing CaM in chick embryos produced the expected elongated beak phenotype [37]. Together, these studies suggested a modular developmental genetic basis for variation in pnc-determined beak morphology in which Bmp4 regulates depth and width, and CaM acts on the length axis. A complementary study that focused on pmx development in the ground finches used the same microarray screen to reveal three candidate genes for pmx patterning: TGFβ receptor type II (TGFβIIr), β-catenin, and Dickkopf-3 (Dkk3) [39]. Further analysis showed that domains of expression of these candidate genes correlated with adult morphology and that the genes interact to determine different axes of growth [39].
Prior to Lamichhaney et al.'s comprehensive genomic analysis [17], two Darwin's finch genomes had been sequenced. In 2012, the genome of a female medium ground finch, Geospiza fortis was published [40] as part of a suite of avian genomes [41-43]. In 2013, Rands et al. [44] published the genome of G. magnirostris and analyzed it in comparison to other vertebrates; zebra finch and G. fortis in particular. An analysis of positive selection by Rands et al. [44], based on patterns of synonymous and non-synonymous substitutions in a filtered set of 1,452 orthologs, yielded 21 genes with putatively adaptive amino acid substitution in the Darwin's finch lineage. At least two of these genes, POU1F1 and IGF2R, have been implicated in craniofacial development, suggesting a potential role in beak morphogenesis.
Integrating genomics of adaptive radiation and field biology
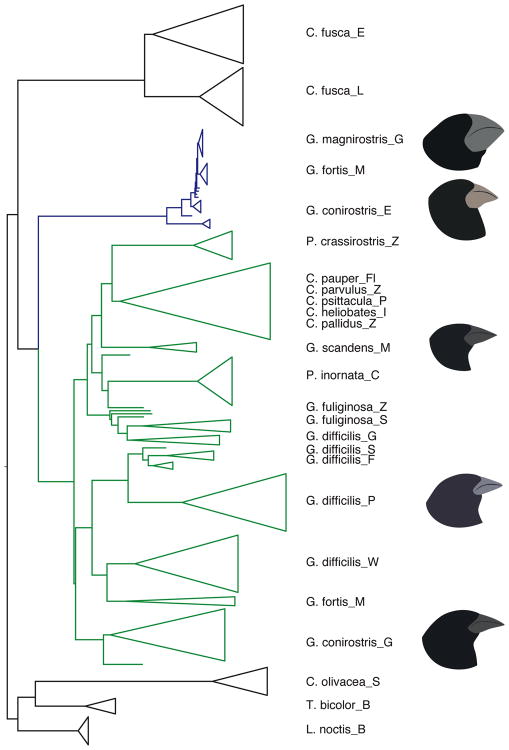
In the latest advance in this historic tale, Lamichhaney et al. [17] present an expanded genomic approach to understanding the evolutionary history of Darwin's finches. They sequence 120 full genomes, representing all the species in the Darwin's finch radiation and incorporating populations from multiple islands, and two closely related tanagers, Tiaris bicolor and Loxigilla noctis. Analysis of this dataset reveals some striking patterns. First, in concordance with prior observations reporting interspecies hybridization and migration between islands, whole-genome comparisons between species reveal high genetic diversity and extensive sharing of genetic variation, especially between ground and tree finches. The autosomal genome-based phylogenetic tree dates the birth of the radiation to circa 900,000 years ago and the radiation of ground and tree finches to 100,000-300,000 years ago. This tree topology generally supports the classical taxonomy based on mtDNA and morphology [45-47], with warbler finches as the first group to branch off and the ground and tree finches as the crown group. However, the phylogeny reveals two polyphyletic species, G. difficilis (also reported in [45]) and G. conirostris, which depart from the existing taxonomy. G. difficilis, which occurs on six islands, is split into three groups, and G. conirostris is split into two groups (Figure 2). The polyphyletic groupings of both species are associated with taxon sampling from multiple islands, emphasizing the importance of geography in the branching order of recently evolved groups.
Figure 2. Maximum-likelihood phylogenies of Darwin's finches.
Dashed arrows indicate gene flow between species. Highlighted bars denote key discordances between the trees. Branches ending with triangles indicate multiple genomes. Letters after species names indicate island sampled: S: Santiago, E: Española, L: San Cristóbal, Z: Santa Cruz, F: Fernandina, C: Cocos, P: Pinta, Fl: Floreana, I: Isabela, M: Daphne, D: Darwin, W: Wolf, G: Genovesa.
Lamichhaney et al. attribute allele sharing between species to introgressive hybridization via multiple lines of evidence, including ABBA-BABA tests [48,49] and discordance between autosomal and sex-linked phylogenetic tree topologies (Figure 2). ABBA-BABA tests show that despite a closer genetic relationship to G. magnirostris on Genovesa, G. difficilis on Wolf shares alleles with G. difficilis on Pinta. Another ABBA-BABA comparison confirms the proximate genetic relationship of G. magnirostris on Genovesa and G. conirostris on Española, but also shows gene flow between G. conirostris on Española and G. conirostris on Genovesa. Introgression of loci affecting phenotypic characters could explain the similarities upon which the two G. difficilis populations and the two G. conirostris populations were grouped in the classical taxonomy. In addition to these cases of recent introgressive hybridization, the authors find ABBA-BABA support for gene flow between the warbler finch C. fusca and the common ancestor of the non-warbler finches.
Sex-linked genes are well known to play a large role in speciation, and hence these loci generally show less interspecific gene flow in comparison to autosomal loci [50]. The discordance between the autosomal and sex-linked tree topologies for Darwin's finches, particularly with respect to the placement of G. difficilis from Pinta, Fernandina, and Santiago, supports the hypothesis of gene flow between this group and the ground and tree finches after the split of the Cocos finch. Phylogenies based on mtDNA and W-linked loci also support this interpretation. In addition, a separate analysis of demographic history within the group shows a large effective population size among the ground finches in comparison to the other taxa, consistent with gene flow among ground finch species.
To explore the genetic architecture of beak shape variation, Lamichhaney et al. perform a genome-wide scan of genetic differentiation (ZFST) on 15-kilobase (kb) windows between two groups of closely related finches distinguished by having blunt (G. magnirostris from Genovesa and G. conirostris from Española) or pointed (G. conirostris from Genovesa and G. difficilis from Wolf) beaks. A number of genomic regions emerge from this analysis, many of which house genes with potential roles in beak development. The highest-scoring window contains the gene ALX homeobox 1 (ALX1), which is known to play a central role in vertebrate craniofacial development [51,52]. A phylogenetic tree of Darwin's finches based on the ALX1 region groups individuals into two clades based on their blunt versus pointed beak morphology (Figure 3). The divergence between blunt and pointed haplotypes is inferred to be quite old, having occurred soon after the split of the warbler finches and other Darwin's finches. The medium ground finch, G. fortis, a species that varies in beak shape on Daphne Major, is also polymorphic for these highly divergent ALX1 haplotypes, and SNP genotyping of G. fortis specimens from this island reveals a significant association between the ALX1 locus and blunt versus pointed beak morphology.
Figure 3. Haplotype tree of beak shape locus ALX1.
Neighbor-joining tree based on ALX1 reveals a deep split between blunt and pointed beak haplotypes. Representative finch heads reflect species grouping by beak morphology as opposed to historical branching order. Branches ending with triangles indicate multiple ALX1 haplotype sequences.
Introgression and adaptation
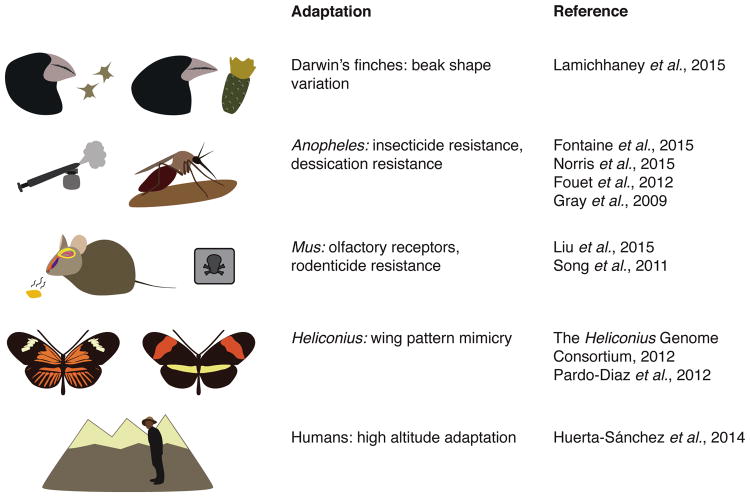
Genomic studies in various systems have uncovered evidence for adaptive allele sharing between closely related species, apparently as a result of introgression [53-60], and several of these investigations point to the exchange of distinct phenotypic traits (Figure 4). For instance, mosquitoes in the Anopheles gambiae complex have experienced such extensive interspecific gene flow that most of the genome no longer reflects the history of species-level diversification [18], and traits such as desiccation resistance and insecticide resistance have been transferred between species as a result [61-63]. Similarly, the house mouse, Mus musculus domesticus, has experienced substantial gene flow from the Algerian mouse, Mus spretus [20], resulting in introgression of alleles at olfactory receptors [20] as well as rodenticide resistance [64]. Heliconius butterflies have a long history of divergence with gene flow [65] and haplotypes at two major loci controlling wing patterning have been transferred among groups of closely related species as a result of natural selection for mimicry [15, 66]. Even modern humans have benefited from introgression: a haplotype at the hypoxia pathway gene EPAS1, associated with high-altitude hemoglobin concentration in Tibetans, appears to have been acquired from a Denisovan-like archaic human lineage [19].
Figure 4. Summary of recent studies of adaptive introgression in animals.
Examples highlight the exchange of distinct adaptive phenotypic traits between species.
It is important to note, however, that two distinct evolutionary processes can produce similar patterns of shared ancestry at focal regions of the genome: introgression and incomplete lineage sorting (ILS) of ancestral variation. For instance, in the case of the finches, it is possible that because the blunt and pointed ALX1 haplotypes diverged long ago, prior to much of the species-level diversification, this may be a long-standing polymorphism that has sorted out over time, resulting in some species becoming fixed for one ancient haplotype (blunt) and other species becoming fixed for the other ancient haplotype (pointed). Under this scenario, species share similar sequences today and group by phenotype on the ALX1 tree as a result of ILS, not introgression. Work in Anopheles [18], Mus [20], Heliconius [67], and humans [19] has explicitly considered ILS as an alternative explanation to introgression at target loci and the data from these systems generally support the introgression hypothesis. Overall, Lamichhaney et al. present compelling genome-wide evidence for a history of divergence with gene flow among Darwin's finches and remarkable allele sharing at a locus responsible for phenotypic diversity and adaptation. In terms of ALX1 specifically, some patterns in the data, such as the very short branch lengths among blunt haplotype sequences on the ALX1 tree, are definitely consistent with introgression (and selection). In contrast, the relatively long branch lengths among pointed haplotype sequences, and the fact that the G. fortis sequences do not group with G. scandens and G. fuliginosa, putative donor species [17], may suggest a potential role for ancestral variation as well. Future work including expanded sampling and sequencing of G. fortis from Daphne Major, and analyses of DNA sequence divergence and ABBA-BABA patterns of allele sharing among putatively introgressed ALX1 haplotypes will help clarify this history.
Conclusions and outlook
Lamichhaney et al.'s discovery of ALX1 raises many fascinating questions. For instance, what is the ultimate source of the two highly divergent, blunt and pointed haplotypes? The authors detect a genome-wide signature of ancient introgression between C. fusca and the common ancestor of the non-warbler finches, which may indicate that ancient hybridization contributed some of the critical genetic variation - perhaps one of these ALX1 haplotypes even - that originally fueled the finch radiation on the Galápagos. Furthermore, Lamichhaney et al. present evidence that these two haplotypes frequently occur together in heterozygotes, both in interspecific hybrids and in polymorphic species such as G. fortis. However, based on the sequence data presented, the two haplotypes do not appear to recombine. This may suggest that structural variation, like a chromosomal inversion polymorphism, is maintaining alternate copies. By reducing recombination between loci, inversions can maintain linkage between co-adapted alleles and this can have profound impacts on adaptation and speciation [68-70]. Moving beyond ALX1, it will be fascinating to explore the evolution of the many other genes that emerged from the genome-wide comparison between finches with blunt and pointed beaks. Given that birds with differing beak morphology are also likely to differ in other aspects of their biology, a detailed analysis of these genes and their evolutionary histories is almost certain to yield insights far beyond beak development.
Finally, we must note that it is not only the incredible dataset, sophisticated analyses, and discovery of ALX1 that make this most recent study of Darwin's finches so remarkable, but the integration of these new insights with detailed knowledge collected over decades about the ecology and evolutionary history of the study system. Genome sequencing technology will continue to advance and the application of these tools will proceed virtually without limit, but the biological context in which we interpret these genomic data is irreplaceable. In our modern exploration of evolutionary biology, natural history and ecology are essential counterparts to genomics because they enable us to establish direct connections between sequence variation and natural selection. For this reason, Darwin's finches have been providing critical insight into the evolutionary process for over 150 years and it seems that they still have plenty more to tell us.
Acknowledgments
We thank Leif Andersson, Peter Grant, Rosemary Grant, and two anonymous reviewers for insightful comments on the manuscript.
References
- 1.Darwin CR. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Darwin CR, Wallace AR. On the tendency of species to form varieties; and on the perpetuation of varieties and species by natural means of selection. Zool J Linn Soc. 1858;3:46–50. [Google Scholar]
- 3.Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Orgogozo V. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–50. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 5.Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution ? Evolution. 2008;62:2155–77. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–51. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rausher MD, Delph LF. Commentary: When does understanding phenotypic evolution require identification of the underlying genes? Evolution. 2015:n/a–n/a. doi: 10.1111/evo.12687. [DOI] [PubMed] [Google Scholar]
- 8.Barrett RD, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nature Rev Genet. 2011;12:767–80. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- 9.Lee YW, Gould BA, Stinchcombe JR. Identifying the genes underlying quantitative traits: a rationale for the QTN programme. Aob Plants. 2014;6 doi: 10.1093/aobpla/plu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockman MV. The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution. 2012;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadeau NJ, Jiggins CD. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 2010;26:484–92. doi: 10.1016/j.tig.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Stapley J, Reger J, Feulner PGD, Smadja C, et al. Adaptation genomics: the next generation. Trends Ecol Evol. 2010;25:705–12. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Brawand D, Wagner CE, Li YI, Malinsky M, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–81. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones FC, Grabherr MG, Chan YF, Russell P, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–8. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan S, Zhang W, Niitepold K, Hsu J, et al. The genetics of monarch butterfly migration and warning colouration. Nature. 2014;514:317–21. doi: 10.1038/nature13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamichhaney S, Berglund J, Almen MS, Maqbool K, et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–5. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 18.Fontaine MC, Pease JB, Steele A, Waterhouse RM, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347:1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huerta-Sanchez E, Jin X, Asan, Bianba Z, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–7. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu KJ, Steinberg E, Yozzo A, Song Y, et al. Interspecific introgressive origin of genomic diversity in the house mouse. Proc Natl Acad Sci USA. 2015;112:196–201. doi: 10.1073/pnas.1406298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulloway F. Darwin and his finches: The evolution of a legend. J Hist Biol. 1982;15:1–53. [Google Scholar]
- 22.Darwin CR. Journal of Researches Into the Natural History and Geology of the Countries Visited During the Voyage of H.M.S Beagle Round the World. London: John Murray; 1845. [Google Scholar]
- 23.Gibbs HL, Grant PR. Oscillating selection on Darwin's finches. Nature. 1987;327:511–3. [Google Scholar]
- 24.Grant BR, Grant PR. Evolution of Darwin's finches caused by a rare climatic event. Proc R Soc Lond B. 1993;251:111–7. [Google Scholar]
- 25.Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin's finches. Science. 2002;296:707–11. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 26.Grant PR. Hybridization of Darwin's Finches on Isla Daphne Major, Galapagos. Phil Trans R Soc Lond B. 1993;340:127–39. doi: 10.1098/rstb.2014.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant PR, Grant BR. Conspecific versus heterospecific gene exchange between populations of Darwin's finches. Phil Trans R Soc Lond B. 2010;365:1065–76. doi: 10.1098/rstb.2009.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant PR, Grant BR. Hybridization, sexual imprinting, and mate choice. Am Nat. 1997;149:1–28. [Google Scholar]
- 29.Grant PR, Grant BR. Mating patterns of Darwin's Finch hybrids determined by song and morphology. Biol J Linn Soc. 1997;60:317–43. [Google Scholar]
- 30.Grant BR, Grant PR. Hybridization and speciation in Darwin's finches: the role of sexual imprinting on a culturally transmitted trait. In: Howard DJ, Berlocher SH, editors. Endless forms: species and speciation. New York: Oxford Univ. Press; 1998. pp. 404–22. [Google Scholar]
- 31.Grant PR, Grant BR, Markert JA, Keller LF, et al. Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution. 2004;58:1588–99. doi: 10.1111/j.0014-3820.2004.tb01738.x. [DOI] [PubMed] [Google Scholar]
- 32.Grant PR, Grant BR. 40 Years of Evolution: Darwin's Finches on Daphne Major Island. Princeton, NJ: Princeton University Press; 2014. [Google Scholar]
- 33.Podos J. Correlated evolution of morphology and vocal signal structure in Darwin's finches. Nature. 2001;409:185–8. doi: 10.1038/35051570. [DOI] [PubMed] [Google Scholar]
- 34.Huber SK, De León LF, Hendry AP, Bermingham E, Podos J. Reproductive isolation of sympatric morphs in a population of Darwin's finches. Proc R Soc Lond B. 2007;274:1709–14. doi: 10.1098/rspb.2007.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendry AP, Grant PR, Rosemary Grant B, Ford HA, Brewer MJ, et al. Possible human impacts on adaptive radiation: beak size bimodality in Darwin's finches. Proc R Soc Lond B. 2006;273:1887–94. doi: 10.1098/rspb.2006.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De León LF, Raeymaekers JAM, Bermingham E, Podos J, Herrel A, et al. Exploring Possible Human Influences on the Evolution of Darwin's Finches. Evolution. 2011;65:2258–72. doi: 10.1111/j.1558-5646.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- 37.Abzhanov A, Kuo WP, Hartmann C, Grant BR, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–7. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 38.Abzhanov A, Protas M, Grant BR, Grant PR, et al. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–5. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 39.Mallarino R, Grant PR, Grant BR, Herrel A, et al. Two developmental modules establish 3D beak-shape variation in Darwin's finches. Proc Natl Acad Sci USA. 2011;108:4057–62. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, Parker P, Li B, Li H, et al. The genome of Darwin's Finch (Geospiza fortis) GigaScience. 2012 http://dx.doi.org/10.5524/100040.
- 41.Jarvis ED, Mirarab S, Aberer AJ, Li B, et al. Phylogenomic analyses data of the avian phylogenomics project. Gigascience. 2015;4:4. doi: 10.1186/s13742-014-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G, Li B, Li C, Gilbert MT, et al. Comparative genomic data of the Avian Phylogenomics Project. Gigascience. 2014;3:26. doi: 10.1186/2047-217X-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Li C, Li Q, Li B, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–20. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rands C, Darling A, Fujita M, Kong L, et al. Insights into the evolution of Darwin's finches from comparative analysis of the Geospiza magnirostris genome sequence. BMC Genomics. 2013;14:95. doi: 10.1186/1471-2164-14-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrington HL, Lawson LP, Clark CM, Petren K. The evolutionary history of Darwin's finches: speciation, gene flow, and introgression in a fragmented landscape. Evolution. 2014;68:2932–44. doi: 10.1111/evo.12484. [DOI] [PubMed] [Google Scholar]
- 46.Lack D. Darwin's Finches. Cambridge Univ. Press; 1947. [Google Scholar]
- 47.Petren K, Grant PR, Grant BR, Keller LF. Comparative landscape genetics and the adaptive radiation of Darwin's finches: the role of peripheral isolation. Mol Ecol. 2005;14:2943–57. doi: 10.1111/j.1365-294X.2005.02632.x. [DOI] [PubMed] [Google Scholar]
- 48.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–52. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green RE, Krause J, Briggs AW, Maricic T, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qvarnström A, Bailey RI. Speciation through evolution of sex-linked genes. Heredity. 2009;102:4–15. doi: 10.1038/hdy.2008.93. [DOI] [PubMed] [Google Scholar]
- 51.Dee CT, Szymoniuk CR, Mills PE, Takahashi T. Defective neural crest migration revealed by a Zebrafish model of ALX1-related frontonasal dysplasia. Hum Mol Genet. 2013;22:239–51. doi: 10.1093/hmg/dds423. [DOI] [PubMed] [Google Scholar]
- 52.Uz E, Alanay Y, Aktas D, Vargel I, et al. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am J Hum Genet. 2010;86:789–96. doi: 10.1016/j.ajhg.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344:1410–4. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- 54.Ellegren H, Smeds L, Burri R, Olason PI, Backstrom N, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–60. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 55.Martin CH, Cutler JS, Friel JP, Dening Touokong C, Coop G, et al. Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution. 2015:n/a–n/a. doi: 10.1111/evo.12674. [DOI] [PubMed] [Google Scholar]
- 56.Joyce DA, Lunt DH, Genner MJ, Turner GF, Bills R, et al. Repeated colonization and hybridization in Lake Malawi cichlids. Current Biology. 2011;21:R108–R9. doi: 10.1016/j.cub.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 57.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vernot B, Akey JM. Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science. 2014;343:1017–21. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 59.Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–6. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 60.Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–46. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 61.Norris LC, Main BJ, Lee Y, Collier TC, et al. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci USA. 2015;112:815–20. doi: 10.1073/pnas.1418892112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fouet C, Gray E, Besansky NJ, Costantini C. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One. 2012;7:e34841. doi: 10.1371/journal.pone.0034841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray EM, Rocca KAC, Costantini C, Besansky NJ. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malaria J. 2009;8:215. doi: 10.1186/1475-2875-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song Y, Endepols S, Klemann N, Richter D, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 2011;21:1296–301. doi: 10.1016/j.cub.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–28. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pardo-Diaz C, Salazar C, Baxter SW, Merot C, et al. Adaptive introgression across species boundaries in Heliconius butterflies. Plos Genet. 2012;8:e1002752. doi: 10.1371/journal.pgen.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith J, Kronforst MR. Do Heliconius butterfly species exchange mimicry alleles? Biol Lett. 2013;9:0130503. doi: 10.1098/rsbl.2013.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–34. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joron M, Frezal L, Jones RT, Chamberlain NL, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–6. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, et al. doublesex is a mimicry supergene. Nature. 2014;507:229–32. doi: 10.1038/nature13112. [DOI] [PubMed] [Google Scholar]