Abstract
Introduction
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was established in 2004 to facilitate the development of effective treatments for Alzheimer’s disease (AD) by validating biomarkers for AD clinical trials.
Methods
We searched for ADNI publications using established methods.
Results
ADNI has (1) developed standardized biomarkers for use in clinical trial subject selection and as surrogate outcome measures; (2) standardized protocols for use across multiple centers; (3) initiated worldwide ADNI; (4) inspired initiatives investigating traumatic brain injury and post-traumatic stress disorder in military populations, and depression, respectively, as an AD risk factor; (5) acted as a data-sharing model; (6) generated data used in over 600 publications, leading to the identification of novel AD risk alleles, and an understanding of the relationship between biomarkers and AD progression; and (7) inspired other public-private partnerships developing biomarkers for Parkinson’s disease and multiple sclerosis.
Discussion
ADNI has made myriad impacts in its first decade. A competitive renewal of the project in 2015 would see the use of newly developed tau imaging ligands, and the continued development of recruitment strategies and outcome measures for clinical trials.
Keywords: Alzheimer’s disease, Data-sharing, Amyloid phenotyping, Clinical trial biomarkers, Tau imaging, AD biomarker signature, Worldwide ADNI
1. Introduction
The overall goal of the Alzheimer’s Disease Neuroimaging Initiative (ADNI), established in 2004, is to facilitate the development of effective treatments for Alzheimer’s disease (AD) by validating biomarkers for AD clinical trials. Although no treatment has yet been shown to slow the progression of AD, the many accomplishments of ADNI have served as a model for other initiatives and programs.
A framework for pathophysiological changes occurring during disease progression was developed in the 1990s which centered on the accumulation of amyloid as a central pathogenic event [1]. However, at the turn of the century, details of the timing of the cascade of antecedent events leading to neurodegeneration and their relationship to clinical phenotypes were lacking [2]. The clinical diagnosis of AD was almost exclusively based on clinical assessment, the apolipoprotein E (APOE) ε4 allele was the primary known genetic AD risk factor, and mild cognitive impairment (MCI) had been recently recognized as a prodromal state of the disease [3,4]. The pharmaceutical industry was developing disease-modifying treatments to be tested, but clinical trials of these treatments were limited because clinical and cognitive outcome measures were the only ways to detect treatment effects. Patient functioning and cognition, especially memory, are extremely important, but brain function is affected by many factors other than AD pathology. Therefore, clinical and cognitive measurements may not be sufficiently powerful to detect the effects of treatments to slow AD progression within time and size constraints of clinical trials. Magnetic resonance imaging (MRI) and positron emission tomography (PET) biomarkers offered more precise alternatives to cognitive tests to assess disease progression, especially early in the disease. If such biomarkers were validated, the cost and length of drug trials could be reduced. Furthermore, the AD field would greatly benefit from surrogate outcome measures, that is, biomarkers of disease progression with greater statistical power than clinical or cognitive measurements used alone. Alternatively, improvement of the ability of cognitive tests to assess disease progression would also benefit clinical trials. The efficacy of these biomarkers could be accurately assessed using a standardized cohort using standardized methods [5,6] and ADNI was established primarily to fill this need.
Designed as a multisite, longitudinal study of normal cognitive aging, MCI, and early AD, the primary goal of ADNI was to develop imaging and other biomarkers for clinical trials [5,6]. To achieve this, ADNI enrolled a large cohort (>800) of participants across the spectrum of the disease [7] and developed optimized and standardized methods for use in a multisite setting to characterize the cohort with clinical, cognitive, MRI, PET, biofluid, and genetics measurements. One aim was to develop biomarkers that could consistently identify the disease with high sensitivity and specificity at an earlier stage and to better monitor disease progression and treatment effects. As the need for effective AD treatments was so pressing and the task of developing them was too great for any one public agency or private company, funding was secured from both the public and private sector, establishing ADNI as a model for public-private partnerships. Initial funding for a 5-year study came from the National Institute on Aging ($40 million), 13 pharmaceutical companies, and 2 not-for-profit foundations ($20 million). After the initial funding of ADNI-1 in 2004, further Foundation and Industry funding allowed the addition of PET amyloid imaging using the radiotracer 11C-Pittsburgh Compound B, genome-wide association studies (GWAS), and additional cerebrospinal fluid (CSF) analysis [8]. A unique feature of the original ADNI grant (now called ADNI-1) was that all clinical, cognitive, imaging, and biomarker data collected by the ADNI database would be immediately available to all scientists in the world who requested it, with no embargo. ADNI-1 was then extended by a Grand Opportunities grant (ADNI-GO). In 2010, ADNI was competitively renewed (termed ADNI-2) with funding through mid-2016. Each study used ongoing advances in imaging and genetics technologies, and ADNI-GO and ADNI-2 included an additional cohort of early MCI patients to study the earlier stages of the disease. Subjects enrolled in ADNI-2 and those continuing from ADNI-1 and ADNI-GO have had amyloid PET scanning with florbetapir, lumbar puncture for CSF analysis, and fluorodeoxyglucose-PET, MRI, and an extensive clinical and cognitive battery.
ADNI is conducted at 57 academic sites across the United States and Canada and comprises eight cores (clinical, MRI, PET, biomarker, neuropathology, genetics, biostatistics, and informatics) under supervision of the Administrative Core, led by Dr Michael W. Weiner [5]. ADNI is governed by Steering Committee including representatives from all funding sources and the principal investigators of ADNI sites. The Industry Scientific Advisory Board provides input from pharmaceutical stakeholders. The structure of the study, detailed in ref. [5], has been integral to the success of this multicenter study, and has served as a model for other similar initiatives.
In 2011, ADNI was identified by the U.S. government as a key player in achieving goals of accelerating the development of treatments that would prevent, halt, or reverse the course of AD and improving early diagnosis in the National Plan to Address Alzheimer’s Disease (U.S. Department of Health and Human Services) developed in response to the National Alzheimer’s Project Act. What specific impacts has ADNI made over the last decade? The pharmaceutical industry has benefitted from the development of standardized biomarkers, the establishment of amyloid phenotyping as a method for selection of subjects for AD trials, and the generation of data to guide trial design. Various companies have benefitted from the use of ADNI data to help validate their products and methods. Investigators worldwide have benefitted from access to ADNI data and samples, resulting in progress often far beyond the original ADNI mandate. ADNI genetics data are now being used in a whole-genome sequencing project in a “big data” approach to finding AD treatments. Our understanding of AD pathophysiology and genetics has benefitted from over 600 publications using ADNI data. In particular, the AD model reported by Jack et al. [9] has provided the field an overall conceptual model that stimulated hypothesis testing and other studies, and ADNI research has contributed to a broadening of the cognitive spectrum to include early MCI and subjective complaint cohorts. The research community has benefitted from the development of a plethora of methods using ADNI data, often applicable to areas outside AD research. ADNI structure and methods are now also being used in studies of the role of depression in AD and of special risk factors for AD in veterans. In addition, the ADNI model has fostered similar projects worldwide and inspired initiatives in other diseases such as Parkinson’s disease and multiple sclerosis (MS).
Three sequential, comprehensive reviews of all studies using ADNI data have been published since 2012 [8,10,11]. In addition to highlighting key ADNI publications, this review details the methodological, organizational, and funding achievements of ADNI in its first decade from 2004 to 2014, and how these have improved clinical trial efficiency and inspired similar initiatives worldwide.
2. Impacts of ADNI
2.1. ADNI has improved clinical trials for AD modifying and preventative treatments
ADNI has provided an important venue for precompetitive public-private interaction around biomarkers and clinical trial methodologies for AD. It has improved clinical trial efficiency by contributing to a better understanding of the pathophysiology of the disease, providing data to guide trial design, and by developing standardized biomarkers and methodologies.
Companies that provide imaging services for clinical trials, such as Bioclinica [12–15] (recently merged with SYNARC [16–19]), IXICO, and Lilly [20] have used ADNI data to develop and validate their image quantification methods. For example, the learning embeddings for atlas propagation technology for repeated automated hippocampal volumetry was developed by ADNI researchers at Imperial College, London, based in part on de novo analyses of ADNI data [21,22], before being licensed to IXICO [23]. This technology was used in the qualification of hippocampal volume for the enrichment of amnestic MCI clinical trial populations by the European Medicines Agency, which was coordinated in a precompetitive fashion by the Coalition Against Major Diseases [24].
Both hippocampal volume and CSF biomarkers remain the focus of ongoing qualification efforts with the Food and Drug Administration (FDA) [24]. Amyloid biomarkers are actively used for subject selection in clinical trials of candidate therapeutics. Amyloid biomarker substudies in the recent bapineuzumab phase III program revealed that even in AD dementia populations, more than 20% of enrolled mild and moderate AD subjects were amyloid negative by CSF amyloid beta (Aβ) or amyloid PET [25]. Subsequent trials of antiamyloid therapeutic candidates are requiring amyloid biomarkers at screening and amyloid positivity as an inclusion criterion. Longitudinal measures of amyloid are also being increasingly used later in the drug development process to assess potential disease-modifying effects.
To date, there have been no successful clinical trials for AD preventive treatments. However, it is now widely believed, in part due to ADNI research, that successful therapies will result from intervention at the very early stages of the disease. Accordingly, investigators have proposed new trial designs for intervention at the prodromal [26] and preclinical [27] stages of disease that have been adopted by academic and industry investigators, contributing to the development of new regulatory guidance [28]. In particular, the A4 trial [29] launched in 2014 as an industry-academia collaboration, represents the first therapeutic trial in preclinical sporadic AD.
2.2. Standardization of methods
At the outset of ADNI, a major obstacle to producing meaningful data for analysis was the development of standardized methods. A major collaborative effort has resulted in a set of protocols (available at http://adni.loni.usc.edu/methods/) that allow the direct comparison of results worldwide [5]. As a result of ADNI’s contributions, pharmaceutical companies developing disease-modifying treatments for AD and studies funded by the National Institutes of Health and private foundations have used ADNI methods in virtually all their clinical trials.
2.2.1. Positron emission tomography
Acquisition methods, quality control standards, and methods for preparing data for FDG-PET and amyloid imaging using Pittsburgh’s Compound and florbetapir were developed by the ADNI PET core [30]. The standardized protocols were designed to be compatible with multiple commercially available scanner hardware and software combinations, which can result in a twofold difference in intrinsic resolution. Raw PET images from all sites undergo quality control processes at the ADNI PET site at the University of Michigan. The gold standard digital Hoffman Phantom is used as a comparison to correct image resolution, and to enhance image uniformity, producing a variety of sets of images such as images that are registered to one another or oriented to a standardized grid. Different ADNI sites are then responsible for a variety of image analysis processes such as SPM5 to examine correlations between changes in glucose metabolism and cognition and to map cross-sectional differences between patient groups, and the determination of the standardized uptake value ratio (SUVR) in multiple regions of interest. These protocols are detailed at http://adni.loni.usc.edu/methods/pet-analysis/ and result in a set of images available at Laboratory of Neuroimaging (LONI) (http://adni.loni.usc.edu), a form that can be readily analyzed by investigators. The development of standardized methods has clearly demonstrated that multicenter PET amyloid imaging is feasible and can produce data sets of great value to investigators.
2.2.2. Magnetic resonance imaging
The development of standardized MRI procedures by the ADNI MRI core for use in the multiple ADNI centers is a major contribution of the initiative to the scientific community. Protocols needed to be compatible with three different vendors of scanners (GE, Siemens, and Philips), a variety of hardware/software configurations within each vendor product line, and two MRI field strengths. Methods were initially developed using technology widely available at the beginning of ADNI with the philosophy that the protocol must maximize scientific utility while minimizing the scan time burden on participants [31]. Pulse sequences were optimized for longitudinal scans to ensure stability and reproducibility [32]. The final protocol could be run in less than 30 minutes, capturing both structural information and detected relevant brain pathologies, and using a phantom to monitor scanner performance. The protocol also included quality control for all images acquired and postacquisition corrections to correct scaling changes and image artifacts such as intensity nonuniformity, and warping because of gradient nonlinearity [33–35]. A total of 38 different vendor- and platform-specific protocols were required to run ADNI MRI sequences at 59 sites with 89 MRI scanners. The final protocol achieved consistent acquisitions across this broad distribution of sites and technologies [33]. After the initial protocols were developed, it became apparent that MRI scans in ADNI also needed to image white-matter disease and so a fluid-attenuated inversion recovery sequence to detect cerebrovascular disease was added to the core sequence for ADNI-GO and ADNI-2. In addition three emerging MRI applications—functional MRI, Arterial Spin Labeling Perfusion Imaging, and Diffusion Tensor Imaging—were added in ADNI-GO and ADNI-2 as vendor-specific protocols to pilot their potential use in multicenter clinical trials [33]. A comparison of sequences used in ADNI-1, ADNI-GO, and ADNI-2 may be found at: http://adni.loni.usc.edu/methods/mri-analysis/mri-acquisition/.
A key factor in the success of ADNI MRI protocols was the use of a high-resolution geometric phantom to assess the reliability of scanner hardware across longitudinal scans. Consisting of polycarbonate spheres filled with water and copper sulfate in a precise geometrical pattern, the ADNI phantom is scanned after each patient to detect linear and nonlinear spatial distortion, signal-to-noise ratio, and image contrast, allowing these artifacts and problems to be identified and subsequently corrected. The ADNI phantom helped correct scanner scaling errors or miscalibrations [36] and to reduce between scanner imaging artifacts in longitudinal studies [37]. Without the monitoring of scanner performance using the ADNI phantom, around 20% of all scans would have been affected by these types of errors [36]. This phantom has been so successful that it has been used in numerous phase 2 and phase 3 treatment trials [5].
With the increasing number of studies published using ADNI data came the realization that the direct comparison of results was hampered by the lack of standardized data sets. To address this, the MRI core developed a series of standardized data sets that have met rigorous quality control standards [38]. Although it is too early to assess the impact of the standardized data sets on the analysis of MRI data, this strategy should facilitate the direct and meaningful comparison and replication of different algorithms and promote consistency in data analysis.
Beyond the standardization of methods and data sets, MRI studies carried out with the ADNI cohort have impacted clinical trials in a number of ways. Fox and coworkers developed improved methods for measuring the rate of atrophy across multiple sites and for reducing required sample sizes [39–41], and also developed automated methods to measure brain and hippocampal volume and rates of atrophy [39,42,43]. These have been incorporated into large commercial clinical trials and submitted to the European Medicines Agency, leading to guidance on hippocampal volume measurement in trials [24].
One challenge in the selection of clinical trial populations is the heterogeneity of individual responses to treatment due to differing underlying pathologies such as vascular brain injury. Effects of white matter hyperintensities on cognition, brain atrophy, and cerebral metabolism are dissociable from the effects of amyloid [44–46] and they likely contribute to the heterogeneity of individual responses to treatment [47,48]. Clinical trials may therefore benefit from reducing heterogeneity by excluding or stratifying individuals with vascular brain injury as measured by MRI.
2.2.3. CSF biomarkers
The ADNI Biomarker Core has developed and improved methods to analyze of CSF biomarkers, initially establishing a flow-cytometry based assay using xMAP technology [49,50] and assessing its within-site and intersite reliability. Best performance was assured by strict attention to standard operating procedures and including appropriate quality control specimens [51]. Their establishment of the predictive ability of the CSF biomarker signature provided support for the lumbar puncture procedure and hastened its acceptance as a valid tool in the AD diagnosis arsenal. More recently, this core has developed an alternative assay to measure CSF Aβ42 using two-dimensional ultra-performance liquid chromatography tandem mass-spectrometry, characterized the diagnostic ability of this assay using receiver operator curves and correlation analyses, and developed a surrogate matrix for calibration purposes [52]. The inclusion of CSF biomarkers in the newly revised National Institute on Aging–Alzheimer’s Association (NIA-AA) criteria for the diagnosis of AD in research settings [53,54] has led to the use of these assays to help select AD patients at the predementia stage, and to improve the statistical power of clinical trial design. Ongoing standardization efforts by the Biomarker Core are aimed at minimizing sources of analytical variability and developing reference methods and standardized reference materials. Assessment of the NIA-AA criteria in the ADNI cohort provided support for their utility and also highlighted possible weaknesses in their classification scheme such as the categorization of patients as “undefined” or “uninformative”. The Biomarker Core has suggested improvements to these criteria to better stratify patients across the AD spectrum [55].
2.3. ADNI has been a model for data sharing without embargo
In recent years, the potential of big data that integrates clinical, scientific, and population level information for use in developing therapies for AD has been increasingly recognized. Databases such as the Global Alzheimer Association Interactive Network (www.gaain.org) seek to organize such information globally and the integration of disparate databases to leverage resources around the world holds much promise. However, when ADNI was established in 2004, the concept that data generated by the initiative would be shared openly and without embargo to all qualified researchers worldwide was a relatively new and radical one. Research data were generally considered to be owned by investigators who guarded it to avoid competition, the possibility of their results of not being duplicated, or from misuse by unqualified persons. The sharing of all data associated with an experiment allows the external duplication of findings and meta-analyses by combining data from multiple experiments, and new experiments to be performed using the same data [56]. The quantity of imaging, clinical, cognitive, biochemical, and genetic data generated throughout ADNI by geographically distributed investigators has required powerful informatics systems and mechanisms of processing, integrating, and disseminating these data. With these goals in mind, the Bioinformatics Core of ADNI, led by Dr Arthur Toga, developed a sophisticated informatics infrastructure based at to the LONI currently at the University of Southern California. This well-curated scientific data repository, owned collectively by ADNI rather than any participating entity, facilitates data integration, access, and sharing of data in a standardized manner with individuals with research credentials [57]. Also included in LONI are data generated by the Australian Imaging Biomarkers and Lifestyle (AIBL) Flagship Study of Ageing, and from new analyses by researchers accessing data.
ADNI is recognized by the medical research community as a leading example of how timely and extensive sharing of well-characterized data can promote further research, improve drug development, and therefore benefit public health [56]. As of July 15, 2014, there have been over 5.6 million downloads of image data, 322,940 downloads of clinical data, and 5867 downloads of genetic data by 3234 separate downloaders (personal communication, Dr Arthur Toga).
The ADNI database also serves as a model for other projects such as the Parkinson’s Progression Markers Initiative (PPMI) and, very recently, the North American Registry for Care and Research in Multiple Sclerosis (NARCRMS). PPMI aims to identify biomarkers for Parkinson’s Disease progression [58] and shares the LONI informatics data repository. NARCRMS, a database to collect MRI and other biomarker information data from patients with MS in the United States, is modeled specifically on ADNI’s database and will provide freely available data on MS patients to clinicians, patients, and pharmaceutical companies [59].
ADNI shared data have also been used in studies beyond the original project mandate, playing a critical role in identifying novel AD genetic risk factors, and contributing to research sometimes completely unrelated to AD for which data from a well-characterized cohort is desirable. These include investigations of stroke, hypertension, depression, and even mapping skull shape gradients in historical population movements [11].
In the mid-2013, whole-genome sequencing data for the entire ADNI cohort were added to the LONI database. Funded by the Alzheimer’s Association and the Brin Wojcicki Foundation, this project added around 165 terabytes of data to the repository and signaled the entry of ADNI into the world of big data. The full impact of this project has yet to be realized, but the combination of whole-genome sequences with existing longitudinal assessments of neuropsychological, imaging, and biological measures will allow investigators worldwide to discover new associations between rare genetic variants and these disease features and to develop novel targets for new disease-modifying or preventative therapies (http://alzforum.org/news/research-news/adni-full-genetic-sequences-now-available-download).
The sum of the ADNI data repository is now being leveraged in a computational challenge jointly run by the Global CEO Initiative for Alzheimer’s Disease, DREAM, and Sage Bionetworks. The Alzheimer’s Disease Big Data DREAM Challenge #1 (https://www.synapse.org/#!Synapse:syn2290704) challenges bioinformatics experts worldwide to predict the best biomarkers for early AD-related cognitive decline and for discordance between high amyloid levels and cognitive decline. Over 200 teams in both the public and private sector accepted the challenge, which are also using data provided by Rush University Medical Center, and the AddNeuroMed Study. The best-performing predictive models will be tested in a similar independent data set, with results expected in early 2015. In a sense, this challenge represents the ultimate in data-sharing in which “crowd-sourcing” of data analysis in a competitive manner is expected to greatly accelerate research in this area for the public good.
2.4. ADNI data have been used in over 600 publications
One measure of the impact of ADNI is more than 600 scientific publications (as of February 2015) that have used data generated by the initiative. ADNI Data and Publications Policy require authors to submit manuscripts using ADNI data to the Data and Publications Committee (DPC) for administrative review before submitting them for peer review and publication. We used lists provided by the DPC in addition to PubMed for searches of the terms “ADNI” and “Alzheimer’s Disease Neuroimaging Initiative” to generate the current list.
Around a third describe methods ranging from the standardization of methods for use in a multicenter setting, to improvements in neuroimaging techniques, to new approaches to classifying patients and predicting their likelihood of future decline, and to methods to improve genetic and statistical analyses. Around a quarter of papers describe disease progression and associations between ADNI measures; many articles relate imaging, genetic and CSF biomarkers, and cognitive measures. Approximately 15% of papers have primarily focused on improving clinical trial efficiency by selecting subpopulations more likely to progress within the time frame of a trial and by developing more sensitive outcome measures, both imaging and clinical. The ADNI data set has been used in another 15% of publications that have identified around 20 AD genetic risk factors beyond the APOE ε4 allele. A smaller number focus on cognitively normal participants, worldwide ADNI (WW-ADNI) and finally, the total includes a number of reviews and perspectives.
Ultimately, the most significant contributions of ADNI data to the scientific community can be distilled to a select group of high impact publications. We chose the following publications based on our assessment of novelty of the concept and the influence of the work on AD research, and were partially guided by number of times the article was cited and the impact rating of the journal of publication. The intent of this section is not to extensively review ADNI literature (this can be found in [8]), but rather to highlight some of the landmark findings of ADNI researchers. Table 1 summarizes significant ADNI findings.
Table 1.
Major findings using ADNI data
| Area of research | Major findings using ADNI data | References |
|---|---|---|
| Relationships between biomarkers | Biomarker “signature” for AD based on levels of Aβ42 and tau found in cognitively normal patients, suggesting AD pathology develops years before manifestation of clinical symptoms | [49,60] |
| Model for temporal ordering of biomarkers in AD pathogenesis largely supported. Biomarkers predicted to become abnormal in following order: CSF Aβ/amyloid PET > CSF tau/FDG-PET glucose metabolism > structural MRI Aβ deposition neuronal damage atrophy |
[9,61–65] | |
| CSF Aβ42, or amyloid PET associated with earlier stage neurodegeneration, but less with cognitive decline | [66–71] | |
| Abnormal tau associated with later-stage neurodegeneration, cognitive decline | [68,70,71] | |
| Abnormal glucose (fluorodeoxyglucose-PET) metabolism develops from parietal and temporal lobes in MCI to frontal and orbitofrontal lobes on AD, is associated with measures of cognitive decline. | [72–76] | |
| Hippocampal atrophy and ventricular expansion associated with decline in cognitive measures, rates of atrophy associated with rates of cognitive decline. | [60,66,69,77] | |
| Patterns of neurodegeneration in disease progression | Neurodegeneration generally occurs in following order: Temporal (hippocampus > entorhinal cortex/[lateral ventricle] >other)>parietal/posterior cingulate > frontal/occipital > anterior cingulate Early MCI late MCI early AD advanced AD |
[78–81] |
| Rate of neurodegeneration increases from cognitively normal to MCI to AD patients, with highest rates at each diagnostic stage in the specific areas outlined previously (e.g., hippocampus in MCI, frontal/occipital in late-AD) | [79,82–85] | |
| Development of summary scores to represent level of AD-like neurodegeneration: STructural Abnormality iNDex (STAND), Spatial Pattern of Abnormality for Recognition of Early Alzheimer’s disease (SPARE-AD). | [86,87] | |
| Neuropathological findings | High percentage of coincident pathologies, including dementia with Lewy bodies, medial temporal lobe pathology, vascular pathology, found in demented patients at autopsy. | [88] |
| Development of novel biomarkers | α-Synuclein strongly correlated with p-tau181, MMSE scores, patient status | [89,90] |
| Blood-based biomarkers show diagnostic potential | [91–94] | |
| White matter changes | Recognition of importance of white matter abnormalities in cognitive decline in AD, independent of amyloid deposition | [95–99] |
| Amyloid imaging | 11C-PiB-PET in agreement with CSF Aβ42, as measure of amyloid deposition | [100,101] |
| 18F-florbetapir PET in agreement with CSF Aβ42, as measure of amyloid deposition | [102,103] | |
| Diagnosis | Optimum diagnostic accuracy from selection of maximally discriminative multimodal features (typically longitudinal MRI, APOE and amyloid status, age) combined with dimensionality reduction: accuracies >95%, and >75% for CN versus AD and CN versus MCI, respectively. | [104–106] |
| Improvement of clinical trial efficiency | Best predictors of MCI to AD conversion combine maximally discriminative multimodal features (typically temporal lobe/entorhinal cortex/hippocampal MRI + t-tau/Aβ). PredictAD software combines modalities in weighted manner. Accuracies over 3 years up to 77%. | [107,108] |
| Lowest N80s with subject selection using baseline MRI atrophy, Aβ and t-tau, and MRI outcome measures (hippocampal or entorhinal cortex atrophy). For example, N80s for MCI (CN) for 24-month trial = 60 (499). | [109–111] | |
| Cognitive | Memory composite score, ADNI-Mem, predicted changes in neuroimaging parameters associated with memory changes. | [112] |
| ADAS-cog improved for increased sensitivity at earlier stages of clinical decline. | [113–115] | |
| Genetics and Genomics | APOE ε4 allele associated with faster hippocampal atrophy | [22,86,116–121] |
| APOE ε4 allele modulates amyloid deposition | [49,86,102,121,122] | |
| Discovery/replication of confirmed AD risk loci: CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, MS4A4E, CD2AP, and the identification of novel risk variants such as TREM2, SPON1. | [123–127] | |
| First uses of quantitative phenotypes in GWAS: CSF Aβ and tau, florbetapir amyloid PET, whole-brain ROIs, longitudinal hippocampal change, memory | [128–131] | |
| Novel approaches as copy number variation, gene pathway analysis, whole-exome sequencing, analysis of transcriptional networks, role of genetic variation in blood biomarker levels | [132–135] | |
| First voxel-wise and gene-wise GWAS, GWAS of structural connectome | [125,136] |
Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; Aβ, amyloid beta; APOE, apolipoprotein E; CSF, cerebrospinal fluid; PET, positron emission tomography; MRI, magnetic resonance imaging; AD, Alzheimer’s disease; MMSE, Mini-Mental State Examination; PiB, Pittsburgh Compound B; CN, cognitively normal; MCI, mild cognitive impairment; GWAS, genome-wide association studies; ROI, region of interest.
2.4.1. Establishing relationships between biomarkers, memory, and APOE genotype
Two early landmark papers examined the relationships between CSF biomarkers, hippocampal atrophy and memory, and the effect of the APOE ε4 allele on these measures. In cognitively normal healthy elderly subjects, Mormino et al. [60] found an inverse relationship between Aβ deposition (as measured by 11C-PiB uptake) and hippocampal volume; episodic memory loss was predicted by hippocampal volume, but not by 11C-PiB uptake. This study suggested that the accumulation of amyloid may reflect the early stages of AD pathogenesis and may subsequently mediate declines in episodic memory and therefore dementia through an effect on hippocampal volume. Likewise, hippocampal atrophy was associated with increased deposition of Aβ in MCI patients by Schuff et al. [66] who also reported that the APOE ε4 allele exacerbated hippocampal loss in AD patients. Together, these studies have been cited more than 500 times and provided evidence that led to the development of a model for how these crucial biomarkers change over the process of AD pathogenesis [61].
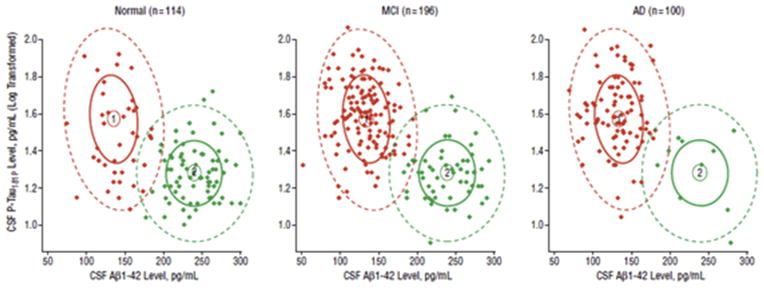
As AD biomarkers were being developed, it was suspected that patients could be cognitively normal but biomarker positive, thereby harboring an increased risk for developing the disease. The question of the level at which CSF biomarkers could be considered abnormal—the cut-point defining this change in risk—was therefore a pressing one. Shaw et al. [49] defined specific cut points for a CSF signature for AD based on an ADNI-independent cohort of autopsy-confirmed AD and cognitively normal patients. This AD signature, which combined low Aβ42 and high t-tau or p-tau181 concentrations, was then applied to the ADNI cohort. De Meyer et al. [137] focused their study of CSF biomarkers on cognitively normal elderly and formulated a CSF biomarker signature almost identical to that of Shaw et al.—for example, their Aβ42 cut-off was 188 pg/mL compared with 192 pg/mL in the former. Unexpectedly, a third of patients possessed the signature which suggested that AD pathology develops at a much earlier stage than previously envisioned (Fig. 2). This discovery would lead eventually to the finding that abnormal changes in some markers can be detected up to 10 years in advance of clinical symptoms and is in accordance with the more recent view of AD being a continuum of disease ending in dementia [138,139]. Aβ cut-offs are robust and show high agreement independently of the platform used to establish the presence of brain amyloid deposition (CSF or amyloid PET scans) or the pipelines and references used to calculate PET summary SUVRs, although biomarker dynamic ranges differ in the extremes of the normal and pathological range [140].
Fig. 2.
A cerebrospinal fluid (CSF) biomarker signature for Alzheimer’s disease (AD). Signature 1 (red) is AD, signature 2 (green) is the healthy signature. From De Meyer et al. [137].
The AD CSF biomarker signature has proved remarkably accurate in diagnosing AD, reaching a sensitivity of 90% to 95% and a specificity of around 90% [141]. Diagnostic accuracy has been further enhanced by the addition of other neuroimaging and clinical measures [11]. These cut point values have become widely accepted as the research standard with these two articles together cited more than 900 times.
2.4.2. A model for biomarker dynamics in AD pathogenesis
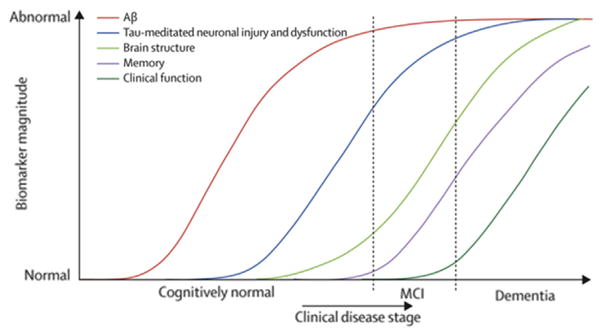
Perhaps the most influential of the ADNI articles was the work of Jack et al. [9] who presented a hypothetical model for biomarker dynamics in AD pathogenesis. The basic tenet of the model was that biomarkers become abnormal in a temporal order, beginning with markers of brain amyloid deposition (CSFAβ and amyloid PET), progressing to markers of neuronal damage (CSF-tau and FDG-PET), and ending with structural MRI which detects atrophy in certain areas typical of AD (Fig. 1). The model proposed that biomarkers become abnormal in a staged but overlapping manner and each follows a sigmoidal shape over time. Critical aspects of the model were based on prior work by the same group. After investigating the relationship between rates of amyloid deposition and ventricular expansion in the ADNI cohort by examining serial 11C-PiB PET and MRI scans [142] and examining relationships between the risk of progression from MCI to AD, and hippocampal atrophy and amyloid load [67], Jack et al. concluded that the deposition of Aβ is decoupled from cognitive decline, whereas neurodegeneration is closely associated with clinical symptoms of the disease. The deposition of Aβ into plaques was proposed to be necessary but not sufficient for clinical manifestation of the disease. Finally, the model suggested that the time frame of disease progression differed between individuals, and that differences in individual cognitive reserve and comorbid non-Alzheimer’s pathologies, in particular, could alter the lag between the appearance of abnormal biomarkers and cognitive decline.
Fig. 1.
A model for biomarker dynamics in Alzheimer’s disease (AD) pathogenesis. From Jack et al. [9].
The fundamental principles of this model have largely stood the test of time and accumulated evidence. The temporal ordering of biomarkers is now well-established and supported by numerous studies. Studies of presymptomatic patients largely support the order of the pathological changes proposed by this model, for example, presymptomatic cerebral amyloid is associated with increased neurodegeneration and may be a harbinger of cognitive decline [45,143,144]. Other studies have supported the acceleration of neurodegeneration from control to MCI to AD patients [78,82]. There is a strong evidence for the sigmoidal trajectory of amyloid biomarkers and some evidence that neurodegenerative biomarkers also follow the same pattern as they rise to abnormal levels, although the steepness of the curve appears to vary between biomarkers [61]. Results from several studies of ADNI biomarkers have diverged in part from the predictions of the model. Mouiha et al. [62] reported nonsigmoidal biomarker trajectories, the work of Yang et al. [145] suggested that Aβ levels may plateau after tau, Jedynak et al. [63] found that the Rey Auditory Verbal Learning Test-30 test of cognition was the first biomarker to become abnormal, and the longitudinal study of Han et al. [146] found that Aβ affected brain structure and function independent of tau, and that tau affected baseline cognition independent of neuroimaging measures. Further longitudinal studies of these preclinical subjects are required to determine whether biomarker trajectories predicted by the model are correct. An updated model by Jack et al. [61] retained the essential elements of the original, primarily adjusting only the horizontal axis from disease stage to years, recognizing the influence of cognitive reserve and other factors on the clinical stage of the disease while acknowledging that the time scale of this axis will vary in every individual. The original model has been cited more than 1200 times and has formed the basis for numerous studies that have substantially deepened our knowledge of AD pathophysiology. The revised model may well prove to have an equal or greater impact.
2.4.3. Diagnosis and prediction of future decline
Diagnostic classification and the prediction of future decline were not original goals of ADNI, but the initiative has generated a rich data set with which to explore new approaches to these challenges. Initially, cross-sectional information was targeted for both classification and prediction and more recently, longitudinal data have been used in the prediction of factors indicating clinical decline. In 2009, twin papers by Vemuri et al. first reported the use of combinations of MRI and CSF biomarkers for AD diagnosis [147] and the prediction of future clinical change [148] in the ADNI data set. The first article reported that although CSF biomarkers were not correlated with cognitive measures in any patient group, they acted to increase the diagnostic accuracy of MRI biomarkers. Likewise in the second article, CSF biomarkers augmented the ability of MRI biomarkers to predict subsequent cognitive decline. Currently cited by over 400 papers, these studies formed the basis for many subsequent diagnosis and prediction papers and ultimately lead to far more refined methods for selecting clinical trial populations likely to show measurable clinical decline within the length of the trial.
As methods were developed for the automatic classification of AD patients using anatomical MR data, the need arose for a standardized side-by-side comparison of different preprocessing strategies on classification accuracy. Cuingnet et al. [149] compared five voxel-based approaches, three cortical approaches, and two methods based on hippocampal shape and volume using ADNI data. This thorough study allowed researchers to directly compare methods that were originally published using different data sets and parameters, and consequently became an essential reference for developing automatic classification strategies.
The selection of AD-like features from imaging data enabled multivariate classification by reducing the “curse of dimensionality”. Likewise, the selection of features that are most AD-like across multiple modalities was a critical step in constructing an accurate classifier. Chen et al. [72] developed a FDG-PET based hypometabolic convergence index that was associated with the hazard for conversion to probable AD. In combination with hippocampal volume measurement, this selected MCI patients with an even higher likelihood of conversion. Zhang et al. [150] selected imaging (MRI and FDG-PET) regions of interest using a linear support vector machine and combined them with levels of CSF biomarkers according to the predefined cut points. This multimodal classifier was highly accurate and marked the beginning of a proliferation of ever more efficient methods that used the full breadth of ADNI data for AD diagnosis and to predict future decline. For instance, one article that quickly followed [104] combined a multitask feature selection with a multimodal support vector machine to integrate disparate imaging and biological data for the estimation of continuous variables such as scores neuropsychological tests. These approaches have produced accuracies in excess of 95% and 75% for the classification of AD and MCI patients, respectively, from cognitively normal controls [105,151]. Likewise, multimodal strategies which combine maximally discriminative multimodal features (typically temporal lobe/entorhinal cortex/hippocampal MRI and t-tau/Aβ) have predicted the conversion of MCI patients to AD within 3 years with accuracies up to 77% [107].
2.4.4. Improvements to clinical trial design
The recognition that emblematic AD disease pathology is present in a subset of cognitively normal patients [137,152], years ahead of any manifestation of clinical symptoms has led to a broadening of the cognitive spectrum of clinical trials of AD therapies to include early MCI and subjective complaint cohorts. The development of subject selection strategies and outcome measures which together reduce N80s to practicable sizes has therefore been an important focus of ADNI. Although several studies have shown Clinical Dementia Rating - sum of boxes (CDR-SB) to be a better outcome measure than ADAS-cog [109,153,154], others have focused on improving the commonly used later test to be more sensitive to cognitive changes earlier in the disease process [113–115]. APOE ε4 status, baseline MRI atrophy, and abnormal tau and Aβ42 have been used as successful stratification strategies [8]. Grill et al. [154] estimated N80s of 258 for MCI patients with enrichment using t-tau and Aβ42 and CDR-SB as an outcome measure. However, in a systematic study, Holland et al. [110] reported that the optimum combination of subject selection strategies and outcome measures was the selection of MCI patients with abnormal MRI, p-tau, and Aβ42, and the use of entorhinal cortex atrophy as an outcome measure. The estimated N80 using this combination was 60 (95% CI: 42 100) compared with 294 (204 456) using no subject selection, 234 (151 455) using CDR-SB as an outcome measure, and 583 (416 894) using no subject selection and CDR-SB as an outcome measure. In cognitively normal ADNI participants, Grill et al. [154] estimated an N80 of 499 (243 1659) using enrichment with APOE ε4 and the AVLT as an outcome measure. Their N80 estimates using other cognitive endpoints had prohibitively high end-points, suggesting that in order for clinical trials in presymptomatic cohorts to be feasible, a biomarker-based outcome measure should be considered.
2.4.5. Genetics and genomics
After a decade, ADNI has made contributions to AD genetics far beyond the original mandate of the initiative. Since the first ADNI genome-wide association study (GWAS) in 2009 [155], over 200 publications using ADNI data alone or in combination with other cohorts have been reported. Genetic variance accounts for approximately 30% of phenotypic variance in AD [156]. ADNI data have repeatedly confirmed the importance of the APOE ε4 allele as the largest genetic risk factor in AD [123], accounting for about 6% of this variance, and a number of ADNI studies have investigated the mechanisms by which the APOE ε4 allele increases AD susceptibility. These have shown that the APOE ε4 allele increases Aβ deposition [86,102], even in presymptomatic patients [122], and that it is associated with increased hippocampal atrophy [116–118,122].
The ADNI Genetics core has been instrumental in pioneering GWAS which leverage the rich array of quantitative phenotypes from multiple imaging and biomarker modalities available in the ADNI data set. Significantly, these have most recently moved toward longitudinal frameworks. ADNI data have also played a vital role in the search for the “missing heritability” of AD by comprising subsets of the very large data sets required to gain sufficient statistical power to identify novel risk variants in these meta-analytic case-control GWAS. Together, these uses of ADNI genetics data are leading to a deeper understanding of the biological pathways involved in disease trajectory and cognitive decline. Selected highlights of ADNI GWAS and related studies in MCI and AD patients are presented later.
In 2009, the publication of a GWAS of MRI hippocampal volume in AD [155] represented the first of many “firsts” for the ADNI Genetics Core; ADNI data was later used in a hippocampal volume analysis by the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium—analyzing over 30,000 people with MRI and GWAS—which discovered common variants that affect hippocampal volume. In the following 2 years, ADNI reported the first GWAS of CSF amyloid and tau markers [157], the first whole-brain ROI-based [128] and voxel-based GWAS [136], the first GWAS of longitudinal hippocampal MRI change [129] and one of the first studies of mitochondrial DNA variations in AD [158]. In 2012, ADNI studies were among the first to report copy number variation in AD or MCI patients [132], and gene pathway analyses of memory impairment in older adults [133]. In 2013, the first MRI study of the recently discovered TREM2 variant [124] reported that carriers of variants in the TREM2 gene showed faster atrophy than noncarriers, and the first GWAS of the healthy human structural connectome implicated the SPON1 gene [125]. ADNI investigators also reported the first whole-exome sequencing study in MCI that identified functional variants for the rate of change in hippocampal volume in MCI [134], and investigated the role of APOE genotype in early MCI [122].
ADNI genetics data continue to enhance the biological understanding of underlying disease mechanisms. Kim et al. [159] examined the influence of genetic variation on plasma protein levels in older adults using a multianalyte panel, and confirmed previously identified gene-protein associations for the interleukin-6 receptor, chemokine CC-4, angiotensin-converting enzyme, and angiotensinogen. In 2014, Ramanan et al. [130] performed the first GWAS of amyloid PET using ADNI florbetapir scans and reported that the APOE and BCHE genes were modulators of cerebral amyloid deposition together accounting for nearly 15% of the variance in amyloid deposition. Swaminathan et al. (2014) reported that the association between plasma Aβ and cortical amyloid deposition is modulated by APOE ε4 status.
Two landmark case-control GWAS of AD, published as companion reports in Nature Genetics [126,127], included the ADNI-1 data in their replication data sets. Hollingworth et al. [126] reported five novel risk variants for AD: ABCA7, MS4A6A/MS4A4E, EPHA1, CD33, and CD2AP, whereas Naj et al. [127] independently reported CD2AP, EPHA1, and CD33 in addition to confirming the previously identified risk variants, in CR1, CLU, BIN1, and PICALM. All variants identified in these reports have now been confirmed and make up a substantial proportion of the over 20 risk variants now identified for the disease [160]. The ADNI cohort was also included in studies of over 30,000 individuals with MRI scans by the ENIGMA and Cohorts for Heart and Aging Research in Genomic Epidemiology consortia ([161,162] Hibar et al. 2014, Nature [under revision]). These studies found common variants influencing hippocampal volume, brain volume, and numerous other subcortical volumes, measured from MRI; carriers and noncarriers of specific small nucleotide polymorphisms differed in hippocampal volume, on average, by an amount equivalent to about 3 years of normal aging. Rhinn et al. [135] used an integrative genomic approach based on the analysis of transcriptional networks in the human brain to identify candidate genes predicted to mediate transcriptional changes in carriers of the APOE ε4 allele. Two genes of interest that affect amyloid deposition and the age of onset in APOE ε4 allele carriers, FYN, and RNF2 19, were subsequently confirmed using a meta-analytic GWAS using ADNI data. Lambert et al. [163] performed a meta-analysis of 74,046 individuals including the ADNI cohort, and identified 11 new susceptibility loci for AD. ADNI also played a prominent role in the largest GWAS of human memory to date including the NIA Health and Retirement Study cohort plus ADNI, the Religious Orders Study and Memory and Aging Project cohort, and other samples (Ramanan et al., in press). This GWAS implicated the FASTKD2 gene for both episodic memory and hippocampal structure on MRI and nominated this gene as a potential neuroprotective target.
Numerous discovery, replication, and methods publications using ADNI genetics data continue to appear from groups around the world at an accelerating pace [131]. Overall, the articles outlined previously along with dozens of other reports using multidimensional phenotypes from several ADNI data sets have confirmed key findings in the genetics of AD and also identified a number of novel candidate genes warranting further investigation in independent cohorts.
2.4.6. ADNI review
The proliferation of articles published using ADNI data is undoubtedly a measure of the success of the initiative. However, these studies represent a sometimes overwhelming volume of information to the average researcher. The review of ADNI papers by Weiner et al. [10] and its update [11] summarized this research and enabled researchers to avoid the unnecessary duplication of efforts and to determine where future directions might lie.
2.5. ADNI is a model for similar neuroimaging projects around the world
ADNI has provided a model for neuroimaging initiatives worldwide run under the direction of the umbrella organization, Worldwide ADNI (WW-ADNI), sponsored by the Alzheimer’s Association. Programs using ADNI methods have been established in Japan, Australia, Argentina, Taiwan, China, Korea, Europe, and Italy [164] with the common goals of harmonizing protocols and results internationally and sharing standardized data across the international research community. It is hoped that WW-ADNI approaches will establish internationally recognized standards to identify and diagnose AD and document cognitive and physical changes throughout disease progression in diverse ethnic groups.
WW-ADNI initiatives share the use of established ADNI protocols for structural MRI, PET, and the collection of cognitive, blood, and genomic data but differ in cohort size and composition, and in the emphasis of some studies. Three international initiatives were established shortly after the North American ADNI. European ADNI (E-ADNI) began as a pilot study and has now expanded to a network of 50 sites across Europe with a particular focus on standardizing protocols for measuring hippocampal volume [165–167]. In conjunction with E-ADNI, the European Union funded the informatics infrastructure, neuGRID and its successor, neuGRID for You (N4U), which have been designed to be interoperable with the LONI data repository. Neuroimaging data from Australian ADNI, also known as the AIBL, established in 2006, is also available through LONI. AIBL is a long-term longitudinal investigation sharing many of the same goals as ADNI but with a particular emphasis on examining various health and lifestyle factors and their effect on cognitive decline [168]. AIBL data have resulted in over 80 publications including a recent work that described a panel of blood-based biomarkers able to accurately predict the conversion of MCI patients to AD [91]. Japan ADNI (J-ADNI) was established in 2007 enrolling 600 participants and using a research protocol designed to maximize compatibility with North American ADNI [169]. Conclusions reached from cognitive, structural MRI, FDG, and amyloid PET data from J-ADNI are largely in agreement with those from North American ADNI. However, J-ADNI has reported a rate of MCI to AD progression nearly double that observed in the North American initiative [164].
Since 2010, four additional initiatives have been established in Taiwan, Korea, China, and Argentina. These projects are in various initial stages of establishing infrastructure and enrolling participants and are modeled largely on the North American initiative. One significant difference in Korean ADNI is the focus on vascular risk factors for AD progression as Subcortical Vascular Dementia is more prevalent in Asian dementia patients [164].
Results from AIBL, E-ADNI, and J-ADNI prove that the ADNI model is highly effective and can be transposed to many settings around the world. It is expected that the initiatives in Korea, Taiwan, China, and Argentina should also make important contributions to painting a global picture of AD disease progression. WW-ADNI is the result of an unprecedented degree of international cooperation. The willingness of scientists worldwide to participate in open data sharing will play a key role in the identification and development of disease-modifying and preventive treatments for AD.
2.6. ADNI has inspired other projects to investigate AD risk factors
The development of ADNI infrastructure, methods, and data collection techniques has facilitated the establishment of additional projects investigating specific risk factors in different populations.
2.6.1. Department of Defense Alzheimer’s Disease Neuroimaging Initiative
Traumatic brain injury (TBI) and post-traumatic stress disorder (PTSD) are well-known risk factors for AD [170–173]. Military veterans in particular have elevated risks of both TBI and PTSD over the course of their service due to combat and other exposures. Funded by the Department of Defense (DOD), a new study termed DOD-ADNI is investigating whether TBI and/or PTSD in veterans increases the risk for AD and decreases cognitive reserve [174]. This longitudinal study uses ADNI methods to obtain baseline and 1-year measurements of AD pathophysiological markers, medial temporal brain atrophy, and cognitive function in three groups of veterans: those with a history of TBI (with or without PTSD), those with ongoing PTSD (without TBI), and control subjects comparable in age, sex, and education [174]. DOD-ADNI is being conducted across a number of established ADNI sites. A future study will examine the same questions in veterans with MCI and TBI/PTSD.
2.6.2. ADNI depression study
One of the most debilitating aspects of Late Life Depression (LLD) is the cognitive impairment suffered by up to 60% of individuals. Accelerated cognitive decline in LLD is likely the result of multiple factors including hypoperfusion, amyloid deposition, cortical atrophy, white matter signal hyperintensities, and genetic susceptibility. In the past, determining specific mechanisms contributing to cognitive impairment in LLD has been challenging due to the co-occurrence of neurodegenerative disease and methodological limitations related to small sample sizes. The ADNI Depression Study (ADNI-D) aims to clarify the degree to which these distinct mechanisms are associated with the accelerated rate of cognitive decline in LLD. This longitudinal study will use standardized ADNI methods and data-sharing protocols, enroll participants who meet the criteria for LLD or Major Depression at two established ADNI sites, and compare these participants to ADNI-2 control subjects.
2.7. ADNI has inspired other initiatives unrelated to AD
As an example of an extremely successful precompetitive public-private partnership in the neurosciences, ADNI has served as an impetus for a coordinated and focused process of biomarker development across multiple therapeutic areas. By proving the feasibility of a multisite study aimed at developing biomarkers to track disease pathophysiology for subsequent use in clinical trials, ADNI has inspired other initiatives focusing on different neurodegenerative diseases.
2.7.1. Parkinson’s Progressive Markers Initiative
The Parkinson’s Progressive Markers Initiative (PPMI) was launched in 2010 to identify biomarkers for Parkinson disease (PD) progression to improve the understanding of disease pathophysiology and to facilitate more efficient PD-modifying therapeutic trials [58]. This observational, international, multicenter study was based largely on ADNI, using a largely similar structure, organization, and funding as a public-private partnership initiated by the Michael J Fox Foundation for Parkinson’s Research. PPMI and ADNI share the same LONI Data Informatics core headed by Arthur Toga, and Fluid Biomarker core headed by John Trojanowski and Leslie Shaw. In addition, ADNI has contributed many of its standardized methods to PPMI, especially for the analysis of certain CSF biomarkers. Like ADNI, PPMI’s data and samples are freely available to qualified researchers. PPMI data are already being downloaded extensively with 192,458, 57,024, and 561 downloads of image, clinical, and genetic data, respectively, by 645 distinct downloaders as of July 2014 (Arthur Toga, personal communication). PPMI has quickly generated significant results with an initial biomarker article reporting the prognostic and diagnostic potential of CSF biomarkers in early stage PD [175].
2.7.2. Frontotemporal Lobar Degeneration Neuroimaging Initiative
ADNI infrastructure forms the basis of the recently established Frontotemporal Lobar Degeneration Neuroimaging Initiative, which aims to determine the optimum methods (MRI, FDG-PET, and biomarker measures) for following the progression of frontotemporal lobar degeneration. This longitudinal study hopes to identify brain regions in which changes in metabolism and structure occur in this common cause of dementia.
2.7.3. North American Registry for Care and Research in MS
ADNI is also the prototype for the NARCRMS, announced in May 2014 and slated to be launched in 2015. This public-private partnership aims to track disease progression in MS, identify new biomarkers, and compare therapeutic outcomes. Participating doctors will use standardized methods to collect and report information on their MS patients including biomarker levels, demographic and clinical data, and imaging test results. Like the ADNI database, the NARCRMS database will offer open access for patients, physicians, and industry [59].
2.7.4. Down Syndrome Biomarker Initiative
Another recent study structured largely on ADNI is the Down Syndrome Biomarker Initiative [176] which aims to investigate the link between Down Syndrome and AD. This 3-year pilot study is currently being run at UC San Diego under the auspices of the Alzheimer’s Disease Cooperative Study with pharmaceutical funding. Twelve participants are undergoing specialized cognitive testing, retinal amyloid imaging, brain PET amyloid imaging, structural MRI, and screening for promising blood biomarkers. It is hoped that this initial investigation, launched in March 2013, will pave the way for a much more extensive study using many of the hallmarks of ADNI structure and standardized methods.
3. Future directions
Future planning for the next decade of ADNI is currently focused on a competitive renewal of the ADNI-2 grant, termed ADNI-3. ADNI-3 would continue to improve clinical trial design by developing strategies for subject selection and validating more sensitive outcome measures. Accordingly, one major focus of ADNI-3 would be the development of fluid, imaging, and genetic biomarkers that effectively identify AD in its earliest stages. These may include biomarkers that reflect the heterogeneity of underlying pathologies evident in AD [88] such as total α-synuclein and phospho-α-synuclein to investigate the role of comorbidities in AD.
A second major focus of ADNI-3 would be the development of surrogate outcome measures. Numerous clinicopathological studies have established that the amount and distribution of tau tangles correlate with cognitive impairment and severity of dementia [177–181]. Several PET ligands have recently been developed that have reasonable sensitivity and specificity to detect tau tangles in the living human brain [182–189]. Preliminary reports with tau PET appear to confirm the view that the extent and location of tau correlates with severity of cognitive impairment [186,190,191]. This suggests that tau PET has the potential to become a “surrogate outcome measure” for AD clinical trials, which would greatly facilitate and accelerate all such trials. A large scale longitudinal observational study of tau PET would be the next step toward the development of a surrogate outcome measure, which could ultimately be approved by the FDA and other regulatory agencies. ADNI has been granted funding from the Department of Defense to conduct tau PET studies at baseline and after 1 year in DOD ADNI subjects in addition to a subset of cognitively normal, MCI, and AD ADNI-2 subjects.
If funded, ADNI-3 would run for 5 years (2016–2021). It would follow subjects currently enrolled in ADNI-2 and enroll additional cohorts with an emphasis on cognitively normal and MCI patients reflective of a change in focus to earlier stages of AD. Subjects would be studied using existing methods and novel additions such as computerized cognitive testing, analysis using advanced MRI techniques (including structural, perfusion, resting state functional magnetic resonance imaging, and diffusion tensor imaging), and tau-PET imaging.
Another promising direction for ADNI is its emerging collaboration with the Dominantly Inherited Alzheimer’s Disease Network (DIAN), which has a great potential for high impact results. ADNI and DIAN investigators have met and developed a plan for data exchange and analysis. It is hoped that this collaboration will lead to more information concerning the similarities and differences in biomarker changes between early onset dominantly inherited AD and late-onset AD.
4. Limitations of ADNI
One limitation of ADNI is that our population represents a primarily amnestic clinical population and not an epidemiologically selected real life population. Our subjects have limited comorbidities, as those with cortical strokes, heart failure, substance abuse, cancer, and other preexisting conditions are excluded from the study. Therefore, it remains to be determined how relevant ADNI findings are to the greater population. The use of ADNI methods in population-based studies such as the Mayo Clinic Study of Aging may help to address this question. A second limitation is the age range of ADNI participants (55–90 years), which may be too old to detect the earliest stages of disease in many subjects. The enrolment of a higher proportion of cognitively normal subjects in ADNI-3 than in ADNI-1 or ADNI-2 is proposed in part to address this issue. However, longitudinal studies of subjects beginning at a young age will be required to gain a full understanding of the pathophysiological sequence of events occurring in AD.
5. Conclusions
The original and continuing goal of ADNI has been to validate biomarkers for AD clinical trials. By all accounts ADNI has accomplished this goal, and helped to establish the critical diagnostic role of amyloid phenotyping. ADNI demonstrates the feasibility and impact of large scale data sharing without embargo and it now serves as the model for other programs wishing to openly share data. ADNI is a model of a successful public-private partnership and this structure combined with ADNI’s development of standardized protocols for use in multicenter settings has inspired other initiatives aimed at evaluating additional AD risk factors, and at developing biomarkers for other diseases. ADNI has also helped to establish a worldwide network of AD clinical trial sites. The economic impact of ADNI, although not quantified, is substantial. Research using ADNI data has generated over 600 publications in a decade and has significantly advanced our knowledge of the progression of AD pathology and of genetic risk factors for the disease. The recent piloting of tau imaging technologies augurs well for a second outstanding decade of innovation and progress.
RESEARCH IN CONTEXT.
Systematic review: The authors reviewed the literature using traditional sources (e.g., PubMed), accessed information from websites of relevant initiatives, which have not yet reached publication stage, and solicited data by personal communication.
Interpretation: Our findings indicate that the Alzheimer’s Disease Neuroimaging Initiative (ADNI) has had wide-ranging and profound impacts on many areas including basic research into Alzheimer’s disease (AD) and other diseases, clinical trials, and data sharing.
Future directions: Imaging studies using tau positron emission tomography (PET) ligands will bring a new dimension to clinicopathological studies of AD and may become a “surrogate outcome measure” for AD clinical trials. The extension of current longitudinal studies will continue to add to the body of data on AD progression. It is likely that ADNI will inspire further initiatives based on its private-public partnership funding structure and model for data sharing.
Acknowledgments
This work was supported by NIH grant 5U01AG024904-10 funded by the National Institute on Aging to Dr Michael Weiner.
Conflicts of Interest
Michael W. Weiner has served on the scientific advisory boards for Lilly, Araclon, and Institut Catala de Neurociencies Aplicades, Gulf War Veterans Illnesses Advisory Committee, VACO, Biogen Idec, and Pfizer; has served as a consultant for Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics LTD, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, SA, Servier, Synarc, Pfizer, and Janssen; has received funding for travel from NeuroVigil, Inc., CHRU-Hopital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California, San Diego–ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, CTAD (Clinical Trials on Alzheimer’s Disease), Pfizer, AD PD meeting, Paul Sabatier University, Novartis, Tohoku University; has served on the editorial advisory boards for Alzheimer’s & Dementia and MRI; has received honoraria from NeuroVigil, Inc., Insitut Catala de Neurociencies Aplicades, PMDA/Japanese Ministry of Health, Labour, and Welfare, and Tohoku University; has received commercial research support from Merck and Avid; has received government research support from DOD and VA; has stock options in Synarc and Elan; and declares the following organizations as contributors to the Foundation for NIH and thus to the NIA funded Alzheimer’s Disease Neuroimaging Initiative: Abbott, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Anonymous Foundation, AstraZeneca, Bayer Healthcare, BioClinica, Inc. (ADNI 2), Bristol-Myers Squibb, Cure Alzheimer’s Fund, Eisai, Elan, Gene Network Sciences, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck, Novartis, Pfizer Inc., Roche, Schering Plough, Synarc, and Wyeth.
Dallas P. Veitch has no conflicts of interest to report.
Paul S. Aisen has served as a consultant to NeuroPhage, Elan, Eisai, Bristol-Myers Squibb, Eli Lilly, Merck, Roche, Amgen, Genentech, Abbott, Pfizer, Novartis, AstraZeneca, Janssen, Medivation, Ichor, Toyama, Lundbeck, Biogen Idec, iPerian, Probiodrug, Somaxon, Biotie, Cardeus, Anavex, Abbvie, Cohbar.; and receives research support from Eli Lilly, Janssen and the NIH (NIA U01-AG10483 [PI], NIA U01-AG024904 [Coordinating Center Director], NIA R01-AG030048 [PI], and R01-AG16381 [Co-I]).
Laurel A. Beckett receives funding from the following NIH grants: 2P30CA093373-11 (deVere White, Ralph), 2P30AG 010129-23 (deCarli, Charles), 5U01AG024904-09 (Weiner), 5R01CA159447-02 (Miller, Lisa), and 5P30AG 043097-02 (Hinton, Ladson). In addition, she has received funding from the following California Breast Cancer Research Program grant: CBCRP # 16BB-1600 (von Friederichs-Fitzwater). She also has received funding from the nonprofit Critical Path Institute (Arizona) for consultation on analysis of potential biomarkers for Alzheimer’s disease clinical trials.
Nigel J. Cairns has no conflicts of interest to report.
Jesse Cedarbaum is an employee of Biogen Idec.
Michael C. Donohue has no conflicts of interest to report.
Robert C. Green has no conflicts of interest to report.
Danielle Harvey is Associate Editor for Statistics for the Journal of Alzheimer Disease and Associated Disorders, on the Statistical Advisory Board for PLoS ONE, and on the Advisory Board for Eye & Contact Lens. She receives funding from NIH (5P30AG010129-21, 2U01AG024904-06, 5R01HD042974-08, 1U54NS079202-01, 1U54HD079 125-01) and DOD (W81XWH-12-2-0012, W81XWH-13-1-0259).
Clifford R. Jack has provided consulting services for Janssen Research & Development, LLC, and Eli Lilly. He receives research funding from the National Institutes of Health (R01-AG011378, U01-HL096917, U01-AG024904, RO1 AG041851, R01 AG37551, R01AG043392, U01-AG06786), and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Foundation.
William Jagust has served as a consultant to Synarc, Genentech, Banner Alzheimer’s Institute, and Merck.
John Morris has participated or is currently participating in clinical trials of antidementia drugs sponsored by the following companies: Janssen Immunotherapy, Pfizer, Eli Lilly/Avid Radiopharmaceuticals, SNIFF (The Study of Nasal Insulin to Fight Forgetfulness) study, and A4 (The Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease) trial. Dr. Morris has served as a consultant for Lilly USA, ISIS Pharmaceuticals, and Charles Dana Foundation. He receives research support from Eli Lilly/Avid Radiopharmaceuticals and is funded by NIH grants #P50AG005681; P01AG003991; P01AG026276; and U19AG032438.
Ronald C. Petersen is the Chair of the Data Monitoring Committee for Pfizer, Inc. and Janssen Alzheimer Immunotherapy and serves as a consultant for Roche Inc., Merck Inc., and as a consultant and member of the adjudication committee for Genentech Inc.
Andrew J. Saykin has received support, in addition to ADNI, from NIA R01 AG19771 and P30 AG10133 and NLM R01 LM011360, and investigator initiated research support from Siemens Healthcare.
Leslie M. Shaw has provided quality control oversight for the use of the AlzBio3 (Fujirebio Europe) immunoassay in the ADNI study and serves as consultant to Eli Lilly and Janssen Research & Development.
Paul M. Thompson has received funding, in addition to ADNI, from the NIA, NIMH, NINDS, and from the NIH Big Data Centers of Excellence program (U54 EB 020403), which is funded by several NIH agencies including NIBIB and NCI.
Arthur W. Toga has no conflicts of interest to report.
John Q. Trojanowski may accrue revenue in the future as coinventor on Aβ amyloid imaging related patents submitted by the University of Pennsylvania and he received revenue from the sale of Avid to Eli Lilly as coinventor on Aβ amyloid imaging related patents submitted by the University of Pennsylvania.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Khachaturian ZS. Toward a comprehensive theory of Alzheimer’s disease—challenges, caveats, and parameters. Ann N Y Acad Sci. 2000;924:184–93. doi: 10.1111/j.1749-6632.2000.tb05577.x. [DOI] [PubMed] [Google Scholar]
- 3.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–8. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 4.Petersen RC, Waring SC, Smith GE, Tangalos EG, Thibodeau SN. Predictive value of APOE genotyping in incipient Alzheimer’s disease. Ann N Y Acad Sci. 1996;802:58–69. doi: 10.1111/j.1749-6632.1996.tb32599.x. [DOI] [PubMed] [Google Scholar]
- 5.Weiner MW, Aisen PS, Jack CR, Jr, Jagust WJ, Trojanowski JQ, Shaw L, et al. The Alzheimer’s Disease Neuroimaging Initiative: progress report and future plans. Alzheimers Dement. 2010;6:202–2117. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer’s Disease Neuroimaging Initiative. Neuroimaging Clin N Am. 2005;15:869–77. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–9. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ. 2014 update of The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement. 2015;11:e1–120. doi: 10.1016/j.jalz.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9:119–28. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8(1 Suppl):S1–68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9:e111–94. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roche F, Svhaerer J, Gouttard S, Istace A, Belaroussi B, Yu HJ, Bracoud L, Pachai C, De Carli C and the Alzheimer’s Neuroimaging Initiative. Accuracy of BMAS hippocampus segmentation using the harmonized hippocampal protocol. Alzheimer’s Association International Conference; Copenhagen, Denmark. 2014. [Google Scholar]
- 13.Bracoud L, Marek K, Cedarbaum J, Schaerer J, Mendick S, Berman F, et al. and the Alzheimer’s Disease Neuroimaging Initiative. Predictive value of baseline hippocampal volumes and brain amyloid burden on atrophy rates for predementia Alzheimer’s disease subjects with data from study CN156-018 and ADNI-1. Clinical Trials on Alzheimer’s Disease (CTAD) conference; San Diego, CA. 2013. [Google Scholar]
- 14.Bracoud L, Coric V, Roche F, Cedarbaum J, Gouttard S, Luo F, et al. Longitudinal volumetric changes in a predementia AD study of Avagacestat (CN156-018), as compared to ADNI-1. Clinical Trials on Alzheimer’s Disease (CTAD) conference; Philadelphia, PA. 2013. [Google Scholar]
- 15.Roche F, Singh J, Schaerer J, Belaroussi B, Gouttard S, Istace A, Yu HL, Fletcher E, Bracoud RL, Pachai C, Decarli C. Reproducibility of intracranial and hippocampal volume quantification at 1.5T and 3T MRI: application to ADNI I. Alzheimer’s Association International Conference; Washington, DC. 2013. [Google Scholar]
- 16.Klein G, Sampat M, Staewen D, Scott D, Landau S, Suhy J. A new look at FDG PET longitudinal analyses in Alzheimer’s studies using a Freesurfer native space method. The 12th international conference on Alzheimer’s and Parkinson’s diseases; Nice, France. 2015. [Google Scholar]
- 17.Klein G, Sampat M, Staewen D, Scott D, Suhy J. Comparative assessment of SUVR methods in amyloid cross-sectional and longitudinal studies. Miami Beach, FL: Human Amyloid Imaging; 2015. [Google Scholar]
- 18.Klein G, Sampat M, Staewen D, Scott D, Suhy J. Hoffman phantom acquisition and analysis methods for qualification of PET centers in Multicenter Neurology Clinical Trials. Clinical Trials on Alzheimer’s Disease (CTAD) conference; Philadelphia, PA. 2014. [Google Scholar]
- 19.Klein G, Landau S, Scott D, Sharoyan V, Koeppe, Suhy J. Scanner resolution effects on quantitative measurements of Pittsburgh compound B and florbetapir. Alzheimer’s Association International Conference; Boston, MA. 2013. [Google Scholar]
- 20.Yu P, Sun J, Wolz R, Stephenson D, Brewer J, Fox NC, et al. Operationalizing hippocampal volume as an enrichment biomarker for amnestic mild cognitive impairment trials: effect of algorithm, test-retest variability, and cut point on trial cost, duration, and sample size. Neurobiol Aging. 2014;35:808–18. doi: 10.1016/j.neurobiolaging.2013.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolz R, Aljabar P, Hajnal JV, Hammers A, Rueckert D. LEAP: learning embeddings for atlas propagation. Neuroimage. 2010;49:1316–25. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolz R, Heckemann RA, Aljabar P, Hajnal JV, Hammers A, Lotjonen J, et al. Measurement of hippocampal atrophy using 4D graph-cut segmentation: application to ADNI. Neuroimage. 2010;52:109–18. doi: 10.1016/j.neuroimage.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Wolz R, Schwarz AJ, Yu P, Cole PE, Rueckert D, Jack CR, Jr, et al. Robustness of automated hippocampal volumetry across magnetic resonance field strengths and repeat images. Alzheimers Dement. 2014;10:430–4382. doi: 10.1016/j.jalz.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Hill DL, Schwarz AJ, Isaac M, Pani L, Vamvakas S, Hemmings R, et al. Coalition Against Major Diseases/European Medicines Agency biomarker qualification of hippocampal volume for enrichment of clinical trials in predementia stages of Alzheimer’s disease. Alzheimers Dement. 2014;10:421–4293. doi: 10.1016/j.jalz.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Salloway S, Sperling R, Fox NC, Blennow K, Klunk W, Raskind M, et al. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer’s disease. N Engl J Med. 2014;370:322–33. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aisen PS, Andrieu S, Sampaio C, Carrillo M, Khachaturian ZS, Dubois B, et al. Report of the task force on designing clinical trials in early (predementia) AD. Neurology. 2011;76:280–6. doi: 10.1212/WNL.0b013e318207b1b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperling RA, Jack CR, Jr, Aisen PS. Testing the right target and right drug at the right stage. Sci Transl Med. 2011;3:111cm33. doi: 10.1126/scitranslmed.3002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozauer N, Katz R. Regulatory innovation and drug development for early-stage Alzheimer’s disease. N Engl J Med. 2013;368:1169–71. doi: 10.1056/NEJMp1302513. [DOI] [PubMed] [Google Scholar]
- 29.Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, et al. The A4 study: stopping AD before symptoms begin? Sci Transl Med. 2014;6:228fs13. doi: 10.1126/scitranslmed.3007941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6:221–9. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–40. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jack CR, Jr, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, et al. Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2010;6:212–20. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarkson MJ, Ourselin S, Nielsen C, Leung KK, Barnes J, Whitwell JL, et al. Comparison of phantom and registration scaling corrections using the ADNI cohort. Neuroimage. 2009;47:1506–13. doi: 10.1016/j.neuroimage.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DL, et al. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage. 2008;39:1752–62. doi: 10.1016/j.neuroimage.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunter JL, Bernstein MA, Borowski BJ, Ward CP, Britson PJ, Felmlee JP, et al. Measurement of MRI scanner performance with the ADNI phantom. Med Phys. 2009;36:2193–205. doi: 10.1118/1.3116776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruggel F, Turner J, Muftuler LT. Impact of scanner hardware and imaging protocol on image quality and compartment volume precision in the ADNI cohort. Neuroimage. 2010;49:2123–33. doi: 10.1016/j.neuroimage.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyman BT, Harvey DJ, Crawford K, Bernstein MA, Carmichael O, Cole PE, et al. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimers Dement. 2013;9:332–7. doi: 10.1016/j.jalz.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung KK, Barnes J, Ridgway GR, Bartlett JW, Clarkson MJ, Macdonald K, et al. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2010;51:1345–59. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schott JM, Bartlett JW, Barnes J, Leung KK, Ourselin S, Fox NC. Reduced sample sizes for atrophy outcomes in Alzheimer’s disease trials: baseline adjustment. Neurobiol Aging. 2010;31:1452–14622. doi: 10.1016/j.neurobiolaging.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox NC, Ridgway GR, Schott JM. Algorithms, atrophy and Alzheimer’s disease: cautionary tales for clinical trials. Neuroimage. 2011;57:15–8. doi: 10.1016/j.neuroimage.2011.01.077. [DOI] [PubMed] [Google Scholar]
- 42.Leung KK, Barnes J, Modat M, Ridgway GR, Bartlett JW, Fox NC, et al. Brain MAPS: an automated, accurate and robust brain extraction technique using a template library. Neuroimage. 2011;55:1091–108. doi: 10.1016/j.neuroimage.2010.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorge Cardoso M, Leung K, Modat M, Keihaninejad S, Cash D, Barnes J, et al. STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcellation. Med Image Anal. 2013;17:671–84. doi: 10.1016/j.media.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Carmichael O, Schwarz C, Drucker D, Fletcher E, Harvey D, Beckett L, et al. Longitudinal changes in white matter disease and cognition in the first year of the Alzheimer disease neuroimaging initiative. Arch Neurol. 2010;67:1370–8. doi: 10.1001/archneurol.2010.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes J, Carmichael OT, Leung KK, Schwarz C, Ridgway GR, Bartlett JW, et al. Vascular and Alzheimer’s disease markers independently predict brain atrophy rate in Alzheimer’s Disease Neuroimaging Initiative controls. Neurobiol Aging. 2013;34:1996–2002. doi: 10.1016/j.neurobiolaging.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haight TJ, Landau SM, Carmichael O, Schwarz C, DeCarli C, Jagust WJ, et al. Dissociable effects of Alzheimer disease and white matter hyperintensities on brain metabolism. JAMA Neurol. 2013;70:1039–45. doi: 10.1001/jamaneurol.2013.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nettiksimmons J, Beckett L, Schwarz C, Carmichael O, Fletcher E, Decarli C. Subgroup of ADNI normal controls characterized by atrophy and cognitive decline associated with vascular damage. Psychol Aging. 2013;28:191–201. doi: 10.1037/a0031063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nettiksimmons J, Decarli C, Landau S, Beckett L Alzheimer’s Disease Neuroimaging I. Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimers Dement. 2014;10:511–5211. doi: 10.1016/j.jalz.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–13. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw LM, Vanderstichele H, Knapik-Czajka M, Figurski M, Coart E, Blennow K, et al. Qualification of the analytical and clinical performance of CSF biomarker analyses in ADNI. Acta Neuropathol. 2011;121:597–609. doi: 10.1007/s00401-011-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang JH, Vanderstichele H, Trojanowski JQ, Shaw LM. Simultaneous analysis of cerebrospinal fluid biomarkers using microsphere-based xMAP multiplex technology for early detection of Alzheimer’s disease. Methods. 2012;56:484–93. doi: 10.1016/j.ymeth.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Korecka M, Waligorska T, Figurski M, Toledo JB, Arnold SE, Grossman M, et al. Qualification of a surrogate matrix-based absolute quantification method for amyloid-beta(4)(2) in human cerebrospinal fluid using 2D UPLC-tandem mass spectrometry. J Alzheimers Dis. 2014;41:441–51. doi: 10.3233/JAD-132489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo LH, Alexopoulos P, Eisele T, Wagenpfeil S, Kurz A, Perneczky R. The National Institute on Aging-Alzheimer’s Association research criteria for mild cognitive impairment due to Alzheimer’s disease: predicting the outcome. Eur Arch Psychiatry Clin Neurosci. 2012;263:325–33. doi: 10.1007/s00406-012-0349-0. [DOI] [PubMed] [Google Scholar]
- 54.Petersen RC, Aisen P, Boeve BF, Geda YE, Ivnik RJ, Knopman DS, et al. Criteria for mild cognitive impairment due to Alzheimer’s disease in the community. Ann Neurol. 2013;74:199–208. doi: 10.1002/ana.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lowe VJ, Peller PJ, Weigand SD, Montoya Quintero C, Tosakulwong N, Vemuri P, et al. Application of the National Institute on Aging-Alzheimer’s Association AD criteria to ADNI. Neurology. 2013;80:2130–7. doi: 10.1212/WNL.0b013e318295d6cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilhelm EE, Oster E, Shoulson I. Approaches and costs for sharing clinical research data. JAMA. 2014;311:1201–2. doi: 10.1001/jama.2014.850. [DOI] [PubMed] [Google Scholar]
- 57.Toga AW, Crawford KL. The informatics core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2010;6:247–56. doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parkinson Progression Marker I. The Parkinson Progression Marker Initiative (PPMI) Prog Neurobiol. 2011;95:629–35. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rammohan KW. Transformation of MS care in the 21st century. How NARCRMS will change the way we practice. 6th Cooperative Meeting of the Consortium of Multiple Sclerosis Centers (CMSC) and the Americas Committee for Treatment and Research In Multiple Sclerosis (ACTRIMS); 2014. [Google Scholar]
- 60.Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, et al. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132(Pt 5):1310–23. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jack CR, Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–16. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mouiha A, Duchesne S Alzheimer’s Disease Neuroimaging I. Toward a dynamic biomarker model in Alzheimer’s disease. J Alzheimers Dis. 2012;30:91–100. doi: 10.3233/JAD-2012-111367. [DOI] [PubMed] [Google Scholar]
- 63.Jedynak BM, Lang A, Liu B, Katz E, Zhang Y, Wyman BT, et al. A computational neurodegenerative disease progression score: method and results with the Alzheimer’s Disease Neuroimaging Initiative cohort. Neuroimage. 2012;63:1478–86. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jack CR, Jr, Vemuri P, Wiste HJ, Weigand SD, Aisen PS, Trojanowski JQ, et al. Evidence for ordering of Alzheimer disease biomarkers. Arch Neurol. 2011;68:1526–35. doi: 10.1001/archneurol.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donohue MC, Jacqmin-Gadda H, Le Goff M, Thomas RG, Raman R, Gamst AC, et al. Estimating long-term multivariate progression from short-term data. Alzheimers Dement. 2014;10(5 Suppl):S400–10. doi: 10.1016/j.jalz.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132(Pt 4):1067–77. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jack CR, Jr, Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010;133:3336–48. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stricker NH, Dodge HH, Dowling NM, Han SD, Erosheva EA, Jagust WJ, et al. CSF biomarker associations with change in hippocampal volume and precuneus thickness: implications for the Alzheimer’s pathological cascade. Brain Imaging Behav. 2012;6:599–609. doi: 10.1007/s11682-012-9171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, et al. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009;45:645–55. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Apostolova LG, Hwang KS, Andrawis JP, Green AE, Babakchanian S, Morra JH, et al. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol Aging. 2010;31:1284–303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beckett LA, Harvey DJ, Gamst A, Donohue M, Kornak J, Zhang H, et al. The Alzheimer’s Disease Neuroimaging Initiative: annual change in biomarkers and clinical outcomes. Alzheimers Dement. 2010;6:257–64. doi: 10.1016/j.jalz.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen K, Ayutyanont N, Langbaum JB, Fleisher AS, Reschke C, Lee W, et al. Characterizing Alzheimer’s disease using a hypometabolic convergence index. Neuroimage. 2011;56:52–60. doi: 10.1016/j.neuroimage.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen K, Langbaum JB, Fleisher AS, Ayutyanont N, Reschke C, Lee W, et al. Twelve-month metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment assessed using an empirically pre-defined statistical region-of-interest: findings from the Alzheimer’s Disease Neuroimaging Initiative. Neuroimage. 2010;51:654–64. doi: 10.1016/j.neuroimage.2010.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu X, Chen K, Yao L, Ayutyanont N, Langbaum JB, Fleisher A, et al. Assessing the reliability to detect cerebral hypometabolism in probable Alzheimer’s disease and amnestic mild cognitive impairment. J Neurosci Methods. 2010;192:277–85. doi: 10.1016/j.jneumeth.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Landau SM, Harvey D, Madison CM, Koeppe RA, Reiman EM, Foster NL, et al. Associations between cognitive, functional, and FDG-PET measures of decline in AD and MCI. Neurobiol Aging. 2011;32:1207–18. doi: 10.1016/j.neurobiolaging.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Habeck C, Risacher S, Lee GJ, Glymour MM, Mormino E, Mukherjee S, et al. Relationship between baseline brain metabolism measured using [(1)(8)F]FDG PET and memory and executive function in prodromal and early Alzheimer’s disease. Brain Imaging Behav. 2012;6:568–83. doi: 10.1007/s11682-012-9208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45(1 Suppl):S3–15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evans MC, Barnes J, Nielsen C, Kim LG, Clegg SL, Blair M, et al. Volume changes in Alzheimer’s disease and mild cognitive impairment: cognitive associations. Eur Radiol. 2010;20:674–82. doi: 10.1007/s00330-009-1581-5. [DOI] [PubMed] [Google Scholar]
- 79.Fennema-Notestine C, Hagler DJ, Jr, McEvoy LK, Fleisher AS, Wu EH, Karow DS, et al. Structural MRI biomarkers for preclinical and mild Alzheimer’s disease. Hum Brain Mapp. 2009;30:3238–53. doi: 10.1002/hbm.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schuff N, Tosun D, Insel PS, Chiang GC, Truran D, Aisen PS, et al. Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol Aging. 2012;33:845–55. doi: 10.1016/j.neurobiolaging.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karow DS, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, Jennings RG, Brewer JB, et al. Relative capability of MR imaging and FDG PET to depict changes associated with prodromal and early Alzheimer disease. Radiology. 2010;256:932–42. doi: 10.1148/radiol.10091402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leung KK, Bartlett JW, Barnes J, Manning EN, Ourselin S, Fox NC, et al. Cerebral atrophy in mild cognitive impairment and Alzheimer disease: rates and acceleration. Neurology. 2013;80:648–54. doi: 10.1212/WNL.0b013e318281ccd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDonald CR, Gharapetian L, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, Holland D, et al. Relationship between regional atrophy rates and cognitive decline in mild cognitive impairment. Neurobiol Aging. 2012;33:242–53. doi: 10.1016/j.neurobiolaging.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chou YY, Lepore N, Avedissian C, Madsen SK, Parikshak N, Hua X, et al. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. Neuroimage. 2009;46:394–410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chou YY, Lepore N, Saharan P, Madsen SK, Hua X, Jack CR, et al. Ventricular maps in 804 ADNI subjects: correlations with CSF biomarkers and clinical decline. Neurobiol Aging. 2010;31:1386–400. doi: 10.1016/j.neurobiolaging.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010;67:308–16. doi: 10.1002/ana.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davatzikos C, Xu F, An Y, Fan Y, Resnick SM. Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain. 2009;132(Pt 8):2026–35. doi: 10.1093/brain/awp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toledo JB, Cairns NJ, Da X, Chen K, Carter D, Fleisher A, et al. Clinical and multimodal biomarker correlates of ADNI neuropathological findings. Acta Neuropathol Commun. 2013;1:65. doi: 10.1186/2051-5960-1-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toledo JB, Korff A, Shaw LM, Trojanowski JQ, Zhang J. CSF alpha-synuclein improves diagnostic and prognostic performance of CSF tau and Abeta in Alzheimer’s disease. Acta Neuropathol. 2013;126:683–97. doi: 10.1007/s00401-013-1148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Korff A, Liu C, Ginghina C, Shi M, Zhang J Alzheimer’s Disease Neuroimaging I. alpha-Synuclein in cerebrospinal fluid of Alzheimer’s disease and mild cognitive impairment. J Alzheimers Dis. 2013;36:679–88. doi: 10.3233/JAD-130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doecke JD, Laws SM, Faux NG, Wilson W, Burnham SC, Lam CP, et al. Blood-based protein biomarkers for diagnosis of Alzheimer disease. Arch Neurol. 2012;69:1318–25. doi: 10.1001/archneurol.2012.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Bryant SE, Xiao G, Barber R, Huebinger R, Wilhelmsen K, Edwards M, et al. A blood-based screening tool for Alzheimer’s disease that spans serum and plasma: findings from TARC and ADNI. PLoS One. 2011;6:e28092. doi: 10.1371/journal.pone.0028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnstone D, Milward EA, Berretta R, Moscato P. Multivariate protein signatures of pre-clinical Alzheimer’s disease in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) plasma proteome dataset. PLoS One. 2012;7:e34341. doi: 10.1371/journal.pone.0034341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Llano DA, Devanarayan V, Simon AJ Alzheimer’s Disease Neuroimaging I. Evaluation of plasma proteomic data for Alzheimer disease state classification and for the prediction of progression from mild cognitive impairment to Alzheimer disease. Alzheimer Dis Assoc Disord. 2013;27:233–43. doi: 10.1097/WAD.0b013e31826d597a. [DOI] [PubMed] [Google Scholar]
- 95.Provenzano FA, Muraskin J, Tosto G, Narkhede A, Wasserman BT, Griffith EY, et al. White matter hyperintensities and cerebral amyloidosis: necessary and sufficient for clinical expression of Alzheimer disease? JAMA Neurol. 2013;70:455–61. doi: 10.1001/jamaneurol.2013.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guzman VA, Carmichael OT, Schwarz C, Tosto G, Zimmerman ME, Brickman AM, et al. White matter hyperintensities and amyloid are independently associated with entorhinal cortex volume among individuals with mild cognitive impairment. Alzheimers Dement. 2013;9:S124–31. doi: 10.1016/j.jalz.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lo RY, Jagust WJ Alzheimer’s Disease Neuroimaging I. Vascular burden and Alzheimer disease pathologic progression. Neurology. 2012;79:1349–55. doi: 10.1212/WNL.0b013e31826c1b9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nir T, Jahanshad N, Jack CR, Weiner MW, Toga AW, Thompson PM, et al. Small world network measures predict white matter degeneration in patients with early-stage mild cognitive impairment. Proc IEEE Int Symp Biomed Imaging; 2012; pp. 1405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, et al. Effectiveness of regional DTI measures in distinguishing Alzheimer’s disease, MCI, and normal aging. Neuroimage Clin. 2013;3:180–95. doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–9. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weigand SD, Vemuri P, Wiste HJ, Senjem ML, Pankratz VS, Aisen PS, et al. Transforming cerebrospinal fluid Abeta42 measures into calculated Pittsburgh Compound B units of brain Abeta amyloid. Alzheimers Dement. 2011;7:133–41. doi: 10.1016/j.jalz.2010.08.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murphy KR, Landau SM, Choudhury KR, Hostage CA, Shpanskaya KS, Sair HI, et al. Mapping the effects of ApoE4, age and cognitive status on 18F-florbetapir PET measured regional cortical patterns of beta-amyloid density and growth. Neuroimage. 2013;78:474–80. doi: 10.1016/j.neuroimage.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Landau SM, Breault C, Joshi AD, Pontecorvo M, Mathis CA, Jagust WJ, et al. Amyloid-beta imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nucl Med. 2013;54:70–7. doi: 10.2967/jnumed.112.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage. 2012;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Westman E, Muehlboeck JS, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. Neuroimage. 2012;62:229–38. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 106.Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-task feature selection for AD/MCI classification. Med Image Comput Comput Assist Interv. 2013;16(Pt 1):308–15. doi: 10.1007/978-3-642-40811-3_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trzepacz PT, Yu P, Sun J, Schuh K, Case M, Witte MM, et al. Comparison of neuroimaging modalities for the prediction of conversion from mild cognitive impairment to Alzheimer’s dementia. Neurobiol Aging. 2014;35:143–51. doi: 10.1016/j.neurobiolaging.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 108.Liu Y, Mattila J, Ruiz MA, Paajanen T, Koikkalainen J, van Gils M, et al. Predicting AD conversion: comparison between prodromal AD guidelines and computer assisted PredictAD tool. PLoS One. 2013;8:e55246. doi: 10.1371/journal.pone.0055246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Holland D, McEvoy LK, Desikan RS, Dale AM Alzheimer’s Disease Neuroimaging I. Enrichment and stratification for predementia Alzheimer disease clinical trials. PLoS One. 2012;7:e47739. doi: 10.1371/journal.pone.0047739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holland D, McEvoy LK, Dale AM. Unbiased comparison of sample size estimates from longitudinal structural measures in ADNI. Hum Brain Mapp. 2012;33:2586–602. doi: 10.1002/hbm.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hua X, Hibar DP, Ching CR, Boyle CP, Rajagopalan P, Gutman BA, et al. Unbiased tensor-based morphometry: improved robustness and sample size estimates for Alzheimer’s disease clinical trials. Neuroimage. 2013;66:648–61. doi: 10.1016/j.neuroimage.2012.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2012;6:502–16. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hobart J, Cano S, Posner H, Selnes O, Stern Y, Thomas R, et al. Putting the Alzheimer’s cognitive test to the test II: Rasch Measurement Theory. Alzheimers Dement. 2013;9(1 Suppl):S10–20. doi: 10.1016/j.jalz.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 114.Hobart J, Cano S, Posner H, Selnes O, Stern Y, Thomas R, et al. Putting the Alzheimer’s cognitive test to the test I: traditional psychometric methods. Alzheimers Dement. 2013;9(1 Suppl):S4–9. doi: 10.1016/j.jalz.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 115.Llano DA, Laforet G, Devanarayan V. Derivation of a new ADAS-cog composite using tree-based multivariate analysis: prediction of conversion from mild cognitive impairment to Alzheimer disease. Alzheimer Dis Assoc Disord. 2010;25:73–84. doi: 10.1097/WAD.0b013e3181f5b8d8. [DOI] [PubMed] [Google Scholar]
- 116.Spampinato MV, Rumboldt Z, Hosker RJ, Mintzer JE. Apolipoprotein E and gray matter volume loss in patients with mild cognitive impairment and Alzheimer disease. Radiology. 2011;258:843–52. doi: 10.1148/radiol.10100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, et al. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31:1401–18. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hostage CA, Roy Choudhury K, Doraiswamy PM, Petrella JR Alzheimer’s Disease Neuroimaging I. Dissecting the gene dose-effects of the APOE epsilon4 and epsilon2 alleles on hippocampal volumes in aging and Alzheimer’s disease. PLoS One. 2013;8:e54483. doi: 10.1371/journal.pone.0054483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008;43:458–69. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Caroli A, Frisoni GB. The dynamics of Alzheimer’s disease biomarkers in the Alzheimer’s Disease Neuroimaging Initiative cohort. Neurobiol Aging. 2010;31:1263–74. doi: 10.1016/j.neurobiolaging.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lo RY, Hubbard AE, Shaw LM, Trojanowski JQ, Petersen RC, Aisen PS, et al. Longitudinal change of biomarkers in cognitive decline. Arch Neurol. 2011;68:1257–66. doi: 10.1001/archneurol.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Risacher SL, Kim S, Shen L, Nho K, Foroud T, Green RC, et al. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Front Aging Neurosci. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Cedarbaum J, et al. Alzheimer’s Disease Neuroimaging Initiative. 2014 update of The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement. 2015;11:e1–120. doi: 10.1016/j.jalz.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rajagopalan P, Hibar DP, Thompson PM. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1565–7. doi: 10.1056/NEJMc1306509#SA3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, et al. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc Natl Acad Sci U S A. 2013;110:4768–73. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage. 2010;53:1051–63. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2011;16:1130–8. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19:351–7. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8:183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Swaminathan S, Huentelman MJ, Corneveaux JJ, Myers AJ, Faber KM, Foroud T, et al. Analysis of copy number variation in Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. PLoS One. 2012;7:e50640. doi: 10.1371/journal.pone.0050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6:634–48. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–7. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE epsilon4 effectors in Alzheimer’s disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- 136.Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS) Neuroimage. 2010;53:1160–74. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.De Meyer G, Shapiro F, Vanderstichele H, Vanmechelen E, Engelborghs S, De Deyn PP, et al. Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67:949–56. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Buchhave P, Minthon L, Zetterberg H, Wallin AK, Blennow K, Hansson O. Cerebrospinal fluid levels of beta-amyloid 1-42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch Gen Psychiatry. 2012;69:98–106. doi: 10.1001/archgenpsychiatry.2011.155. [DOI] [PubMed] [Google Scholar]
- 139.Randall C, Mosconi L, de Leon M, Glodzik L. Cerebrospinal fluid biomarkers of Alzheimer’s disease in healthy elderly. Front Biosci. 2013;18:1150–73. doi: 10.2741/4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Toledo J, Bjerke M, Da X, Landua S, Foster N, Jagust W, et al. CSF and florbetapir Aβ measures are not interchangeable across the spectrum of Alzheimer’s disease. JAMA Neurol. 2015;72:571–81. doi: 10.1001/jamaneurol.2014.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K, et al. Advancing research diagnostic criteria for Alzheimer’s disease: the IWG-2 criteria. Lancet Neurol. 2014;13:614–29. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 142.Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain. 2009;132(Pt 5):1355–65. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schott JM, Bartlett JW, Fox NC, Barnes J. Increased brain atrophy rates in cognitively normal older adults with low cerebrospinal fluid Abeta1-42. Ann Neurol. 2010;68:825–34. doi: 10.1002/ana.22315. [DOI] [PubMed] [Google Scholar]
- 144.Young AL, Oxtoby NP, Daga P, Cash DM, Fox NC, Ourselin S, et al. A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain. 2014;137(Pt 9):2564–77. doi: 10.1093/brain/awu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang X, Tan MZ, Qiu A. CSF and brain structural imaging markers of the Alzheimer’s pathological cascade. PLoS One. 2012;7:e47406. doi: 10.1371/journal.pone.0047406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Han SD, Gruhl J, Beckett L, Dodge HH, Stricker NH, Farias S, et al. Beta amyloid, tau, neuroimaging, and cognition: sequence modeling of biomarkers for Alzheimer’s disease. Brain Imaging Behav. 2012;6:610–20. doi: 10.1007/s11682-012-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009;73:287–93. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, et al. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2010;56:766–81. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 150.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;55:856–67. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification. Neuroimage. 2014;84:466–75. doi: 10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nettiksimmons J, Harvey D, Brewer J, Carmichael O, Decarli C, Jack CR, Jr, et al. Subtypes based on cerebrospinal fluid and magnetic resonance imaging markers in normal elderly predict cognitive decline. Neurobiol Aging. 2010;31:1419–28. doi: 10.1016/j.neurobiolaging.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cedarbaum JM, Jaros M, Hernandez C, Coley N, Andrieu S, Grundman M, et al. Rationale for use of the Clinical Dementia Rating Sum of Boxes as a primary outcome measure for Alzheimer’s disease clinical trials. Alzheimers Dement. 2013;9(1 Suppl):S45–55. doi: 10.1016/j.jalz.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 154.Grill JD, Di L, Lu PH, Lee C, Ringman J, Apostolova LG, et al. Estimating sample sizes for predementia Alzheimer’s trials based on the Alzheimer’s Disease Neuroimaging Initiative. Neurobiol Aging. 2013;34:62–72. doi: 10.1016/j.neurobiolaging.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ridge PG, Mukherjee S, Crane PK, Kauwe JS Alzheimer’s Disease Genetics C. Alzheimer’s disease: analyzing the missing heritability. PLoS One. 2013;8:e79771. doi: 10.1371/journal.pone.0079771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Han MR, Schellenberg GD, Wang LS. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lakatos A, Derbeneva O, Younes D, Keator D, Bakken T, Lvova M, et al. Association between mitochondrial DNA variations and Alzheimer’s disease in the ADNI cohort. Neurobiol Aging. 2010;31:1355–63. doi: 10.1016/j.neurobiolaging.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim S, Swaminathan S, Inlow M, Risacher SL, Nho K, Shen L, et al. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS One. 2013;8:e70269. doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–61. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–51. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carrillo MC, Bain LJ, Frisoni GB, Weiner MW. Worldwide Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2012;8:337–42. doi: 10.1016/j.jalz.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 165.Boccardi M, Bocchetta M, Ganzola R, Robitaille N, Redolfi A, Duchesne S, et al. Operationalizing protocol differences for EADC-ADNI manual hippocampal segmentation. Alzheimers Dement. 2015;11:184–94. doi: 10.1016/j.jalz.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 166.Frisoni GB. Alzheimer’s disease neuroimaging Initiative in Europe. Alzheimers Dement. 2010;6:280–5. doi: 10.1016/j.jalz.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 167.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement. 2011;7:171–4. doi: 10.1016/j.jalz.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 168.Ellis KA, Rowe CC, Villemagne VL, Martins RN, Masters CL, Salvado O, et al. Addressing population aging and Alzheimer’s disease through the Australian imaging biomarkers and lifestyle study: collaboration with the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2010;6:291–6. doi: 10.1016/j.jalz.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 169.Arai H, Okamura N, Furukawa K, Kudo Y. Geriatric medicine, Japanese Alzheimer’s disease neuroimaging initiative and biomarker development. Tohoku J Exp Med. 2010;221:87–95. doi: 10.1620/tjem.221.87. [DOI] [PubMed] [Google Scholar]
- 170.Yaffe K, Vittinghoff E, Lindquist K, Barnes D, Covinsky KE, Neylan T, et al. Posttraumatic stress disorder and risk of dementia among US veterans. Archives of general psychiatry. 2010;67:608–13. doi: 10.1001/archgenpsychiatry.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fleminger S, Oliver DL, Lovestone S, Rabe-Hesketh S, Giora A. Head injury as a risk factor for Alzheimer’s disease: the evidence 10 years on; a partial replication. Journal of neurology, neurosurgery, and psychiatry. 2003;74:857–62. doi: 10.1136/jnnp.74.7.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Qureshi SU, Kimbrell T, Pyne JM, Magruder KM, Hudson TJ, Petersen NJ, et al. Greater prevalence and incidence of dementia in older veterans with posttraumatic stress disorder. Journal of the American Geriatrics Society. 2010;58:1627–33. doi: 10.1111/j.1532-5415.2010.02977.x. [DOI] [PubMed] [Google Scholar]
- 173.Khachaturian AS, Khachaturian ZS. Military risk factors for Alzheimer’s dementia and neurodegenerative disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014;10(3 Suppl):S90–1. doi: 10.1016/j.jalz.2014.05.1085. [DOI] [PubMed] [Google Scholar]
- 174.Weiner MW, Veitch DP, Hayes J, Neylan T, Grafman J, Aisen PS, et al. Effects of traumatic brain injury and posttraumatic stress disorder on Alzheimer’s disease in veterans, using the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2014;10(3 Suppl):S226–35. doi: 10.1016/j.jalz.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kang JH, Irwin DJ, Chen-Plotkin AS, Siderowf A, Caspell C, Coffey CS, et al. Association of cerebrospinal fluid beta-amyloid 1-42, T-tau, P-tau181, and alpha-synuclein levels with clinical features of drug-naive patients with early Parkinson disease. JAMA Neurol. 2013;70:1277–87. doi: 10.1001/jamaneurol.2013.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ness S, Rafii M, Aisen P, Krams M, Silverman W, Manji H. Down’s syndrome and Alzheimer’s disease: towards secondary prevention. Nat Rev Drug Discov. 2012;11:655–6. doi: 10.1038/nrd3822. [DOI] [PubMed] [Google Scholar]
- 177.Arriagada PV, Marzloff K, Hyman BT. Distribution of Alzheimer-type pathologic changes in nondemented elderly individuals matches the pattern in Alzheimer’s disease. Neurology. 1992;42:1681–8. doi: 10.1212/wnl.42.9.1681. [DOI] [PubMed] [Google Scholar]
- 178.Bierer LM, Hof PR, Purohit DP, Carlin L, Schmeidler J, Davis KL, et al. Neocortical neurofibrillary tangles correlate with dementia severity in Alzheimer’s disease. Arch Neurol. 1995;52:81–8. doi: 10.1001/archneur.1995.00540250089017. [DOI] [PubMed] [Google Scholar]
- 179.Gomez-Isla T, Hollister R, West H, Mui S, Growdon JH, Petersen RC, et al. Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer’s disease. Ann Neurol. 1997;41:17–24. doi: 10.1002/ana.410410106. [DOI] [PubMed] [Google Scholar]
- 180.Giannakopoulos P, Herrmann FR, Bussiere T, Bouras C, Kovari E, Perl DP, et al. Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology. 2003;60:1495–500. doi: 10.1212/01.wnl.0000063311.58879.01. [DOI] [PubMed] [Google Scholar]
- 181.Ingelsson M, Fukumoto H, Newell KL, Growdon JH, Hedley-Whyte ET, Frosch MP, et al. Early Abeta accumulation and progressive synaptic loss, gliosis, and tangle formation in AD brain. Neurology. 2004;62:925–31. doi: 10.1212/01.wnl.0000115115.98960.37. [DOI] [PubMed] [Google Scholar]
- 182.Villemagne VL, Okamura N. In vivo tau imaging: obstacles and progress. Alzheimers Dement. 2014;10(3 Suppl):S254–64. doi: 10.1016/j.jalz.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 183.Shin J, Lee SY, Kim SH, Kim YB, Cho SJ. Multitracer PET imaging of amyloid plaques and neurofibrillary tangles in Alzheimer’s disease. Neuroimage. 2008;43:236–44. doi: 10.1016/j.neuroimage.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 184.Fodero-Tavoletti MT, Furumoto S, Taylor L, McLean CA, Mulligan RS, Birchall I, et al. Assessing THK523 selectivity for tau deposits in Alzheimer’s disease and non-Alzheimer’s disease tauopathies. Alzheimers Res Ther. 2014;6:11. doi: 10.1186/alzrt240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Okamura N, Furumoto S, Harada R, Tago T, Yoshikawa T, Fodero-Tavoletti M, et al. Novel 18F-labeled arylquinoline derivatives for noninvasive imaging of tau pathology in Alzheimer disease. J Nucl Med. 2013;54:1420–7. doi: 10.2967/jnumed.112.117341. [DOI] [PubMed] [Google Scholar]
- 186.Okamura N, Furumoto S, Fodero-Tavoletti MT, Mulligan RS, Harada R, Yates P, et al. Non-invasive assessment of Alzheimer’s disease neurofibrillary pathology using 18F-THK5105 PET. Brain. 2014;137(Pt 6):1762–71. doi: 10.1093/brain/awu064. [DOI] [PubMed] [Google Scholar]
- 187.Chien DT, Bahri S, Szardenings AK, Walsh JC, Mu F, Su MY, et al. Early clinical PET imaging results with the novel PHF-tau radioligand [F-18]-T807. J Alzheimers Dis. 2013;34:457–68. doi: 10.3233/JAD-122059. [DOI] [PubMed] [Google Scholar]
- 188.Chien DT, Szardenings AK, Bahri S, Walsh JC, Mu F, Xia C, et al. Early clinical PET imaging results with the novel PHF-tau radioligand [F18]-T808. J Alzheimers Dis. 2014;38:171–84. doi: 10.3233/JAD-130098. [DOI] [PubMed] [Google Scholar]
- 189.Maruyama M, Shimada H, Suhara T, Shinotoh H, Ji B, Maeda J, et al. Imaging of tau pathology in a tauopathy mouse model and in Alzheimer patients compared to normal controls. Neuron. 2013;79:1094–108. doi: 10.1016/j.neuron.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Villemagne VL, Furumoto S, Fodero-Tavoletti MT, Mulligan RS, Hodges J, Harada R, et al. In vivo evaluation of a novel tau imaging tracer for Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2014;41:816–26. doi: 10.1007/s00259-013-2681-7. [DOI] [PubMed] [Google Scholar]
- 191.Mintun M, Schwarz A, Joshi A, Shcherbinin S, Chien D, Elizarov A, et al. Exploratory analyses of regional human brain distribution of the PET tau tracer F18-labeled T807 (AV-1541) in subjects with normal cognitive function or cognitive impairment thought to be due to AD. Alzheimers Dement. 2013;9:P842. [Google Scholar]