Abstract
The aim of this work was to compare the effects of electrosynthesis on different bacterial species. The effects of neutral red-mediated electrosynthesis on the metabolite profiles of three microorganisms: Escherichia coli, Klebsiella pneumoniae, and Zymomonas mobilis, were measured and compared and contrasted. A statistically comprehensive analysis of neutral red-mediated electrosynthesis is presented using the analysis of end-product profiles, current delivered, and changes in cellular protein expression. K. pneumoniae displayed the most dramatic response to electrosynthesis of the three bacteria, producing 93% more ethanol and 76% more lactate vs. control fermentation with no neutral red and no electron delivery. Z. mobilis showed no response to electrosynthesis except elevated acetate titers. Stoichiometric comparison showed that NAD+ reduction by neutral red could not account for changes in metabolites during electrosynthesis. Neutral red-mediated electrosynthesis was shown to have multifarious effects on the three species.
Keywords: Neutral red, Electron transfer, Bioelectrochemical system, Electrosynthesis
1. Introduction
The delivery or removal of electrons during fermentation can alter the terminal metabolite profiles of microorganisms fermenting a reduced substrate. This process, termed “electrosynthesis” or “electro-fermentation” (Rabaey and Rozendal, 2010), involves the mediated transfer of electrons between electrodes and cells in order to shift the terminal metabolite profile toward more reduced or oxidized end-products by delivering or removing electrons, respectively. Microbial electrosynthesis could provide a marked benefit vs. traditional fermentation if the electrons were able to reduce NAD(P)+ to NAD(P)H, because yields in many industrial fermentations are limited by the ratio of energy carriers in the cytoplasm (Leonardo et al., 1996). If these mediated cathodic electrons could be made available to the bacterial metabolism to do useful work, the result would be a shift toward more valuable reduced metabolic end products like ethanol or 1,3-propanediol, at the expense of less valuable, more oxidized products like acetate.
A number of reports have used reduced redox dyes or quinone analogues as the electron mediators in microbial electrosynthesis with the goal of higher yields of reduced fermentation products or the reduction of environmental contaminants. Among the mediators employed, which include benzyl and methyl viologen (Peguin and Soucaille, 1996; Rao and Mutharasan, 1986, 1987), anthraquinone disulfate (AQDS) (Clark et al., 2012; Emde and Schink, 1990; Sasaki et al., 2013), and neutral red (Girbal et al., 2006; Hongo and Iwahara, 1979; Park and Zeikus, 1999; Park et al., 2005), the latter has shown promise because it does not form a radical, undergoes two-electron reduction and oxidation (Azariah et al., 1998), binds to the cell membrane and is less toxic (Park and Zeikus, 1999), and has a standard reduction potential below that of the NADH/NAD+ couple (−525 mVAg/AgCl vs. −520 mVAg/AgCl) (McKinlay and Zeikus, 2004). It has been known for over 100 years that certain bacteria biochemically reduce neutral red anaerobically. This “neutral red reaction” was briefly used to identify E. coli contamination in water sources (Makgill, 1901) before it was shown to be non-specific with the identification of several genera that are capable of reducing neutral red (Gage and Phelps, 1902). It was also shown that this was related to the ability of a bacterium to attain low reduction potentials near the hydrogen electrode (Clark and Perkins, 1932). This reaction has been more recently applied to microbial fuel cells, where neutral red was employed as a mediator to transfer electrons from E. coli to electrodes (Park and Zeikus, 2000).
The reversal of this process, where reduced neutral red is reduced at a cathode (Eo = −525 mVAg/AgCl) to influence the metabolism of bacteria during fermentation, was first reported by Hongo and Iwahara (1979), with the enhancement of l-glutamic acid formation in a process they called “electro-energizing”. Since then, cathodically reduced neutral red has been shown to drive fumarate reduction in Actinobacillus succinogenes (Park and Zeikus, 1999), act as the sole electron donor for growth and metabolite formation (Park et al., 1999), generate a proton motive force in combination with a proton-translocating fumarate reductase (Park and Zeikus, 1999), chemically reduce NAD+ (Park and Zeikus, 1999), enhance ethanol production in Clostridium thermocellum and Saccharomyces cerevisiae (Shin et al., 2002), enhance reduced pathway flux in Weissella kimchii (Park et al., 2005), modify carbon flow in Clostridium acetobutylicum (Girbal et al., 2006), and when immobilized on a graphite electrode, enhance ethanol formation in Zymomonas mobilis (Jeon et al., 2009; Jeon and Park, 2010) and Ralstonia Eutropha (Jeon et al., 2013). There is confusion from some of these reports where high voltage drops were involved, which may have produced hydrogen via electrolysis in addition to reducing neutral red. Some of these voltage drops include: 2 V (Jeon et al., 2012), 3 V (Jeon et al., 2009, 2013), and 1-10 V variable (Jeon and Park, 2010; Shin et al., 2002). Very few studies employed the use of a potentiostat to monitor and control working electrode potential while simultaneously quantifying charge transfer in order to compare electron delivery with metabolite production, with at least one recent exception (Choi et al., 2014). Additionally, where electrolysis occurred in these systems, there may have been induced local temperature effects from Joule heating. The confusion in the literature and the lack of a standard, controlled process for neutral red-mediated microbial electrosynthesis has made it difficult to interpret its effects and the mechanisms involved.
Assuming that the prevailing explanation for the effects of neutral red-mediated electrosynthesis – that neutral red reduces NAD+ (Park and Zeikus, 1999) – was true, the hypothesis for this work was that microbial electrosynthesis with neutral red would alter all fermentative metabolisms towards more reduced products, in turn indicating where those products were limited by NADH availability. However, since metabolism is specific to every individual organism, with different bottlenecks, regulatory mechanisms, and redox balances, it was expected that the effect of neutral red cathodic electrosynthesis would affect specific bacteria in distinct ways. The goal was to test the effects of neutral red-mediated microbial electrosynthesis under well-controlled conditions by using a potentiostat to compare current delivery to fermentation end-product titers. Electrosynthesis conducted in bioelectrochemical reactors were compared with control fermentations (without electrodes or neutral red) using three different bacteria with different metabolisms: Escherichia coli, which uses branched pathways to convert d-glucose to pyruvate and then to mixed acids (Lee et al., 2005), Klebsiella pneumoniae, which uses different branched pathways to convert glycerol to several products including the industrially important 1,3-propanediol (Huang et al., 2002), and Zymomonas mobilis, a highly efficient ethanol producer, which converts d-glucose to ethanol in a near-completely balanced linear pathway with only minor alternate metabolite products (Jeffries, 2005). A comprehensive statistical analysis of the effect of neutral red-mediated microbial electrosynthesis is presented, and suggests a mechanism of action in bacteria.
2. Methods
2.1. Bacterial strains
Escherichia coli K-12 (ATCC 10,798), Klebsiella pneumoniae NCTC 418 (ATCC 15,380), and Zymomonas mobilis AX (PTA-1797) were used to perform fermentations.
2.2. Fermentation media
All chemicals were purchased from Fisher unless otherwise noted. E. coli was grown in media composed of the following, L−1: M-9 salts 5×, 200 mL; MgSO4 (1 M), 2 mL; CaCl2 (1M), 100 μL; yeast extract, 5 g; D-glucose, 20 g (111 mM), pH 7.0. K pneumoniae was grown in a media composed of the following, L−1: CaCl2, 0.2 g; MgSO4·7H2O, 0.4 g; K2HPO4, 1 g; KH2PO4, 1 g; yeast extract, 5 g; peptone, 10 g; glycerol, 20 g (217 mM), pH 7.0. Z. mobilis was grown in a media composed of the following, L−1: KH2PO4, 2 g; MgSO4·7H2O, 0.5 g; (NH4)2SO4, 5 g; yeast extract, 10 g; D-glucose, 50 g (278 mM), pH 6.0. Electrosynthesis media additionally included 200 μM neutral red (CAS: 553-24-2, Sigma) to serve as a redox mediator to deliver electrons to growing cells, which was added separately from a 20× stock solution after sterile-filtering both the media and neutral red solution.
2.3. Bioelectrochemical reactors (BERs)
Adams and Chittenden 100 mL microbial fuel cells were employed as bioelectrochemical reactors. VICI-JOUR bottle caps (JR-S-11003) and bulkhead fittings (Eldon James – BH-1428-2-2NK) provided ports for nitrogen gas sparging. Side arms allowed for media sampling via syringe. One of the two chambers was filled with 100 mL of medium. Control fermentations included a rubber separator between the two chambers to isolate a single 100 mL chamber, contained no electrodes, and were not connected to a potentiostat. Electrosynthesis reactors included a cation exchange membrane (Membranes International – CMI-7000) to separate the working electrode (fermentation) chamber from the counter electrode chamber, which was filled with 100 mM sodium phosphate buffer of similar pH to the medium to prevent any initial pH gradient across the membrane. A graphite felt coupon (HP Materials Solutions, Inc., diameter = 2.2 cm, thickness = 0.6 cm) connected to a titanium wire (Keego Technologies, LLC) was submerged into the fermentation media to act as the working electrode during electrosynthesis, while a graphite rod counter electrode (Sigma – 496,553) and Ag/AgCl reference electrode were submerged in the counter electrolyte. The working electrode was polarized to –650 mVAg/AgCl overnight prior to inoculation, through the end of the experiment to ensure near-complete and continuous reduction of the neutral red. Cells were only inoculated to electrosynthesis reactors that were drawing a steady-state background current of between −50 and 0 μA, which indicated that neutral red reduction was complete. The initial reductive current when the potentiostat was connected, oxygen was removed, and neutral red was present was consistently between −1.6 and −1.9 mA, whereas no neutral red control experiments showed only background current when the working electrode potential was controlled at −650 mVAg/AgCl (Harrington et al., 2015 – accompanying manuscript). This indicated that there were no electron mediators present in the background media (once oxygen was removed) in sufficient quantity to deliver electricity from the cathode to the planktonic cells. Pre-inoculation cyclic voltammograms were smooth, displaying no significant peaks or waves. For these reasons, controls where a polarized electrode was included during fermentation without the addition of neutral red were excluded in favor of no electrode, no neutral red fermentations. Completely reduced media exhibited a yellow color with a green fluorescence (Makgill, 1901) at the surface prior to inoculation as well as during the course of fermentation. Current delivered to the fermentation media was monitored by a custom-built potentiostat (Renslow et al., 2011), where negative current indicates delivery of electrons to the fermentation media (cathodic current). Starter cultures (50 mL, neutral red-free media) were grown anaerobically to OD650 = 0.55–0.6, and 2.5 mL from the starter cultures were inoculated to each reactor at t = 0 h. Reactors were operated in incubators set to 37, 37, and 30 °C for E. coli, K. pneumoniae, and Z mobilis fermentations, respectively. Samples were taken at regular intervals throughout the course of the batch electrosynthesis and control fermentations. Nitrogen gas was continuously sparged through the fermentation media chambers. Stir bars were controlled at 150 rpm.
2.4. Analytical techniques
Fermentation samples (0.7 mL) were assayed for absorbance (OD650) to track cell growth using an Agilent 8453 UV–Vis spectrophotometer. 650 nm was chosen because it is outside the absorption spectrum window of neutral red. After assaying cell growth, samples were centrifuged, and the supernatants were analyzed for metabolite concentrations with an Agilent 1100 HPLC equipped with a diode array and refractive index detector.
Current delivery profiles were integrated after using the Interpolation function in Mathematica with “InterpolationOrde r” = 7 to calculate total charge transferred to the growing cells during the course of the fermentations. Averages were calculated from four replicate electrosyntheses and converted to the equivalent of μM of electron pairs to enable a simple comparison to the terminal metabolites.
2.5. Protein extraction
Samples were collected during the mid-to-late-exponential phase of growth (3 × 1 mL samples taken at six hours post-inoculation, four hours post-inoculation, and 10 h post-inoculation for E. coli, K. pneumoniae, and Z mobilis, respectively) from each of four separate fermentations and four separate electrosyntheses for each of the three species of bacterium (three technical replicates of four biological replicates per organism, per condition).
Protein extraction was adapted from published protocols (Wu et al., 2014). First, (1 mL) frozen fermentation samples were sonicated in an ice water bath for 30–45 s. Each sample was extracted using a series of 1% SDS, Phenol-HCl and centrifuged at 16,000×g for 5 min at 4 °C. Each pellet was precipitated overnight at 4 °C by adding 0.1 M ammonium acetate in methanol at a ratio of 5:1 methanol/phenol mixed thoroughly. The resulting pellet was then washed with 80% acetone in methanol and centrifuged at 16,000×g for 5 min at 4 °C. Finally, the pellet was washed with 70% methanol and pelleted at 16,000×g for 5 min at 4 °C. After air-drying the samples to remove residual methanol, protein was resuspended in 50 mM ammonium bicarbonate, pH 7.5. Protein was then quantified with a Qubit Protein Assay Kit (Life Technologies, Carlsbad, CA) in compliance with the manufacturer's protocol. Disulfide bonds were reduced using 100 mM dithiothreitol (DTT) at a ratio of 1:10 DTT/sample volume and incubated at 60 °C for 30 min. Cysteine bonds were then alkylated with 300 mM iodoacetamide at the same volume ratio for 30 min at room temperature. Finally, protein was digested with trypsin (Promega V511A) at a 1:50 ratio of trypsin/protein, and incubated at 37 °C for 8 h.
2.6. LC-MS/MS parameters and processing
Nano ultra performance liquid chromatography-ion mobility time of flight tandem mass spectrometry (UPLC-IMS-TOFMS/MS) was used to evaluate the relative abundance of protein within each sample. Chromatographic separation was achieved by injecting 50 or 100 ng of protein into the Nanoacquity (Waters, Milford MA) reverse phase C18 column at a flow rate of 0.45 μL min−1 with a linear gradient over 85 min of 5% ACN to 95% ACN in H2O with 0.1% formic acid. The Nanoacquity was in line with the Synapt G2S (Waters, Milford MA) equipped with a nano-electrospray ionization (nESI) source in positive mode. High energy MS/MS function included a transfer collision energy ramp from 15 to 50 eV. The lock mass Leu-enkephaline was acquired every 30 s. Comprehensive databases for peptide identification were constructed using UniProtKB, Swissprot-Prot, and TrEMBL for E. coli, K. pneumoniae, and Z mobilis (The UniProt Consortium, 2012). Data processing was performed using TransOmics V1.1 (Waters, Milford MA). Data filtering required that peaks appear in at least two of three biological replicates, i.e., 66.6% consistency and that only proteins with >2 unique peptides be counted. Peak intensities were normalized by total ion chromatogram (Wang et al., 2003). The TTest function in the Mathematica software package was used to test the null hypothesis that the mean protein expression under electrosynthesis conditions was no different from the mean expression under control fermentation conditions. The means of the three technical replicates for each sample were calculated, and these means were used to calculate the T-test statistic. Degrees of freedom were adjusted when normality or equivalent variances could not be assumed (Welch's T-test). All proteins with P<0.05 were considered significant and the null hypothesis was rejected. The proteomics raw data and statistical analysis are presented in the Supplementary Information. The same method for data processing was used on the metabolite data.
3. Results and discussion
3.1. Escherichia coli electrosynthesis
Electrosynthesis during fermentation of glucose with neutral red by E. coli K-12 resulted in the current and product profile changes shown in Fig. 1. Fig. 1A shows that the current increased after inoculation from 0 to 2 h, then dropped in correspondence with cell growth from 2 to 6 h, before slowly increasing back towards background after the cells stopped growing. Electrosynthesis resulted in slightly lower biomass concentration. Electrosynthesis also resulted in the consumption of 3.35 mM more glucose (1B), and the production of 2.49 mM more ethanol (1C), 3.07 mM more acetate (1D), 0.6 mM more succinate (1E), 5.52 mM more lactate (1G) and 0.24 mM more pyruvate (1F) after 15.5 h. Formate concentration peaked at 6.5 h, and was dissimilated to H2 and CO2 faster during electrosynthesis vs. control. Proteomic analysis (Supplementary Information) indicated that formate hydrogen lyase was significantly overexpressed during electrosynthesis, which corresponds with the faster dissimilation of formate shown in 1F. Several hydrogenase subunits and hydrogenase maturation proteins (hyaA, hybA, hycE, hycF, hycI, hypB) were moderately to significantly overexpressed. 13 ribosomal subunits (obgE, rplB, rplN, rplX, rplY, rpmD, rpmF, rpsQ, rbfA, rpsC, rpsL, rpsO, rsfS), 10 proteins involved in tRNA synthesis (aspS, fmt, glyQ, lysS, metG, mnmA, selD, tufA, tusE, ybaK), and 8 proteins involved in nucleotide synthesis (aroK, deoD, gmk, mrp, prs, purC, pyrD, rsuA) were overexpressed during electrosynthesis, and none from these categories were repressed. Seven proteins involved in sugar metabolism (galM, glf, glk, tktA, tktB, tpiA, yqaB) were overexpressed, and two were repressed (araA, galK). Transcription regulating proteins were the largest group of proteins that were differentially expressed, accounting for 16 proteins that were overexpressed (allR, arcA, galR, metJ, mhpR, narL, narP, nusA, nusB, nusG, pspA, qseB, stpA, trpR, yebC, yeeN), and one that was repressed (exuR).
Fig. 1.
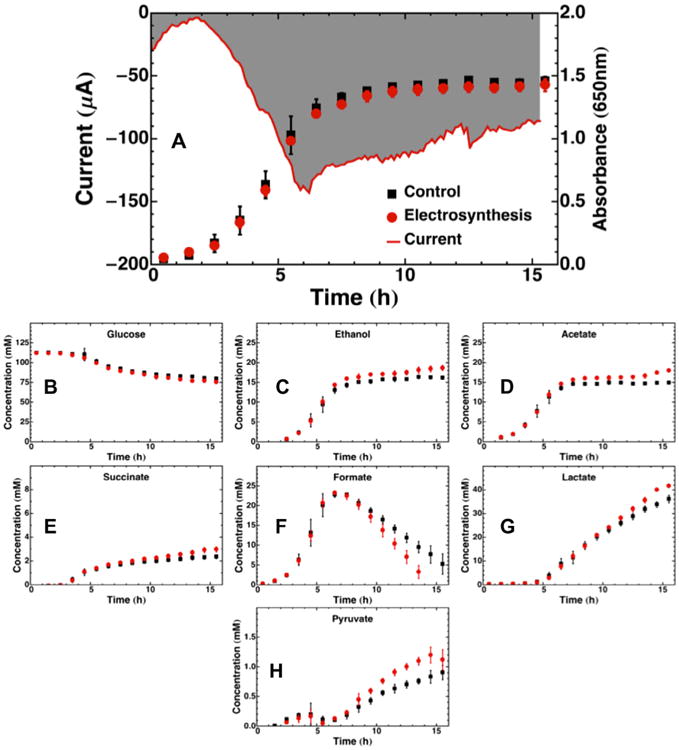
Batch control and electrosyntheses or fermentation profiles for E. coli growing on glucose over 15.5 h. (A) Chronoamperometry plot of current delivered to fermentation media plotted together with cell growth tracked by OD650. The gray shaded area represents the area integrated to obtain the total charge transferred. Substrate and product metabolite profiles during the same 16 h period are shown for (B) glucose, (C) ethanol, (D) acetate, (E) succinate, (F) formate, (G) lactate, and (H) pyruvate. All data points are averages of four replicate fermentations or electrosynthesis, with error bars representing one standard deviation. The chronoamperometry plot of current delivered in (A) is from a single representative reactor, not an average.
3.2. Klebsiella pneumoniae electrosynthesis
Electrosynthesis during fermentation of glycerol with neutral red by K. pneumoniae resulted in the current and product profile changes shown in Fig. 2. Fig. 2A shows that electrosynthesis resulted in higher final biomass concentration. Similarly to E. coli, the current increased initially for the first 1.5 h, then dropped sharply from 1.5 to 4 h during the exponential growth phase. After 4 h, the current rose back toward zero more quickly than the current in the E. coli reactors. Electrosynthesis resulted in the consumption of 22.78 mM more glycerol (1B), and the production of 12.73 mM more ethanol (1C), 1.17 mM less acetate (1D), 0.26 mM more succinate (1E), 6.01 mM more 1,3-propanediol (1F), and 2.98 mM more lactate (1E). The final titers of ethanol and lactate, in particular, were nearly doubled during electrosynthesis vs. control. Proteomic analysis (Supplementary Information) of K. pneumoniae electrosynthesis vs. control revealed only eight significant (P<0.05) differentially-expressed proteins (budB, dhaF, KPR_1933, KPR_4061, potD, topA, tyrS, uspA) out of the 686 identified proteins, of which one could be correlated with the metabolomics data. Glycerol dehydratase reactivating factor (dhaF, Log2 = −0.56), which is required to regenerate an enzyme that converts glycerol to 1,3-propanediol, was repressed during electrosynthesis. This correlates with and could possibly explain the 10% lower yield of 1,3-propanediol on glycerol seen during electrosynthesis vs. control fermentation.
Fig. 2.
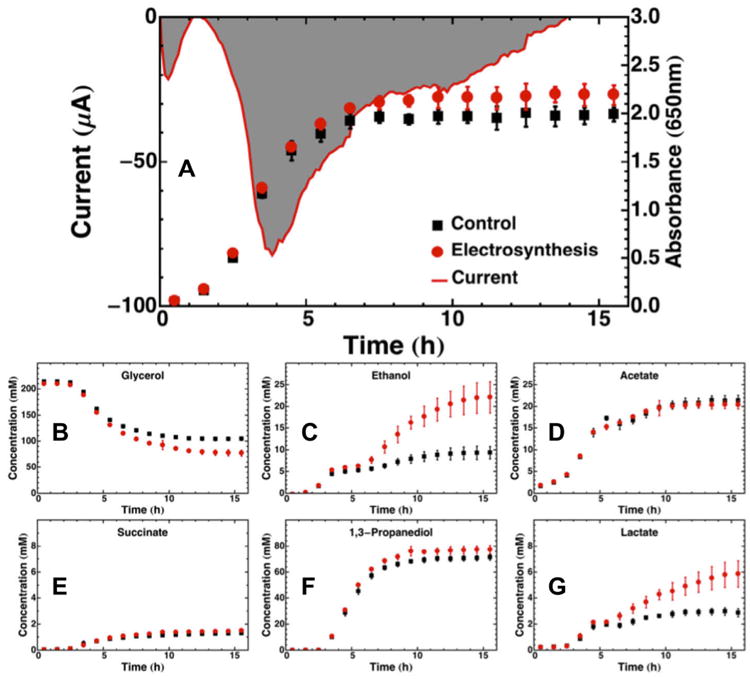
Batch control and electrosynthesis fermentation profiles for K. pneumoniae growing on glycerol over 15.5 h. (A) Chronoamperometry plot of current delivered to fermentation media plotted together with cell growth tracked by OD650. The gray shaded area represents the area integrated to obtain the total charge transferred. Substrate and product metabolite profiles during the same 16 h period are shown for (B) glycerol, (C) ethanol, (D) acetate, (E) succinate, (F) 1,3-propanediol, and (G) lactate. All data points are averages of four replicate experiments, with error bars representing one standard deviation. The chronoamperometry plot of current in delivered in (A) is from a single representative reactor, not an average.
3.3. Zymomonas mobilis electrosynthesis
Electrosynthesis during fermentation of glucose with neutral red by Z. mobilis resulted in the cathodic current and product profiles shown in Fig. 3. Fig. 3 shows that the current drop had a very late onset that trailed the onset of exponential growth by 5 h. The peak current was reached at 15.5 h and then dropped off toward background thereafter. Interestingly, despite numerous reports of higher ethanol yields from Z. mobilis with electrically-reduced neutral red (Jeon et al., 2009; Jeon and Park, 2010), no significant difference was found in final titer for any metabolites except for a slightly higher titer of acetate (5.48 mM) during electrosynthesis vs. control. Z. mobilis had 54 proteins that were identified as differentially-expressed, out of 620 identified (Supplementary Information). The only statistically significant difference in end product formation for Z. mobilis was in the acetate profile, and no changes in protein expression were found that could be correlated to this difference. However, there were several proteins involved in central metabolism that were over-expressed (glk, pgi, ZZ6_0880, ZZ6_1055, ZZ6_1712), suggesting that these enzymes were not rate-limiting. For instance, pyruvate decarboxylase was slightly overexpressed during electrosynthesis (Log2 = 0.78), but this did not manifest as an increase in ethanol productivity. Glucokinase, Glucose-6-phosphate isomerase, 6-phosphogluconate dehydratase, and pyruvate kinase were also up-regulated. Six ribosomal subunits were moderately up-regulated (rplJ, rplM, rplN, rplY, rpsB, rpsD, rpsM), as well as five proteins involved in amino acid metabolism (argG, carB, hisC, ilvC, proB, ZZ6_1462).
Fig. 3.
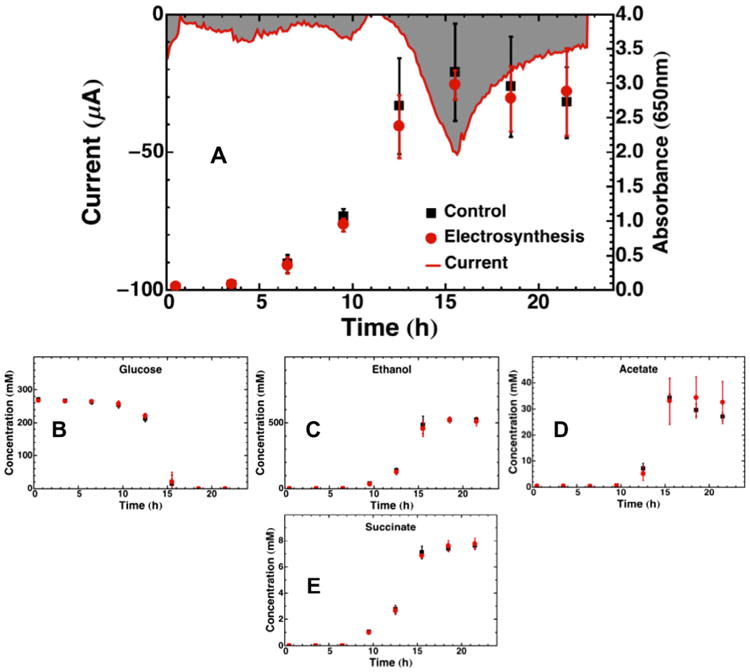
Batch control and electrosynthesis fermentation profiles for Z. mobilis growing on glycerol over 21.5 h. (A) Chronoamperometry plot of current delivered to fermentation media plotted together with cell growth tracked by OD650. The gray shaded area represents the area integrated to obtain the total charge transferred. Substrate and product metabolite profiles during the same 24 h period are shown for (B) glucose, (C) ethanol, (D) acetate, and (E) succinate. All data points are averages of four replicate experiments, with error bars representing one standard deviation. The chronoamperometry plot of current delivered in (A) is from a single representative reactor, not an average.
3.4. Comparison of fermentations
Central metabolism in bacteria often contains branch points where a single substrate can be converted to two or more products. Conversion to the different products at these metabolic “nodes” or “branch points” are sometimes influenced by the availability of NADH or NAD+, which drive intracellular metabolites to more reduced or oxidized products, respectively. The central metabolisms of the three chosen organism/substrate combinations are quite different from each other, and represent an archetype for their respective genres of central metabolism branching patterns. E. coli performs a mixed acid fermentation in which glucose is converted to pyruvate in a linear manner, followed by branched conversion to several different organic acids (Lee et al., 2005). K. pneumoniae performs a branched fermentation in which the initial electron donor, glycerol, is itself a metabolic branch point like pyruvate, and the subsequent oxidized pathway is further branched (Huang et al., 2002). Z. mobilis performs a nearly linear conversion of glucose to ethanol, with the production of only a minor amount of byproducts (Jeffries, 2005). As expected from the numerous differences in their metabolisms, these organisms demonstrated different responses to neutral red-mediated electrosynthesis. E. coli and K. pneumoniae both produced higher titers of reduced products during electrosynthesis vs. control, whereas Z mobilis only produced higher titers of acetate. The current profiles were also different: E. coli and K. pneumoniae current decreased in correspondence with the exponential phase of growth, while Z mobilis current profiles lagged the exponential growth phase by a period of 5–6 h, and peaked to a lower magnitude of current (−50 μA vs. −75 to −150 μA), even with a higher number of cells available to accept electrons.
Table 1 displays tabulated values for the endpoint (15.5 h for E. coli and K. pneumoniae, and 21.5 h for Z. mobilis) changes in concentration of each terminal metabolite present for all three sets of fermentations. Standard deviation of four biological replicates for metabolite data and student's T-test values are also displayed, as well as yield changes for each metabolite. Significant differences (P < 0.05) in final concentrations of several metabolites were found between electrosynthesis and control fermentation conditions with E. coli and K. pneumoniae. However, the only significant difference in Z mobilis was an increase in final acetate concentration of 5.48 mM, or a 1.1% increase in yield. K. pneumoniae experienced the largest percent changes in a metabolite concentration out of the three organisms, producing 93% higher ethanol yield and 75% higher lactate yield on glycerol vs. control. The effect on E. coli was more moderate, but still significant. E. coli experienced 14% higher succinate yield, 13% higher pyruvate yield, 9.4% higher acetate yield, and 5% higher lactate yield on glucose during electrosynthesis vs. control. At first glance, the E. coli and K. pneumoniae results appear consistent with the hypothesis that reduced neutral red reduces NAD+ to NADH, and drives carbon flux down reductive pathways. E. coli experienced higher yields of the reduced products succinate and lactate, and K. pneumoniae experienced higher titers of the reduced products ethanol and lactate. However, close inspection revealed a more complicated story. Electrosynthesis resulted in higher consumption of the electron donor by both organisms, with E. coli consuming 3.35 mM more glucose, and K. pneumoniae consuming 22.78 mM more glycerol. Though E. coli consumed more glucose, it produced less biomass per glucose consumed, which corresponded with a 6.6% increase in total metabolite yield on glucose. In effect, electrosynthesis diverted glucose slightly away from biomass formation and toward metabolite formation by E. coli. K. pneumoniae overall metabolite yield on glycerol was unchanged by electrosynthesis, and the increase in biomass during electrosynthesis was accounted for by the increased glycerol consumption. A further result that could not have been predicted by the NAD+ reduction hypothesis is that electrosynthesis with K. pneumoniae resulted in a lower yield of the reduced metabolite 1,3-propanediol on glycerol. If electrosynthesis resulted in NAD+ reduction, then the higher NADH/NAD+ ratio should have driven glycerol flux down the very first reduced pathway branch, resulting in higher 1,3-propanediol yields vs. control. That this was not the case may possibly be explained by the reduced expression of glycerol dehydratase reactivating factor, which may have lowered reductive glycerol flux. Glycerol dehydratase does not involve a nicotinamide cofactor, and it was previously shown to be the limiting step in 1,3-propanediol production by a Clostridium strain (Abbad-Andaloussi et al., 1996). Further support that NAD+ reduction does not explain the metabolite shifts comes from an analysis of the current delivery data as represented by the current profiles in Figs. 1A, 2A, and 3A.
Table 1.
Stoichiometric comparison of metabolites and electrons delivered.
| Metabolite | Control (mM) t = 15.5 h | σ | Electrosynthesis (mM) t = 15.5 h | σ | T | df | P | Control % yield | NR % yield | % change | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | Glucose | −32.99 | 1.21 | −36.34 | 1.57 | 3.38 | 6 | 0.0149 | |||
| Ethanol | 16.15 | 0.37 | 18.64 | 0.61 | 6.98 | 6 | 0.0004 | 24.5% | 25.6% | 4.8% | |
| Acetate | 14.92 | 0.19 | 17.99 | 0.34 | 15.76 | 6 | 0.0001 | 22.6% | 24.7% | 9.4% | |
| Succinate | 2.36 | 0.16 | 2.96 | 0.22 | 4.41 | 6 | 0.0045 | 3.6% | 4.1% | 13.9% | |
| Formate | 22.52 | 0.67 | 22.96 | 0.53 | 1.03 | 6 | 0.3427 | 34.1% | 31.6% | −7.4% | |
| Lactate | 35.78 | 1.29 | 41.30 | 0.67 | 7.59 | 6 | 0.0003 | 54.2% | 56.8% | 4.8% | |
| Pyruvate | 0.98 | 0.35 | 1.22 | 0.43 | 0.87 | 6 | 0.4199 | 1.5% | 1.7% | 12.7% | |
| Total metabolites* | 35.09 | 1.56 | 41.05 | 1.20 | 6.28 | 6 | 0.0008 | 106.4% | 113.0% | 6.2% | |
| Electron pairs delivered (μM) | 271.4 | 44.12 | |||||||||
|
| |||||||||||
| K. pneumoniae | Metabolite | Control (mM) t = 15.5 h | σ | Electrosynthesis (mM) t = 15.5 h | σ | T | df | P | Control % yield | NR % yield | % change |
| Glycerol | −110.07 | 3.75 | −132.85 | 5.86 | 6.55 | 6 | 0.0006 | ||||
| Ethanol | 9.61 | 1.43 | 22.34 | 3.59 | 6.59 | 6 | 0.0006 | 8.7% | 16.8% | 92.7% | |
| Acetate | 19.76 | 1.17 | 18.59 | 1.02 | 1.51 | 6 | 0.1824 | 18.0% | 14.0% | −22.1% | |
| Succinate | 1.22 | 0.05 | 1.48 | 0.14 | 3.50 | 6 | 0.0129 | 1.1% | 1.1% | 0.5% | |
| Propanediol | 71.31 | 2.24 | 77.32 | 3.24 | 3.05 | 6 | 0.0225 | 64.8% | 58.2% | −10.2% | |
| Lactate | 2.66 | 0.58 | 5.64 | 1.01 | 5.12 | 6 | 0.0022 | 2.4% | 4.2% | 75.7% | |
| Total metabolites | 104.56 | 2.96 | 125.37 | 5.05 | 7.11 | 6 | 0.0004 | 95.0% | 94.4% | −0.7% | |
| Electron pairs delivered (μM) | 68.77 | 38.82 | |||||||||
|
| |||||||||||
| Z. mobilis | Metabolite | Control (mM) t = 21.5 h | σ | Electrosynthesis (mM) t = 21.5 h | σ | T | df | P | Control % yield | NR % yield | % change |
| Glucose | −270.32 | 6.87 | −266.74 | 3.59 | 0.92 | 6 | 0.3913 | ||||
| Ethanol | 521.89 | 10.22 | 509.06 | 33.09 | 0.74 | 6 | 0.4867 | 96.5% | 95.4% | −1.1% | |
| Acetate | 27.02 | 1.26 | 32.50 | 2.84 | 3.53 | 6 | 0.0124 | 5.0% | 6.1% | 21.9% | |
| Succinate | 7.62 | 0.23 | 7.78 | 0.44 | 0.64 | 6 | 0.5431 | 1.4% | 1.5% | 3.5% | |
| Total metabolites | 278.26 | 10.30 | 274.67 | 33.21 | 0.21 | 6 | 0.8432 | 102.9% | 103.0% | 0.0% | |
| Electron pairs delivered (μM) | 57.91 | 19.19 | |||||||||
(σ: standard deviation, T = T-statistic, df = degrees of freedom, P = P-value, NR = neutral red,
formate was excluded from total yield calculations because it is a byproduct of the conversion of pyruvate to acetate).
3.5. Electron balance
Before inoculation of the cultures to the electrosynthesis BERs, neutral red was completely reduced, which was indicated by a constant low background current (≥ −50 μA, background-subtracted in Figs. 1A, 2A, and 3A). Oxidation of the neutral red by growing cells resulted in the working electrode passing larger cathodic (negative) currents in order to re-reduce the neutral red that had been oxidized. Because of this, integration of the current profiles delivered to the electrosynthesis cathode chambers approximates the amount of neutral red that was oxidized by the cells during electrosynthesis. Integration of the current profiles from Figs. 1A, 2A, and 3A revealed that during 15.5 h electrosynthesis, an average of 271.40 μM of electron pair equivalents were delivered to E. coli cells, while 68.77 μM were delivered to K. pneumoniae cells. Z. mobilis cells received the least amount of electrons; an average of 57.91 μM electron pairs during a longer electrosynthesis time period of 21.5 h. Current delivery only happened in the presence of both neutral red and cells (control data not shown). The tally of electron pair equivalents that were delivered to the electrosynthesis reactors was not at all comparable to the changes in final titers of the products. The electron pairs delivered to the broth were from one to three orders of magnitude smaller when compared to the increases in concentration seen in several of the reduced metabolites at the end of the E. coli and K. pneumoniae electrosynthesis.
Taken together, this data led to the conclusion that the assumption behind the hypothesis – that neutral red reduces NAD+ – was either an insignificant part of the whole effect of neutral red-mediated microbial electrosynthesis, or does not occur at all in the studied model systems. These results demonstrate that the assumed mechanism behind the hypothesis was incorrect, even though significant changes were observed during electrosynthesis with K. pneumoniae and E. coli vs. control. The data presented here is by no means exhaustive with respect to the large number of organisms and fermentation pathways where neutral red-mediated microbial electrosynthesis has been previously applied. However, the analysis of the three presented-organisms suggests that previous studies may have erroneously assumed that larger amounts of electron delivery by neutral red had occurred in order to explain the observed higher yields of reduced metabolites seen during electrosynthesis. This misattribution may have been partly due to the lack of potentiostatic control to account for charge delivered, and partly due to a lack of accounting for hydrogen production via electrolysis, which was avoided in this study as evidenced by the steady-state currents pre-inoculation. Because the amount of current delivered does not support the NAD+ reduction hypothesis, a plausible mechanism behind the dramatic response of E. coli and K. pneumoniae to electrosynthesis is a regulatory response, perhaps a stress response or a redox-potential sensing response (Liu et al., 2013), resulting in altered protein expression and fermentation profiles. In contrast, electrosynthesis with Z. mobilis results in fewer product changes, despite large changes in expression of several cellular proteins. This suggests that a different mechanism may operate in Z. mobilis, rendering electrosynthesis ineffective at altering metabolite profiles toward more reduced metabolites like ethanol.
3.6. The special case of Zymomonas mobilis
The lack of response of Z. mobilis to electrosynthesis was a surprise considering previous studies that showed as much as a 50% increase in ethanol production when fermented in a chamber with a cathodically charged electrode with covalently-immobilized neutral red (Jeon et al., 2009). However, the results make sense considering the Z. mobilis metabolism. What makes Z. mobilis such a great ethanol producer is its nearly linear conversion of one glucose molecule to two ethanol molecules, with only minor byproducts succinate and acetate (Kalnenieks, 2006). For each glucose molecule metabolized to two ethanol molecules, Z. mobilis consumes and generates exactly 2 NAD(P)+ and 2 NAD(P)H, respectively. Additional NADH entering the system would stop or slow glycolysis by lowering the availability of NAD+, unless it was subsequently removed by re-oxidation to NAD+. However, producing more ethanol would not solve the problem because there is nowhere to borrow carbon from in a linear fermentation pathway. Succinate is ethanol's only competition for reducing equivalents in Z. mobilis, and requires two equivalents to be produced via reduction of pyruvate, whereas ethanol only requires one. Succinate production is thus stoichiometrically limited in Z. mobilis by NADH because oxidation of glucose gives two NAD(P)H, and reduction of the two resulting pyruvate molecules to succinate would require four NAD(P)H. As opposed to the ethanol pathway, the succinate pathway could sustain an influx of NAD(P)H by borrowing carbon flux from the ethanol pathway. NAD(P)+ reduction by neutral red during electrosynthesis would therefore have been easily observed as an increase in succinate production at ethanol's expense, as it would have been the only option to relieve the excess electron pressure. That this did not occur is further evidence that NAD(P)+ reduction did not occur. Perhaps the most interesting difference seen in Z. mobilis when compared to E. coli and K. pneumoniae is the overall lower current delivery to the fermentation broth, despite the fact that the cells grew to a much larger cell density during a 39% longer fermentation time, while converting at least 8× more D-glucose than E. coli.
The current delivery to growing cells of each of the three species presented in this study did not stoichiometrically account for the changes in end-product metabolite concentrations. The observed changes in behavior for E. coli and K. pneumoniae may be explained by the involvement of a regulatory mechanism, similar or identical in effect to low-redox potential fermentations, rather than the previously hypothesized chemical reduction of NAD+ to NADH. Further, the neutral red mechanism appears to be species-specific, and cannot be equally applied to any fermenting organism with the expectation of the same effect.
4. Conclusions
Neutral red-mediated microbial electrosynthesis by E. coli, K. pneumoniae, and Z. mobilis yielded distinct results. E. coli overexpressed formate hydrogen lyase, dissimilated formate faster, and produced higher titers of reduced metabolites. K. pneumoniae consumed more glycerol and achieved statistically significant higher lactate and ethanol yields. Z mobilis experienced no significant changes except for acetate. Current delivery did not stoichiometrically account for metabolite changes. The effects of electrosynthesis are organism specific, and are not caused by NAD+ reduction. The observed metabolite and protein changes in E. coli and K. pneumoniae are consistent with the involvement of a low-redox sensing mechanism.
Supplementary Material
Highlights.
Neutral red-mediated electrosynthesis was applied to three different bacteria.
Electrosynthesis altered metabolite profiles in E. coli and K. pneumoniae.
No effect was observed on Z. mobilis metabolites.
Hypothesized NAD+ reduction did not account for metabolite changes.
Effects of electrosynthesis are consistent with a regulatory response.
Acknowledgments
This work was supported by National Science Foundation (NSF) – USA Career Award 0954186, and TDH was supported by NIH Training Grant 5T32GM008336-24. Proteomic analyses were performed on an instrument acquired through the National Science Foundation MRI Program Grant No. DBI-1229749 to DRG. HB acknowledges additional support from the Fundamental and Applied Chemical and Biological Catalysts to Minimize Climate Change, Create a Sustainable Energy Future, and Provide a Safer Food Supply” and it is Project #WNP00807.
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.biortech.2015.06.005.
References
- Abbad-Andaloussi S, Guedon E, Spiesser E, Petitdemange H. Glycerol dehydratase activity: the limiting step for 1,3-propanediol production by Clostridium butyricum DSM 5431. Lett Appl Microbiol. 1996;22:311–314. http://dx.doi.org/10.1111/j.1472-765X.1996.tb01168.x. [Google Scholar]
- Azariah AN, Sheela B, Yegnaraman V. Electrochemical behaviour of neutral red. B Electrochem. 1998;14:309–314. [Google Scholar]
- Choi O, Kim T, Woo HM, Um Y. Electricity-driven metabolic shift through direct electron uptake by electroactive heterotroph Clostridium pasteurianum. Sci Rep. 2014;4:6961. doi: 10.1038/srep06961. http://dx.doi.org/10.1038/srep06961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IC, Carlson HK, Iavarone AT, Coates JD. Bioelectrical redox cycling of anthraquinone-2,6-disulfonate coupled to perchlorate reduction. Energy Environ Sci. 2012;5:7970. http://dx.doi.org/10.1039/c2ee21594b. [Google Scholar]
- Clark WM, Perkins ME. Studies on oxidation-reduction XVII neutral red. J Am Chem Soc. 1932;54:1228–1248. [Google Scholar]
- Emde R, Schink B. Enhanced propionate formation by Propionibacterium-freudenreichii subsp freudenreichii in a three-electrode amperometric culture system. Appl Environ Microbiol. 1990;56:2771–2776. doi: 10.1128/aem.56.9.2771-2776.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage SD, Phelps EBE. Notes on B. coli and allied forms, with special reference to the neutral-red reaction. Public Health Pap Rep. 1902;28:402–412. [PMC free article] [PubMed] [Google Scholar]
- Girbal L, Vasconcelos I, Saint-Amans S, Soucaille P. How neutral red modified carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH. FEMS Microbiol Rev. 2006;16:151–162. http://dx.doi.org/10.1111/j.1574-6976.1995.tb00163.x. [Google Scholar]
- Harrington TD, et al. The mechanism of neutral red-mediated microbial electrosynthesis in Escherichia coli: menaquinone reduction. Bioresour Technol. 2015;192:689–695. doi: 10.1016/j.biortech.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo M, Iwahara M. Application of electro-energizing method to L-glutamic acid fermentation. Agric Biol Chem. 1979;43:2075–2081. [Google Scholar]
- Huang HH, Gong CSC, Tsao GTG. Production of 1,3-propanediol by Klebsiella pneumoniae. Appl Biochem Biotechnol. 2002;98–100:687–698. http://dx.doi.org/10.1385/ABAB:98-100:1-9:687. [PubMed] [Google Scholar]
- Jeffries TWT. Ethanol fermentation on the move. Nat Biotechnol. 2005;23:40–41. doi: 10.1038/nbt0105-40. http://dx.doi.org/10.1038/nbt0105-40. [DOI] [PubMed] [Google Scholar]
- Jeon BY, Hwang TS, Park DH. Electrochemical and biochemical analysis of ethanol fermentation of Zymomonas mobilis KCCM11336. J Microbiol Biotechnol. 2009;19:666–674. doi: 10.4014/jmb.0809.509. http://dx.doi.org/10.4014/jmb.0809.509. [DOI] [PubMed] [Google Scholar]
- Jeon BY, Jung IL, Park DH. Conversion of carbon dioxide to metabolites by Clostridium acetobutylicum KCTC1037 cultivated with electrochemical reducing power. Adv Microbiol. 2012;2:332–339. http://dx.doi.org/10.4236/aim.2012.23040. [Google Scholar]
- Jeon BY, Park DH. Improvement of ethanol production by electrochemical redox combination of Zymomonas mobilis and Saccharomyces cerevisiae. J Microbiol Biotechnol. 2010;1:94–100. http://dx.doi.org/10.4014/jmb.0904.04029. [PubMed] [Google Scholar]
- Jeon BY, Yi JY, Jung IL, Park DH. Activation of ethanol production by combination of recombinant Ralstonia eutropha and electrochemical reducing power. Adv Microbiol. 2013;3:42–45. http://dx.doi.org/10.4236/aim.2013.31006. [Google Scholar]
- Kalnenieks U. Physiology of Zymomonas mobilis: some unanswered questions. Adv Microb Physiol. 2006;51:73–117. doi: 10.1016/S0065-2911(06)51002-1. http://dx.doi.org/10.1016/S0065-2911(06)51002-1. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee DY, Kim TY, Kim BH, Lee J, Lee SY. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl Microbiol Biotechnol. 2005;71:7880–7887. doi: 10.1128/AEM.71.12.7880-7887.2005. http://dx.doi.org/10.1128/AEM.71.12.7880-7887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo MR, Dailly Y, Clark DP. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Xue C, Lin YH, Bai FW. Redox potential control and applications in microaerobic and anaerobic fermentations. Biotechnol Adv. 2013;31:257–265. doi: 10.1016/j.biotechadv.2012.11.005. http://dx.doi.org/10.1016/j.biotechadv.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Makgill RH. The neutral-red reaction as a means of detecting Bacillus colin in water supplies. J Hyg (Lond) 1901;1:430–436. doi: 10.1017/s0022172400000383. http://dx.doi.org/10.1017/S0022172400000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinlay JB, Zeikus JG. Extracellular iron reduction is mediated in part by neutral red and hydrogenase in Escherichia coli. Appl Microbiol Biotechnol. 2004;70:3467–3474. doi: 10.1128/AEM.70.6.3467-3474.2004. http://dx.doi.org/10.1128/AEM.70.6.3467-3474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Laivenieks M, Guettler MV, Jain MK, Zeikus JG. Microbial utilization of electrically reduced neutral red as the sole electron donor for growth and metabolite production. Appl Environ Microbiol. 1999;65:2912–2917. doi: 10.1128/aem.65.7.2912-2917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Zeikus JG. Utilization of electrically reduced neutral red by Actinobacillus succinogenes: physiological function of neutral red in membranedriven fumarate reduction and energy conservation. J Bacteriol. 1999;181:2403–2410. doi: 10.1128/jb.181.8.2403-2410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Zeikus JG. Electricity generation in microbial fuel cells using neutral red as an electronophore. Appl Microbiol Biotechnol. 2000;66:1292–1297. doi: 10.1128/aem.66.4.1292-1297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Kang HS, Park DW, Park DH. Electrochemical control of metabolic flux of Weissella kimchii sk10: neutral red immobilized in cytoplasmic membrane as electron channel. J Microbiol Biotechnol. 2005;15:80–85. [Google Scholar]
- Peguin S, Soucaille P. Modulation of metabolism of Clostridium acetobutylicum grown in chemostat culture in a three-electrode potentiostatic system with methyl viologen as electron carrier. Biotechnol Bioeng. 1996;51:342–348. doi: 10.1002/(SICI)1097-0290(19960805)51:3<342::AID-BIT9>3.0.CO;2-D. http://dx.doi.org/10.1002/(SICI)1097-0290(19960805)51:3<342∷AID-BIT9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Rabaey K, Rozendal RA. Microbial electrosynthesis — revisiting the electrical route for microbial production. Nat Rev Microbiol. 2010;8:706–716. doi: 10.1038/nrmicro2422. http://dx.doi.org/10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- Rao G, Mutharasan R. Alcohol production by Clostridium acetobutylicum induced by methyl viologen. Biotechnol Lett. 1986;8:893–896. [Google Scholar]
- Rao G, Mutharasan R. Altered electron flow in continuous cultures of Clostridium acetobutylicum induced by viologen dyes. Appl Microbiol Biotechnol. 1987;53:1232–1235. doi: 10.1128/aem.53.6.1232-1235.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renslow R, Donovan C, Shim M, Babauta J, Nannapaneni S, Schenk J, Beyenal H. Oxygen reduction kinetics on graphite cathodes in sediment microbial fuel cells. Phys Chem Chem Phys. 2011;13:21573–21584. doi: 10.1039/c1cp23200b. http://dx.doi.org/10.1039/c1cp23200b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Tsuge Y, Sasaki D, Kondo A. Increase in lactate yield by growing Corynebacterium glutamicum in a bioelectrochemical reactor. J Biosci Bioeng. 2013;117:598–601. doi: 10.1016/j.jbiosc.2013.10.026. http://dx.doi.org/10.1016/j.jbiosc.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Shin HS, Zeikus JG, Jain MK. Electrically enhanced ethanol fermentation by Clostridium thermocellum and Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2002;58:476–481. doi: 10.1007/s00253-001-0923-2. http://dx.doi.org/10.1007/s00253-001-0923-2. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2012;41:43–47. doi: 10.1093/nar/gks1068. http://dx.doi.org/10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. http://dx.doi.org/10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- Wu X, Xiong E, Wang W, Scali M, Cresti M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat Prot. 2014;9:362–374. doi: 10.1038/nprot.2014.022. http://dx.doi.org/10.1038/nprot.2014.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


