ABSTRACT
Modern molecular technology, and particularly high-throughput sequencing (HTS), has revolutionized virus discovery and expanded the depth and breadth of the virome. Recent HTS was used to identify and discover a previously undescribed member of the family Flaviviridae that has genomic features characteristic of both hepaciviruses and pegiviruses. This virus, designated human hepegivirus-1 (HHpgV-1), may represent a previously undescribed new genus in the Flaviviridae family with implications for public health and blood supply safety. Detecting uncharacterized viruses such as HHpgV-1 in clinical samples requires an unbiased screening method that is as sensitive as PCR, while simultaneously detecting multiple rare viral sequences. The virome-capture-sequencing platform for vertebrate viruses (VirCapSeq-VERT) uses positive-selection oligonucleotide capture to sensitively detect sequences from every known vertebrate virus, even in high-background specimens with low-abundance viruses. VirCapSeq-VERT can also detect uncharacterized viruses with sequence homology to known viruses, enabling a new paradigm for virus detection.
COMMENTARY
Modern molecular biology has accelerated the pace of virus discovery, allowing pathogen identification based on genome characterization to proceed much more rapidly than traditional virologic, microscopic, or serologic methods. In particular, high-throughput sequencing (HTS) has revolutionized the unbiased discovery of novel viruses, often in unexpected places that are highly relevant to human health. This is exemplified by a recent article by Kapoor and colleagues from W. Ian Lipkin’s research laboratory (1), in which a novel human virus was found in serum samples from blood transfusion recipients and hemophilia patients treated with plasma-derived clotting factors. Further analysis indicated that this virus combines features of hepaciviruses and pegiviruses, two distinct genera within the family Flaviviridae. This virus, named human hepegivirus-1 (HHpgV-1), may represent a new genus within the Flaviviridae and a potential human pathogen.
Previous work from the Lipkin laboratory has enhanced our understanding of hepacivirus and pegivirus diversity in the wild. Hepatitis C virus (HCV) was long thought to be the sole member of the genus Hepacivirus, until the discovery of a novel canine hepacivirus (2) suggested that there may be greater hepacivirus diversity in nonhuman mammalian species. Since then, numerous hepaciviruses have been characterized in horses, bats, rodents, and nonhuman primates (3–7). A similar host range diversity has been observed for pegiviruses (8). As HTS-based methods increase for viromics study applications, we also expect to discover additional novel members of Flaviviridae and develop a more comprehensive understanding of flavivirus biology.
The HHpgV-1 sequence shows that it has a type IV internal ribosome entry site (IRES) in its 5′ untranslated region (UTR) with considerable structural homology to that found in HCV. This suggests that like hepaciviruses, translation of the viral polyprotein occurs via a common internal initiation mechanism. HHpgV-1 also contained a distinguishing sequence in the pseudoknot domain that is highly conserved across the genus Hepacivirus. However, analysis of two relatively conserved nonstructural genes, the viral helicase (NS3) and RNA-dependent RNA polymerase (NS5B), demonstrated that HHpgV-1 is phylogenetically grouped as a distinct branch within the pegivirus clade. Thus, HHpgV-1 shares characteristics of both genera, suggesting either a common ancestor or recombination between two prototypic viruses. This raises the possibility that HHpgV-1 may cause chronic infections like other hepaciviruses and pegiviruses. It is currently unknown whether HHpgV-1 has the potential to cause a pathogenic infection, like HCV, or merely a silent, nonpathogenic infection similar to human pegivirus (HpgV), which persists for years without causing disease in healthy people.
HHpgV-1 may have substantial implications for public health, as Kapoor and colleagues detected the virus in posttransfusion samples, strongly suggesting transmission via transfused blood products. The blood-borne HCV was spread extensively by contaminated blood products prior to identification of the virus as the etiologic agent of the then-called non-A, non-B hepatitis and the subsequent development of tests to ensure the safety of the blood supply. Indeed, the observation that HHpgV-1 persisted in both hemophilia patients—in one case for more than 5 years—demonstrates that this virus can establish chronic infections. While the consequences for human health are unknown, a more detailed assessment of HHpgV-1 prevalence in the population will start to yield answers. For this purpose, a precise, highly sensitive means of screening clinical specimens is essential. Ideally, this would require an unbiased method for detecting multiple HHpgV-1 variants, in addition to other related persistent viruses, with a sensitivity equal to or greater than the sensitivity of quantitative PCR.
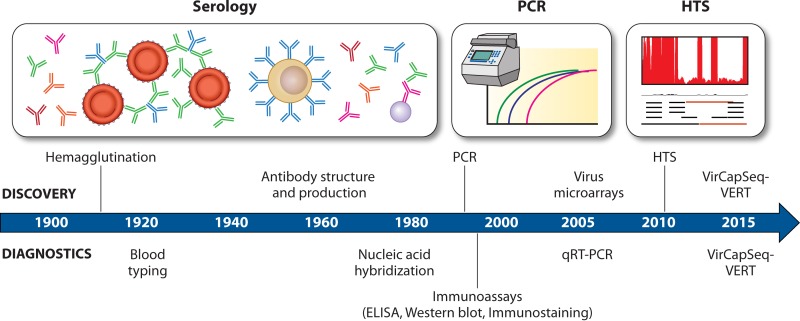
A decade ago, microarray-based technologies showed great promise for allowing the simultaneous detection of multiple viruses (9, 10); however, low sensitivity has hindered these technologies from becoming standard for clinical diagnostics. While HTS can be used to perform unbiased pathogen detection and discovery, this technology is still insufficiently sensitive for patient samples, in which viral sequences may be very rare (Fig. 1). Efforts to enrich pathogen sequences by depleting host nucleic acids has not sufficiently improved sensitivity to warrant using HTS as a standard diagnostic assay. Furthermore, the high cost and practical complexity of obtaining and analyzing data have delayed the implementation of HTS in clinical diagnostics, compared to relatively simpler, less-expensive virus-specific PCR-based methods. While PCR-based methods are highly sensitive, they cannot identify unknown pathogens and have been used only to confirm diagnosis of a suspected pathogen. Another recent article from the Lipkin laboratory by Briese et al. (11) upends this paradigm. The virome-capture-sequencing platform for vertebrate viruses (VirCapSeq-VERT) represents a novel means of detecting every known vertebrate virus, even from limited clinical samples with high abundance of host background sequences.
FIG 1 .
History of virus diagnostics. Timeline showing how the development of different technologies (shown above the timeline) enabled standard methods for diagnosing virus infection (shown below the timeline). Serology and PCR subsequently enabled detection of hundreds of viruses, largely individually, while modern HTS technology platforms, such as VirCapSeq-VERT, allow detection of thousands of viruses simultaneously.
The VirCapSeq-VERT method dramatically enhances HTS-based virus detection by positively selecting, or capturing, viral sequences using a library of oligonucleotide probes estimated to cover 88 to 98% of target sequences. This library was designed by extracting coding sequences from all nonbacteriophage viruses from the EMBL Coding Domain Sequence database and cross-referencing these with the Master Species List of the International Committee on the Taxonomy of Viruses. Probe sequences were optimized to account for variant sequences and assay chemistry, resulting in a library of nearly 2 million 50-mer to 100-mer nucleotide probes. This library can effectively detect any virus reported to infect vertebrate animals.
Compared experimentally with conventional HTS methods, the VirCapSeq-VERT system showed 100- to 1,000-fold-greater on-target viral reads in lung and whole-blood samples spiked with various quantities of mixed viral nucleic acids, as well as substantial reductions in background host reads. Coverage was drastically improved, with nearly complete genome sequences detected for all viruses. Compared with conventional methods for virus sequence enrichment such as nuclease digestion and rRNA depletion, the VirCapSeq-VERT platform was clearly superior, demonstrating 10,000-fold relative increases in mapped read counts for multiple, diverse RNA and DNA viruses across multiple sample types.
Importantly, the VirCapSeq-VERT method showed high sensitivity in clinical samples (such as nasal swabs and fecal specimens) that may contain viral sequences in very low abundance relative to background host sequences. VirCapSeq-VERT recovered partial sequences even from samples with extremely low copy numbers of input virus, indicating that this method is suitable for detecting virus even from the most limited clinical samples. Furthermore, the assay can be readily multiplexed, and it efficiently detects multiple viruses from the same specimen. The exciting implication of this breakthrough method is that HTS technology may now be applied to clinical diagnostics.
Although the VirCapSeq-VERT method is not completely unbiased, as the oligonucleotide probes were generated based on known viral sequences deposited in the EMBL or NCBI database, Briese and colleagues have shown that this system may be useful for detecting novel or poorly characterized viruses. A novel rodent hepacivirus isolate with less than 65% nucleotide sequence identity from the VirCapSeq-VERT probes was enriched in conserved NS3 helicase and NS5B polymerase genes, and similar enrichment of conserved regions was observed for uncharacterized rotaviruses in bat fecal specimens. Thus, VirCapSeq-VERT may also have some utility for virus discovery by identifying novel viruses through enrichment of conserved sequences.
The VirCapSeq-VERT platform represents the first example of HTS technology with true clinical applicability, capable of achieving a sensitivity equal to that of PCR without the limitations imposed by primer-dependent amplification. Not only does this represent a turning point for clinical diagnostics, in which multiple viruses can be detected even from very spare samples despite high host background, but it can also be used to more accurately characterize the true burden of many viruses in the human population. For example, VirCapSeq-VERT can be applied to determine the prevalence of HHpgV-1 in the blood supply, possibly uncovering disease associations. The VirCapSeq-VERT platform heralds a new era of virus detection, in which any sample can be tested for every known virus simultaneously.
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Footnotes
Citation Rasmussen AL. 2015. Probing the viromic frontiers. mBio 6(6):e01767-15. doi:10.1128/mBio.01767-15.
REFERENCES
- 1.Kapoor A, Kumar A, Simmonds P, Bhuva N, Singh Chauhan L, Lee B, Sall AA, Jin Z, Morse SS, Shaz B, Burbelo PD, Lipkin WI. 2015. Virome analysis of transfusion recipients reveals a novel human virus that shares genomic features with hepaciviruses and pegiviruses. mBio 6:e01466-15. doi: 10.1128/mBio.01466-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. 2011. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A 108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons S, Kapoor A, Sharp C, Schneider BS, Wolfe ND, Culshaw G, Corcoran B, McGorum BC, Simmonds P. 2012. Nonprimate hepaciviruses in domestic horses, United Kingdom. Emerg Infect Dis 18:1976–1982. doi: 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol 86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drexler JF, Corman VM, Müller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C. 2013. Evidence for novel hepaciviruses in rodents. PLOS Pathog 9:e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MOV, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A 110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauck M, Sibley SD, Lara J, Purdy MA, Khudyakov Y, Hyeroba D, Tumukunde A, Weny G, Switzer WM, Chapman CA, Hughes AL, Friedrich TC, O’Connor DH, Goldberg TL. 2013. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old world primate. J Virol 87:8971–8981. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheel TKH, Kapoor A, Nishiuchi E, Brock KV, Yu Y, Andrus L, Gu M, Renshaw RW, Dubovi EJ, McDonough SP, Van de Walle GR, Lipkin WI, Divers TJ, Tennant BC, Rice CM. 2015. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc Natl Acad Sci U S A 112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. 2002. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A 99:15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palacios G, Quan PL, Jabado OJ, Conlan S, Hirschberg DL, Liu Y, Zhai J, Renwick N, Hui J, Hegyi H, Grolla A, Strong JE, Towner JS, Geisbert TW, Jahrling PB, Büchen-Osmond C, Ellerbrok H, Sanchez-Seco MP, Lussier Y, Formenty P, Nichol MS, Feldmann H, Briese T, Lipkin WI. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis 13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briese T, Kapoor A, Mishra N, Jain K, Kumar A, Jabado OJ, Lipkin WI. 2015. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio 6:e01491-15. doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]