Abstract
Background
Ubiquitination is a post-translation modification where ubiquitin is attached to a substrate. Ubiquitin-conjugating enzymes (E2s) play a major role in the ubiquitin transfer pathway, as well as a variety of functions in plant biological processes. To date, no genome-wide characterization of this gene family has been conducted in maize (Zea mays).
Methodology/Principal Findings
In the present study, a total of 75 putative ZmUBC genes have been identified and located in the maize genome. Phylogenetic analysis revealed that ZmUBC proteins could be divided into 15 subfamilies, which include 13 ubiquitin-conjugating enzymes (ZmE2s) and two independent ubiquitin-conjugating enzyme variant (UEV) groups. The predicted ZmUBC genes were distributed across 10 chromosomes at different densities. In addition, analysis of exon-intron junctions and sequence motifs in each candidate gene has revealed high levels of conservation within and between phylogenetic groups. Tissue expression analysis indicated that most ZmUBC genes were expressed in at least one of the tissues, indicating that these are involved in various physiological and developmental processes in maize. Moreover, expression profile analyses of ZmUBC genes under different stress treatments (4°C, 20% PEG6000, and 200 mM NaCl) and various expression patterns indicated that these may play crucial roles in the response of plants to stress.
Conclusions
Genome-wide identification, chromosome organization, gene structure, evolutionary and expression analyses of ZmUBC genes have facilitated in the characterization of this gene family, as well as determined its potential involvement in growth, development, and stress responses. This study provides valuable information for better understanding the classification and putative functions of the UBC-encoding genes of maize.
Introduction
Protein ubiquitination is mediated by the sequential action of an E1 activating enzyme, an E2 conjugating enzyme, and a range of E3 proteins, which are thought to confer substrate specificity [1]. Ubiquitin-conjugating enzymes (E2s) comprise a class of eukaryotic enzymes that function at an intermediate step in the reaction pathway, leading to protein ubiquitination. E2 accepts thioester-linked ubiquitin from E1 to their own active-site cysteine via a transthiolation reaction. E2 then transfers ubiquitin either to a substrate directly aided by an E3 or to a cysteine of an ubiquitin protein ligase (E3s) through a second transthiolation reaction that then transfers ubiquitin to the substrate. E2s for ubiquitin and most UBLs contain a conserved catalytic domain of approximately 140 to 200 amino acids, called the UBC domain [2]. The required conserved cysteinyl residue for thioester formation is contained within this region. This conservation is observed in both sequence and structural comparisons of E2s. For example, the 11 E2s and two ubiquitin-like conjugating enzymes (UBC9 and UBC12) of Saccharomyces cerevisiae exhibit 20%–92% identity [3]. The UBC domains of different E2s have a high degree of sequence homology and adopt similar structures comprised of four α-helices, an anti-parallel β-sheet formed by four strands, and a short 310–helix [4–6]. The highly conserved active-site cysteine (Cys) is located in a shallow groove formed by a short loop connecting α-helix 2 with α-helix 3 and a long loop proximal to the active site [7]. It is thought that the requirement of E2s to interact with E1 and ubiquitin (or its cognates) during E2-ubiquitin thiol ester formation places considerable evolutionary constraints on the E2 structure, resulting in the observed conservation [8].
The protein family has expanded during evolution. Lower eukaryotes have lower numbers of E2 enzymes than higher ones. All eukaryotes possess several E2s, ranging from no more than 20 in algae to a little over 40 in certain multicellular plants and animals. For example, in S. cerevisiae, 16 UBC proteins are found, whereas in human, 40 E2s (including both active E2 proteins and inactive E2 variants) are described [9]. In sequenced algal genomes, 12, 18, and 19 E2 enzymes are encoded in Ostreococcus tauri, Micromonas sp. RCC299, and Chlamydomonas reinhardtii, respectively (data not shown). A total of 48 UBC domain-containing proteins have been identified in Arabidopsis [10], of which three carry thioester-linked UBLs, 2 are Related Ubiqutin (RUB) conjugating enzymes (RCE1, At4g36800 and RCE2, At2g18600), and one is a SUMO-conjugating enzyme (AtSCE1, At3g57870), so while these UBL-specific enzymes function as E2s, they are not ubiquitin E2s. Eight other UBC proteins lack the Cys active site, referred to as ubiquitin-conjugating enzyme variants (UEVs), are not active by themselves, leaving 37 potential E2s [11]. Kraft et al. [12] classified 37 Arabidopsis E2s into 14 groups based on detailed sequence homology analyses. In the rice genome, 48 genes encoding Ub-conjugating (UBC) fold-containing putative E2 proteins were classified into 15 groups [13]. The number of E2s increases with the developmental complexity of the organisms during evolution [14]. Indeed, eukaryotic evolution has also been associated with an increase in the number and diversity of E3s and DUBs [15]. It is now generally believed that genetic expansion and subsequent acquisition of novel molecular functions are the fundamental processes that drive biological diversity [16].
Compared to the extensive studies on E3s, functional studies on E2s in higher plants are limited. Polyubiquitylation through a noncanonical Lys63 chain has been reported, and is required for error-free DNA damage tolerance (or postreplication repair) in plants. Wen et al. [17] cloned and functionally characterized Arabidopsis UBC13 genes, which functionally complement the yeast UBC13 null mutant for spontaneous mutagenesis and sensitivity to DNA damaging agents, suggesting the existence of an error-free DNA damage tolerance pathway in plants. Lately, the same research group isolated and characterized four Arabidopsis UEV1 genes that form a stable complex with AtUBC13, indicating that the UCB13-UEV complex may be involved in DNA repair and damage tolerance [18]. In addition, AtUBC13 has been implicated in Arabidopsis epidermal cell differentiation and iron deficiency responses [17, 19]. COP10 is an E2 enzyme variant lacking a Cys residue that is important for ubiquitin conjugation and necessary for COP10-mediated protein degradation in Arabidopsis [20]. COP10 forms protein complex with other E2s, UV-damaged DNA-binding protein 1A (DDB1A), and de-etiolated 1 (DET1), and the resulting complex plays a crucial role in COP1-mediated photo-morphogenesis and is involved in plant tolerance response against UV-B [21]. AtUBC2 is involved in tolerance response to UV stress, as well as activation of a floral repressor gene [22]. Ectopic expression of E2 gene from wild rice OgUBC1 confers resistance to UV-B radiation and Botrytis infection in Arabidopsis [23]. A tomato UBC13-type homologous protein, FNI3, is involved in the regulation of the immune response [24]. Recent reports have shown that tomato-specific E2 regulates fruit ripening [25]. Two classes of E2s (E2-C and UBC4) capable of contributing to APC-dependent protein ubiquitylation in vivo during the cell cycle have been reported [26]. Arabidopsis encodes two genes belonging to the E2-C gene family (i.e., UBC19 and UBC20), and these proteins functionally replace its yeast ortholog for protein degradation during mitosis, indicating that AtUBC19/20 E2s play a key function in the cell cycle [26]. AtUBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance [27], whereas AtUBC21 (AtPEX4) is specialized for ubiquitination in peroxisome maintenance [28]. While ubiquitination promotes protein degradation, SUMOylation largely regulates protein interactions [29]. There are at least two distinct and functional SUMO E2 conjugases in Caenorhabditis reinhardtii. One (CrUBC9) is involved in essential stress-induced SUMOylations, and one (CrUBC3) is involved in housekeeping SUMOylation [30]. RUB is an evolutionary conserved 76-amino-acid protein that is closely related to ubiquitin, and its conjugating enzyme 1 (RCE1) has been identified in Arabidopsis based on its homology to human UBC12 [31]. RUB conjugation of AtCUL1 affects the function of SCF E3s, which are required for auxin response [32]. PHO2 encodes a putative E2 conjugase, UBC24, which interacts with NLA protein, an E3 ubiquitin ligase, and the NLA-UBC24 complex polyubiquitinates and disrupts the high-affinity Pi transporter, PT2, to regulate Pi homeostasis/signaling in plants [33]. These results suggest that plant E2s play various important roles during plant growth and response to stress.
Maize (Zea mays L.) is an important cereal crop and is the staple food for most people around the world. Under natural conditions, high salinity, drought, and cold are the major environmental stresses experienced by maize plants [34, 35]. The completion of the maize genome sequencing project in 2009 has facilitated in the generation of maize models that have been employed in investigating basic biological processes [36]. The complex history of genome duplications and chromosomal rearrangements in maize provides an opportunity to study gene family expansion patterns over the course of genome evolution [37]. To date, information on the UBC genes of maize has not been report. The present study provides comprehensive information on the genomic structures, chromosomal locations, sequences homologies, and expression patterns of 75 UBC-containing proteins related to maize development and its response to abiotic conditions (drought, cold, or salt stress). The distinct spatiotemporal expression patterns of ZmUBC genes and their differential responses to abiotic stress provides clues for the functional characterization of UBC genes involved in maize development and stress response. The expression patterns of each gene, together with the information of orthologs identified from other species, were used in generating inferences on the potential function of UBC genes in maize. Our study on UBC protein families in maize has provided valuable genomic resources for future evolutionary, biochemical, and physiological studies on maize.
Materials and Methods
2.1 Identification and bioinformatics analysis of candidate genes
To identify potential members of the maize UBC protein family, published Arabidopsis, rice, and yeast UBC protein sequences were retrieved and used as queries in BLASTP searches against the database of the Maize Genetics and Genomics Database (MaizeGDB, http://www.maizegdb.org/) and Phytozome (http://bioinformatics.psb.ugent.be/plaza/versions/plaza/) [38]. The identified putative ZmUBC proteins were designated as ZmUBC1 to ZmUBC75 for convenient discussion of the results. All candidate maize UBC protein sequences were examined using domain analysis programs, Simple Modular Architecture Research Tool (SMART; http://smart.emblheidelberg.de/) [39], Protein family (Pfam; http://pfam.sanger.ac.uk/) [40], and INTERPRO (http://www.ebi.ac.uk/interpro/) [41]. Information on ZmUBC genes, including chromosomal location, coding sequence (CDS), exons and introns number, open reading frame (ORF) and amino acid (AA) lengths, was obtained from the maize B73 sequencing database. The molecular weight and theoretical isoelectric point (PI) of the maize UBC proteins were investigated using ExPASy online tools (http://expasy.org/tools/). The exon-intron organization of ZmUBC genes was determined by comparing the CDS of its corresponding genomic sequences using the Gene Structure Display Server (GSDS) software (http://gsds.cbi.pku.edu.cn/) [42]. The putative localization of all candidate ZmUBC proteins was analyzed by using TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) [43], WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html), and Plant-mPLoc (http://www.csbio.sjtu.edu.cn/cgi-bin/PlantmPLoc.cgi) [44].
2.2 Analysis of phylogenetic relationships and gene duplication
All DNA and amino acid sequence alignments were performed with the MUSCLE program using default parameters [45]. We excluded ambiguously aligned sequences to produce an alignment of 139 amino acid characters for subsequent maximum likelihood (ML) and neighbor joining (NJ) analyses. Phylogenetic trees were constructed with MEGA (Version 6.0; http://www.megasoftware.net/) using the neighbor-joining (NJ) method, and a bootstrap test conducted with 1,000 iterations to test the significance of the nodes [46]. ML analysis was performed on the PhyML web server [47] using the LG substitution model with discrete gamma and four categories. Representations of the calculated trees were constructed using TreeView (Version 1.6.6; http://taxonomy.zoology.gla.ac.uk/rod/treeview). For detection of segmental and tandem duplications, paralogs were regarded as tandem duplicated genes provided two ZmUBCs were separated by eight or fewer gene loci according to the maize B73 genome annotation [48]. Paralogs were designated as segmentally duplicated genes when these were located on duplicated chromosomal blocks as previously described by Wei et al.[49].
2.3 Plant material and stress treatment
Seeds of maize inbred line B73 were used in the present study. The B73 seeds were surface sterilized, washed with sterile water, and germinated in Petri plates in a greenhouse at 28 ± 2°C, with a photoperiod of 14 h light and 10 h dark. Then, the seedlings were transferred to a nutrient solution (half-strong modified Hoagland’s), the pH of the nutrient solution was adjusted to 5.6, and the nutrient solution was changed every 3 days. All environmental treatments were conducted when uniform-sized seedlings developed three fully opened trifoliate leaves (approximately three weeks after sowing). For cold treatment, seedlings were placed at 4°C for 1 h, 6 h, and 24 h, and maize seedlings were collected at different time points for RNA isolation. For drought treatment, three-week-old maize seedlings were treated with 20% PEG6000 and harvested at time points of 1 h, 6 h, and 24 h after treatment. For salt stress treatment, seedlings were subjected to 200 mM NaCl and harvested at time points of 1 h, 6 h, and 24 h after treatment. In all cases, parallel and untreated plants at the same stage were used as controls. The experiments were performed in triplicate. Student’s t-test analysis between untreated seedlings and stress-inoculated plants was performed to identify differential expression patterns of the ZmUBC family genes. The leaves of the seedlings were harvested after treatment, flash-frozen in liquid nitrogen, and stored at -80°C until RNA isolation.
2.4 RNA isolation and expression analysis
Total RNA was extracted from 0.1 g of tissue using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer's instructions. RNA concentrations were determined by using a NanoDrop ND-1000 UV-vis spectrophotometer (NanoDrop Technologies, Inc., Wilmington, USA), and RNA integrity was assessed on a 1% (w/v) agarose gel. Reverse transcription reactions were performed using 5 μg of RNA by M-MLV reverse transcriptase (Takara Bio Inc., Otsu, Japan), following the manufacturer's instructions, after incubation with RNase-free DNase I. Semi-quantitative reverse transcription PCR (RT-PCR) was performed as described elsewhere [50], the PCR conditions were as follows: 94°C for 4 min, followed by 26 cycles of a stepped program (94°C for 40 s, 58°C for 40 s, and 72°C for 30 s), and terminated by an extension at 72°C for 10 min. The maize actin 1 gene was used as reference. For quantitative real-time PCR, total RNAs were first extracted from various maize samples, following the instructions provided with the TRIzol reagent (Takara). Reverse transcription reactions were performed using 5 μg of RNA with the PrimeScript RT reagent kit with gDNA Eraser (Takara), according to the supplier’s manual. Real-time PCR was performed with a Bio-Rad real-time thermal cycling system (LightCycler 480) using SYBR-green to assess gene expression levels. Each reaction consisted of 10 μL of 2 ×SYBR Premix Ex Taq™ II (Takara), 2.0 μL of each cDNA sample, and 400 nM of a gene-specific primer, in a final volume of 20 μL. The primers used are listed in S1 Table. The reaction conditions were as follows: pre-incubation at 94°C for 5 min, followed by 40 cycles at 94°C for 10 s, 58°C–63°C for 20s, 72°C for 30 s. After amplication was complete, a melting curve was obtained by holding at 95°C for 5 s and then at 65°C for 15 s, followed by heating slowly at 0.1°C/s to 95°C. Quantification of results was obtained by using a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., USA). The specificity of the reactions was verified by melting curve analysis. The relative mRNA level for each gene was calculated as △△C T values relative to that of untreated seedlings. Maize actin 1 gene was used as internal control for normalization. cDNAs from three biological samples were used for analysis, and all the reactions were run in triplicate. In the comparative expression analysis of ZmUBCs, genes that were up- or downregulated by at least two-fold were considered differentially expressed.
Results and Discussion
Identification of maize genes encoding UBCc-E2 proteins
In the present study, we used the previously reported E2 proteins from Arabidopsis, rice, and yeast as BLAST queries to search for maize E2s in the MaizeGDB and Phytozome10.3 databases. A total of 82 genes encoding UEVs were detected in maize. E2s for conjugating Rub (family UBC12) and SUMO (family UBC9) were also identified. After the redundant sequences were removed, a total of 80 sequences were obtained. Further scanning of the 80 sequences for the UBCc domain by motif scan using SMART, Pfam, or INTERPRO searches with filter off by default setting reduced the total number of maize UBC protein family members to 75. For convenience, the 75 ZmUBC genes were named ZmUBC-01 to ZmUBC-75 based on their order on the chromosomes, from chromosomes 1 to 10. The information of these 75 UBC-encoding genes, including TIGR locus, chromosome location, ORF, intron, size, PI, MW (kDa), and putative localization of each protein is shown in Table 1. The size of the deduced UBC proteins varied markedly, ranging from 133 amino acids (ZmUBC-29) to 1,102 amino acids (ZmUBC-56), its corresponding molecular mass ranged from 12.49 kDa to 121.98 kDa, and its predicted isoelectric point was between 4.09 (ZmUBC-02) and 11.07 (ZmUBC-07). In addition, the subcellular localization analysis showed that most of the ZmUBC proteins were located in the nucleus, only a few ZmUBC proteins had functions related to the Cytoplasm, chloroplast, mitochondrion, or endoplasmic reticulum. These findings suggested that different UBC proteins functioned in different micro-environments. These result were partially confirmed by previous reports involving the AtPEX4 protein, which was localized in the peroxisome, AtUBC32 in the ER, and AtPHO2 in the endomembrane [27, 28, 51], GmUBC2 in both the cytosol and nucleus, and AtCOP10, a UEV that has been localized to the nucleus [52].
Table 1. The information of ZmE2 gene family.
| Gene name | Gene locus | Chromosome Location | ORF(bp) | Size(aa) | PI | MW(KDa) | Intron | Putative localizatin |
|---|---|---|---|---|---|---|---|---|
| ZmUBC-01 | GRMZM2G070047 | Chr1: 4,981,301–4,984,643 | 483 | 160 | 8.43 | 18.03 | 4 | Nucleus |
| ZmUBC-02 | GRMZM2G150867 | Chr1: 10,590,399–10,593,290 | 552 | 183 | 4.09 | 19.16 | 4 | Nucleus |
| ZmUBC-03 | GRMZM5G824629 | Chr1: 49,614,813–49,617,825 | 945 | 314 | 5.52 | 34.45 | 4 | Nucleus |
| ZmUBC-04 | GRMZM2G312693 | Chr1: 83,176,776–83,179,023 | 483 | 160 | 8.43 | 17.99 | 4 | Nucleus |
| ZmUBC-05 | GRMZM2G007381 | Chr1: 180,012,788–180,017,472 | 486 | 161 | 7.72 | 18.25 | 5 | Nucleus |
| ZmUBC-06 | GRMZM2G053764 | Chr1: 236,088,923–236,128,384 | 1113 | 370 | 4.60 | 41.69 | 10 | Nucleus |
| ZmUBC-07 | GRMZM2G116840 | Chr1: 256,367,147–256,407,916 | 417 | 138 | 11.07 | 15.44 | 1 | Nucleus |
| ZmUBC-08 | GRMZM2G022859 | Chr1:267,922,012–267,928,571 | 486 | 161 | 5.13 | 18.36 | 4 | Nucleus |
| ZmUBC-09 | GRMZM2G120674 | Chr1: 286,845,415–286,850,582 | 459 | 152 | 5.37 | 17.30 | 4 | Nucleus |
| ZmUBC-10 | GRMZM2G102471 | Chr2: 2,803,488–2,806,329 | 447 | 148 | 7.72 | 16.51 | 3 | Nucleus |
| ZmUBC-11 | GRMZM2G038851 | Chr2: 15,245,261–15,247,000 | 480 | 159 | 9.12 | 17.73 | 4 | Nucleus |
| ZmUBC-12 | GRMZM2G341089 | Chr2: 15,269,254–15,273,461 | 480 | 159 | 5.25 | 12.57 | 2 | Nucleus |
| ZmUBC-13 | GRMZM2G016176 | Chr2: 161,497,123–161,501,579 | 762 | 253 | 10.16 | 28.54 | 10 | Nucleus |
| ZmUBC-14 | GRMZM2G000601 | Chr2: 172,703,543–172,707,425 | 447 | 148 | 6.40 | 16.66 | 1 | Nucleus |
| ZmUBC-15 | GRMZM2G113396 | Chr2: 206,899,426–206,903,057 | 573 | 190 | 9.58 | 20.84 | 3 | Nucleus |
| ZmUBC-16 | GRMZM2G022206 | Chr2: 233,793,739–233,798,436 | 459 | 152 | 5.64 | 17.34 | 3 | Nucleus |
| ZmUBC-17 | GRMZM2G010460 | Chr3: 1,684,241–1,688,895 | 1122 | 373 | 8.39 | 41.93 | 6 | Nucleus |
| ZmUBC-18 | GRMZM2G123519 | Chr3: 1,726,294–1,729,668 | 1155 | 384 | 4.91 | 43.51 | 4 | Nucleus |
| ZmUBC-19 | GRMZM2G086583 | Chr3: 24,458,611–24,460,536 | 804 | 267 | 6.71 | 30.29 | 3 | Nucleus |
| ZmUBC-20 | GRMZM2G002830 | Chr3: 41,472,011–41,473,369 | 552 | 183 | 5.17 | 19.57 | 2 | Nucleus |
| ZmUBC-21 | GRMZM2G434519 | Chr3: 55,353,651–55,355,715 | 525 | 174 | 6.96 | 19.60 | 3 | Nucleus |
| ZmUBC-22 | GRMZM2G018447 | Chr3: 93,187,188–93,205,169 | 486 | 161 | 7.72 | 18.23 | 5 | Nucleus |
| ZmUBC-23 | GRMZM5G866947 | Chr8: 170,631,089–170,636,009 | 447 | 148 | 7.71 | 16.49 | 3 | Nucleus |
| ZmUBC-24 | GRMZM2G007300 | Chr3: 181,446,796–181,451,566 | 510 | 169 | 5.05 | 18.98 | 5 | Nucleus |
| ZmUBC-25 | GRMZM5G862131 | Chr3: 209,846,496–209,850,012 | 462 | 153 | 6.74 | 17.20 | 7 | Nucleus |
| ZmUBC-26 | GRMZM5G814314 | Chr3: 212,436,396–212,440,285 | 444 | 147 | 7.72 | 16.55 | 3 | Nucleus |
| ZmUBC-27 | GRMZM2G157605 | Chr3: 223,745,568–223,748,681 | 1734 | 577 | 8.57 | 63.23 | 5 | Nucleus |
| ZmUBC-28 | GRMZM2G106143 | Chr4: 50,730,408–50,737,299 | 954 | 317 | 9.74 | 35.90 | 11 | Cytoplasm |
| ZmUBC-29 | GRMZM2G461533 | Chr4: 145,324,353–145,325,491 | 402 | 133 | 8.42 | 14.90 | 4 | Nucleus |
| ZmUBC-30 | GRMZM2G161545 | Chr4: 145,368,230–145,371,847 | 474 | 157 | 8.69 | 17.80 | 3 | Nucleus |
| ZmUBC-31 | GRMZM2G027378 | Chr4: 220,508,652–220,512,101 | 447 | 148 | 6.42 | 16.77 | 3 | Nucleus |
| ZmUBC-32 | AC233922.1_FGT008 | Chr4: 238,041,057–238,042,180 | 447 | 148 | 7.70 | 16.51 | 2 | Nucleus |
| ZmUBC-33 | GRMZM2G007260 | Chr5: 4,495,065–4,499,934 | 618 | 205 | 6.08 | 13.54 | 4 | Nucleus |
| ZmUBC-34 | GRMZM2G090172 | Chr5: 10,931,101–10,936,807 | 501 | 166 | 5.40 | 18.93 | 4 | Cytoplasm. Nucleus |
| ZmUBC-35 | GRMZM2G148130 | Chr5: 13,646,747–13,649,731 | 495 | 164 | 6.02 | 14.73 | 3 | Nucleus |
| ZmUBC-36 | GRMZM2G146374 | Chr5: 31,648,146–31,651,787 | 624 | 207 | 4.49 | 23.32 | 3 | Nucleus |
| ZmUBC-37 | GRMZM2G115939 | Chr5: 56,740,173–56,751,799 | 555 | 184 | 8.67 | 20.66 | 4 | Nucleus |
| ZmUBC-38 | GRMZM2G466265 | Chr5: 70,756,337–70,760,939 | 1149 | 382 | 9.11 | 43.33 | 6 | Nucleus |
| ZmUBC-39 | GRMZM5G828302 | Chr5: 188,118,457–188,127,477 | 474 | 157 | 8.69 | 17.80 | 3 | Nucleus |
| ZmUBC-40 | GRMZM2G411771 | Chr5: 193,685,680–193,722,957 | 984 | 327 | 4.86 | 36.65 | 5 | Cytoplasm |
| ZmUBC-41 | GRMZM2G147579 | Chr5:217,470,848–217,472,628 | 1437 | 478 | 9.65 | 52.13 | 0 | Nucleus |
| ZmUBC-42 | GRMZM2G110983 | Chr6: 66,201,455–66,205,271 | 537 | 178 | 4.82 | 12.82 | 4 | Nucleus |
| ZmUBC-43 | GRMZM2G102421 | Chr6: 87,278,503–87,284,723 | 552 | 183 | 8.36 | 20.63 | 4 | Nucleus |
| ZmUBC-44 | GRMZM5G895435 | Chr6: 94,713,649–94,717,995 | 762 | 253 | 8.98 | 27.47 | 4 | Nucleus |
| ZmUBC-45 | GRMZM2G116919 | Chr6:107,362,893–107,368,461 | 720 | 239 | 9.01 | 26.91 | 8 | Nucleus |
| ZmUBC-46 | GRMZM2G156517 | Chr6: 110,386,088–110,389,342 | 447 | 148 | 7.68 | 16.52 | 3 | Nucleus |
| ZmUBC-47 | GRMZM2G012052 | Chr6: 151,770,259–151,778,529 | 510 | 169 | 5.04 | 18.97 | 4 | Nucleus |
| ZmUBC-48 | GRMZM2G085849 | Chr6: 163,835,366–163,840,407 | 1524 | 507 | 5.61 | 56.87 | 7 | Nucleus |
| ZmUBC-49 | GRMZM2G381709 | Chr6: 163,843,112–163,847,600 | 2616 | 871 | 4.77 | 96.37 | 7 | Nucleus |
| ZmUBC-50 | GRMZM2G007122 | Chr6: 164,260,122–164,263,444 | 465 | 154 | 6.67 | 17.34 | 7 | Nucleus |
| ZmUBC-51 | GRMZM2G433968 | Chr7: 12,311,212–12,312,732 | 534 | 177 | 4.73 | 19.29 | 2 | Nucleus |
| ZmUBC-52 | GRMZM2G173756 | Chr7: 79,006,728–79,011,032 | 447 | 148 | 6.40 | 16.68 | 1 | Nucleus |
| ZmUBC-53 | GRMZM2G056501 | Chr7: 82,007,091–82,010,522 | 486 | 161 | 6.96 | 18.30 | 3 | Nucleus |
| ZmUBC-54 | GRMZM2G072506 | Chr7: 160,242,028–160,246,175 | 555 | 184 | 8.53 | 19.84 | 3 | Nucleus |
| ZmUBC-55 | GRMZM2G007057 | Chr8: 3,195,388–3,196,814 | 765 | 254 | 5.36 | 19.5 | 2 | Nucleus |
| ZmUBC-56 | GRMZM2G078360 | Chr8: 10,926,318–10,939,316 | 3309 | 1102 | 4.55 | 121.98 | 6 | Nucleus |
| ZmUBC-57 | GRMZM2G122003 | Chr8: 28,846,608–28,852,930 | 1317 | 438 | 5.14 | 49.63 | 5 | Nucleus |
| ZmUBC-58 | GRMZM2G027546 | Chr8: 67,380,589–67,387,931 | 1563 | 520 | 5.42 | 58.37 | 7 | Nucleus |
| ZmUBC-59 | GRMZM2G015287 | Chr8: 68,928,768–68,932,536 | 462 | 153 | 6.74 | 17.21 | 7 | Nucleus |
| ZmUBC-60 | GRMZM2G007276 | Chr8: 116,776,837–116,781,561 | 510 | 169 | 6.28 | 12.49 | 3 | Nucleus |
| ZmUBC-61 | GRMZM2G132759 | Chr8: 119,039,106–119,043,451 | 510 | 169 | 7.71 | 16.50 | 3 | Nucleus |
| ZmUBC-62 | GRMZM2G086088 | Chr8: 148,200,019–148,203,135 | 444 | 147 | 7.72 | 16.55 | 3 | Nucleus |
| ZmUBC-63 | GRMZM2G115828 | Chr8: 150,010,557–150,014,289 | 468 | 155 | 6.74 | 17.19 | 7 | Nucleus |
| ZmUBC-64 | GRMZM2G134176 | Chr8: 160,396,937–160,401,812 | 594 | 197 | 4.77 | 21.39 | 4 | Nucleus |
| ZmUBC-65 | GRMZM2G085600 | Chr8: 169,009,649–169,013,585 | 510 | 169 | 4.40 | 16.43 | 5 | Nucleus |
| ZmUBC-66 | GRMZM2G177276 | Chr9: 11,703,081–11,707,342 | 720 | 239 | 9.01 | 26.99 | 8 | Nucleus |
| ZmUBC-67 | GRMZM2G464572 | Chr9: 56,902,708–56,907,334 | 2616 | 871 | 4.74 | 96.37 | 7 | Nucleus |
| ZmUBC-68 | GRMZM2G153924 | Chr9: 105,689,843–105,694,388 | 759 | 252 | 9.22 | 27.38 | 4 | Nucleus |
| ZmUBC-69 | GRMZM2G163398 | Chr9: 125,296,709–125,299,946 | 483 | 160 | 7.76 | 18.07 | 4 | Nucleus |
| ZmUBC-70 | GRMZM2G121303 | Chr9: 135,149,922–135,154,003 | 552 | 173 | 5.40 | 18.72 | 2 | Nucleus |
| ZmUBC-71 | GRMZM2G440918 | Chr9: 143,394,526–143,397,331 | 957 | 318 | 9.03 | 35.37 | 3 | Nucleus |
| ZmUBC-72 | AC149818.2_FGT006 | Chr9: 152,442,986–152,445,781 | 552 | 183 | 4.27 | 20.87 | 5 | Nucleus |
| ZmUBC-73 | GRMZM2G063931 | Chr9: 153,779,802–153,784,716 | 483 | 160 | 8.42 | 18.01 | 4 | Nucleus |
| ZmUBC-74 | GRMZM2G109582 | Chr10: 21,820,190–21,821,376 | 1053 | 350 | 6.16 | 38.01 | 0 | Nucleus |
| ZmUBC-75 | GRMZM2G146142 | Chr10: 62,900,930–62,907,699 | 1146 | 381 | 9.99 | 42.86 | 8 | Nucleus |
Chromosomal distribution and gene duplication of maize UBCc-E2 genes
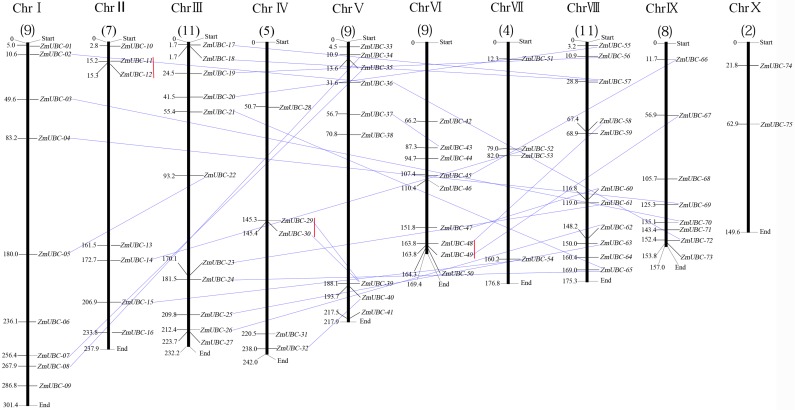
Chromosomal location analyses showed that 75 ZmUBC genes were distributed across 10 chromosomes (Fig 1). The distribution of the ZmUBC genes was highly variable; only 2 genes were detected on chromosome X, 4 genes was found on chromosome VII, 5 genes was found on chromosome IV, 7 genes were localized to chromosome II, 8 genes on chromosome IX, whereas chromosomes I, V andVI harbored 9 ZmUBC genes. The largest number of ZmUBC genes was identified on chromosomes III and VIII (11 genes). According to the definition of gene clusters [53], out of 75 maize homeobox genes, 21 gene pairs were assigned to 10 maize linkage groups, whereas the rest were located on scaffolds. Some ZmUBC genes were sparsely situated on maize chromosomes, whereas some were densely distribution in other chromosomes.
Fig 1. Chromosomal and gene duplication events involving ZmUBC genes.
The chromosomal position of each UBC-encoding gene was mapped to the maize genome. The chromosomal number is indicated at the top of each chromosome. The number below represented the number of UBC-encoding genes in each chromosome. The segmental duplication genes are connected by a blue straight line. The tandemly duplicated genes are linked by a red straight line. Chromosomal distances are in Mbp.
Gene duplication plays an extremely important role in gene family expansion and protein functional diversification [54]. To explore the contribution of duplication events to this family, we analyzed the occurrence of tandem duplications and large-scale segmental duplications during the evolution of this gene family. Phylogenetic analysis and chromosomal localization of ZmUBC genes determined that a total of 24 duplicated maize gene pairs (48 of 75 ZmUBC genes, 64%) underwent segmental duplication, and three gene pairs (6 of 75 ZmUBC genes, 8%) were involved in tandem duplication (ZmUBC-11 and ZmUBC-12, ZmUBC29 and ZmUBC30, ZmUBC48 and ZmUBC49) (Fig 1). A recent report showed that 15 of 39 (38.5%) OsUBCs resulted from segmental duplication events, and four genes (10.2%) from tandem duplication, indicating the different roles of gene duplication in the evolution of E2s between the maize and rice genomes [55]. The three tandemly duplicated ZmUBC genes were localized to chromosomes 2, 4, and 6 and grouped into subfamilies UBC9, UBC10, and UBC15, respectively. These findings suggested that gene duplication events, particularly segmental duplications, contributed to the expansion of the ZmUBC family in maize.
Phylogenetic analysis of the ZmUBC gene family
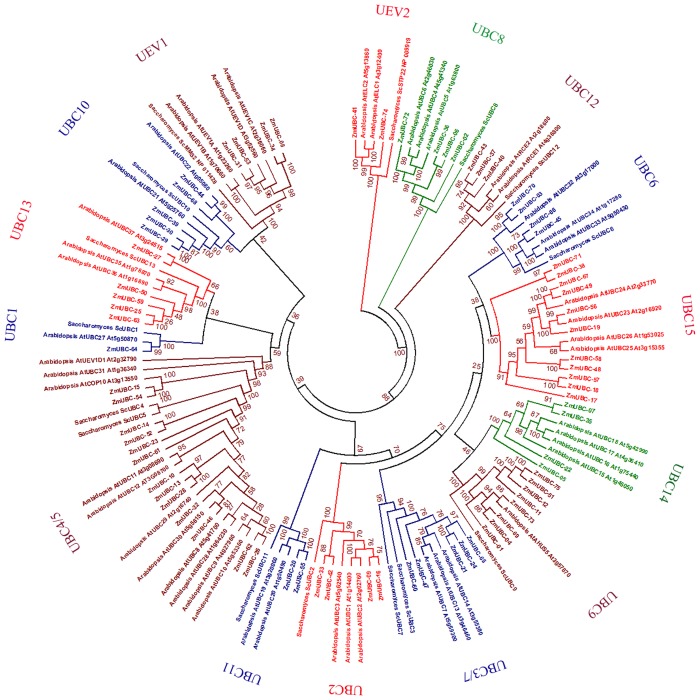
To evaluate the evolutionary relationship of the ZmUBC proteins, full-length amino acid sequences of 75 ZmUBC genes, 15 UBC-encoding genes from Saccharomyces cerevisiae, and 48 UBC-encoding genes from A. thaliana were subjected to a multiple sequence alignment using MUSCLE and later manually optimized. The alignment was then used for the construction of an unrooted phylogenetic tree using the ML and NJ methods. Fig 2 and S1 Fig shows that based on the consistent topology between ML and NJ trees, except for some tiny differences, the UBC proteins could be further classified into 13 E2 groups and two independent UEV groups as monophyletic clades with at least 50% bootstrap support, with groups UBC9 and UBC12 functioning in SUMO and RUB1 conjugation pathways and COP10. The third UEV group was nested in the UBC4/5 subfamily. The subfamilies were named according to their identity to S. cerevisiae UBC proteins: UBC1, UBC2, UBC3, UBC4/5, UBC6, UBC3/7, UBC8, UBC9, UBC10, UBC11, UBC12, UBC13, UBC14, UBC15, UEV1 and UEV2. The subfamilies UBC4/5 and UBC3/7 shared two highly identical paralogous yeast genes respectively, according to previous reports we named this two subclades as subfamily UBC4/5 and UBC3/7 [14, 56]. The result is consistent with the findings of previous studies [12]. The analysis shows that all maize UBC proteins have orthologous Arabidopsis proteins and green algae proteins (data not shown). These findings also indicated that the highly complex extant genomes and the expansion of Arabidopsis UBC-encoding genes were derived from a common ancestor.
Fig 2. Phylogenetic relationship of various ZmE2s.
The ML tree includes 75 UBC proteins from Zea mays, 15 from Saccharomyces cerevisiae, and 48 from Arabidopsis thaliana. The tree shows 15 phylogenetic subgroups depicted in various colors to distinguish diversification of subfamilies into clusters.
Comparison of the number of UBC-encoding genes and its corresponding mRNAs within and between species indicated that both gene duplication and alternative splicing contributed to the genomic complexity and proteomic diversity of the ZmUBC genes. The phylogenetic tree shows that UBC4/5 and UBC15 underwent considerable expansion of 13 and 11 E2s, respectively, which comprised the largest subfamily, whereas the clades of UEV2, UBC1, and UBC11 only have one or two UBC proteins in maize. The remaining subfamilies have an intermediate number E2s, ranging from 3 in UBC12 to 8 in UBC9. These findings indicated that each group has a different evolutionary history that finally formed the E2 family in the extant species. Alternative splicing can generate more transcripts from a single gene than the number of genes in an entire genome. The observed high percentages [(human: 19/50 (38%), Arabidopsis: 21/48 (43.8%), maize: 52/75 (69.3%)] of alternatively spliced transcripts indicated that maize E2s underwent extensive expansion at the transcriptional level.
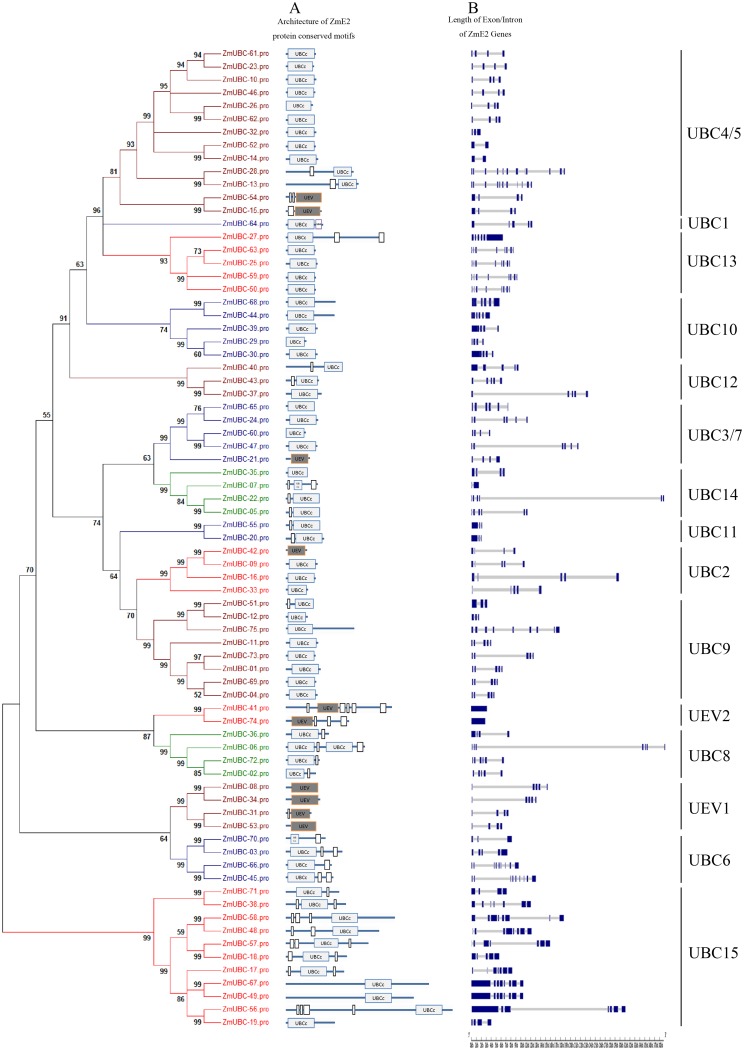
Gene structure and conserved motif distribution analysis
We then used the SMART program to analyze the conserved motifs in UBC proteins. Fig 3A shows that according to the UBC/UEV domain and the N- or C-terminal structure, the ZmUBC proteins could be further divided into four classes [2, 12, 57]. Class I proteins mainly comprised the UBC/UEV domain, which contained 42 ZmUBC proteins, and those genes contained in all groups except UBC 1, 6, 8, and 11 and UEV 2. Class II proteins harbored the UBC/UEV domain plus a C-terminal extension, which contained 15 UBC proteins, and those genes contained in UBC 4/5, 9, 11, 12, 14, 15, and UEV 1. Class III proteins have the UBC/UEV domain plus an N-terminal extension, which contained 12 ZmUBC proteins, and those genes contained in UBC 1, 6, 8, 13, 15, and UEV2. Class IV proteins have the UBC/UEV domain plus both the N-terminal and C-terminal extension, which consisted of 6 ZmUBC proteins, and those genes contained in UBC 14, UBC 15, and UEV 2.
Fig 3. The architecture of conserved protein motifs and intron pattern of ZmE2s.
(A) Architecture of conserved protein motifs of UBC proteins. (B) The gene structure is presented by blue exon(s) and spaces between the blue box corresponding to intron (s). The sizes of the exons and intron can be estimated using the horizontal lines.
To better understand the structural diversity of ZmUBC genes, its intron/exon arrangements and conserved motifs were also compared. We obtained each gene structure by comparing their ORFs with their corresponding genomic sequences. Table 1 and Fig 3B shows that all the ZmUBC genes possessed at least 1 intron. The most common pattern contained 3–4 introns (63% of ZmUBCs). In addition, most members within the same subfamily shared the same exon/intron structure. For example, in subfamily UBC 2, ZmUBC-09, ZmUBC-16, ZmUBC-33, and ZmUBC-42 consisted of four introns, ZmUBC-20 and ZmUBC-55 in subfamily UBC11 harbored two introns, and UBC 11, 12, UEV 1, and UEV 2 presented the same exon/intron structure. The members of UBC9, 10, and 13 within the same subfamily shared two kinds of exon/intron structures. For example, in UBC 10, ZmUBC-29, ZmUBC-30, and ZmUBC-39 consisted of three introns, and two genes (ZmUBC-44 and ZmUBC-68) harbored four introns. In contrast to the highly conserved structural patterns in the above groups, the members of UBC 4/5, 3/7, 6, 8, 14, and 15 showed a complex distribution of exons and introns, including different pattern subsets within the same phylogenetic group. For instance, in group 4/5, 10 genes harbored less than 3 introns, whereas two genes consisted of more than 10 introns. Overall, most closely related members in the same subfamilies shared a similar exon/intron structure in terms of intron number and exon length.
The expression pattern of ZmUBC genes
Because gene expression patterns have been examined in relation to gene function [58], the expression of the ZmUBC gene in different tissues was investigated. Using a semi-quantitative RT-PCR approach, the mRNA accumulation of each gene in the root, stem, leaf, tassel, young seed (YS), and silk tissues was assessed. Fig 4 shows that among the 75 UBC-encoding genes, 10 genes were constitutively expressed in different tissues at similar levels, and 61 genes were expressed in at least one tissue, whereas the transcript levels of 4 genes were undetectable, suggesting these might be pseudogenes or might be expressed at specific developmental stages, or under special conditions.
Fig 4. Tissue-specific expression of ZmUBC genes.
(A) Semi-quantitative RT-PCR analysis of ZmUBC genes. (B) RT-qPCR analysis of representative ZmUBC genes. The letter R above the column of the expression data refers to root, St represents stem, L indicates leaf, T refers to tassel, Ys indicates young seed, and Si represents silk.
By combining these results, we determined that among the 61 ZmUBC genes, 4 genes were only detected in one tissue, 3 genes were detected in two tissues, 8 genes in three tissues, 5 genes in four tissues, 11 genes in five tissues, and 30 genes in all tested tissues with tissue-specific expression profiles. Intriguingly, the expression of ZmUBC-12, ZmUBC-20, and ZmUBC-72 was only detected in YSs, suggesting that those genes might be involved in the formation and development of fruit. To validate the results of semi-quantitative PCR analysis, RT-qPCR analysis was performed on representative genes. The results indicated that the expression patterns of these genes were in general agreement with the findings of RT-PCR analysis. Six tested genes (ZmUBC-29/31/45/53/60/75) showed high expression levels in YS, with an approximately 10-40-fold higher level of expression than that observed in the root. Similar results were also reported in rice. Five rice UBCs (OsUBC13/17/18/26/35) exhibited either predominant or tissue-specific expression in YR, YL, or ML, and four genes (OsUBC13/32/33/34) were highly expressed in leaves [55]. In Arabidopsis, AtUBC1 and AtUBC2 were ubiquitously expressed in the roots, leaves, flowers, and seedlings, and the double mutant atubc1-1atubc2-1 showed a dramatically reduced number of rosette leaves and an early-flowering phenotype [22]. However, OsUBC7 and OsUBC8, orthologs of AtUBC1 and AtUBC2, were upregulated in most organs, particularly in YR and ML. Collectively, our results indicated that ZmUBC genes played multiple roles in the development of maize.
Expression profiles of ZmUBC genes in response to various stressors
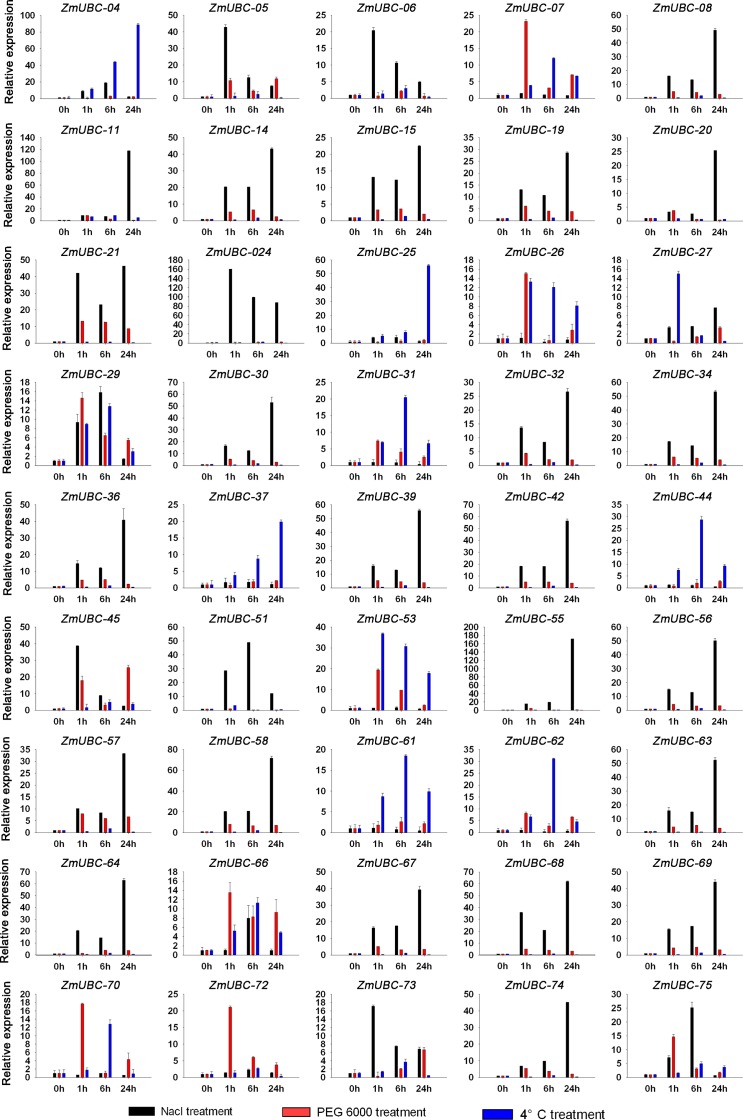
Plants and crops are frequently challenged by abiotic stressors such as salt, drought, and low temperature. Recent studies have suggested that UBC proteins are widely involved in signaling and response to abiotic stimuli [23, 27, 55], although information on the involvement of UBC proteins in stress responses in maize is limited. To investigate the potential roles of ZmUBC genes in response to environmental stresses, we examined their expression patterns under salt (200 mM NaCl), drought (20% PEG 6000), and cold (4°C) treatment as described in the Materials and Methods section. Untreated seedlings grown in nutrient solution were used as control seedlings.
Fig 5 and S2 Fig shows that under NaCl treatment, all of the ZmUBCs tested, except 14 genes (ZmUBC-01/07/16/26/31/37/44/46/47/53/61/62/70), were upregulated by salt stress. Twenty-six genes (ZmUBC-02/03/04/05/06/09/10/12/18/22/24/25/28/29/33/35/45/48/49/50/51/52/54/65/73/75) were upregulated at early time points during treatment, downregulated at later time points. Thirty-five genes (ZmUBC-08/11/13/14/15/17/19/20/21/23/24/27/30/32/34/36/38/39/40/41/42/43/55/56/57/58/59/60/63/64/66/67/68/69/71) were upregulated throughout the NaCl treatment. The ZmUBC-08/11/24/30/34/39/42/58/63/64 genes were upregulated by >50-fold at the 24 h time point of the NaCl treatment. By comparing the expression data of each pair of duplicated ZmUBC genes, 14 pairs of duplicated genes (ZmUBC-5/22; ZmUBC-08/34; ZmUBC-19/56; ZmUBC-20/55; ZmUBC29/30/39; ZmUBC21/24/65; ZmUBC48/58; ZmUBC25/63; ZmUBC26/62; ZmUBC15/54; ZmUBC14/52; ZmUBC-04/69; ZmUBC49/67; and ZmUBC45/66) in salt stress showed a similar expression pattern. However, 8 pairs of duplicated genes (ZmUBC-02/36/72, ZmUBC-03/70; ZmUBC07/35; ZmUBC-17/18/57; ZmUBC-23/61; ZmUBC-32/40; ZmUBC37/43; and ZmUBC47/60) showed completely differently expression profiles during salt stress, which can be attributed to lack of intense selection pressure and need for diversification.
Fig 5. Differential expression levels of ZmUBC genes following NaCl, PEG 6000, and 4°C treatment.
The X-axis indicates time course/treatment and the Y-axes are scales of relative expression levels. The maize actin gene was used as internal control. The presented data are representative of three independent experiments.
For PEG6000 treatment, 34 genes (ZmUBC-06/08/11/14/15/18/22/26/28/29/30/31/32/34/36/39/40/41/42/47/53/55/56/57/58/62/63/65/67/68/69/70/72/74/75) were upregulated during early time points, and then downregulated at later time points. The expression levels of 22 genes (ZmUBC-02/03/05/07/09/10/13/16/17/19/20/21/23/27/35/44/45/46/48/64/66/73) were continuously increased throughout the time course. On the other hand, 19 genes (ZmUBC-01/04/12/17/ 24/25/33/37/38/43/49/50/51/53/54/59/60/61/71) were not responsive to PEG-monitored drought stress. For the 4°C treatment, 28 genes (ZmUBC-03/07/09/10/11/16/18/26/27/28/29/31/40/41/44/45/50/51/52/53/57/61/62/66/70/75) were upregulated during the early time points of the treatment, then downregulated at the later time points. The expression level of 42 genes(ZmUBC-01/02/05/06/08/12/13/14/15/17/19/20/21/22/23/24/30/32/34/35/36/38/39/42/43/47/54/55/56/58/59/60/63/65/67/68/69/71/72/73/74) did not change with the 4°C treatment, whereas five genes (ZmUBC-04/25/33/37/46) were upregulated with the 4°C treatment. ZmUBC-25 was upregulated by >50-fold at the 24 h time point during cold treatment.
Similar to salt stress, 18 and the 14 pairs of duplicated genes under drought and cold stress, respectively, showed a similar expressional pattern, whereas 8 and 14 pairs of duplicated genes under drought and cold stress, respectively, showed drastically differential expression profiles. In addition, a total of 6 ZmUBCs (ZmUBC-07, ZmUBC-26, ZmUBC-45, ZmUBC-53, ZmUBC-70, and ZmUBC-75) were markedly upregulated at least by two stimuli, and two genes, ZmUBC29 and ZmUBC65, was strongly induced by three stimuli, indicating that these genes played a complex role in response to abiotic stress. We concluded that ZmUBCs played an essential role in response to abiotic stresses. This conclusion was supported by the previously reported plant homologous genes such as AtUBC32, AtUBC24, OgUBC1, CmUBC2, and VrUBC1 [23, 27, 51, 55], which were identified as salt-responsive genes and regulate salt tolerance in plants.
Discussion
Ubiquitin-conjugating enzymes are an integral component of the ubiquitin proteasome system that is involved in very important roles during plant development and growth. Despite the potential functional significance of UBC members, only a few UBC family members have been described in higher plants. Maize (Z. mays L.) is one of the most important cereal crops and a plant model for investigations in genetics, evolution, and domestication. Drought, cold, and salt stresses can all be harmful to plants by causing cellular desiccation, and plants combat these three stresses using a common mechanism [55]. This background knowledge prompted us to identify the full complement and expression profile of this important gene family during development and under abiotic stresses in maize.
In the present study, a comprehensive set of 75 nonredundant UBC-encoding genes (including 69 E2s and 6 UEVs) were identified and characterized from the current version of the maize (B73) genome. Exploration for the full complement of UBC proteins in Arabidopsis and rice genomes resulted in the identification of 45 and 48 genes, respectively [12, 55]. A total of 52 nonredundant E2 genes have also recently been identified in tomato [25]. The observed higher number of UBC-encoding genes in maize can not only be explained by its larger genome size (~2,300 Mb) compared to that of Arabidopsis and rice (~125 Mb and ~389 Mb), as a similar number of UBC-encoding genes exist in these species (48 and 45 genes, respectively), although the size of the rice genome is ~3.7 times larger than that of Arabidopsis. Gene duplication events play a significant role in the amplification of gene family members in the genome [55]. Research has estimated that the fraction of retained paralogs is 72% in maize, which occurred over the course of the past 11 million years [59]. The expansion mechanism of the ZmUBC gene family was analyzed to understand gene duplication events. In our study, a total of 24 sister gene pairs of maize UBC proteins were determined to be involved in segmental duplications by shared phylogenetic clade combinations within the same groups and by locations within the segmental duplicated blocks. Three maize UBC genes involved in tandem duplication were localized to chromosomes 2, 3, and 4, respectively, forming a single cluster. These results suggest that segmental duplications are the main contributor to the expansion of the maize UBC family. Therefore, it can be inferred that the expansion of the ZmUBC gene family might not solely rely on the independent duplication of individual sequences and thus might also be the consequence of segmental chromosomal duplication and rearrangement events. An increasing number of studies has shown that segmental duplications are largely responsible for the expansion of maize gene families, which include the CCCH, HD-Zip HSF, bZIP, and PRX gene families [60–63].
The phylogenetic analysis categorized all the ZmUBCs into 13 discrete E2 groups and 2 UEV groups. Our classification resulted in similar clusters, similar to that described in previous studies involving Arabidopsis [12], tomato [25], and rice [55], although with minimal differences. For example, two genes, AtUBC21 and AtUBC22, were grouped as a subfamily with weak bootstrap support (60%) in our study, whereas their orthologs were divided into two subfamilies in previous results [12, 25, 55]. The congruity of tree topology in different studies indicated the reliability of our clade- and subgroup designations. The phylogenetic tree showed obvious differences in numbers among subfamilies. Subfamilies UBC4/5, UBC15, and UBC9 have 13, 11, and 8 members, respectively, whereas less than 3 members comprised subfamilies UBC1, UBC11, and UBC12. The change in the number of ZmUBC members suggested that the ZmUBC family had undergone lineage-specific expansion and functional divergence during the course of evolution. Phylogenetic data also suggested that UBC9 and UBC15 expanded in monocots but not in Arabidopsis. In subclasses UBC9 and UBC15, one and four members were detected in Arabidopsis, whereas eight and three members were present in maize and rice genome, respectively, indicating that UBC-encoding genes of these subclasses expanded in a species-specific manner from common ancestral genes that were present prior to the diversification of the monocot and dicot lineages. Relationship analysis of intragroup members with well-supported bootstrap values revealed that all of the 13 subfamilies included both proteins from eudicots and monocots, which indicated that the defined subfamilies were already present in the common ancestor of both groups, and evolved prior to the divergence of monocots and dicots. Arabidopsis and maize have at least one close homolog to the 15 UBC consensus enzymes of yeast. However, there are three UBC gene families (UBC14,UBC15,UBC16) in Arabidopsis and maize for which no probable orthologs exist in the budding yeast, whereas all three groups have potential orthologs in animals [64], which was indicative of possible gene loss during yeast evolution.
Previous reports have shown that plant E2 genes are involved in developmental and physiological processes, as well as biotic/abiotic stress responses under normal and stressed growth conditions [23]. Therefore, these are important factors for various plants to withstand adverse environmental conditions. The expression of UBC E2 genes in a number of plant species are regulated by tissues and/or developmental stage, as well as by environmental conditions [23, 27, 65]. In a previous report, three rice genes (OsUBC2/5/18) and five Arabidopsis genes (AtUBC13/17/20/26/31) in the UBC family were significantly downregulated, whereas only three rice genes (OsUBC13/15/45) were significantly upregulated under salt and drought stresses [55]. In maize, however, ZmUBCs showed different expression profiles in Arabidopsis and rice. Our data showed that over half of the ZmUBC genes (48 genes) and 16 ZmUBC genes were significantly upregulated under salt and drought condition, respectively. In Arabidopsis, the expression of AtUBC24 (PHO2) and AtUBC32 was upregulated by salt stress, and their mutants showed a reduction in the uptake and accumulation of Na+, leading to enhanced salt tolerance [27, 51]. Our data showed that ZmUBC56, ZmUBC60, and ZmUBC71, orthologs of AtUBC24 and AtUBC32, respectively, had high expression levels under salt stress. Similar expression patterns suggested that these genes might play important roles under salt-stressed conditions during maize development. Contrary to ZmUBC71, another orthologous gene of AtUBC32, ZmUBC62, was highly expressed under cold stress. The observed differences in expression profiles between duplicated gene pairs, ZmUBC62 and ZmUBC71, indicated that these genes might have undergone significant diversification after the duplication of the respective genomic segments, leading to neo-functionalization of the paired partners. These data indicated that ZmUBCs might play different roles under abiotic stresses compared to rice and Arabidopsis.
Conclusions
It is essential to systematically analyze the members of the ubiquitin-conjugating enzyme family to elucidate its functions in plant development and stress response. In the present study, we present the genome-wide identification and analysis of the ubiquitin-conjugating enzyme families in maize. A total of 75 ZmUBC genes were identified in the maize genome. Phylogenetic analysis of maize, Arabidopsis, and yeast indicated that these ubiquitin-conjugating enzymes could be divided into 13 E2 and 2 UEV groups. Comparison of the number of UBC-encoding genes and mRNAs within and between species indicated that both gene duplication and alternative splicing contributed to the genomic complexity and proteomic diversity of ZmUBC genes. Analysis of exon-intron junctions and the conserved motif of each candidate gene has revealed high levels of conservation within and between phylogenetic groups. Clear orthologous relationships were established for the majority of UBC-encoding genes, facilitating in the generation of functional inference of the maize ubiquitin-conjugating enzymes. Although nearly all the ZmUBC genes were expressed in the examined organs, some genes were upregulated in one or several specific organs. The expression of some E2s could be up- or downregulated by different abiotic stress treatments, indicating the critical roles of this gene family in maintaining maize normal growth under stress conditions. However, additional studies on the detailed functions of each gene are warranted.
Supporting Information
The NJ tree includes 75 UBC proteins from Zea mays, 15 from Saccharomyces cerevisiae, and 48 from Arabidopsis thaliana. The tree shows 15 phylogenetic subgroups depicted in various colors to distinguish diversification of subfamilies into clusters.
(TIF)
The X-axis indicates time course/treatment and the Y-axes are scales of relative expression levels. The maize actin gene was used as internal control. The presented data are representative of three independent experiments.
(TIF)
(DOC)
Acknowledgments
This research was supported by the grants from the National Natural Science Foundation of China (Grant No. 30900784) and supported by the Fundamental Research Funds of the National Non profit Research Institute for South Subtropical crops Rearch Institute, Chinese Academy of Tropical Agricultural Sciences (No. 1630062015003, 1630062013001, 1630062015017).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the grants from the National Natural Science Foundation of China (Grant No. 30900784) and supported by the Fundamental Research Funds of the National Nonprofit Research Institute for South Subtropical crops Research Institute, Chinese Academy of Tropical Agricultural Sciences (No. 1630062015003, 1630062013001, 1630062015017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome research. 2009;19(10):1905–11. 10.1101/gr.093963.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schumacher FR W G, Day CL. The N-Terminal Extension of UBE2E Ubiquitin-Conjugating Enzymes Limits Chain Assembly. Journal of Molecular Biology. 2013;425(22):4099–111. 10.1016/j.jmb.2013.06.039 [DOI] [PubMed] [Google Scholar]
- 3. Ptak C, Gwozd C, Huzil JT, Gwozd TJ, Garen G, Ellison MJ. Creation of a pluripotent ubiquitin-conjugating enzyme. Molecular and cellular biology. 2001;21(19):6537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Engin O, Hongtao Y, Johann D. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):18890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature Structural & Molecular Biology. 2006;13(10):915–20. [DOI] [PubMed] [Google Scholar]
- 6. Lin Y, Hwang WC, Basavappa R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. Journal of Biological Chemistry. 2002;277(24):21913–21. [DOI] [PubMed] [Google Scholar]
- 7. Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature Reviews Molecular Cell Biology. 2009;10(11):755–64. 10.1038/nrm2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. Embo Journal. 1992;11(8):3091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheng Y, Hong JH, Doherty R, Srikumar T, Shloush J, Avvakumov GV, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Molecular & cellular proteomics: MCP. 2012;11(8):329–41. 10.1074/mcp.O111.013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michelle C, Vourc’H P, Mignon L, Andres CR. What Was the Set of Ubiquitin and Ubiquitin-Like Conjugating Enzymes in the Eukaryote Common Ancestor?. Journal of Molecular Evolution. 2009;68(6):616–28. 10.1007/s00239-009-9225-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Callis J. The Ubiquitination Machinery of the Ubiquitin System. Arabidopsis Book. 2014;12:e0174–e. 10.1199/tab.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, et al. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiology. 2005;139(4):1597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bae H, Kim WT. The N-terminal tetra-peptide (IPDE) short extension of the U-box motif in rice SPL11 E3 is essential for the interaction with E2 and ubiquitin-ligase activity. Biochemical & Biophysical Research Communications. 2013;433(2):266–71. [DOI] [PubMed] [Google Scholar]
- 14. Jones D, Crowe E, Stevens TA, Candido EPM. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biology. 2002;3(1):129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Semple CAM, Group RG, Members G. The Comparative Proteomics of Ubiquitination in Mouse. Genome research. 2003;13(6B):1389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang S, Arguello JR, Li X, Ding Y, Zhou Q, Chen Y, et al. Repetitive Element-Mediated Recombination as a Mechanism for New Gene Origination in Drosophila. Plos Genetics. 2008;4(1):63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wen R, Newton L, Li G, Wang H, Xiao W. Arabidopsis thaliana UBC13: implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Molecular Biology. 2006;61(1–2):241–53(13). [DOI] [PubMed] [Google Scholar]
- 18. Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W. Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell. 2008;20(1):213–27. 10.1105/tpc.107.051862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li W, Schmidt W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant Journal. 2010;volume 62(2):330–43(14). [DOI] [PubMed] [Google Scholar]
- 20. Lau OS, Deng XW. Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochemical Journal. 2009;418(3):683–90. 10.1042/BJ20081943 [DOI] [PubMed] [Google Scholar]
- 21. Yuki Y, Sullivan JA, Setsuko K, Giuliana G, Genki S, Jianning Y. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004;18(17):2172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu L,Ménard R,Berr A,Fuchs J,Cognat V, Meyer D, et al. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant Journal. 2009;57(2):279–88. 10.1111/j.1365-313X.2008.03684.x [DOI] [PubMed] [Google Scholar]
- 23. En Hee J, Jung Hun P, Jin KM, Hye Jeong K, Hyun SS, Jai Heon L, et al. Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochemical & Biophysical Research Communications. 2012;427(2):309–14. [DOI] [PubMed] [Google Scholar]
- 24. Mural RV, Liu Y, Rosebrock TR, Brady JJ, Hamera S, Connor RA, et al. The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell. 2013;25(9):3615–31. 10.1105/tpc.113.117093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Wang W, Cai J, Zhang Y, Qin G, Tian S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biology. 2014;15(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Criqui MC, DAE J, Camasses A, Capron A, Parmentier Y, Inzé D, et al. Molecular characterization of plant ubiquitin-conjugating enzymes belonging to the UbcP4/E2-C/UBCx/UbcH10 gene family. Plant Physiology. 2002;130(3):1230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng C, Lijing L, Qingzhen Z, Zhonghui Z, Qingliang L, Baoying L, et al. Arabidopsis Ubiquitin Conjugase UBC32 Is an ERAD Component That Functions in Brassinosteroid-Mediated Salt Stress Tolerance. Plant Cell. 2012;24(1):233–44. 10.1105/tpc.111.093062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zolman BK, Melanie MA, Silva ID, Bonnie B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17(12):3422–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pichler A KP, Oberhofer E, van Dijk WJ, Körner R, Olsen JV, Olsen JV, et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nature Structural & Molecular Biology. 2005;12(3):264–9. [DOI] [PubMed] [Google Scholar]
- 30. Knobbe AR, Horken KM, Plucinak TM, Balassa E, Cerutti H, Weeks DP. SUMOylation by a Stress-Specific Small Ubiquitin-Like Modifier E2 Conjugase Is Essential for Survival of Chlamydomonas reinhardtii under Stress Conditions. Plant Physiology. 2015;167(3):753–65. 10.1104/pp.114.256081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pozo JCD, Estelle M. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proceedings of the National Academy of Sciences. 1999;96(26):15342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mergner J, Schwechheimer C. The NEDD8 modification pathway in plants. Frontiers in Plant Science. 2014;5(2):113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bong Soo P JS S, Nam-Hai C. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell. 2014;26(1):454–64. 10.1105/tpc.113.120311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jie S, Xu G, Hui QZ. Apoplastic barrier development and water transport in Zea mays seedling roots under salt and osmotic stresses. Protoplasma. 2014;252(1):173–80. 10.1007/s00709-014-0669-1 [DOI] [PubMed] [Google Scholar]
- 35. Zongliang X, Quanjun L, Jianyu W, Junqiang D. ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene. 2012;495(2):146–53. 10.1016/j.gene.2011.12.028 [DOI] [PubMed] [Google Scholar]
- 36. Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112-. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- 37. Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, Pontaroli AC, et al. Uneven chromosome contraction and expansion in the maize genome. Genome research. 2006;16(10):1241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sebastian P, Michiel VB, Lieven S, Kenny B, Thomas VP, Yves VD P, et al. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21(12):3718–31. 10.1105/tpc.109.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz Jr, et al. Smart 4.0: towards genomic data integration. Nucleic Acids Research. 2004;32(22):142–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer ELL. The Pfam Protein Families Database. Nucleic Acids Research. 2000;28(1):263–6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: the integrative protein signature database. Nucleic Acids Research. 2009;37(suppl 1):D211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo AY, Zhu QH, Chen X. [GSDS: a gene structure display server]. Hereditas. 2007;29(8):1023–6. [PubMed] [Google Scholar]
- 43. Emanuelsson O, Brunak S, Von HG, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocol. 2007;2(4):953–71. [DOI] [PubMed] [Google Scholar]
- 44. Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One. 2010;5(6):e11335 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(3):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura K S G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology & Evolution. 2013;30(12):2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59(3):307–21. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 48. Kaifa W, Juan C, Yanmei W, Yanhui C, Shaoxiang C, Yina L, et al. Genome-wide analysis of bZIP-encoding genes in maize. Dna Research An International Journal for Rapid Publication of Reports on Genes & Genomes. 2012;19(6):463–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E. et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. Plos Genetics. 2007;3(7):: e123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hou H, Li J, Gao M, Singer SD, Wang H, Mao L, et al. Genomic Organization, Phylogenetic Comparison and Differential Expression of the SBP-Box Family Genes in Grape. Plos One. 2013;8(3):e59358 10.1371/journal.pone.0059358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kenji M, Aiko S, Masaru O, Jun F. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta. 2011;234(6):1191–9. 10.1007/s00425-011-1476-y [DOI] [PubMed] [Google Scholar]
- 52. Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes & Development. 2002;16(5):554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nature Reviews Genetics. 2001;2(7):516–27. [DOI] [PubMed] [Google Scholar]
- 54. Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 55. E ZG, Zhang ZY, Li TT, Wang L, Zhao HM. Characterization of the Ubiquitin-Conjugating Enzyme Gene Family in Rice and Evaluation of Expression Profiles under Abiotic Stresses and Hormone Treatments. PLoS One. 2015;10(4):e0122621 10.1371/journal.pone.0122621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Winn PJ, Religa TL, Battey JND, Banerjee A, Wade RC. Determinants of Functionality in the Ubiquitin Conjugating Enzyme Family. Structure. 2004;12(9):1563–74. [DOI] [PubMed] [Google Scholar]
- 57. Papaleo E, Casiraghi N, Arrigoni A, Vanoni M, Coccetti P, Gioia LD. Loop 7 of E2 Enzymes: An Ancestral Conserved Functional Motif Involved in the E2-Mediated Steps of the Ubiquitination Cascade. Plos One. 2012;7(7):398-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, Depamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant Journal. 2007;50(5):873–85. [DOI] [PubMed] [Google Scholar]
- 59. Ahn S, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proceedings of the National Academy of Sciences. 1993;90(17):7980–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. Bmc Genomics. 2011;12(1):: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang Y, Wang Q, Zhao Y, Han G, Zhu S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene. 2015;566:95–108. 10.1016/j.gene.2015.04.041 [DOI] [PubMed] [Google Scholar]
- 62. Xiaojian P, Yang Z, Jiangang C, Wei Z, Haiyang J, Xiaoyu L, et al. CCCH-Type Zinc Finger Family in Maize: Genome-Wide Identification, Classification and Expression Profiling under Abscisic Acid and Drought Treatments. Plos One. 2012;7(7):: e40120 10.1371/journal.pone.0040120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang Z, Yuqiong Z, Haiyang J, Xiaoyu L, Defang G, Xiaojian P, et al. Systematic Analysis of Sequences and Expression Patterns of Drought-Responsive Members of the HD-Zip Gene Family in Maize. Plos One. 2011;6(12):: e28488 10.1371/journal.pone.0028488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends in Plant Science. 2001;6(10):463–70. [DOI] [PubMed] [Google Scholar]
- 65. Zhou G, Chang R, Qiu L. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Molecular Biology. 2010;72(4–5):357–67. 10.1007/s11103-009-9575-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The NJ tree includes 75 UBC proteins from Zea mays, 15 from Saccharomyces cerevisiae, and 48 from Arabidopsis thaliana. The tree shows 15 phylogenetic subgroups depicted in various colors to distinguish diversification of subfamilies into clusters.
(TIF)
The X-axis indicates time course/treatment and the Y-axes are scales of relative expression levels. The maize actin gene was used as internal control. The presented data are representative of three independent experiments.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.