Abstract
Berberine has been identified with anti-proliferative effects on various cancer cells. Many researchers have been trying to elucidate the anti-cancer mechanisms of berberine based on differentially expressed genes. However, differentially alternative splicing genes induced by berberine might also contribute to its pharmacological actions and have not been reported yet. Moreover, the potential functional cross-talking between the two sets of genes deserves further exploration. In this study, RNA-seq technology was used to detect the differentially expressed genes and differentially alternative spliced genes in BEL-7402 cancer cells induced by berberine. Functional enrichment analysis indicated that these genes were mainly enriched in the p53 and cell cycle signalling pathway. In addition, it was statistically proven that the two sets of genes were locally co-enriched along chromosomes, closely connected to each other based on protein-protein interaction and functionally similar on Gene Ontology tree. These results suggested that the two sets of genes regulated by berberine might be functionally cross-talked and jointly contribute to its cell cycle arresting effect. It has provided new clues for further researches on the pharmacological mechanisms of berberine as well as the other botanical drugs.
Introduction
Alternative splicing is a tightly regulated process during gene expression that can produce different forms of a protein from the same gene. Moreover, it has been proved that alternative splicing can also determine binding properties, intracellular localization, enzymatic activity, protein stability and posttranslational modifications of a large number of proteins [1]. In recent years, more and more pharmacological researchers have found that small molecular drugs could exert their pharmacological effects not only through regulating transcription levels of genes but also through changing gene alternative splicing [2].
Berberine, an isoquinoline quaternary alkaloid isolated from Berberis species [3], has a wide spectrum of pharmacological effects such as anti-microbe, anti-diabetes and anti-inflammation [4]. Clinically, it has been used to treat a range of disorders, including coronary artery disease, diabetes, non-alcoholic fatty liver disease, hyperlipidaemia, metabolic syndrome, obesity and polycystic ovary syndrome (https://www.clinicaltrials.gov/). Recently, accumulating studies have found that berberine also possessed potent anticancer activity, with few or minimal undesired toxic effects [5, 6]. Therefore, many researchers have been trying to elucidate the anti-cancer mechanisms of berberine based on gene differential expression. For instance, it is reported that berberine could induce apoptosis in HepG2 cells through AMPK-mediated mitochondrial pathway by increasing the ratio of Bax/Bcl-2 [7]. In our previous study, we also have found that berberine could induce G1 cell cycle arrest in BEL-7402 cells partially via disturbing the interaction of calmodulin with CaMKII and blocking subsequent p27 protein degradation. [8]
Although these works have provided some fundamental knowledge about the anticancer mechanism of berberine, alternative splicing in cancer cells after treated with berberine has not been reported yet. Firstly, it has been implied that many anticancer drugs could inhibit the growth of cancer cells by altering gene alternative splicing [9–11]. In addition, it is reported that gene transcription and alternative splicing might be physically and functionally coupled [12, 13], thereby jointly contribute to the pharmacological actions of these drugs. Therefore, it is necessary to simultaneously measure the differentially expressed genes (DEGs) and the differentially alternatively spliced genes (DASGs) in cancer cells after treatment with berberine and systematically examine whether there is any kind of functional cross-talking between these two sets of genes.
For the question mentioned above, the latest next-generation sequencing technology (RNA-seq) was used to obtain the transcriptomic profiling of BEL-7402 cells after berberine treatment. Then, the DEGs and DASGs induced by berberine were carefully detected by a suite of sequence analysis tools. Finally, the functional crosstalk between these DEGs and DASGs was statistically analysed and their possible contributions to the anticancer effect of berberine were discussed.
Materials and Methods
Cell culture
The established human liver cancer cell line (BEL-7402, Cat. No.: TCHu_10) [8, 14, 15] was bought from and deposited in the cell bank of the institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Science (http://www.cellbank.org.cn/) and kindly provided by Dr. Xuan Liu (Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, P.R.China). Cells were maintained in a humidified 37°C atmosphere containing 5% CO2 and cultured in RPMI-1640 medium (GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum, 2 mM/L-glutamine, 50 units/mL penicillin, and 50 mg/mL streptomycin. In this study, we have pooled 3~5 biological repeats into one RNA-seq experiment to achieve the same amount of total RNA content. More specifically, at the cell culturing section, cancer cells in 8~10 different petri dishes were divided into two equal groups. One group was used as berberine treated group, the other was used as control/untreated group.
RNA isolation
For extraction of total RNA, cells were plated on 10-cm plates (Corning, Acton, MA, USA) in complete medium for 12 h. Then, 50 μM berberine (purity ≥ 98%, Tauto Biotech, Shanghai, China, IC50 according to Ref.[8]) was added and left in contact for 24 h. After that, cells were rinsed in PBS and lysed in TRIzol reagent (Invitrogen, Carlsbad, CA). Residual contaminating genomic DNA was removed from the total RNA fraction using Turbo DNA-free kit (Ambion, Austin, TX, USA). mRNA was isolated from DNA-free total RNA using the Dynabeads mRNA Purification Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Finally, to obtain enough quantity of mRNA content for sequencing, the total RNA samples from each group were pooled together as one pooled sample.
Transcriptome sequencing
mRNA selection, library preparation and sequencing was performed by Shanghai Ceneter for Bioinformation Technology (SCBIT) on an Illumina Hiseq 2000 sequencing platform according to manufacturer specifications. Briefly, mRNA was selected using oligo (dT) probes and then fragmented using divalent cations. cDNAs were synthesized using random primers, modified and enriched for attachment to the Illumina flow cell. We sequenced two 60-cycle 100 bp × 2 paired-end lanes, generating ~83.1 million reads.
Read alignment
Raw sequencing reads were trimmed out Ns and low quality regions (average base quality score in a 5-mer slide-window less than 20, that is, sequencing error rate ≤1%). Read-pairs with at least 36 bp left in each read were kept as clean reads. Sequencing quality was assessed using FastQC [16]. Then, clean reads were mapped to human reference genome (Ensembl GRCh37.66 = hg19) using aligner Tophat [17] (version 2.0.6) with parameters (-r/—inner-mate-distance: 70, -g/—max-multihits: 1,—no-coverage-search,—no-novel-junc, the others as default). The outputs of Tophat were used as the inputs for HTSeq [18]and AltAnalyze [19] to perform gene differential expression or alternative splicing analysis respectively.
Differential gene expression analysis
DEGs (Differentially Expressed Genes) refer to those genes with significant changes in their expression level. Firstly, the number of reads mapped to the genomic region of each gene for each sample was estimated with HTSeq [18]. Then DESeq [20] was used to assess the difference in gene expression between different samples. Finally, DEGs were filtered by the following rules: 1) the absolute value of log2 [fold-change] was greater than 1. 2) The p-value was less than 0.05.
Differentially gene alternative splicing analysis
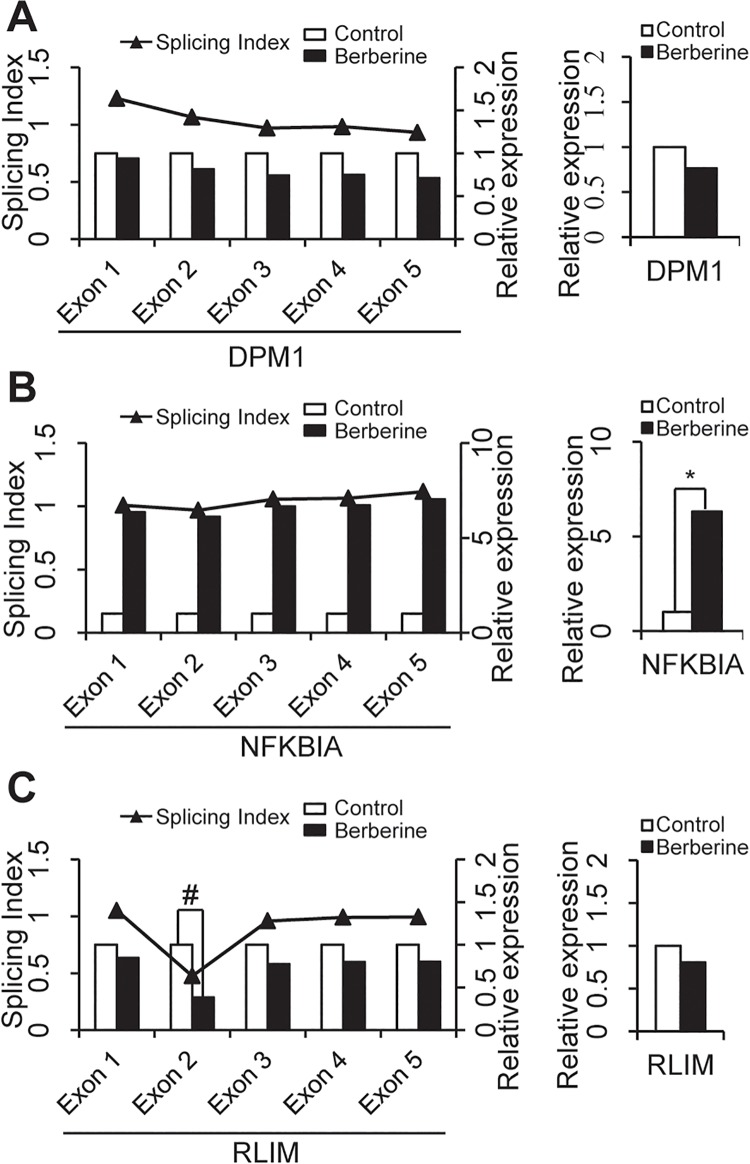
DASGs (Differentially Alternative Spliced Genes) refer to those genes with no or small variations in their whole-gene expression level but significant changes in their exon usage, especially those alternative exons (Fig 1). Differential splicing analysis describes the differences in alternative splicing site usage between two samples, which is critical for studies involving mechanisms of alternative splicing and its regulation, and able to uncover functional diversity that is missed by differential gene expression analysis [21]. Dozens of available software tools and packages, such as Cuffdiff2 [22], MISO [23], DEXseq [24], DEGseq [25], DiffSplice [26], Splicing compass [27], AltAnalyze [19] and etc., took conceptually different approaches that could identify differential splicing at the level of isoform/transcript, exon, or both. However, the choice of a suitable approach for a study depends on the experimental objective and expected outcome. Among these approaches and packages, only AltAnalyze provides a graphical user interface, while the rest are run on the commend line. Besides, AltAnalyze provides a convenient workflow to extract the significant alternative splicing regulation events and subsequently investigate the potential biological implications of such splicing events and related mechanisms in the context of molecular interactions and binding sites [19]. Multiple algorithms are available in AltAnalyze to identify individual features (exons or junctions) or reciprocal junctions that are differentially regulated relative to gene expression changes. In this work, single feature analysis (Splicing Index, SI) was performed immediately after reciprocal junction analyses (Analysis of Splicing by Isoform Reciprocity, ASPIRE) [28] on the same list of expressed features, which allows us to examine alternative exons predicted by pairs of reciprocal junctions in addition to those predicted by a single regulated exons/junctions (see AltAnalyze-Manual). Splicing index is one measurement of differential splicing between two samples. Exons with SI value larger than 1 means more inclusion of the exon in the matured mRNA during splicing process in the treated sample compared with the control sample, while those with SI value less than 1 means less inclusion but more skipping of the exon, and those with SI value equal to 1 means there is no change in the usage of the exon.
Fig 1. Representation of differentially expressed and alternatively spliced genes after berberine treatment.
A. The expression or alternative splicing of DPM1 gene remained unchanged after berberine treatment. B. The splicing pattern of NFKBIA gene remained unchanged after berberine treatment as indicated by the splicing index (SI) value of each of its exons. However, the expression of NFKBIA gene was upregulated by more that 2-fold after berberine treatment (p<0.05) since each of its exons were overexpressed. C. The splicing pattern of RLIM gene was altered by berberine treatment as indicated by the SI value of exon 2 (SI < 0.5) within RLIM gene. But the expression of RLIM gene did not changed significantly after berberine treatment. Relative expression, the relative expression level of the gene or all its exons compared to the control sample. *, DEG; #, DSAG.
Differentially alternative splicing analysis was carried out using AltAnalyze with default parameters. Firstly, there were three criteria of exons to perform differentially gene alternative splicing analysis: 1) At least 5 reads were mapped to the exon; 2) the RPKM (Reads Per Kilo-base of exon model per Million mapped reads)value of the exon was larger than 0.3; 3) the expression change of the corresponding gene was less than 2-fold. Then, two exon-centric algorithms were used to detect the differentially alternative splicing exons: Splicing Index (SI) and Analysis of Splicing by Isoform Reciprocity (ASPIRE) [28]. The SI value for each exon or junction was calculated as below.
| (1) |
In which, RPKM(exon i) and RPKM(gene) were the expression value of the i-th exon and the gene respectively. In SI algorithm, exons with SI>2-fold were called AS-exons_SI. In ASPIRES algorithm, exons with ΔI>0.2 were called AS-exons_ASPIRE. The details of these two algorithms have been described in AltAnalyze workflow (http://www.altanalyze.org/). Only overlapped AS-exons in both algorithms were chosen for further functional analysis. And the genes containing these exons were called differentially alternative spliced genes (DASG).
Gene Set Functional Enrichment Analysis
Gene set enrichment analyses were performed for the functional annotation of the DEGs and DASGs separately. Functional Annotation Tools in DAVID Bioinformatics Resources [29] were used to carry out these analyses. Those gene ontology (GO) [30] biological process terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) [31] pathways with enrichment p-value less than 0.05 (modified Fisher’s exact test) and more than two genes were considered as significantly enriched functions for further analysis.
Local co-enrichment analysis along chromosome sequences
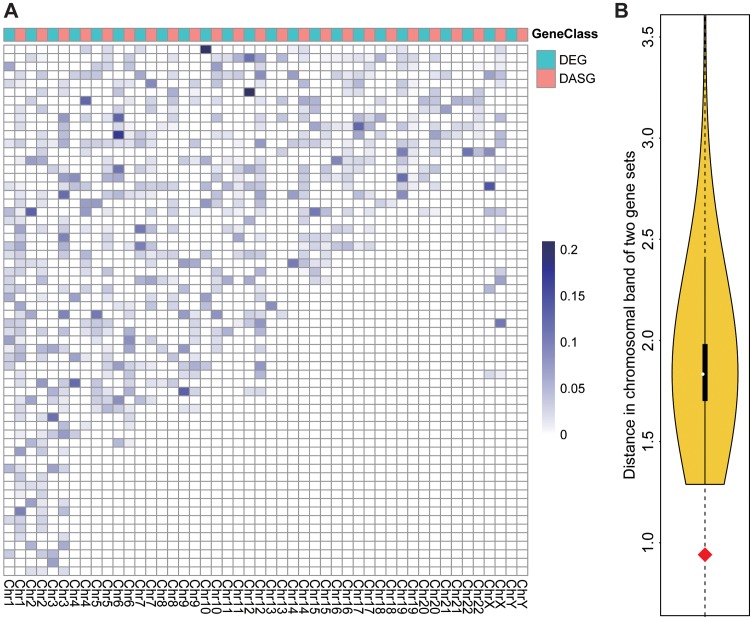
To test whether the DEGs and DASGs showed similar enrichment distribution along the chromosomes, local co-enrichment analysis was performed. Firstly, an enrichment score (ES) was calculated for a gene set at each chromosome band, which was also displayed as the intensity of that region as shown in Fig 2A [32]. Given gene set X:{x 1,x 2,…x N }, chromosome band B:{b 1,b 2,…b M }, the number of genes in each band G:{g 1,g 2,…g M }, then ES score was calculated as following.
Fig 2. Positional co-enrichment of DEGs and DASGs along 24 chromosomes.
A. Global intensity plot of positional enrichment for DEGs and DASGs. Each cell represents a chromosomal band and the intensity of its color is proportional to the corresponding enrichment of DEGs or DASGs in this band, which is normalized by both the total amount of genes in the band and the number of either DEGs or DASGs. Each column represents the chromosome containing all the bands and the color, either blue or red, ahead of the column distinguishing the class of genes from DEGs and DASGs. B. The violin plot for the positional co-enrichment score (distance) between two gene sets. The curve covering the yellow region represents the distance distribution of two random gene sets in the same size of DEGs and DASGs respectively. A shorter distance means a higher co-enrichment of these two gene sets. The red diamond point refers to the distance between DEGs and DASGs.
| (2) |
Then, a ES score vector EX:{ex 1,ex 2,…ex M } was got for gene set X across all bands in B. Similarly, another vector EY:{ey 1,ey 2,…ey M } could be got for another gene set Y. Finally, the similarity (LCE) between the two color bands of X and Y was measured as the Euclidian distance between them.
| (3) |
Global closeness in protein-protein interaction (PPI) network
To evaluate the topological closeness between DEGs and DASGs in the PPI network, average shortest path length between them was used as the measure. Average shortest path length is a concept in network topology that is defined as the average number of steps along the shortest paths for all possible pairs of network nodes, which is a measure of the efficiency of information transport in the network [33]. This measure is also applicable on two-subnetwork issues by calculating the average path distance for all possible pairs of nodes from these two networks, x and y.
| (4) |
Where dis(i, j) is the shortest path distance between the i-th gene of subset x and the jth gene of subset y. In this analysis, the background network was built using the whole PPI data from online database Human Protein Reference Database (HPRD).
Functional semantic similarity on Gene Ontology (GO) tree
On the tree of Gene Ontology, genes are categorized into different functional groups named as GO terms based on the biological processes in which they are involved. Based on the graph structure of GO, semantic similarity between any two GO terms was calculated as described in Wang et.al [34] using GOSemSim package [35]. Then the semantic similarity between two gene sets such as DEGs and DASGs could be calculated as the average of all possible pairs of GO terms involving genes in these two gene sets. Given two lists of GO terms, go 1 and go 2 involving genes in two gene sets, X and Y, the semantic similarity SemSim was defined as:
| (5) |
Where N GO is the total amount of GO terms used for semantic similarity calculation.
Results
Summary of RNA-seq results for BEL-7402 cells treated with berberine
The whole transcriptomic profiling of BEL-7402 cells treated with or without berberine was assessed at base-pair resolution via RNA sequencing. After low-quality reads (as mentioned in Read alignment section, Materials and Methods) were cleaned, approximately 30 million reads out of ~41.5 million raw reads (reads retention rate: >70%, GC content: 45~46%, sequence length:36~100 bp) were mapped to reference genome for each sample (Table 1). According to the “Standards, Guidelines and Best Practices for RNA-Seq” adopted by ENCODE [36], this sequencing depth is good for differential expression analysis and is acceptable for alternative splicing analysis in human transcriptome.
Table 1. General Statistics of Reads Alignment Process.
| Ctrl | Berberine | |||
|---|---|---|---|---|
| Count(left+right) | % | Count(left+right) | % | |
| raw reads | 41671690 | 100.00 | 41434030 | 100.00 |
| clean reads | 33260886 | 79.81 | 32645126 | 78.78 |
| mapped reads | 29934588 | 71.83 | 29109042 | 70.25 |
Differentially expressed and alternatively spliced genes induced by berberine treatment
After analyzed by using DESeq software, RNA-seq results returned 343 DEGs in BEL-7402 cells after treated with berberine, with 111 up-regulated and 232 down-regulated (Table A in S1 File). Interestingly, biological process response to bacterium (GO: 0009617) was found to be significantly enriched by up-regulated DEGs (Table 2), which is consistent with the clinical usage of berberine as a broad-spectrum anti-microbial medicine [3]. And we found the genes related to positive regulation of angiogenesis were significantly down-regulated by berberine, which accounts for the anti-angiogenetic activity of berberine reported in previous works. [37–39] According to KEGG pathway enrichment analysis, the DEGs were significantly enriched in p53 signalling pathway (hsa04115) and several cancer-related pathways such as Jak-STAT signalling pathway (hsa04630), Apoptosis (hsa04210) and Pathways in cancer (hsa05200) (Table 3). The Jak-STAT signalling pathway is a major signalling involved in regulation of the immune system, suggesting the immunity-regulating activity of berberine may contribute to its anti-cancer effect.
Table 2. Gene Ontology Enrichment Result of DEGs and DASGs.
| GeneClass | GO_ID | GO term | Count # | p-value |
|---|---|---|---|---|
| DEG_up | GO:0006334 | nucleosome assembly | 8 | <0.001 |
| DEG_up | GO:0043066 | negative regulation of apoptosis | 11 | <0.001 |
| DEG_up | GO:0009617 | response to bacterium | 8 | <0.001 |
| DEG_up | GO:0045595 | regulation of cell differentiation | 11 | <0.001 |
| DEG_up | GO:0045637 | regulation of myeloid cell differentiation | 5 | <0.001 |
| DEG_up | GO:0002521 | leukocyte differentiation | 6 | <0.001 |
| DEG_up | GO:0051174 | regulation of phosphorus metabolic process | 10 | <0.010 |
| DEG_up | GO:0030097 | hemopoiesis | 7 | <0.010 |
| DEG_up | GO:0051726 | regulation of cell cycle | 8 | <0.010 |
| DEG_up | GO:0031667 | response to nutrient levels | 6 | <0.010 |
| DEG_up | GO:0006986 | response to unfolded protein | 4 | <0.010 |
| DEG_up | GO:0008284 | positive regulation of cell proliferation | 8 | <0.010 |
| DEG_down | GO:0006350 | transcription | 58 | <0.001 |
| DEG_down | GO:0051252 | regulation of RNA metabolic process | 52 | <0.001 |
| DEG_down | GO:0045766 | positive regulation of angiogenesis | 5 | <0.001 |
| DEG_down | GO:0031328 | positive regulation of biosynthetic process | 18 | <0.010 |
| DEG_down | GO:0051174 | regulation of phosphorus metabolic process | 14 | <0.010 |
| DEG_down | GO:0042127 | regulation of cell proliferation | 19 | <0.010 |
| DEG_down | GO:0043067 | regulation of programmed cell death | 19 | <0.010 |
| DEG_down | GO:0007050 | cell cycle arrest | 6 | <0.010 |
| DEG_down | GO:0008284 | positive regulation of cell proliferation | 12 | <0.010 |
| DEG_down | GO:0006915 | apoptosis | 15 | <0.010 |
| DASG | GO:0007049 | cell cycle | 48 | <0.001 |
| DASG | GO:0051276 | chromosome organization | 35 | <0.001 |
| DASG | GO:0065003 | macromolecular complex assembly | 41 | <0.001 |
| DASG | GO:0008104 | protein localization | 47 | <0.001 |
| DASG | GO:0006259 | DNA metabolic process | 30 | <0.001 |
| DASG | GO:0045184 | establishment of protein localization | 40 | <0.001 |
| DASG | GO:0006468 | protein amino acid phosphorylation | 36 | <0.001 |
| DASG | GO:0033554 | cellular response to stress | 32 | <0.001 |
| DASG | GO:0006350 | transcription | 85 | <0.001 |
| DASG | GO:0045926 | negative regulation of growth | 11 | <0.010 |
| DASG | GO:0051640 | organelle localization | 10 | <0.010 |
| DASG | GO:0016044 | membrane organization | 23 | <0.010 |
| DASG | GO:0043407 | negative regulation of MAP kinase activity | 6 | <0.010 |
| DASG | GO:0008219 | cell death | 34 | <0.010 |
| DASG | GO:0008361 | regulation of cell size | 14 | <0.010 |
| DASG | GO:0042770 | DNA damage response, signal transduction | 8 | <0.010 |
#the number of DEGs or DASGs in each Gene Ontology Biological Process term.
Table 3. KEGG pathway enrichment result of DEGs and DASGs.
| GeneClass | KEGG_ID | KEGG pathway | Count # | p-value |
|---|---|---|---|---|
| DEG | hsa04115 | p53 signaling pathway | 7 | <0.001 |
| DEG | hsa05020 | Prion diseases | 5 | <0.010 |
| DEG | hsa04060 | Cytokine-cytokine receptor interaction | 12 | <0.010 |
| DEG | hsa05322 | Systemic lupus erythematosus | 7 | <0.010 |
| DEG | hsa04640 | Hematopoietic cell lineage | 6 | <0.050 |
| DEG | hsa04630 | Jak-STAT signaling pathway | 8 | <0.050 |
| DEG | hsa04210 | Apoptosis | 6 | <0.050 |
| DEG | hsa04710 | Circadian rhythm | 3 | <0.050 |
| DEG | hsa05200 | Pathways in cancer | 11 | <0.050 |
| DASG | hsa04150 | mTOR signaling pathway | 6 | <0.050 |
| DASG | hsa04144 | Endocytosis | 12 | <0.050 |
| DASG | hsa04810 | Regulation of actin cytoskeleton | 13 | <0.050 |
| DASG | hsa00310 | Lysine degradation | 5 | <0.050 |
| DASG | hsa04360 | Axon guidance | 9 | <0.050 |
#the number of DEGs or DASGs in each pathway.
Meanwhile, alternative splicing analysis was carried out using AltAnalyze software. The results showed that 1083 differentially alternatively spliced exons, corresponding to 867 DASGs (Table B in S1 File) were identified in BEL-7402 cancer cells after berberine treatment. Taken RLIM gene as an example (Fig 1), the inclusion level of exon 2 in the matured mRNA was significantly changed (splicing index < 0.5) after berberine treatment, while there was only a small change in its whole gene expression level. In comparison with RLIM, there was dramatically variation (fold-change > 6) in the gene-expression level of NFKBIA (as an example of DEG) but no significant changes (splicing index ≈ 1) in the inclusion of its exons (Fig 1). As shown in Table 2, the DASGs were involved in a variety of biological processes such as cell cycle (GO: 0007049), chromosome organization (GO: 0051276), macromolecular complex assembly (GO: 0065003) and cellular response to stress (GO: 0033554). Pathway enrichment analysis represented that these DASGs were enriched in PI3K-AKT-mTOR signalling pathway (has: 04150) and regulation of actin cytoskeleton (has: 04810) (Table 3), which all highly related with cell cycle process and cell proliferation [40]. Clearly, these results suggested that these DASGs might also be related with the anticancer action of berberine.
Statistical analysis of the functional cross-talking between DEGs and DASGs
To explore whether there is any kind of functional cross-talking between DEGs and DASGs, statistical tests were carried out from three primary but different perspectives.
Local co-enrichment among chromosome sequences
Intuitively, it should be tested firstly whether those DEGs and DASGs are enriched together at the same or nearby regions on the chromosomes. To carry out this exam, an enrichment score was assigned to each band on all the chromosomes for DASGs and DEGs respectively [32]. As shown in Fig 2A, those DEGs and DASGs exhibited quite different enrichment distributions globally. However, patterns could be found in each local region. Most of the DASG-enriched sites are at or near the sites enriched by DEGs. To quantify the co-enrichment pattern between DEGs and DASGs, Euclidean distance (0.941) was calculated between the enrichment scores on all chromosome bands of DEGs and DASGs. To test the statistical significance of the distance between DEGs and DASGs, simulations by randomly sampling two gene sets in the same size of DEGs and DASGs were carried out for 10000 times to produce the distribution of the Euclidean distance between any two gene sets on the chromosomes. As shown in Fig 2B, the distance between DEGs and DASGs was significantly shorter than those between randomly sampled sets. These results clearly demonstrated that the DEGs and DASGs are physically co-enriched on the corresponding chromosomes.
Global closeness based on protein-protein interaction (PPI) network
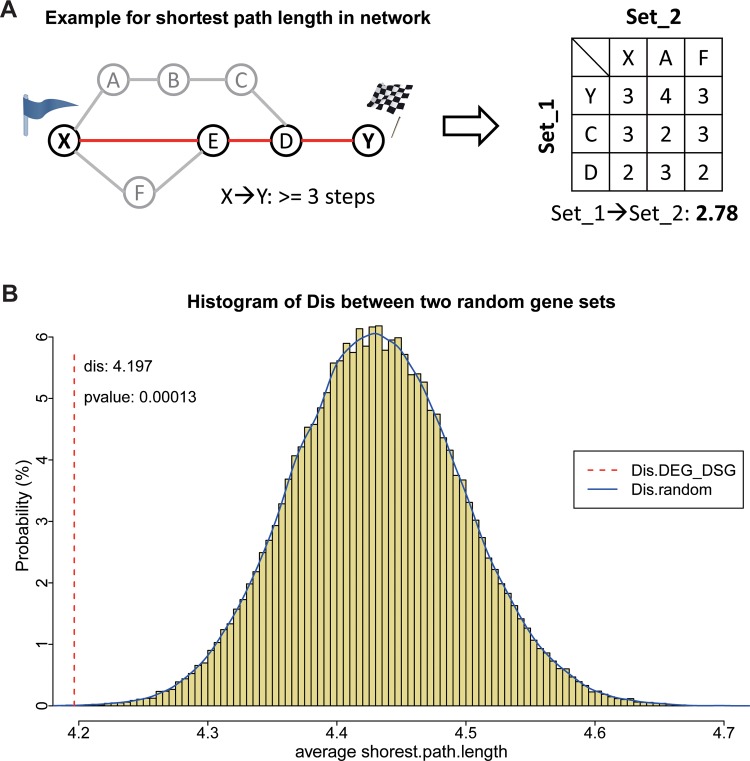
Following the above result, additional efforts were taken to measure the functional connection between these two gene sets based on PPI network. At first, 100000-time random simulations were performed to build the distribution of averaged shortest path distance between any two gene sets in the same scales as DEGs and DASGs [33]. Then, the averaged distance between DEGs and DASGs were calculated and compared with the distribution. As shown in Fig 3, the former is far away from the mean of the simulated distribution at a statistical significance of p-value < 0.05. The results indicated that these DEGs and DASGs might be closely connected through protein-protein interaction.
Fig 3. The PPI closeness (averaged shortest path distance) between DEGs and DASGs.
A. An example illustration of the closeness between two gene sets. Each circled point represents a node in the network and each line represents the edge between two nodes. The red-colored path is the shortest path between X and Y in this network, which is with 3 steps. The right part is the matrix of shortest path length between all possible pairs from two gene sets (set_1{Y, C, D}, set_2{X, A, F}). And their averaged shortest path length is 2.78. B. The blue curve above the yellow histogram fits the distribution of closeness between two random gene sets in the same sizes of DEGs and DASGs respectively. The red dotted line refers to the distance between DEGs and DASGs. The p-value represents the proportion of gene set pairs with shorter distance than that between DEGs and DASGs.
Functional semantic similarity on Gene Ontology (GO) tree
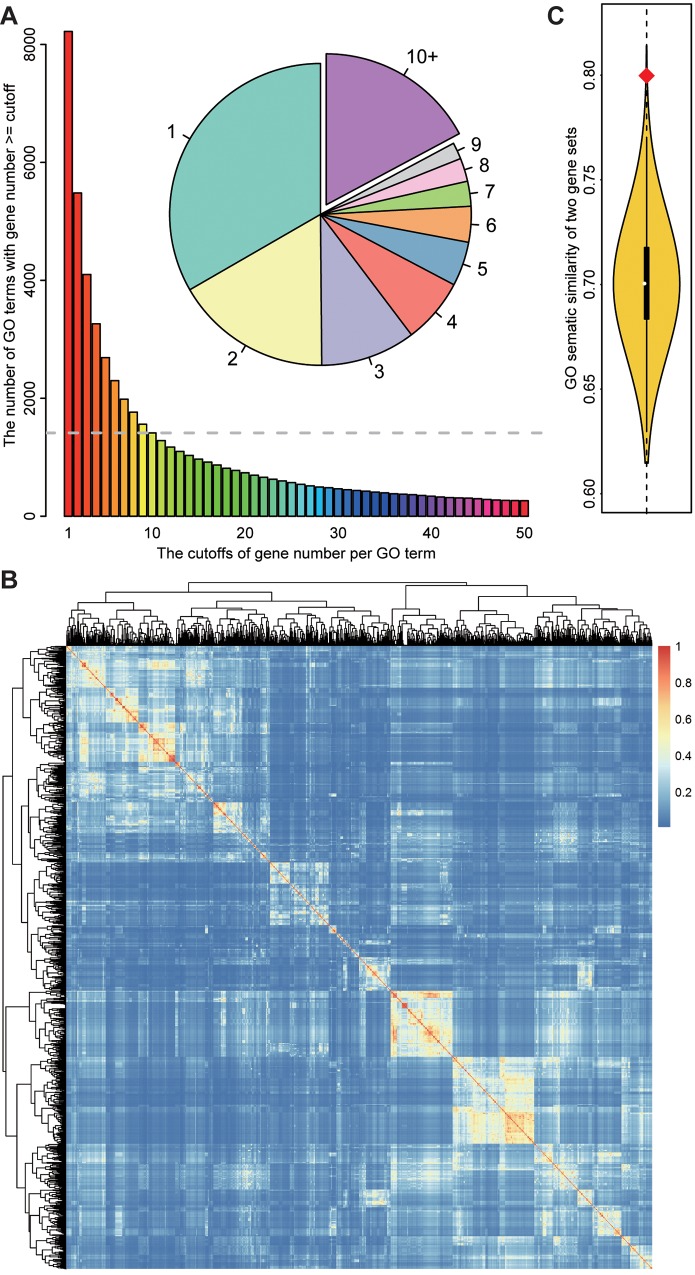
While DEGs and DASGs were found to have a significantly shorter distance compared with random-sampled gene sets according to PPI network, this kind of functional correlation is indirect and not with strong biological significance. Thus, a more straightforward method was employed to measure the functional correlation between DEGs and DASGs based on the biological processes affected by them [35]. To balance the computational efficiency and measurement robustness, those GO biological process terms with at least 10 genes were select as the reference biological process set (Fig 4A). A semantic similarity score was assigned to each two GO terms based on the graph structure of GO, as illustrated in Fig 4B. Then an averaged semantic similarity score was calculated over all pairs of GO terms respectively from the biological processes affected by DEGs and DASGs using the best-match average strategy. And the procedures were performed on two randomly sampled gene sets in the same scale as DEGs and DASGs for 10000 times to get the distribution of the averaged semantic similarity. As shown in Fig 4C, the averaged semantic similarity between DEGs and DASGs is extremely higher than two random-pickup sets, indicating that these two set genes might be functionally connected.
Fig 4. The Gene Ontology semantic similarity between DEGs and DASGs.
A. The pie plot shows the proportion of GO terms with the corresponding gene number. The barplot represents the number of GO terms with genes more than each cutoff of gene number. And the part of GO terms with more than 10 genes were selected for further calculation. B. the heatmap of GO semantic similarity between each two selected GO terms. The warmer the color is, the higher the similarity is between the two terms. C. The curve represents the distribution of semantic similarity between two randomly sampled gene sets in the same size of DEGs and DASGs respectively. The red diamond point refers to the semantic similarity between DEGs and DASGs.
Discussion
Transcription and alternative splicing are two important biological processes during gene expression, which together determine the proteome of cells under a certain condition. In this study, for exploring the functional connection between differentially expressed genes and differentially alternative spliced genes in cancer cells treated with berberine, the mRNA profiling of BEL-7402 cells was collected via RNA-seq. Then, both the DEGs and DASGs induced by berberine were carefully identified by a suite of sequence analysis software. Based on the DEGs functional enrichment analysis, two potential mechanisms underlying the cell cycle arrest property for berberine were suggested as below.
Berberine might induce p53-dependent G1 phase arrest of BEL-7402 cancer cells. After BEL-7402 cells were treated with berberine, five genes (BBC3, FAS, CCNG2, GADD45B, and IGFBP3) were up-regulated and two genes (CASP9, THBS1) were down-regulated (Table C in S1 File) in p53 signaling pathway. Notably, the five up-regulated genes are downstream targets of p53 [41–45]. Meanwhile, p53 was found to be expressed at a modest level in BEL-7402 cells before and after berberine treatment. The wild-type p53 status in BEL-7402 cell line was also confirmed by other researchers [46, 47]. These results suggested that p53 signaling pathway might be influenced under berberine perturbation. As is known, p53 is a crucial cell cycle checkpoint protein that will cause cell cycle arrest responding to DNA damage [48]. Two cell cycle-related genes (CDKN2D, GADD45B), which are in the downstream pathway of p53, were found to be up-regulated after berberine treatment. They could inhibit the activity of CDKs that regulate the G1-S phase transition. And Cell cycle arrest at G1 phase in BEL-7402 cells induced by berberine was also found in our previous work through flow cytometric analysis.[8] Taken together, our results indicated that berberine might inhibit BEL-7402 cell proliferation by inducing G1 cell cycle arrest in a p53-dependent manner. Additionally, p53-dependent cell cycle arrest at G1 phase was also reported in several other cancer cell lines after berberine treatment.[37–39] Our previous studies reported the cell cycle arrest effect of berberine by targeting calmodulin, which might be the upstream signal of p53 pathways [8].
Berberine might suppress the production of inflammatory cytokines through NFκB inhibition. RNA-seq results indicated that NFKBIA (IκB) was significantly up-regulated by berberine treatment (Table D in S1 File). As an inhibitor of NFκB, IκB can hinder the translocation of NFκB from cytoplasm to nucleus [49]. Previous studies have proved that the inhibition of NFκB may influence the expression of many inflammatory factors which could promote tumor cell growth and facility tumor invasion and metastasis [50]. In this study, five cytokines (IL-6, IL-8, IL1A, IL-33 and IL11) (Table D in S1 File) were found to be down-regulated after berberine treatment according to the RNA-seq results. Notably, all these cytokines have been reported as pro-inflammatory factors. For example, IL-6 and IL-8 were reported to be significantly high expressed and play an important role in migration and sustainable proliferation of human liver cancer cells [51–53]. In addition, the silencing of IL-8 by siRNA could inhibit proliferation and delay the G1 to S cell cycle progression in breast cancer cells or prostate cancer cells lines [54]. This result suggested that suppressing the production of inflammatory cytokines through NFκB inhibition might also contribute to the anti-proliferative effect of berberine in BEL-7402 cells.
Meanwhile, according to the functional annotation on those DASGs, the splicing pattern of a group of cell cycle related genes were also interfered with berberine treatment (Table 2). In addition, the connections between the DEGs and DASGs were measured by statistically analyzing local co-enrichment among chromosomes, network distance and functional semantic similarity on GO trees. The results indicated that these DEGs and DASGs might be connected and functionally cross-talked.
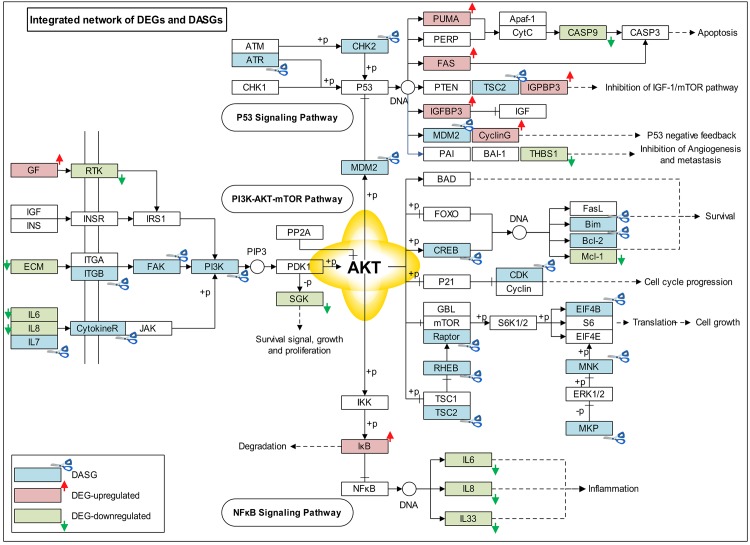
In addition to overall statistically analysis of the connection between DEGs and DASGs, an integrated network included 15 DEGs and 18 DASGs was re-constructed based on p53 signaling pathway, PI3K-AKT-mTOR pathway, and NFκB signaling pathway (Fig 5). In light of this network, two potential functional connections between DEGs and DASGs were suggested as below.
Fig 5. The integrated network affected by DEGs and DASGs.
This plot shows an integrated gene-regulation network which is built based on those KEGG pathways affected by DEGs and DASGs. In the plot, red boxes represent those DEGs which are up-regulated; the green ones represent those DEGs which are down-regulated. Those DASGs are displayed as blue-colored boxes.
DEGs and DASGs synergically regulate p53 pathway. In the upper part in Fig 5, two genes (CHK2, ATR) were differentially alternative spliced, which are responsible for the phosphorylation and activation of p53 [55, 56]. Meanwhile, another four genes (PUMA, FAS, IGF-BP3, CyclinG) were all significantly overexpressed, which are the transcriptional targets of p53 [41–45]. These results highly suggested that these DEGs and DASGs might act synergistically on p53 pathway, thereby inhibit cell cycle progression in cancer cells.
DEGs and DASGs synergically regulate PI3K-AKT-mTOR pathway. Considering the PI3K-AKT-mTOR part in the integrated network (middle part in Fig 5), DASGs were widely distributed both at the upstream and downstream of this pathway. In addition, this pathway seems to play a linking role between p53 pathway and NFκB pathway (Fig 5). For example, AKT is involved in the regulation of NFκB activation through the phosphorylation of IκB kinase (IKK), which could phosphorylate IκB and induce its degradation [57]. Based on DEGs functional analysis, both p53 signalling pathway and NFκB signalling pathway were found to contribute to the anti-proliferative effect of berberine. Moreover, via the DASGs-perturbed PI3K-AKT-mTOR pathway, these two pathways might be linked together for generating berberine’s anticancer effect.
Thus, based on the findings in the current study, we would like to draw attention that in addition to differential expression, differential splicing is very important as well to regulate the cellular activities. The biological outcome of differential splicing is actually the collaborative effects of multiple DASGs coupling with DEGs [13]. Preferably, further experimental validation is desirable when key factors can be identified or the multi-targeting interference could be achieved in one experiment.
Conclusion
In this study, the genome-wide profiling of transcription and alternative splicing of BEL-7402 cells treated with berberine were analysed by RNA-seq. The results showed that the DEGs and DASGs induced by berberine were functionally enriched in the p53 and cell cycle signalling pathway. Importantly, our results indicated that these two sets of genes might be functionally cross-talked and jointly contribute to the anticancer effect of berberine. This RNA-seq study gave us a better understanding of the molecular mechanisms of anti-cancer effect of berberine. In addition, it has provided new clues for further researches of pharmacological activity of berberine as well as other drugs.
Supporting Information
There are four supplementary tables. Table A, information about all the DEGs. Table B, information about all the DASGs. Table C, expression data of DEGs in p53 signaling pathway. Table D, expression data of DEGs in NFκB pathway.
(XLSX)
Acknowledgments
We would like to thank Dr. Qi Liu from Department of Bioinformatics, Tongji University for his constructive suggestions. We are indebted to Ms. Yan Liu, the administrator of High-Performance Computing Center at School of Life science and technology, for her technical assistance.
Abbreviations
- DASG
Differentially Alternative Spliced Genes between two samples
- DEG
Differentially Expressed Genes between two samples
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported partially by the National High Technology Research and Development Program (“863” Program) of China (2012AA020405), and the Funding of Ministry of Health (2012ZX10005001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene 2005. January 3;344:1–20. [DOI] [PubMed] [Google Scholar]
- 2. Tang JY, Lee JC, Hou MF, Wang CL, Chen CC, Huang HW, et al. Alternative splicing for diseases, cancers, drugs, and databases. ScientificWorldJournal 2013;2013:703568 10.1155/2013/703568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berberine. Altern Med Rev 2000. April;5(2):175–7. [PubMed] [Google Scholar]
- 4. Tillhon M, Guaman OL, Lombardi P, Scovassi AI. Berberine: new perspectives for old remedies. Biochem Pharmacol 2012. November 15;84(10):1260–7. 10.1016/j.bcp.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 5.Wang Yi HJLH. Selective Effect of Berberine on Tumor and Normal cell. Focus on Microscopy; 2010; Shanghai, China.
- 6. Ortiz LM, Lombardi P, Tillhon M, Scovassi AI. Berberine, an epiphany against cancer. Molecules 2014;19(8):12349–67. 10.3390/molecules190812349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang X, Huang N. Berberine induces selective apoptosis through the AMPKmediated mitochondrial/caspase pathway in hepatocellular carcinoma. Mol Med Rep 2013. August;8(2):505–10. 10.3892/mmr.2013.1506 [DOI] [PubMed] [Google Scholar]
- 8. Ma C, Tang K, Liu Q, Zhu R, Cao Z. Calmodulin as a potential target by which berberine induces cell cycle arrest in human hepatoma Bel7402 cells. Chem Biol Drug Des 2013. June;81(6):775–83. 10.1111/cbdd.12124 [DOI] [PubMed] [Google Scholar]
- 9. Shkreta L, Froehlich U, Paquet ER, Toutant J, Elela SA, Chabot B. Anticancer drugs affect the alternative splicing of Bcl-x and other human apoptotic genes. Mol Cancer Ther 2008. June;7(6):1398–409. 10.1158/1535-7163.MCT-08-0192 [DOI] [PubMed] [Google Scholar]
- 10. Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, et al. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol 2010. November;17(11):1358–66. 10.1038/nsmb.1912 [DOI] [PubMed] [Google Scholar]
- 11. Dutertre M, Chakrama FZ, Combe E, Desmet FO, Mortada H, Polay EM, et al. A recently evolved class of alternative 3'-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nat Commun 2014;5:3395 10.1038/ncomms4395 [DOI] [PubMed] [Google Scholar]
- 12. Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3'-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol 2008. January;28(2):849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montes M, Becerra S, Sanchez-Alvarez M, Sune C. Functional coupling of transcription and splicing. Gene 2012. June 15;501(2):104–17. 10.1016/j.gene.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 14. Chen JM, Chu FH, Yeh HJ, Shen TW. The establishment and some characteristics of a human liver carcinoma cell line (BEL-7402) in vitro. Acta Biologicae Experimentalis Sinica;11:37–47. [Google Scholar]
- 15. Liu LX, Jiang HC, Liu ZH, Zhu AL, Zhou J, Zhang WH, et al. Gene expression profiles of hepatoma cell line BEL-7402. Hepatogastroenterology 2003. Sep-Oct;50(53):1496–501. [PubMed] [Google Scholar]
- 16. Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 17. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14(4):R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015. January 15;31(2):166–9. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res 2010. July;38(Web Server issue):W755–62. 10.1093/nar/gkq405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hooper JE. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Hum Genomics 2014;8:3 10.1186/1479-7364-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013. January;31(1):46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 2010. December;7(12):1009–15. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 2012. October;22(10):2008–17. 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010. January 1;26(1):136–8. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 26. Hu Y, Huang Y, Du Y, Orellana CF, Singh D, Johnson AR, et al. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res 2013. January;41(2):e39 10.1093/nar/gks1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aschoff M, Hotz-Wagenblatt A, Glatting KH, Fischer M, Eils R, Konig R. SplicingCompass: differential splicing detection using RNA-seq data. Bioinformatics 2013. May 1;29(9):1141–8. 10.1093/bioinformatics/btt101 [DOI] [PubMed] [Google Scholar]
- 28. Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 2005. August;37(8):844–52. [DOI] [PubMed] [Google Scholar]
- 29. Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4(5):P3 [PubMed] [Google Scholar]
- 30. The Gene Ontology project in 2008. Nucleic Acids Res 2008. January;36(Database issue):D440–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014. January;42(Database issue):D199–205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Preter K, Barriot R, Speleman F, Vandesompele J, Moreau Y. Positional gene enrichment analysis of gene sets for high-resolution identification of overrepresented chromosomal regions. Nucleic Acids Res 2008. April;36(7):e43 10.1093/nar/gkn114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mao G, Zhang N. Analysis of Average Shortest-Path Length of Scale-Free Network. Journal of Applied Mathematics;2013:5. [Google Scholar]
- 34. Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. A new method to measure the semantic similarity of GO terms. Bioinformatics 2007. May 15;23(10):1274–81. [DOI] [PubMed] [Google Scholar]
- 35. Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010. April 1;26(7):976–8. 10.1093/bioinformatics/btq064 [DOI] [PubMed] [Google Scholar]
- 36.ENCODE. Standards, Guidelines and Best Practices for RNA-Seq. v1.0 ed; 2011.
- 37. Liu Z, Liu Q, Xu B, Wu J, Guo C, Zhu F, et al. Berberine induces p53-dependent cell cycle arrest and apoptosis of human osteosarcoma cells by inflicting DNA damage. Mutat Res 2009. March 9;662(1–2):75–83. 10.1016/j.mrfmmm.2008.12.009 [DOI] [PubMed] [Google Scholar]
- 38. Choi MS, Oh JH, Kim SM, Jung HY, Yoo HS, Lee YM, et al. Berberine inhibits p53-dependent cell growth through induction of apoptosis of prostate cancer cells. Int J Oncol 2009. May;34(5):1221–30. [PubMed] [Google Scholar]
- 39. Choi MS, Yuk DY, Oh JH, Jung HY, Han SB, Moon DC, et al. Berberine inhibits human neuroblastoma cell growth through induction of p53-dependent apoptosis. Anticancer Res 2008. Nov-Dec;28(6A):3777–84. [PubMed] [Google Scholar]
- 40. Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol 2008. August;8(4):393–412. 10.1016/j.coph.2008.08.004 [DOI] [PubMed] [Google Scholar]
- 41. Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ, et al. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci U S A 2001. September 25;98(20):11318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hernandez-Vargas H, Rodriguez-Pinilla SM, Julian-Tendero M, Sanchez-Rovira P, Cuevas C, Anton A, et al. Gene expression profiling of breast cancer cells in response to gemcitabine: NF-kappaB pathway activation as a potential mechanism of resistance. Breast Cancer Res Treat 2007. April;102(2):157–72. [DOI] [PubMed] [Google Scholar]
- 43. Rozan LM, El-Deiry WS. p53 downstream target genes and tumor suppression: a classical view in evolution. Cell Death Differ 2007. January;14(1):3–9. [DOI] [PubMed] [Google Scholar]
- 44. Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev 2000. March 15;14(6):704–18. [PMC free article] [PubMed] [Google Scholar]
- 45. Gutierrez-Mariscal FM, Yubero-Serrano EM, Rangel-Zuniga OA, Marin C, Garcia-Rios A, Perez-Martinez P, et al. Postprandial activation of p53-dependent DNA repair is modified by Mediterranean diet supplemented with coenzyme Q10 in elderly subjects. J Gerontol A Biol Sci Med Sci 2014. July;69(7):886–93. 10.1093/gerona/glt174 [DOI] [PubMed] [Google Scholar]
- 46. Zeng Y, Prabhu N, Meng R, Eldeiry W. Adenovirus-mediated p53 gene therapy in nasopharyngeal cancer. Int J Oncol 1997. August;11(2):221–6. [PubMed] [Google Scholar]
- 47. Bressac B, Galvin KM, Liang TJ, Isselbacher KJ, Wands JR, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci U S A 1990. March;87(5):1973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev 1994. November 1;8(21):2540–51. [DOI] [PubMed] [Google Scholar]
- 49. Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell 1998. December 11;95(6):749–58. [DOI] [PubMed] [Google Scholar]
- 50. Diamant G, Dikstein R. Transcriptional control by NF-kappaB: elongation in focus. Biochim Biophys Acta 2013. September;1829(9):937–45. 10.1016/j.bbagrm.2013.04.007 [DOI] [PubMed] [Google Scholar]
- 51. Wong VW, Yu J, Cheng AS, Wong GL, Chan HY, Chu ES, et al. High serum interleukin-6 level predicts future hepatocellular carcinoma development in patients with chronic hepatitis B. Int J Cancer 2009. June 15;124(12):2766–70. 10.1002/ijc.24281 [DOI] [PubMed] [Google Scholar]
- 52. Liu Y, Fuchs J, Li C, Lin J. IL-6, a risk factor for hepatocellular carcinoma: FLLL32 inhibits IL-6-induced STAT3 phosphorylation in human hepatocellular cancer cells. Cell Cycle 2010. September 1;9(17):3423–7. [DOI] [PubMed] [Google Scholar]
- 53. Ren Y, Poon RT, Tsui HT, Chen WH, Li Z, Lau C, et al. Interleukin-8 serum levels in patients with hepatocellular carcinoma: correlations with clinicopathological features and prognosis. Clin Cancer Res 2003. December 1;9(16 Pt 1):5996–6001. [PubMed] [Google Scholar]
- 54. Singh RK, Lokeshwar BL. Depletion of intrinsic expression of Interleukin-8 in prostate cancer cells causes cell cycle arrest, spontaneous apoptosis and increases the efficacy of chemotherapeutic drugs. Mol Cancer 2009;8:57 10.1186/1476-4598-8-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 1999. January 15;13(2):152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ou YH, Chung PH, Sun TP, Shieh SY. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell 2005. April;16(4):1684–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ozes ON, Mayo LD, Gustin JA, Pfeffer SR, Pfeffer LM, Donner DB. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature 1999. September 2;401(6748):82–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
There are four supplementary tables. Table A, information about all the DEGs. Table B, information about all the DASGs. Table C, expression data of DEGs in p53 signaling pathway. Table D, expression data of DEGs in NFκB pathway.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.