Abstract
The alarmone (p)ppGpp is involved in regulating growth and several different stress responses in bacteria. In recent years, substantial progress has been made in our understanding of the molecular mechanisms of (p)ppGpp metabolism and (p)ppGpp-mediated regulation. In this Review, we summarize these recent insights, with a focus on the molecular mechanisms governing the activity of the RelA/SpoT Homologue (RSH) proteins, which are key players that regulate the cellular leves of (p)ppGpp, the structural basis of transcriptional regulation by (p)ppGpp and the role of (p)ppGpp in GTP metabolism and in the emergence of bacterial persisters.
Bacteria use an array of sensory systems to monitor their environment to enable adaptation to stressful conditions. Most of these systems convert external stimuli into changes in the intracellular concentration of a secondary messenger molecule, which functions as a pleiotropic regulator of key molecular targets. There are three common nucleotide-based secondary messengers in bacteria: cAMP, c-di-GMP and (p)ppGpp1.
(p)ppGpp is a collective term that refers to two related alarmone nucleotides, ppGpp and pppGpp, which are formed by the addition of a pyrophosphate moiety to the 3′ position of GDP and GTP, respectively (Fig. 1a). (p)ppGpp – or ‘magic spot’ as it is often referred to – has several important roles in bacterial physiology, in particular through the coordination of cellular responses on exposure to stress (Fig. 1a, Box 1). During exponential growth, (p)ppGpp is present at basal levels and functions as one of the major modulators of bacterial growth rate2 and a fine-tuner of general metabolism3, 4. (p)ppGpp contributes to growth rate control by inhibiting ribosomal RNA production5. This occurs, for example, when nutrient limitation slows down the growth rate, and cellular resources and energy are driven from ribosome biosynthesis towards the upkeep of general metabolism6. Regulation of general metabolism by (p)ppGpp is exerted by its action on multiple processes: first, at the level of transcription, (p)ppGpp controls the expression of genes involved in amino acid biosynthesis5; second, it regulates nucleotide metabolism by binding directly to the enzymes involved in nucleotide biosynthesis7 and uptake8.
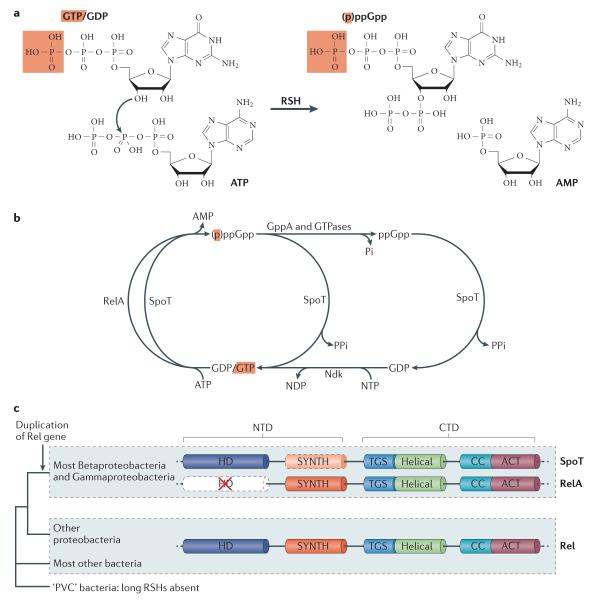
Figure 1. (p)ppGpp metabolism by RelA , SpoT and Rel.
(a) (p)ppGpp synthesis by RSH enzymes. The products of the RSH-catalyzed reaction guanosine 3′-diphosphate 5-′diphosphate (ppGpp) and guanosine 3′-diphosphate 5-'triphosphate (pppGpp) are commonly referred to as (p)ppGpp. The γ-phosphate moiety of GTP and pppGpp are highlighted in red.
(b) (p)ppGpp metabolism in Escherichia coli. The ‘long’ RSHs RelA and SpoT synthesize (p)ppGpp from GTP and GDP, generating AMP as a by-product. Interconversion of pppGpp to ppGpp is catalyzed by guanosine pentaphosphate phosphatase (GppA) and translational GTPases, such as the translocase EF-G15. SpoT catalyzes the degradation of pppGpp and ppGpp to form GTP and GDP, respectively38. Interconversion of GDP to GTP is catalyzed by nucleoside diphosphate kinase (Ndk)75. Adapted from ref Cashel, M., et al., The stringent response, in Escherichia coli and Salmonella: cellular and molecular biology. 1996, ASM Press: Washington DC. p. 1458–1496.
(c) The distribution of ‘long’ RSHs and RSH domain structure. The domains of the long RSHs SpoT, RelA and Rel are shown, along with the distribution of these proteins in bacteria. The coloured boxes representing each domain show their approximate location along the length of the proteins, with dashed borders indicating domains with reduced or absent functional activity. In the case of the SpoT (p)ppGpp synthesis (SYNTH) domain, its synthetic activity is weak, whereas hydrolytic activity is absent in the RelA (p)ppGpp hydrolysis (HD) domain. The HD and SYNTH domains comprise the N-terminal domain (NTD), whereas the ThrRS, GTPase and SpoT (TGS), helical, Conserved Cysteines (CC) and Aspartokinase, Chorismate mutase and TyrA (ACT) domains together comprise the C-terminal domain (CTD). The phylogenetic tree summarizes the evolutionary relationships among bacteria that contain or lack long RSHs. The red arrow indicates the duplication event that led to the emergence of RelA and SpoT from an ancestral Rel protein in the lineage of the Gamma- and Beta-proteobacteria. The Planctomycete, Verrucomicrobia and Chlamidiale superphylum (PVC) of bacteria do not encode any long RSHs. In the absence of a reliable root of the bacterial tree of life, it is not known whether long RSHs evolved after the divergence of PVC bacteria, or whether they were lost in this lineage.
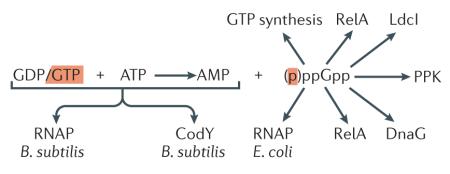
Box 1. Molecular targets of (p)ppGpp.
(p)ppGpp exerts its regulatory effects using both direct and indirect mechanisms (see the figure). The indirect mechanisms includean alteration in the nucleoside triphosphate pool due to depletion of GTP during the stringent response. This strategy is the main pathway of RNAP regulation in Bacillus subtilis: depletion of GTP alters the balance of initiating nucleotides available, which results in changes in the promoter preferences of RNAP62. Another important target of indirect regulation is the transcriptional repressor CodY, which is inhibits expression of more then 100 target genes involved in adaptation to stress and sporulation106. CodY is activated by the binding of GTP and branched amino acids in B. subtilis106, and depletion of GTP during the stringent response causes de-repression of the CodY-regulated ergulatory network. Direct binding of the alarmone to the target enzyme, in addition to the regulation of transcription in E. coli via its effects on RNAP60, 61, leads to activation of (p)ppGpp synthesis by RelA28, in addition to the inhibition of protein biosynthesis (by inhibiting translational GTPases, such as Initiation Factor 2 (IF2)107, 108), replication via DNA primase (DnaG)109, polyphosphate metabolism via polyphosphate kinase (PPK)110 and acid stress response via lysine decarboxylase (Ldcl/CadA)111. Recent sequence analysis and docking studies have further expanded this list to include potential targets, such as orotate phosphoribosyltransferase and glutamine phosphoribosylpyrophosphate amidotransferase, that are awaiting experimental validation112. For an overview of (p)ppGpp-mediated regulation of its molecular targets and the implications of this regulation for bacterial virulence, we refer the reader to an excellent review104.
On exposure to stresses, such as nutrient deprivation or heat shock, the cellular concentration of (p)ppGpp increases, and the alarmone orchestrates concentration-dependent reprogramming of many processes, including transcription9. These physiological changes are commonly referred to as the stringent response. Several classes of enzymes regulate the intracellular concentration of (p)ppGpp (Fig. 1b). RelA/SpoT Homologue (RSH) bifunctional proteins are the key players, synthesizing (p)ppGpp from ATP and GTP/GDP and degrading (p)ppGpp to GTP/GDP and pyrophosphate5, 10. Degradation of (p)ppGpp is also carried out by non-RSH enzymes called Nudix hydrolases11, 12 – a widely distributed class of enzymes capable of hydrolyzing a variety of nucleoside diphosphate compounds13. Rapid conversion of pppGpp to ppGpp is specifically catalyzed by the guanosine pentaphosphate phosphohydrolase, GppA14, as well as unspecifically by GTPase enzymes, such as the translational GTPase EF-G15, which catalyzes in translocation of the ribosome during protein synthesis.
The stringent response and (p)ppGpp have important roles in the regulation of bacterial virulence16, survival during host invasion17, antibiotic resistance18 and persistence19. In this Review, we focus on recent progress in the understanding of the ‘nuts and bolts’ of the bacterial stringent response machinery: the structural and functional studies of RelA/SpoT Homologue (RSH) proteins that modulate cellular (p)ppGpp levels, the molecular mechanism of RNAP regulation by (p)ppGpp, the recently identified role of (p)ppGpp in GTP biosynthesis and progress in the dissection of the long-speculated connection between the stringent response and bacterial persistence.
RSHs: (p)ppGpp homeostasis
The enzymes that synthesize and degrade (p)ppGpp are highly conserved in bacteria10. RSH genes are only absent in Planctomycetes, Verrucomicrobia, and Chlamydiae (the PVC superphylum), as well as a few other species, mainly obligate intracellular endosymbionts such as Buchnera aphidicola and Candidatus Carsonella ruddii, and pathogens with reduced genomes, such as Treponema pallidum and species of Mycoplasma and the order Rickettsiales10.
There are two types of RSH enzymes: ‘short’ enzymes that consist of a single domain (Box 2) and ‘long’ multi-domain RSH enzymes10. Historically, the study of the stringent response initially focused on the γ-proteobacterium E. coli, the workhorse of bacterial genetics. In this organism the stringent response is orchestrated by two multi-domain RSH enzymes: RelA20 and SpoT21. These two proteins originated via gene duplication in the evolutionary lineage to β-proteobacteria and γ-proteobacteria and share the same six-domain structure, comprising a (p)ppGpp hydrolysis domain (HD), a (p)ppGpp synthesis (SYNTH) domain, a ThrRS, GTPase and SpoT (TGS) domain, a domain containing helical, conserved cysteines (CC) and an Aspartokinase, Chorismate mutase and TyrA (ACT) domain10 (Fig. 1c). The exact molecular functions of the latter four domains, collectively referred to as the C-terminal domain (CTD), are unclear but they are suggested to regulate the enzymatic activities of the N-terminal domain (NTD), which contains HD and SYNTH22.
Box 2. Diversity and nomenclature of RSHs.
The “long” multi-domain RSHs have been known for decades and have been used as prototypes for studying the stringent response. Recently, ‘short’ single-domain monofunctional RSH proteins – Small Alarmone Synthases (SAS) and Small Alarmone Hydrolases (SAHs) – have been discovered in various bacterial species, such as Mycobacterium smegmatis113, Bacillus subtilis114 and Streptococcus mutans115. Phylogenetic analysis has identified twenty subgroups of these enzymes, which are typically only present in organisms that encode a ‘long’ RSH10. With the exception of the Mycobacterium smegmatis SAS (MS_RHII-RSD, which has additional RNase activity113), SASs and SAHs are single-domain enzymes. Although biochemical data suggest that these proteins do not require additional interaction partners for the regulation of their enzymatic activities, it is impossible to rule out the existence of unidentified partners, similar to those involved in the regulation of ‘long’ RSHs114. The activity of SAS enzymes seems to be primarily regulated at the transcriptional level: expression of the B. subtilis SAS RelQ (also known as YwaC) is upregulated as part of the σ W regulon in response to cell wall damage116, and transcription of a second B. subtilis SAS RelP (also known as YjbM) is up-regulated by alkaline shock114. Several X-ray structures of putative SAS enzymes have been solved and deposited in the Protein Data Bank (for example Streptococcus mutans, PDB 3L9D, and Streptococcus pneumoniae, 2BE3), but the functionality of these enzymes has not been tested and the structures are not yet published.
An SAH, Mesh1, which is structurally similar to bacterial SAHs, has been identified in humans and other animals10, 117. Mesh1 is capable of hydrolyzing ppGpp in vitro117, but its physiological role is unclear given the absence of (p)ppGpp-synthesizing RSH enzymes10 and the absence of detectable (p)ppGpp in these organisms117, 118. Although several chloroplast RSHs are widely distributed in plants119, the only other eukaryotes that have so far been found to encode cytoplasmic (p)ppGpp synthetase homologues are the amoebae Dictyostelium discoideum and Dictyostelium purpureum, the fungi Aspergillus nidulans, Aspergillus fumigatus and Gibberella zeae, and the heterokont algae Thalassiosira pseudonana and Phaeodactylum tricornutum10. The potential functional roles of (p)ppGpp in these organisms are unknown; however D. discoidium uses another secondary messenger cAMP as a secreted signalling molecule for cellular aggregation120, which raises the question of whether (p)ppGpp has a similar role in extracellular communication.
The use of standardized nomenclature is particularly important as RSH proteins from different organisms are often named in an idiosyncratic manner. An extensive bioinformatic analysis of RSH enzymes has provided a foundation for the rational terminology of RSHs10. This phylogenetic analysis identified thirty distinct RSH subgroups that comprise eleven long, multidomain RSHs (Rel, SpoT, RelA, RshA, RshB, RshC, RshD and Rsh1-4), seven SAHs (paSpo, pbcSpo, pbcSpo2, Mesh1, Mesh1L, rickSpo, divSpo) that carry only the (p)ppGpp hydrolase (HD domain) and twelve SASs (actRel, bdRel, cloRel, fpRel, fpRel2, gRel, capRel, rickRel, RelP, RelQ, RelV and divRel) that only carry the (p)ppGpp syntetase (SYNTH) domain. Not all of these groups are monophyletic; Rel is paraphyletic to the other long RSHs as these evolved from the more ancient Rel subfamily. divRel and divSpo do not define co-clustering clades, but refer to miscellaneous and divergent sequences that contain only the SYNTH or HD domain, respectively.
RelA
The activities of RelA and SpoT are regulated by different stress signals. The bifunctional RSH SpoT senses the limitation of several nutrients: carbon sources, phosphate, iron and fatty acids, and has both (p)ppGpp synthetic and hydrolytic activities23-25. RelA has only (p)ppGpp synthetic activity, which is induced by amino acid starvation26 and heat shock27. These functional differences between RelA and SpoT are reflected in the domain conservation of the proteins (Fig. 1c): although RelA retains a relic of the HD domain, it is highly divergent from that of SpoT and lacks hydrolytic activity10, 28
RelA is a ribosome-associated protein that senses amino acid starvation by directly monitoring the translational capacity of the cell. During normal growth, i.e. exponential growth in the absence of amino acid limitation, amino acids are delivered in the form of aminoacylated tRNA molecules to the ribosomal acceptor site (A-site) to be added to the nascent poypeptide as it is synthesized. During amino acid starvation, deacylated tRNAs, which are not conjugated to amino acids, accumulate and enter the A-site, which strongly activates the (p)ppGpp synthetic activity of RelA29 (Fig. 2a). The 3′-OH group of the terminal adenosine of the tRNA molecule is crucial for the activation of RelA, which suggests that the aminoacylation state of tRNAs is directly inspected by RelA30. In the absence of nucleotide substrates for synthesis of (p)ppGpp, i.e. ATP and GTP (GDP), it has been shown that the presence of a deacylated tRNA in the A-site promotes the binding of enzymatically idle RelA to the ribosome. However, stabilization of deacylated tRNA binding to the ribosome in the presence of enzymatically idle RelA has not been detected31, 32. This is surprising since regardless of the order of association events (whether the tRNA or RelA binds first), the same ternary complex is formed, RelA:deacylated tRNA:70S ribosome, and one would expect mutual stabilization of RelA and tRNA binding to the ribosome. On synthesis of (p)ppGpp, RelA was initially proposed to dislodge the tRNA from the ribosome33, but this was not supported by a subsequent study32 in which a ‘hopping’ model was proposed. This model suggests that the process of (p)ppGpp synthesis by ribosome-bound RelA dislodges RelA from the complex, and by ‘hopping’ between ribosomes, RelA can monitor the translational status of the cell32. Recent single-molecule in vivo analysis suggests a modified model34: activation of RelA during amino acid starvation was shown to induce its dissociation from the ribosome; however, multiple rounds of (p)ppGpp synthesis occurred off the ribosome, rather than on the ribosome. This model implies the existence of ‘molecular memory’, such that RelA remains in an active state after its dissociation from the ribosome. Prolonged disengagement of the auto-inhibitory CTD domain of RelA22 could render it enzymatically active by preventing inactivation of the enzyme off the ribosome, but this hypothesis regarding the nature of RelA’s molecular memory remains to be tested. In addition to deacylated tRNA, the other allosteric activator of RelA is its product ppGpp28, resulting in a conditional positive feedback loop. The presence of ppGpp alone is insufficient to induce maximum catalytic activity of RelA, but it potentiates the activation induced by the ribosome and deacylated tRNA in the A-site.
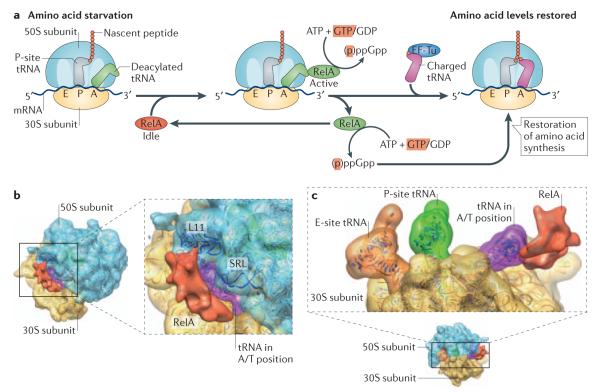
Figure 2. Mechanism of action of RelA.
(a) Amino acid starvation leads to the accumulation of deacylated tRNAs in the cytosol that bind to the ribosomal A-site. This ribosomal state is recognized by RelA, which binds to the 50S subunit and adopts an active conformation. Activation of RelA and consequent synthesis of (p)ppGpp leads to its dissociation from the ribosome, followed by numerous rounds of (p)ppGpp synthesis in the dissociated state32, 34. Increased (p)ppGpp levels direct cellular metabolic resources to amino acid synthesis, which restores normal levels of tRNA aminoacylation. Aminoacylated tRNA is delivered to the ribosome by elongation factor Tu (EF-Tu) in direct competition with binding of RelA and deacylated tRNA.
(b-c) Cryo-EM structure of RelA in complex with the ribosome. On binding to the ribosomal A-site, RelA interacts with the Sarcin-Ricin Loop (SRL) and ribosomal protein L11, as well as deacylated tRNA in the A-site, which adopts a highly distorted conformation (referred to as A/T-like tRNA). The RelA cryo-EM structure (3D-EM database e accession code EMD-2373) is reproduced from35, with permission.
Our understanding of the structural aspects of RelA enzymatic activity is based on a recent low-resolution cryo-EM structure35. On binding to the A-site, RelA interacts with the Sarcin-Ricin Loop (SRL) and ribosomal protein L11 (Fig. 2b). In support of the cryo-EM structure, L11 has been shown to be crucial for activation of RelA36. The enzyme interacts directly with the A-site tRNA, which adopts an unusual conformational state that resembles the A/T state of the aminoacyl tRNA in complex with EF-Tu on the ribosome37 (Fig. 2c). Despite lacking in molecular detail, the current structure provides a framework for understanding well-documented RelA inhibition by antibiotic thiostrepton26 that is targeting translation by intercalating between L11 and helices 43 and 44 of the 23S rRNA (PIMD 18406324), and for the mutual exclusion between active protein synthesis and RelA activation on the ribosome.
SpoT
RelA seems to respond specifically to amino acid starvation, whereas SpoT is a hub protein that integrates various stress signals including fatty acid23, iron24 and carbon source25 starvation. Unlike RelA, E. coli SpoT has both weak (p)ppGpp synthetic activity and strong hydrolytic activity25, 38 (Fig. 1c). Interestingly, patterns of sequence conservation of SpoT in the Moraxellaceae family of γ-proteobacteria suggest that its (p)ppGpp synthetic activity is gradually being lost, indicating a partitioning of functions for RelA and SpoT in this lineage10, but requires further experimental validation. The hydrolysis function of SpoT is crucial for balancing cellular (p)ppGpp concentrations in the presence of RelA, and therefore disruption of the spoT gene in E. coli is lethal25. Several molecular partners of SpoT have been identified; direct interactions with the GTPase Obg (also known as CgtA) has been suggested to repress the (p)ppGpp-synthetic activity of SpoT under nutrient-rich conditions39, whereas the Acyl Carrier Protein (ACP) activates SpoT during fatty acid starvation40. Simultaneous disruption of spoT and the β-Ketoacyl-ACP Synthase III gene (fabH) in E. coli leads to synthetic lethality41, underscoring the intimate connection between SpoT and fatty acid metabolism. However, the molecular details of these regulatory mechanisms are unknown as all attempts to purify full-length recombinant SpoT from E. coli have so far been unsuccessful. This has limited biochemical investigations to the characterization of the crude, partially purified protein38, 42, 43 and structural information is lacking altogether.
Rel
Similarly to SpoT, Rel has both synthetic and hydrolytic activities, and is the sole regulator of (p)ppGpp concentration in the majority of bacteria. Biochemical investigations of Rel have mostly focused on the Mycobacterium tuberculosis enzyme, owing to its importance for long-term survival of non-replicating persister bacteria during chronic infection44, 45. In a reconstituted in vitro system, the synthetic activity of M. tuberculosis Rel was strongly stimulated by ribosomes that contained a deacylated tRNA in the A-site, whereas no effect on its hydrolytic activity46 was observed, suggesting that Rel is the functional counterpart of RelA in terms of its role in acute (p)ppGpp synthesis in response to amino acid starvation. However, experiments with live M. tuberculosis showed that amino acid starvation induced by serine hydroxamate (SHX), a competitive inhibitor of seryl-tRNA synthetase that leads to the accumulation of deacylated tRNASer in E. coli and B. subtilis, did not lead to (p)ppGpp accumulation, and complete starvation (the absence of all nutrients induced by transferring bacteria to a saline buffer) was required for induction of the stringent response45. Similarly, the stringent response is not induced in Helicobacter pylori following exposure to SHX47, but it is induced after a nutritional downshift48 and CO2 deprivation49 . However, since amino acid analogues such as SHX have been shown to be ineffective in eliciting the stringent response in some bacterial species, including H. pylori48, the sensitivity of tRNA synthetases to this analogue and the efficiency of analogue uptake should be considered in such experiments. However, regardless the possible technical caveats, it is clear that the precise triggers for the stringent response do seem to vary between species. For example, a Caulobacter crescentus requires additional stimuli such as carbon or nitrogen starvation – not just amino acid starvation alone – to elicit the Rel-mediated stringent response50. This contrasts with reports of amino acid starvation being sufficient to induce (p)ppGpp synthesis by Rel in other organisms, such as Enterococcus faecalis51, Myxococcus xanthus52 and Streptomyces coelicolor A3(2)53. Such differences in the triggers of the stringent response have been suggested to reflect adaptations to the distinct lifestyles of each species 54.
Unlike SpoT, Rel does not seem to be regulated by ACP55, and whether Obg-mediated regulation occurs is unknown (thus far this has only been tested for SpoT and RelA in E. coli56, 57 and Vibrio cholerae39). An X-ray structure of a truncated Rel from Streptococcus dysgalactiae subsp. equisimilis is available, but unfortunately the regulatory domains of the enzyme are unresolved, which limits structural interpretation of the available biochemical data58.
Regulation of transcription by (p)ppGpp
Accumulation of (p)ppGpp during stress conditions results in alterations in gene expression owing to changes in RNA polymerase (RNAP) activity. For example, during periods of amino acid starvation, (p)ppGpp inhibits transcription from the rRNA and ribosomal protein promoters, but it activates transcription from promoters for amino acid biosynthesis5, 59. In the case of E. coli RNAP, (p)ppGpp interacts directly with RNAP to destabilize the short-lived open complexes that form at certain promoters, such as the promoters of rRNA genes, thereby directly inhibiting transcription initiation 60, 61. In other bacteria, including Bacillus subtilis, (p)ppGpp downregulates transcription without directly interacting with RNAP. This indirect inhibition occurs as production of the alarmone leads to the consumption of GTP and it also inhibits the enzymes responsible for GTP synthesis3, which together reduce the cellular pool of GTP resulting in a decline in the GTP:ATP ratio. This in turn modulates the expression of genes that are governed by promoters sensitive to the concentration of the initiating nucleotide, such that transcription of genes beginning with guanine nucleotides are downregulated, whereas of those beginning with adenosine – activated3, 62.
A crystal structure of Thermus thermophilus RNAP in complex with ppGpp showed that the nucleotide binds near the active site of RNAP63. However, subsequent studies with E. coli RNAP mutants harbouring amino acid substitutions in the vicinity of the putative ppGpp binding site proposed based on the T. thermophilus RNAP structure had no effect on ppGpp-dependent transcriptional regulation of either of E. coli or T. thermophiles enzyme64. Therefore, the relevance of the structural data for predicting the role of ppGpp in the regulation of transcription in E. coli was unclear. In 2013, two studies reported the crystal structure of the E. coli RNAP in complex with ppGpp65, 66. These structures revealed that the ppGpp binding site is located in a cavity surrounded by the α, β’ and ω subunits of RNAP, and it binds to the surface of a double-psi β-barrel (DPBB) domain in the β’subunit and also the N-terminus of the ω subunit (Fig. 3a), which is ~30 Å away from the active site of RNAP. pppGpp binds to the same site as ppGpp on RNAP, but with lower potency65. This newly located (p)ppGpp binding site is consistent with previous observations that the presence of the ω subunit is required for ppGpp-dependent transcription inhibition in vitro67, 68. Amino acid substitutions in the (p)ppGpp binding region of the ω and β’ subunits makes RNAP unresponsive to (p)ppGpp, indicating that this binding site is indeed required for modulating the activity of E. coli RNAP during the stringent response69.
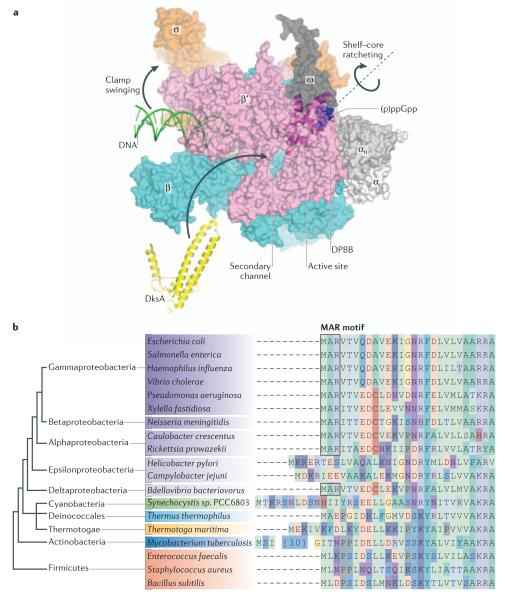
Figure 3. Direct regulation of RNAP activity by (p)ppGpp.
(a) The E. coli RNAP holoenzyme (αI: white, αII, light gray, β, cyan; β’, pink; ω, dark gray; σ70, orange) and promoter DNA (green) complex model showing how the (p)ppGpp and DksA interact with RNAP. The model shows the (p)ppGpp (blue) binding site on RNAP, which is located in the DPBB domain of the β’ subunit (magenta) and the N-termini of the ω subunit. Binding of (p)ppGpp to the DPBB domain may induce an allosteric signal to the catalytic Mg2+ (red sphere) for regulating the catalytic efficiency of RNAP. Alternatively, the (p)ppGpp binding at the shelf-core ratcheting axis may influence the shelf-core ratcheting and/or the DNA binding clamp swinging. The black dashed line indicates the shelf-core ratcheting axis and black arrows show directions of the ratcheting and the DNA binding clamp swinging. DksA is shown as a yellow cartoon model and a blue arrow shows its binding to the E. coli RNAP secondary channel, which may influence the orientation of the core and shelf modules for enhancing the potency of (p)ppGpp. This model was constructed by combining the X-ray crystal structures of E. coli RNAP-ppGpp complex (PDB: 4JK1/4JKR), T. thermophilus RNAP-promoter DNA complex (PDB: 4G7H) and E. coli DksA (PDB: 1TJL).
(b) Taxonomic distribution of the MAR motif. Alignment of the N-terminal region of the ω subunit shows that the MAR motif (shaded box) that (p)ppGpp binds to is conserved only in the α-, β-, δ- and γ-proteobacteria. This suggests that the ancestor of Proteobacteria carried the MAR motif, and it was subsequently lost in the lineage that gave rise to the ε-proteobacteria. The phylogenetic tree on the left shows the evolutionary relationships among the groups of bacteria sampled, according to127.
Models for (p)ppGpp-mediated regulation of transcription
Residues at the (p)ppGpp binding site are not directly involved in RNAP enzymatic activity or in the binding of the DNA template, which suggests an allosteric mechanism for (p)ppGpp-dependent modulation of RNAP activity. As mentioned above, the (p)ppGpp binding site is located on the surface of a double-psi β -barrel (DPBB) domain of the β’ subunit (Fig. 3a) and the other side of the DPBB domain faces the RNAP active center cleft and coordinates the catalytic Mg2+, which is central to RNAP activity. There are two proposed working models to explain how (p)ppGpp binding regulates RNAP. According to the first model, binding of (p)ppGpp to the surface of the DPBB domain induces an allosteric signal, which is transmitted to the other side of the DPBB domain that coordinates the catalytic Mg2+ for the nucleotidyl transfer reaction, thereby regulating the catalytic efficiency of RNAP65. In the second model, (p)ppGpp binding is suggested to influence the shelf and core domains of RNAP (add reference 74 here for define shelf and core domains of RNAP) as it binds at the interface of these two mobile modules. The coordinated motion of these modules, which is known as “shelf-core ratcheting”, causes global and local conformational changes in RNAP, including the swinging of the DNA binding clamp70 (Fig. 3a). Thus, binding of (p)ppGpp at the junction of these mobile modules of RNAP is proposed to determine the opening and closing of the DNA binding clamp, thereby affecting the stability of RNAP-promoter complexes66, 69. However, neither of these models have been tested experimentally yet, so the molecular mechanism of (p)ppGpp regulation remains elusive.
As mentioned above, (p)ppGpp binds to a cavity surrounded by the α, β’ and ω subunits (Fig. 3a). The amino acid residues of the β’and ω subunits are involved in guanine base recognition and interactions with the phosphate backbone, respectively. The N-terminal Met residue of the ω subunit is cleaved by methionine aminopeptidase, which is required for accommodating the phosphate backbone of (p)ppGpp65. Methionine aminopeptidase preferentially cleaves the N-terminal Met if the second amino acid residue has a short side chain, such as Ala, as is the case for the E. coli ω subunit. The third amino acid residue, Arg, is important for mediating interactions with the phosphate group of (p)ppGpp. Thus, the ω subunit N-terminal sequence, MAR, represents a signature for its ability to interact with (p)ppGpp.
An alignment of the N-terminal sequence of the ω subunit69 (Fig. 3b) shows that the MAR motif is found in γ-proteobacteria and the closely related α-, β- and δ-proteobacteria, which suggests that the RNAPs encoded by these classes of bacteria accommodate (p)ppGpp at the ω subunit and probably use direct inhibition by (p)ppGpp for the modulation of transcription. By contrast, other classes of bacteria do not contain the MAR motif, which indicates that they do not have the same (p)ppGpp binding site and may use the indirect mechanism of RNAP regulation as observed in B. subtilis, or they may have an as yet unidentified (p)ppGpp binding site, which has been observed in the RNAPs of Mycobacterium tuberculosis71 and Thermus thermophilus 72. The N-terminal sequence of the ω subunit can be used to predict direct (p)ppGpp–dependent transcriptional regulation, but biochemical experiments are needed to clarify the mechanisms involved.
The role of DksA
In addition to (p)ppGpp, the transcription factor DksA has a crucial role in the stringent response as it amplifies the effect of (p)ppGpp-dependent transcription regulation. DksA binds close to the RNAP secondary channel (which is proposed to transport nucleotide substrates to the active site) and inserts its coiled-coil domain into this channel to reach the active site73 (Fig. 3a). Since (p)ppGpp and DksA bind to RNAP on opposite surfaces, there is no physical interaction between them. DksA binds the RNAP secondary channel in a manner similar to that of the Thermus thermophilus transcription factor Gfh1, which widens this channel, thereby influencing the orientation of the core and shelf modules74. The RNAP and DksA interaction probably makes RNAP more sensitive to the shelf-core ratcheting that is induced by (p)ppGpp, thereby amplifying the signal from (p)ppGpp during transcriptional regulation.
Regulation of GTP biosynthesis by (p)ppGpp
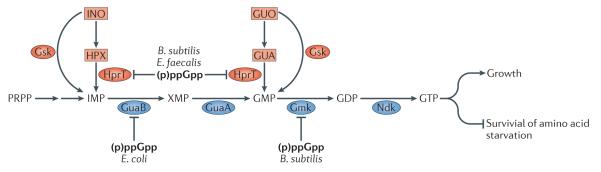
In most bacteria, there are two pathways responsible for GTP biosynthesis: the de novo pathway and the salvage pathway. Both pathways converge on the production of inosine 5′-monophosphate (IMP) – a key intermediate in the synthesis of the purine nucleoside triphosphates ATP and GTP. (Fig. 4)75. The de novo pathway uses phosphoribosyl pyrophosphate (PRPP) as a starting compound for the multi-step pathway of IMP synthesis. IMP is first converted into xanthosine monophosphate (XMP) by the IMP dehydrogenase GuaB, and the GMP synthase GuaA then converts XMP into guanosine monophosphate (GMP). GMP is transformed into the final product, GTP, via sequential rounds of phosphorylation: the GMP kinase Gmk catalyzes the conversion of GMP into guanosine diphosphate (GDP), which is then converted to guanosine triphosphate (GTP) by the nucleoside diphosphate kinase Ndk. By contrast, the salvage pathway uses purines as the starting compounds for GTP synthesis. The nucleobase guanine (GUA) and the corresponding ribonucleoside guanosine (GUO) are converted directly to GMP, whereas the nucleobase hypoxanthine (HPX) and the corresponding ribonucleoside inosine (INO) serve as substrates for the formation of IMP. Guanine and hypoxanthine are very similar chemically: the two compounds differ only in the presence of an amino group on the GUO C-2 carbon atom. As a result, they are recognized by the same proteins: both are substrates of hypoxanthine phosphoribosyltransferase, HprT, which converts these compounds to GMP and IMP, respectively75. Guanine and hypoxanthine are imported from the environment via the same transporters76 and both function as direct repressors of the gua operon, which encodes the enzymes (GuaA and GuaB) for catalyzing the conversion of IMP to GMP77. Moreover, the same enzyme – guanosine kinase, Gsk – converts the nucleosides GUO and INO to GMP and IMP, respectively75.
Figure 4. Role of (p)ppGpp in GTP homeostasis.
In E. coli, the salvage pathway (red) utilizes either guanosine (GUO), guanine (GUA), inosine (INO) and hypoxanthine (HPX) as substrates. Guanosine kinase (Gsk) converts nucleosides GUO and INO to GMP and IMP, respectively. The de novo pathway uses phosphoribosyl pyrophosphate (PRPP) as a starting compound for the multi-step synthesis of inosine 5′-monophosphate (IMP), which is further converted to GTP. The transformation is achieved in four steps: IMP is first converted into xanthosine monophosphate (XMP) by the IMP dehydrogenase GuaB, and the GMP synthase GuaA then converts XMP into guanosine monophosphate (GMP). GMP is transformed into the final product, GTP, via sequential rounds of phosphorylation: the GMP kinase Gmk catalyzes the conversion of GMP into guanosine diphosphate (GDP), which is then converted to guanosine triphosphate (GTP) by the nucleoside diphosphate kinase Ndk. Cellular GTP concentrations have dual effect on bacterial physiology. Until a certain threshold concentration, increasing the GTP level increases the growth rate. However, further increase leads to cytotoxic effects, and at high concentrations GTP inhibits growth, negatively affecting bacterial survival upon amino acid starvation. The specific targets of (p)ppGpp-mediated control vary according to species and differ in E. coli7, 80_, B. subtilis3 and E. faecalis4. In E. coli, (p)ppGpp inhibits the IMP dehydrogenase GuaB; in B. subtilis and E. faecalis (p)ppGpp inhibits hypoxanthine phosphoribosyltransferase (HprT, the enzyme that catalyzes both conversion of HPX to IMP and of GUA to GMP); in B. subtilis (p)ppGpp inhibits the GMP kinase Gmk.
The importance of (p)ppGpp for nucleotide metabolism was first recognized in E. coli more than four decades ago78, and the GTP biosynthesis pathway was subsequently shown to be directly regulated by (p)ppGpp7. The IMP dehydrogenase GuaB was initially identified as the main target of (p)ppGpp in E. coli7, 79; however, recent work has shown that the molecular targets of (p)ppGpp-mediated regulation vary according to the species examined. In B. subtilis, GuaB is only weakly inhibited by (p)ppGpp3, whereas HprT and Gmk are strongly inhibited3. In E. faecalis (p)ppGpp targets HprT, but not Gmk4; similarly, Gmk is insensitive to (p)ppGpp in E. coli and Synechococcus elongatus PCC 794280. These differences in the primary targets of (p)ppGpp are expected to translate into differences in the regulation of GTP biosynthesis in different bacterial species. Regulation is also likely to be further complicated by the effects of (p)ppGpp on nucleotide uptake8, 81, the mode of inhibition (competitive or non-competitive), so further investigations are necessary to establish the precise effects of (p)ppGpp-mediated regulation in distinct species. An overproduction of GTP is observed in a mutant B. subtilis strain that lacks functional RSH genes and is therefore unable to synthesize (p)ppGpp – known as a ppGpp0 strain3. In media containing guanosine, which is incorporated via the salvage pathway, the GTP levels increase even further 3. This imbalance in GTP metabolism leads to decreased survival during starvation3, and B. subtilis82, E. faecalis4 and E. coli25 ppGpp0 strains are auxotrophic for specific amino acids., In E. coli it has been suggested that this auxotrophy is caused by insufficient transcription of amino acid biosynthesis genes61. In ppGpp0 B. subtilis, in addition to the unbalanced regulation on transcriptional level, effects mediate by transcriptional repressor CodY are contributing to the amino acid auxotrophy phenotype. This global metabolic regulator inhibits expression of the target genes (which include several amino synthetic operons) when associated with branched amino acids and GTP106. Reducing the cellular concentration of GTP in the ppGpp0 B. subtilis (through mutations in the promoters or open reading frames of guaB, guaA and gmk) leads to a substantial increase in survival and partially relieves the auxotrophy 83, most likely relieved by amino acid production upon partial de-repression of the CodY regulatory cascade. Surprisingly, a Pseudomonas putida ppGpp0 strain is not auxotrophic for amino acids, which highlights the relevance of species-specific differences in (p)ppGpp-associated physiology84.
(p)ppGpp: TA systems and persistence
Bacterial persisters arise stochastically in bacterial populations and are tolerant to multiple antibiotics, which has led to growing interest in understanding their physiology given the current global problems with antibiotic resistance 19, 85. However, the molecular underpinnings of this phenotype have been poorly understood and it has been suggested that several redundant mechanisms may contribute to the phenomenon (REF). As slow growth is associated with recalcitrance to killing by antibiotics and other environmental insults, by inference, slow growth was initially suggested to be crucial for the drug tolerance phenotype86. Considering that (p)ppGpp modulates bacterial growth rate2 and metabolism87, this raised the possibility that the stringent response has a key role in persistence. However, until recently, direct evidence for this conjecture has been lacking, primarily because persisters are rare in growing populations and are therefore difficult to study.
A decade ago, an elegant study showed that an exponentially growing mutant of E. coli, in which the hipA gene was mutated, generated persisters at a high rate. These cells grew slowly and survived treatment with ampicillin. Importantly, they also formed stochastically and independently of the drug88. A first hint of the underlying mechanism responsible for their emergence came from the realization that hipA encodes the toxin component of a toxin – antitoxin (TA) module (hipAB) and that the persistence phenotype of the hipA mutant was dependent on (p)ppGpp89. This rather enigmatic connection between (p)ppGpp, a TA module and persistence became clearer with the discovery that hipA encodes a kinase that phosphorylates and inhibits glutamyl-tRNA synthetase90, 91. Inhibition of the synthetase leads to the accumulation of uncharged glutamyl-tRNA that, when loaded at the ribosomal A-site, leads to activation of RelA and increased (p)ppGpp levels90, 92. These findings provide a rationale for the observed dependency of hipA-mediated persistence on (p)ppGpp89 and raised the possibility that HipA induces persistence by provoking an increase in (p)ppGpp levels. However, these observations did not exclude the opposite possibility, in which (p)ppGpp might function as the main regulator of persistence. This model predicts that (p)ppGpp should vary stochastically in single cells and that HipA activity should be stimulated by (p)ppGpp in a positive feedback loop.
Indications of a more general involvement of TA genes in persistence came from the observation that the transcription of TA operons encoding mRNA endonucleases (mRNases) was increased in persisters compared to wild-type cells in the same population 93, 94. TA-encoded mRNases are stable inhibitors of bacterial cell growth, whereas their corresponding antitoxins are metabolically unstable because they are readily degraded by the Lon protease95, 96. Thus, the activity of Lon determines the levels of antitoxins and thereby the activities of the toxins. Ectopic expression of TA-encoded toxins was shown to cause a dramatic increase in persistence, consistent with the idea that the toxins are inducers of persistence90, 93, 94, 97. Further strong support for this proposal came from the observation that progressive deletion of ten type II TA operons in E. coli led to a progressive reduction in persistence97. Together, these data suggested that TA modules encode cell growth inhibitors that are pivotal to bacterial persistence.
The link between (p)ppGpp, the TA systems and persistence was illuminated by a study in which (p)ppGpp was found to be the master regulator of persistence and that the toxins of TA systems encode persistence “effectors” 98. This study showed that that both (p)ppGpp levels and transcription of TA operon vary stochastically in single cells98. By using an rpoS::mCherry fluorescent protein fusion as a reliable proxy of the (p)ppGpp level in single cells, it was found that persisters exhibited high levels of (p)ppGpp and high transcript levels of the hipA toxin. The authors propose that the hierarchical signalling pathway involves an increase in (p)ppGpp levels, which inhibits exopolyphosphatase activity, which leads to the accumulation of polyphosphate that binds to and activates the Lon protease. In turn, Lon degrades the antitoxin causing activation of the mRNase toxin, ultimately inhibiting cell growth and inducing persistence. It will be interesting to learn if inhibition of cell growth per se is sufficient to induce persistence or if the TA-encoded toxins are required to induce the transient phenotypic drug tolerance.
The alarmone (p)ppGpp is present in the vast majority of bacterial species, including major pathogens10, thus raising an important question: does (p)ppGpp also control persistence during infection and, if so, can this knowledge be exploited to devise improved treatment strategies? It is well known that, in almost all cases, (p)ppGpp is required for bacterial pathogens to be virulent, which is consistent with the hostile and stress-inducing environments that bacteria encounter during infection16. It is not yet known if (p)ppGpp and the TAs contribute to persistence during infection; however, two recent studies of Salmonella enterica serovar Typhimurium persisters support this view. During infection, Salmonella spp. reside in a niche known as a Salmonella-containing vacuole in macrophages, which are that are relatively acidic and nutrient poor. As a result, the stringent response is induced, which stimulates the formation of persisters99. Moreover, S. Typhimurium mutants that have an impaired stringent response were found to produce fewer persisters in macrophages. Interestingly, persister formation in macrophages depended on both Lon and TA genes, which together with the findings for E. coli, provide strong support for the notion that similar mechansims are used by E. coli and Salmonella spp. to produce persisters99. Another study found that moderately to slow-growing S. Typhimurium survive antibiotic treatment better than fast-growing cells in host tissues100. The slow-growing cells had a higher level of (p)ppGpp synthesizing enzymes, supporting the conjecture that (p)ppGpp controls persistence following antibiotic treatment in an infection context. Similarly, persistence of Pseudomonas aeruginosa, a γ-proteobacterium distantly related to E. coli and Salmonella spp., depends on (p)ppGpp during amino acid starvation, in biofilms and under oxidative stress101, 102. Future studies should investigate whether the involvement of (p)ppGpp in persistence is a widespread phenomenon and whether it could constitute the master regulator of persistence in more diverse bacterial pathogens19, 98.
Conclusions
In the past four decades since the discovery of (p)ppGpp by Cashel and Gallant20, our understanding of its physiological roles has gradually evolved. Initially, the term ‘stringent response’ was used to describe the RelA-mediated stress response to amino acid starvation, with a focus on the inhibition of rRNA transcription103. The concept of the stringent response has since been reformulated to refer to an integrated response to several different types of stress, such as carbon and iron limitation, which is sensed by the other E. coli RSH enzyme, SpoT5. In recent years, there has been a greater appreciation of (p)ppGpp as a crucial component of normal physiology in unstressed conditions, with regulation of growth rate2, amino acid biosynthesis4 and GTP metabolism3 being amongst the key roles of this versatile molecule.
Species-specific downstream effects of (p)ppGpp-mediated regulation are well-recognized104. Prominent examples include its involvement in virulence, such as the induction of cytotoxic phenol-soluble modulins (PSMs) following phagocytosis of Staphylococcus aureus17, as well as the regulation of antibiotic production in Streptomycetes105, and A-factor production and development of fruiting bodies in Myxococcus xanthus52. Importantly, species-specific variations in the core molecular machinery of the stringent response are apparent: the cellular repertoire of RSH proteins differs greatly among different bacterial lineages10; the regulation of transcription can be achieved via either direct binding of (p)ppGpp to RNAP65, 66 or changes in the nucleotide pools62, and lastly, regulation of metabolic targets, such as GTP metabolism, varies widely amongst bacterial species3, 80. As such, the regulatory circuits of (p)ppGpp-mediated sensory systems has been suggested to be intimately linked to bacterial lifestyle54. A direct analysis of this conjecture requires a holistic characterization of all the elements of the stringent response regulatory network, and great caution should be exercised when using biochemical data obtained for one of the more popular model organisms, such as E. coli, to rationalize the in vivo results obtained for any other species. A holistic understanding of the role of the stringent response in normal bacterial physiology is crucial for determining whether it might constitute a viable target for novel antibacterial agents (Box 3). Such inhibitors are likely to also be invaluable molecular tools for dissecting the mechanisms of the stringent response with biochemical, structural and microbiological approaches.
Box 3. Targeting the stringent response as an antibacterial strategy?
As opposed to antibiotics, which generally target essential cellular processes, anti-virulence compounds are expected to exert less selective pressure for resistance due to the non-essentiality of the target121. Inhibition of the stringent response is a promising approach for disarming pathogens without killing them, since bacterial strains incapable of (p)ppGpp production are viable, though metabolically compromised82. There have been several important advances in the development of molecular tools for inhibition of the stringent response pathway, demonstrating its potential as a drug target. First, nucleotide-based inhibitors – the (p)pGpp-based nucleotide Relacin and its derivatives – were developed to directly target the RSH enzymes122, 123 Second, the anti-biofilm peptide 1018 promotes hydrolysis of (p)ppGpp formed in the cell124. However, our growing understanding of the importance of (p)ppGpp during normal physiology, such as regulation of amino acid biosynthesis82 and growth rate2, suggests that inhibition of (p)ppGpp production would have a detrimental effect on bacterial survival, not just virulence. Thus, in clinical settings potential stringent response inhibitors may not strictly function as anti-virulence compounds, but rather as antibiotics. Regardless the exact physiological consequences, selective inhibitors of the stringent response hold great promise as novel therapeutics.
Online summary.
The stringent response is a regulatory mechanism that is controlled by members of the RelA/SpoT Homologue (RSH) protein family in response to stress in bacteria. It is mediated by two related alarmone nucleotides, ppGpp and pppGpp, which are commonly referred to as (p)ppGpp.
The RSH enzymes can be divided into two categories: ‘long’ multi-domain RSHs (RelA, Rel and SpoT) and ‘short’ single-domain enzymes (Small Alarmone Synthases, SAS, and Small Alarmone Hydrolases, SAHs). The enzymatic activity of ‘long’ enzymes is regulated by interactions with molecular effectors, such as ‘starved’ ribosomes in the case of RelA, and the activity of ‘short’ RSHs is regulated on the transcriptional level.
The regulatory role of (p)ppGpp in general metabolism is exerted by direct and indirect mechanisms. The direct mechanisms rely on the alarmone binding to and regulating its molecular target, whereas indirect mechanisms rely on changes in the concentrations of GTP and ATP nucleotides elicited by (p)ppGpp production.
The primary target of (p)ppGpp-mediated regulation is transcription. In Escherichia coli this regulation relies on direct binding of the alarmone to the β’ and ω subunits of RNAP, whereas in Bacillus subtilis, indirect regulation relies on changes in the concentration of the initiator nucleotide.
In addition to transcription, (p)ppGpp production regulates multiple additional molecular targets, such as protein biosynthesis, replication, the acid stress response, polyphosphate metabolism, biosynthesis and uptake of nucleotides, and, via direct interaction with RelA, the stringent response itself.
The effects of (p)ppGpp span a continuum from acute survival responses elicited by stress such as nutrient deprivation or heat shock mediated by high (p)ppGpp levels, to the ‘house-keeping’ role of basal (p)ppGpp levels in normal bacterial metabolism, such as the production of amino acids and nucleotides.
The stringent response has a key role in bacterial virulence and persistence (the formation of antibiotic-tolerant cells). This has prompted the recent development of specific inhibitors of this process, which serve as a starting point for the future development of novel anti-virulence compounds.
Subject categories.
Biological sciences / Microbiology / Bacteria / Bacterial physiology
[URI /631/326/41/1969]
Biological sciences / Microbiology / Bacteria / Cellular microbiology
[URI /631/326/41/88]
Biological sciences / Microbiology / Bacteria / Bacterial structural biology
[URI /631/326/41/2536]
Biological sciences / Microbiology / Bacteria / Bacterial transcription
[URI /631/326/41/2532]
Acknowledgments
The authors are grateful to Victoria Shingler for her insightful comments on the manuscript and Mikel Valle for providing RelA cryoEM images. This work was supported by the funds from European Regional Development Fund through the Centre of Excellence in Chemical Biology (VH and TT), Estonian Science Foundation grants (ETF9012 and PUT37 to VH, ETF9020 to GCA); Umeå University, Swedish Research council, Ragnar Söderberg and Kempe foundations (VH); National Institutes of Health Grant GM087350 (KSM); European Research Council Advanced Investigator Grant (294517; ‘‘PERSIST’’) (KG) and a Novo Nordisk Foundation Laureate Research Grant (KG).
Glossary terms
- Ribosomal translocation
A step in the stepwise addition of amino acids to the growing protein chain by the ribosome, elongation cycle. During translocation mRNA–tRNA moiety advances for one coden on the ribosome, thus allowing the next codon to move into the decoding center and to accept next incoming aminoacylated tRNA.
- Phenol-soluble modulins (PSMs)
a family of amphipathic alpha-helical peptides with surfactant-like properties, that have multiple roles in staphylococcal virulence, contributing in immune evasion and biofilm development. PSMs induce lysis of neutrophils as well as hemolysis, production of proinflammatory cytokines, as well as contribute to biofilm formation.
- A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone)
a signaling molecule regulating that triggers secondary metabolism and morphogenesis in Streptomyces bacteria.
- Fruiting body
an multicellular aggregate state of Myxobacteria developed upon by nutrient deprivation.
- Lon protease
ATP-dependent peptidase that degrades aberrant and short-lived polypeptides.
- Salvage pathway
a pathway in which nucleotides are synthesized from intermediates (nucleobases and ribonucleosides) that are the product of degradation or are imported from the extracellular milieu, rather than constructing them de novo from phosphoribosyl pyrophosphate as a starting compound.
- σW regulon
an operon that is regulated by sigma W transcription factor and is induced in response to various stresses.
- Acyl Carrier Protein (ACP)
an essential component of the fatty acid synthesis pathway that stabilizes and transports the growing lipid chain. Specialized ACPs are also involved in other processes that require acyl transfer, such as polyketide antibiotic synthesis.
- GTPase
enzymes that bind and hydrolyse guanosine-triphosphate (GTP) to produce guanosine diphosphate (GDP).
- Sarcin-Ricin Loop
an essential structural element of the 23S rRNA that interacts with various protein factors during translation.
- EF-Tu
A GTPase that delivers aminoacyl-tRNA to the ribosomal A-site during protein synthesis.
- Toxin – Antitoxin (TA) system
bacterial operons that encode two proteins, a toxin and an antitoxin that combines with and neutralizes the cognate toxin. TA system inhibit various vital cellular functions, most often targeting protein biosynthesis.
- A/T state
a structural conformation in which the aminoacyl-tRNA becomes distorted as it occupies the ribosomal A-site while interacting with EF-Tu.
- Deacylated tRNA
tRNA that is uncharged as it lacks an aminoacyl group at the 3′ CCA end.
- A-site
The ribosome “acceptor” site that accommodates tRNA and various ribosome-interacting protein factors during the translation cycle.
- Nudix hydrolase
phosphohydrolase enzymes that are unrelated to RSH hydrolases but can nevertheless degrade (p)ppGpp.
- ThrRS, GTPase and SpoT domain (TGS domain)
a protein domain with a suggested ligand-binding function.
- Aspartokinase, Chorismate mutase and TyrA domain(ACT domain)
a protein domain involved in ligand binding, often present in metabolic enzymes.
- Monophyletic
in a phylogenetic tree, a monophyletic group of sequences share a single common ancestor.
- Paraphyletic
a paraphyletic clade in a phylogenetic tree contains sequences that share a single common ancestor, but excludes some descendent sequences. ppGpp0 strain: a bacterial strain that is unable to produce (p)ppGpp as it lacks functional ‘large’ and ‘small’ RSHs.
Biography
Vasili Hauryliuk received his MSc degree from Moscow State University, Russia, in 1997, followed by a PhD in Molecular Biology from Uppsala University, Sweden in 2008. He established his research group at University of Tartu, Estonia in 2008, and in 2013 he was recruited as an associate professor at the Laboratory for Molecular Infection Medicine Sweden, Umeå University. In 2014 he became a Ragnar Söderberg Fellow in medicine. The Hauryliuk lab studies the molecular mechanisms of translation and bacterial stress responses.
Gemma C. Atkinson was awarded a BSc in Genetics from Newcastle University, UK in 2003, an MRes in Bioinformatics from the University of Leeds, UK in 2004, and a PhD in Biology from The University of York, UK in 2009. She holds research associate positions at Umeå University, Sweden and the University of Tartu, Estonia. Her research focuses on molecular evolution of function of ribosome-associated proteins and antibiotic resistance elements.
Katsuhiko Murakami received his PhD from the Graduate University of Advanced Studies in Japan in 1997. He engaged in postdoctoral study at the Rockefeller University, and then joined the Pennsylvania State University in 2003. His research interest is centered on understanding the mechanism of prokaryotic RNA polymerase transcription.
Tanel Tenson received his BSci (1992), MSci (1994) and PhD (1997) from University of Tartu, Estonia. The PhD and postdoctoral research was done partly at University of Illinois at Chicago, USA and Uppsala University, Sweden. He is currently professor of the technology of antimicrobial compounds at the University of Tartu. His research interests are related to the mechanisms of antibiotic action and antibiotic resistance. Currently his research focus is on the inhibitors of protein synthesis and mechanisms of persister cell formation.
Kenn Gerdes received his PhD in molecular genetics from the Technical University of Denmark in 1986. In 2002 he became a full professor at the University of Southern Denmark. In 2006 he moved his research group to Newcastle University, UK, where he joined the Centre for Bacterial Cell Biology. In 2014 he moved his group to the University of Copenhagen where he established the Danish National Research and Novo Nordisk Foundations Centre of Bacterial Stress Response and Persistence. The Gerdes lab studies bacterial stress responses, (p)ppGpp and toxin – antitoxins.
References
- 1.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–6. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol. 2011;13:563–75. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriel A, et al. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell. 2012;48:231–41. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaca AO, et al. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio. 2013;4:e00646–13. doi: 10.1128/mBio.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 6.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 7.Gallant J, Irr J, Cashel M. The mechanism of amino acid control of guanylate and adenylate biosynthesis. J Biol Chem. 1971;246:5812–6. [PubMed] [Google Scholar]
- 8.Hochstadt-Ozer J, Cashel M. The regulation of purine utilization in bacteria. V. Inhibition of purine phosphoribosyltransferase activities and purine uptake in isolated membrane vesicles by guanosine tetraphosphate. The Journal of biological chemistry. 1972;247:7067–72. [PubMed] [Google Scholar]
- 9.Traxler MF, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the 'feast to famine' gradient in Escherichia coli. Mol Microbiol. 2011;79:830–45. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito D, et al. Enzymatic and molecular characterization of Arabidopsis ppGpp pyrophosphohydrolase, AtNUDX26. Biosci Biotechnol Biochem. 2012;76:2236–41. doi: 10.1271/bbb.120523. [DOI] [PubMed] [Google Scholar]
- 12.Ooga T, et al. Degradation of ppGpp by nudix pyrophosphatase modulates the transition of growth phase in the bacterium Thermus thermophilus. J Biol Chem. 2009;284:15549–56. doi: 10.1074/jbc.M900582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraszewska E. The plant Nudix hydrolase family. Acta Biochim Pol. 2008;55:663–71. [PubMed] [Google Scholar]
- 14.Keasling JD, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc Natl Acad Sci U S A. 1993;90:7029–33. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamel E, Cashel M. Role of guanine nucleotides in protein synthesis. Elongation factor G and guanosine 5′-triphosphate,3′-diphosphate. Proc Natl Acad Sci U S A. 1973;70:3250–4. doi: 10.1073/pnas.70.11.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–99. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiger T, et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 2012;8:e1003016. doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother. 2012;67:2069–89. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 19.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–48. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 20.Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–41. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 21.Laffler T, Gallant J. spoT, a new genetic locus involved in the stringent response in E. coli. Cell. 1974;1:27–30. [Google Scholar]
- 22.Mechold U, Murphy H, Brown L, Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184:2878–88. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seyfzadeh M, Keener J, Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:11004–8. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinella D, Albrecht C, Cashel M, D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–70. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–90. [PubMed] [Google Scholar]
- 26.Haseltine WA, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238:381–4. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 27.Gallant J, Palmer L, Pao CC. Anomalous synthesis of ppGpp in growing cells. Cell. 1977;11:181–5. doi: 10.1016/0092-8674(77)90329-4. [DOI] [PubMed] [Google Scholar]
- 28.Shyp V, et al. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 2012;13:835–9. doi: 10.1038/embor.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A. 1973;70:1564–8. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprinzl M, Richter D. Free 3′-OH group of the terminal adenosine of the tRNA molecule is essential for the synthesis in vitro of guanosine tetraphosphate and pentaphosphate in a ribosomal system from Escherichia coli. Eur J Biochem. 1976;71:171–6. doi: 10.1111/j.1432-1033.1976.tb11103.x. [DOI] [PubMed] [Google Scholar]
- 31.Payoe R, Fahlman RP. Dependence of RelA-mediated (p)ppGpp formation on tRNA identity. Biochemistry. 2011;50:3075–83. doi: 10.1021/bi1015309. [DOI] [PubMed] [Google Scholar]
- 32.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–88. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 33.Richter D. Stringent factor from Escherichia coli directs ribosomal binding and release of uncharged tRNA. Proc Natl Acad Sci U S A. 1976;73:707–11. doi: 10.1073/pnas.73.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.English BP, et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci U S A. 2011;108:E365–73. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agirrezabala X, et al. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14:811–6. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker J, Watson RJ, Friesen JD. A relaxed mutant with an altered ribosomal protein L11. Mol Gen Genet. 1976;144:111–4. doi: 10.1007/BF00277313. [DOI] [PubMed] [Google Scholar]
- 37.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyltRNA. Science. 2009;326:688–94. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An G, Justesen J, Watson RJ, Friesen JD. Cloning the spoT gene of Escherichia coli: identification of the spoT gene product. J Bacteriol. 1979;137:1100–10. doi: 10.1128/jb.137.3.1100-1110.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raskin DM, Judson N, Mekalanos JJ. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:4636–41. doi: 10.1073/pnas.0611650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–63. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 41.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:E2561–8. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sy J. In vitro degradation of guanosine 5′-diphosphate, 3′-diphosphate. Proc Natl Acad Sci U S A. 1977;74:5529–33. doi: 10.1073/pnas.74.12.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hara A, Sy J. Guanosine 5′-triphosphate, 3′-diphosphate 5′-phosphohydrolase. Purification and substrate specificity. J Biol Chem. 1983;258:1678–83. [PubMed] [Google Scholar]
- 44.Dahl JL, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100:10026–31. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Primm TP, et al. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000;182:4889–98. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avarbock D, Avarbock A, Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000;39:11640–8. doi: 10.1021/bi001256k. [DOI] [PubMed] [Google Scholar]
- 47.Scoarughi GL, Cimmino C, Donini P. Helicobacter pylori: a eubacterium lacking the stringent response. J Bacteriol. 1999;181:552–5. doi: 10.1128/jb.181.2.552-555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wells DH, Gaynor EC. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J Bacteriol. 2006;188:3726–9. doi: 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park SA, Ko A, Lee NG. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol. 2011;11:96. doi: 10.1186/1471-2180-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol. 2011;80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaca AO, Abranches J, Kajfasz JK, Lemos JA. Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiology. 2012;158:1994–2004. doi: 10.1099/mic.0.060236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris BZ, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in myxococcus xanthus. Genes Dev. 1998;12:1022–35. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauch E, Takano E, Baylis HA, Bibb MJ. The stringent response in Streptomyces coelicolor A3(2) Mol Microbiol. 1991;5:289–98. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 54.Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013;21:174–80. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Battesti A, Bouveret E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J Bacteriol. 2009;191:616–24. doi: 10.1128/JB.01195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wout P, et al. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J Bacteriol. 2004;186:5249–57. doi: 10.1128/JB.186.16.5249-5257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Persky NS, Ferullo DJ, Cooper DL, Moore HR, Lovett ST. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2009;73:253–66. doi: 10.1111/j.1365-2958.2009.06767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 59.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A. 2005;102:7823–8. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul BJ, et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–22. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 61.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. Journal of molecular biology. 2001;305:673–88. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 62.Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–83. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Artsimovitch I, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 64.Vrentas CE, et al. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. Journal of molecular biology. 2008;377:551–64. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41:6175–89. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50:430–6. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Igarashi K, Fujita N, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: omega factor is responsible for the ppGpp sensitivity. Nucleic acids research. 1989;17:8755–65. doi: 10.1093/nar/17.21.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–87. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–9. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tagami S, Sekine SI, Yokoyama S. A novel conformation of RNA polymerase sheds light on the mechanism of transcription. Transcription. 2011;2:162–167. doi: 10.4161/trns.2.4.16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tare P, Mallick B, Nagaraja V. Co-evolution of specific amino acid in sigma 1.2 region and nucleotide base in the discriminator to act as sensors of small molecule effectors of transcription initiation in mycobacteria. Mol Microbiol. 2013;90:569–83. doi: 10.1111/mmi.12384. [DOI] [PubMed] [Google Scholar]
- 72.Kasai K, et al. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J Bacteriol. 2006;188:7111–22. doi: 10.1128/JB.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lennon CW, et al. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–46. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tagami S, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–82. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 75.Jensen K, Dandanell G, Hove-Jensen B, Willemoës M. In: EcoSal Plus. Stewart V, editor. ASMscience; 2008. 2008. [DOI] [PubMed] [Google Scholar]
- 76.Papakostas K, Botou M, Frillingos S. Functional identification of the hypoxanthine/guanine transporters YjcD and YgfQ and the adenine transporters PurP and YicO of Escherichia coli K-12. J Biol Chem. 2013;288:36827–40. doi: 10.1074/jbc.M113.523340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meng LM, Nygaard P. Identification of hypoxanthine and guanine as the co-repressors for the purine regulon genes of Escherichia coli. Mol Microbiol. 1990;4:2187–92. doi: 10.1111/j.1365-2958.1990.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 78.Irr J, Gallant J. The control of ribonucleic acid synthesis in Escherichia coli. II. Stringent control of energy metabolism. J Biol Chem. 1969;244:2233–9. [PubMed] [Google Scholar]
- 79.Pao CC, Dyess BT. Effect of unusual guanosine nucleotides on the activities of some Escherichia coli cellular enzymes. Biochim Biophys Acta. 1981;677:358–62. doi: 10.1016/0304-4165(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 80.Nomura Y, et al. Diversity in Guanosine 3′,5′-Bisdiphosphate (ppGpp) Sensitivity among Guanylate Kinases of Bacteria and Plants. J Biol Chem. 2014;289:15631–15641. doi: 10.1074/jbc.M113.534768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beaman TC, et al. Specificity and control of uptake of purines and other compounds in Bacillus subtilis. J Bacteriol. 1983;156:1107–17. doi: 10.1128/jb.156.3.1107-1117.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kriel A, et al. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J Bacteriol. 2014;196:189–201. doi: 10.1128/JB.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bittner AN, Kriel A, Wang JD. Lowering GTP level increases survival of amino acid starvation but slows growth rate for Bacillus subtilis cells lacking (p)ppGpp. J Bacteriol. 2014;196:2067–76. doi: 10.1128/JB.01471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bernardo LM, Johansson LU, Skarfstad E, Shingler V. sigma54-promoter discrimination and regulation by ppGpp and DksA. J Biol Chem. 2009;284:828–38. doi: 10.1074/jbc.M807707200. [DOI] [PubMed] [Google Scholar]
- 85.Lewis K. Persister cells. Annu.Rev.Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 86.Bigger JW. Treatment of staphyloccal infections with penicillin by intermittent sterilisation. Lancet. 1944;ii:497–500. [Google Scholar]
- 87.Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell. 2013;50:475–87. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 88.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 89.Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 90.Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol Cell. 2013;52:248–54. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 91.Kaspy I, et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyltRNA-synthetase. Nat Commun. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 92.Bokinsky G, et al. HipA-triggered growth arrest and beta-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J Bacteriol. 2013;195:3173–82. doi: 10.1128/JB.02210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. Journal of Bacteriology. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shah D, et al. Persisters: a distinct physiological state of E-coli. Bmc Microbiology. 2006;6 doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–23. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 96.Hansen S, et al. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS One. 2012;7:e39185. doi: 10.1371/journal.pone.0039185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc.Natl.Acad.Sci.U.S.A. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Maisonneuve E, Castro-Camargo M, Gerdes K. p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–50. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 99.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–8. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Claudi B, et al. Phenotypic variation of salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 2014;158:722–33. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 101.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol. 2013;195:2011–20. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–6. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cashel M. Regulation of bacterial ppGpp and pppGpp. Annu Rev Microbiol. 1975;29:301–18. doi: 10.1146/annurev.mi.29.100175.001505. [DOI] [PubMed] [Google Scholar]
- 104.Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–12. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 105.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–15. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 106.Geiger T, Wolz C. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int J Med Microbiol. 2014;304:150–5. doi: 10.1016/j.ijmm.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 107.Milon P, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci U S A. 2006;103:13962–7. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitkevich VA, et al. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J Mol Biol. 2010;402:838–46. doi: 10.1016/j.jmb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 109.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–75. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–3. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 111.Kanjee U, et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–44. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kanjee U, Ogata K, Houry WA. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol. 2012;85:1029–43. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- 113.Murdeshwar MS, Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J Bacteriol. 2012;194:4003–14. doi: 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nanamiya H, et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 115.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65:1568–81. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 116.Cao M, et al. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol. 2002;316:443–57. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- 117.Sun D, et al. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat Struct Mol Biol. 2010;17:1188–94. doi: 10.1038/nsmb.1906. [DOI] [PubMed] [Google Scholar]
- 118.Thammana P, Buerk RR, Gordon J. Absence of ppGpp production in synchronised Balb/C mouse 3T3 cells on isoleucine starvation. FEBS Lett. 1976;68:187–90. doi: 10.1016/0014-5793(76)80433-4. [DOI] [PubMed] [Google Scholar]
- 119.Tozawa Y, Nomura Y. Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol (Stuttg) 2011;13:699–709. doi: 10.1111/j.1438-8677.2011.00484.x. [DOI] [PubMed] [Google Scholar]
- 120.O'Day DH, Keszei A. Signalling and sex in the social amoebozoans. Biol Rev Camb Philos Soc. 2012;87:313–29. doi: 10.1111/j.1469-185X.2011.00200.x. [DOI] [PubMed] [Google Scholar]
- 121.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–28. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 122.Wexselblatt E, et al. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 2012;8:e1002925. doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wexselblatt E, Kaspy I, Glaser G, Katzhendler J, Yavin E. Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur J Med Chem. 2013;70:497–504. doi: 10.1016/j.ejmech.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 124.de la Fuente-Nunez C, Reffuveille F, Haney EF, Straus SK, Hancock RE. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014;10:e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 126.Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–40. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 127.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]