Summary
The strychnine-sensitive glycine receptor (GlyR) mediates inhibitory synaptic transmission in the spinal cord and brainstem and is linked to neurological disorders including autism and hyperekplexia. Understanding of molecular mechanisms and pharmacology of GlyRs has been hindered by a dearth of high-resolution structures. Here we report electron cryo-microscopy structures of the α1 GlyR with strychnine, glycine, or glycine/ivermectin. Strychnine arrests the receptor in an antagonist-bound, closed ion channel state, glycine stabilizes the receptor in an agonist-bound open channel state, and the glycine/ivermectin complex adopts a potentially desensitized or partially open state. Relative to the glycine-bound state, strychnine expands the agonist-binding pocket via outward movement of the C loop, promotes rearrangement of the extracellular and transmembrane domain ‘wrist’ interface, and leads to rotation of the transmembrane domain toward the pore axis, occluding the ion conduction pathway. These structures illuminate GlyR mechanism and define a rubric to interpret structures of Cys-loop receptors.
Introduction
Neurotransmitter-gated ion channels mediate fast excitatory and inhibitory signal transduction in the central nervous system (CNS) by controlling ion flux through neuronal cell membranes in response to the binding of a wide-range of neurotransmitters1. Glycine, a major inhibitory transmitter in the CNS2, exerts its inhibitory effect on the glycine receptor (GlyR), a postsynaptic ligand-gated channel receptor, opening a chloride-permeable pore that, in turn, leads to hyperpolarization of the membrane potential and inhibition of neuronal firing3–6. GlyRs mediate neurotransmission throughout the spinal cord and brain stem and control a wide range of motor and sensory functions including vision and audition6–8. Heritable mutations of human GlyR are the major cause of a rare neurological disorder, hyperekplexia or startle disease9,10.
Strychnine, the notorious and complex alkaloid11, is a potent competitive GlyR antagonist that locks the receptor in a closed state, precluding chloride permeation. Employed to facilitate receptor isolation12, and exploited to disentangle glycine-induced synaptic currents, strychnine acts at the intersubunit, canonical neurotransmitter site13. Glycine binds at the same site yet promotes channel opening, allowing permeation of chloride ions through an anion conductive pathway with an estimated diameter of 5.2–6.0 Å14,15. Small molecules and ions, including the macrocyclic lactones ivermectin and related avermectins, modulate the gating activity of GlyRs by potentiating glycine-induced currents by an allosteric mechanism16. Despite the profound roles of GlyRs in the CNS and their prominence in neuroscience, mechanisms to describe the action of strychnine, glycine and ivermectin in terms of atomic structure have proven elusive.
GlyRs belong to the superfamily of Cys-loop receptors that includes the cation-selective nicotinic acetylcholine receptor (nAChR) and the serotonin type-3 receptor (5-HT3R), as well as the anion-selective GABA type A receptor (GABAAR)17,18. Since the landmark studies of Katz and colleagues1, Cys-loop receptors have been studied because of their prominent roles in the nervous system and because they are targets of scores of natural products and synthetic agents, from curare to valium. More recently, high-resolution structures of the prokaryotic pentameric ligand-gated ion channels, GLIC and ELIC19–22, and Lily23, as well as the eukaryotic nAChR24,25, GluCl26,27, 5-HT3R28 and GABAA receptors29 have been elucidated. Molecular understanding of eukaryotic Cys-loop receptors, however, is largely based on comparisons between different receptors30,31 due to the challenge of capturing a single receptor in multiple functional states. To reveal the molecular interplay between competitive antagonists, agonists or allosteric modulators and ion channel gating, we determined GlyR structures in complex with strychnine (str), glycine (gly) or glycine and ivermectin (ivm) using single particle electron cryo-microscopy (cryo-EM).
Structure determination and refinement
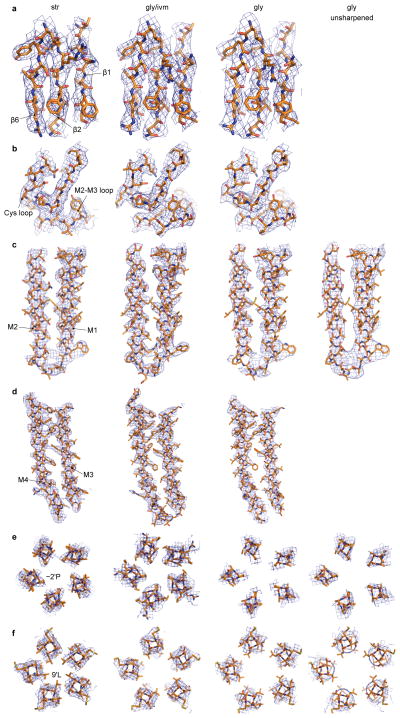
The 3D reconstructions of the str-, gly-, and gly/ivm-bound structures have estimated resolutions of 3.9, 3.9 and 3.8 Å, respectively (Extended Data Fig. 1–3), and are of sufficient quality to allow modeling of almost the entire receptor (Fig. 1; Extended Data Fig. 4). The density for strychnine can be recognized in the str-bound form, and is located at the intersubunit neurotransmitter binding pocket. By contrast, density for glycine is not discernable in either the gly- or the gly/ivm bound forms. Characterized by the distinctive triangular shape of its macrocyclic lactone, the density of ivermectin is unambiguous, and is found wedged between TMD subunit interfaces. The pore-lining M2 helix is best resolved in the gly/ivm- and str-bound structures (Extended Data Fig. 4c), although three (Ala326-Thr328) and two (Gly327-Thr328) residues are not visible in the M3-M4 loop of the str- and gly-bound reconstructions, respectively (Extended Data Fig. 4d). The final structures have excellent stereochemistry (Extended Data Table 1) and correlate well with respective density maps (Extended Data Fig. 1–3).
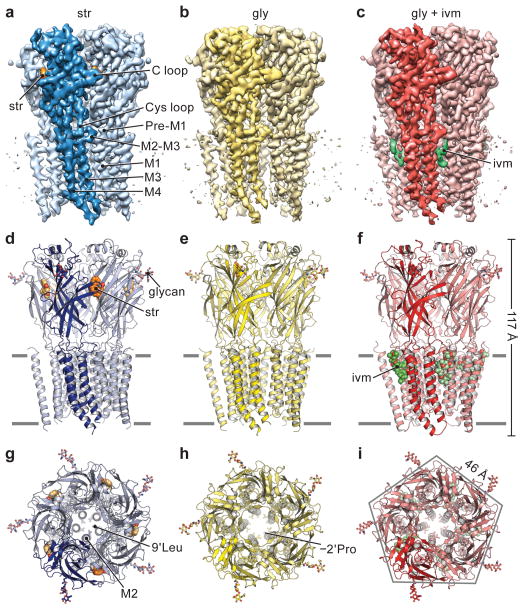
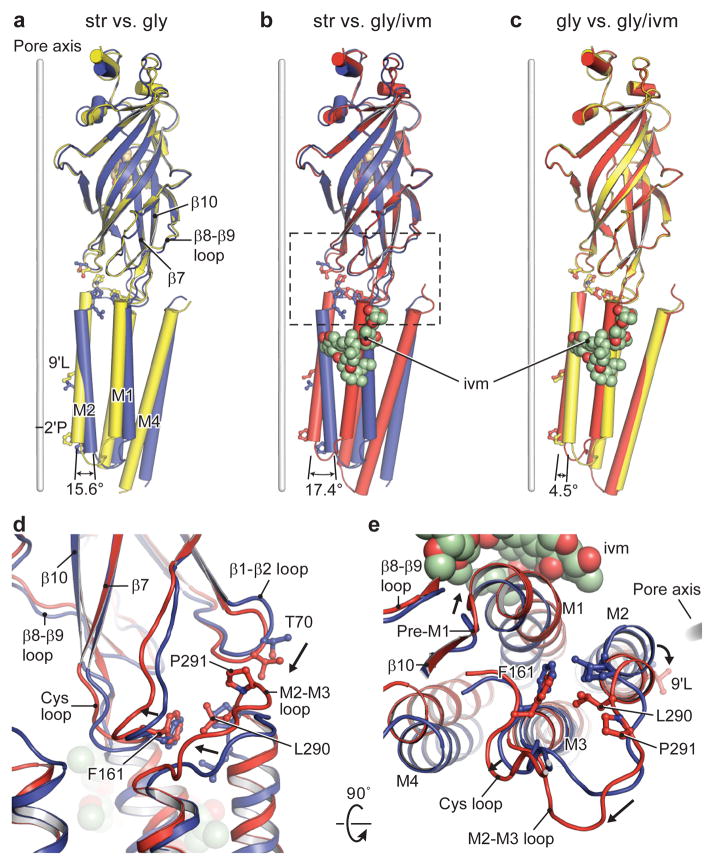
Figure 1. Receptor architecture.
a–c, The 3D reconstruction maps, viewed parallel to the membrane (str-bound in blue, gly-bound in yellow and gly/ivm-bound in red). One subunit is highlighted. The densities for strychnine and ivermectin are orange and green, respectively. d–f, Cartoon representations of the corresponding models of reconstructions shown in a–c, viewed in parallel to the membrane plane. The Asn-linked carbohydrate and associated Asn54 residue are in stick representation. g–i, Views of the structures from the extracellular side of the membrane. Residues −2′Pro (Pro266) and 9′Leu (Leu277) reside on the pore-lining M2 helix.
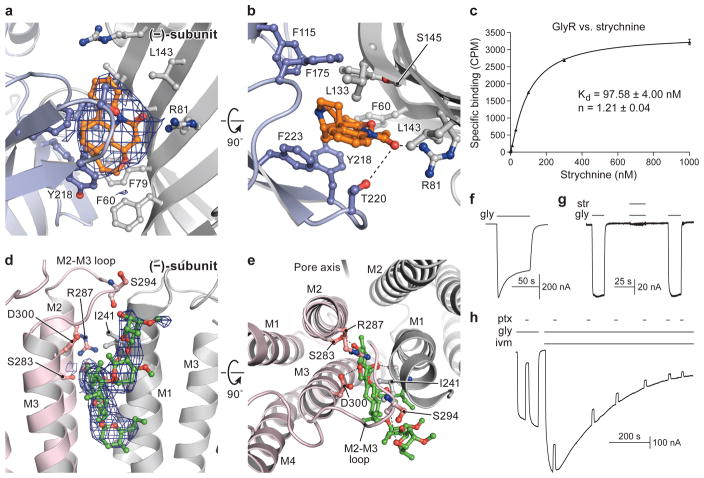
Figure 3. Strychnine and ivermectin bind at subunit interfaces.
Strychnine binding site (+; light blue) (−; gray); view is parallel to the membrane (a) or from the extracellular side (b). Density for strychnine (blue mesh) is contoured at 7 σ. c, Saturation binding of 3H strychnine to the GlyREM construct. Results are the mean of three biological replicates and the error bars represent s.e.m. d–e, Ivermectin binds at TMD intersubunit interface. Views are parallel to the membrane (d) or the extracellular side (e). f, Activation of GlyR currents by 10 mM glycine determined by TEVC. g, Strychnine (1 μM) inhibits glycine-induced (0.3 mM) currents. h, Effect of picrotoxinin (ptx, 1 mM) on glycine (0.3 mM) induced current. Ptx inhibits ~80% of glycine-induced current. Ivermectin (5 μM) potentiates the GlyR current and causes a slow desensitization. The shown recordings are representative of three independent experiments.
Overall architecture
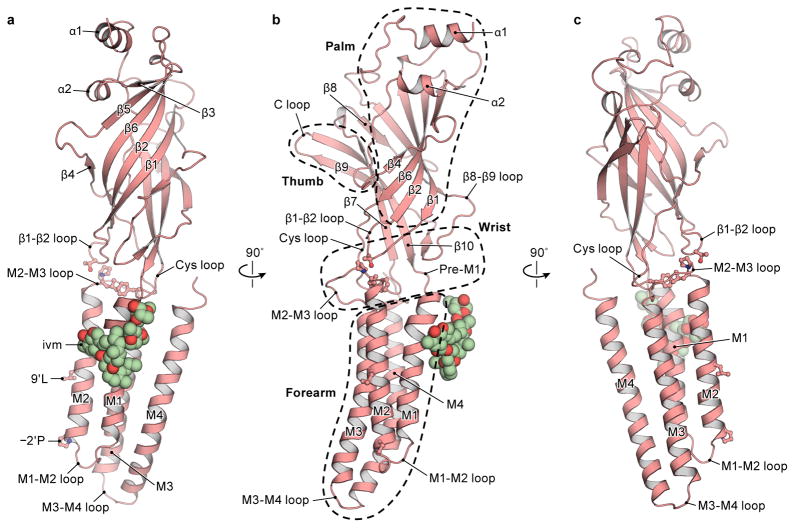
The three GlyR structures have urn-like architectures, akin to the AChR25 and other Cys-loop receptors, with the pentameric assemblage of subunits surrounding a central, 5-fold symmetric pore axis. Each subunit has the shape of an upright left forearm, clad with a mitten (Extended Data Fig. 5). The β strands of the ECD form the palm and the hallmark C loop emerges as the thumb, poised on the back of the hand of an adjacent subunit. A ‘wrist-like’ cuff connects the ECD and TMD and is buttressed by ‘ligaments’ that include the pre-M1 linker and loops of the ECD and TMD. We envision the TMD as the ‘forearm’, consisting of helices M1-M4. With the open palm and thumb of one mitten packing against the back of the hand of the neighbor, together with multiple interactions via the TMD ‘forearms’, these interactions knit the pentamer together (Fig. 1d–f).
The outline of the str-GlyR TMD is approximately perpendicular to the membrane (Fig. 1a, d). By contrast, in the gly-GlyR complex, the extracellular half of TMD is wider than the intracellular half (Fig. 1b, e), reminiscent of a truncated cone. In the gly/ivm-GlyR structure, the intracellular halves of the TMs move closer to each other (Fig. 1c, f). Thus, the pore in the str-bound form is constricted throughout and the pore-lining M2 helices are perpendicular to the membrane, in comparison with the other two forms (Fig. 1g–i). In the gly-bound form, the extracellular opening is wider than in the strychnine complex because the M2 helices tilt and undergo a counterclockwise rotation around the pore axis, viewed from the extracellular side. The binding of ivermectin promotes a clockwise rotation of the intracellular half of the M2, relative to the gly-bound form, constricting the intracellular opening of the pore but leaving the extracellular entry as expanded as in the gly-bound structure.
Ion channel pore
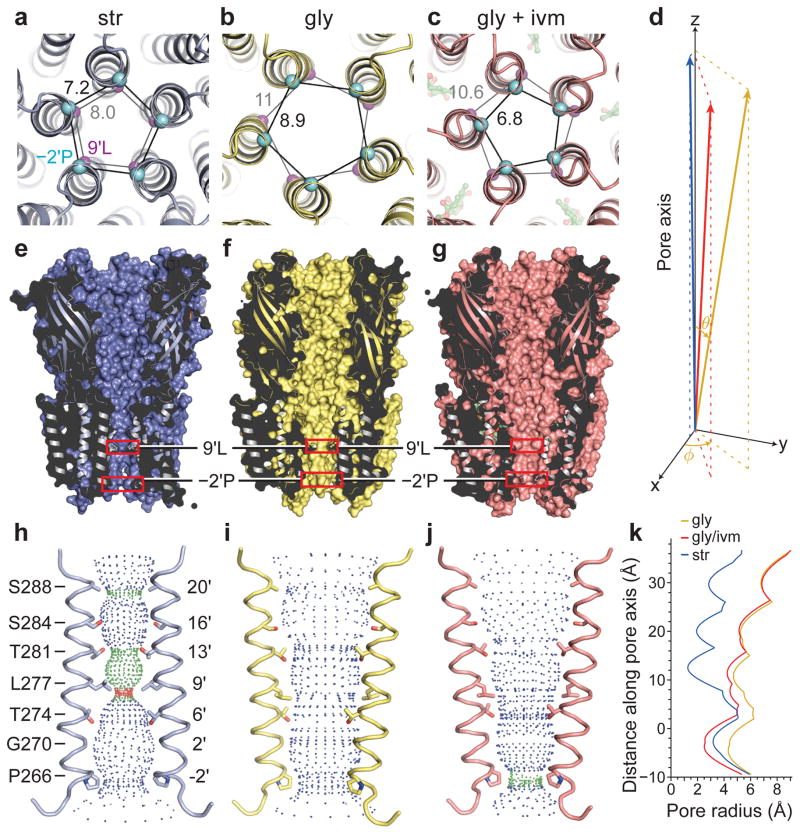
Inspection of the three GlyR structures demonstrate that there are two physical sites of constriction within the ion channel pore located at −2′Pro (Pro266) and 9′Leu (Leu277), highly conserved residues that reside on the M2 helix and are located near the cytoplasmic side and middle of the helix, respectively. The descending order of pore cross-sectional area, as estimated by the distance between adjacent Cα atoms of −2′Pro residues, is gly, str, gly/ivm, while at the position of the 9′Leu, the order is gly, gly/ivm, str (Fig. 2a–c). The ion channel in the str-bound form is the narrowest, with a radius of ~1.4 Å at 9′Leu, too narrow for passage of a dehydrated chloride ion with an ionic radius of 1.8 Å32. In the gly- and gly/ivm-bound structures, 9′Leu rotates away from the 5-fold axis, expanding the ion channel pore to a radius of 4–5 Å. In the gly-bound form, the smallest radius is 4.4 Å at −2′Pro, allowing permeation of hydrated chloride ions, with an ionic radius of 3.3 Å. Thus the str-bound form is an antagonist-bound closed, non-conducting state and the gly-bound form is an agonist-bound open, conducting state (Fig. 2e–j).
Figure 2. The ion channel.
a–c, Two sites of pore constriction are at −2′Pro and 9′Leu, viewed from the cytoplasmic side, with their Cα in cyan and magenta spheres, respectively. Distances are in Ångstrom. d, Plot of the pore-lining M2 as represented by the vector connecting −2′ProCα and 9′LeuCα, with −2ProCα as the origin, the tilt angle θ and the rotation angle φ relative to the pore axis. The φ of str-bound GlyR is set to zero. e–g, Sagittal ‘slice’ views along the pore axis. h–j, Shape and size of the ion permeation pathway. M2 of two subunits are shown as ribbon representation, where the side chains of the pore-lining residues are shown in sticks. Blue, green and red spheres define radii of >3.3Å, 1.8–3.3 Å and <1.8 Å, respectively. k, Plot of pore radii as a function of distance along the pore axis. The Cα position of 0′Arg is set to zero.
To define the structural mechanism underlying the changes in dimension and shape of the ion channel pore, we measured the ‘tilt’ (θ) and rotation angles (φ) of the pore-lining M2 helices relative to the pore axis (Fig. 2d). The tilt angle θ is approximately the same in the gly and gly/ivm complexes and larger in the str form, thus opening the extracellular ‘halves’ of the ion channel pores in the gly- and gly/ivm-bound forms nearly two-fold the width of the str-bound state (Fig. 2h–k). Furthermore, by measuring the rotation angle φ of the M2 helix, we see that the M2 helices have undergone counterclockwise rotations of 49° and 22° in the gly- and gly/ivm-bound states, respectively relative to the str-bound form, when viewed from the extracellular side of the membrane. Considering the tilt and rotation angles together, the M2 helix undergoes an outward rotation from the str- to gly-bound forms, enlarging both constriction sites. By contrast, the relatively smaller rotation in the gly/ivm-bound state leads to a distinct repositioning of the cytoplasmic end of the M2 helix, as defined by measurements at −2′Pro, resulting in a constriction of the pore to a radius of 2.5 Å, which is smaller than a hydrated chloride ion (Fig. 2k).
To augment our interpretations of the GlyR structures, we performed two-electrode voltage clamp electrophysiology (TEVC) experiments. At intermediate (Fig. 3g) and saturating (Fig. 3f) glycine concentrations, current decays are small and slow, consistent with the notion that the GlyREM construct is not prone to desensitization and that the gly-bound structure is an open, conducting state. Strychnine antagonizes the glycine currents (Fig. 3g), in accord with the conclusion that the str-bound structure is a closed, non-conducting state. In the presence of glycine and ivermectin, we observe initial potentiation followed by pronounced decay (Fig. 3h). In addition, block of the gly/ivm current by picrotoxinin, an open channel blocker, is only ~20% on peak or steady state currents. Thus, even though ivermectin initially enhances glycine-induced currents, we propose that the gly/ivm-bound form represents an agonist/allosteric modulator-bound desensitized state or a partially open state with reduced susceptibility to picrotoxinin block. This is in agreement with recent studies which suggest that desensitization in ELIC33 involves constriction at the intracellular entrance of the pore and that the picrotoxinin binding site in GlyR spatially overlaps the desensitization gate34.
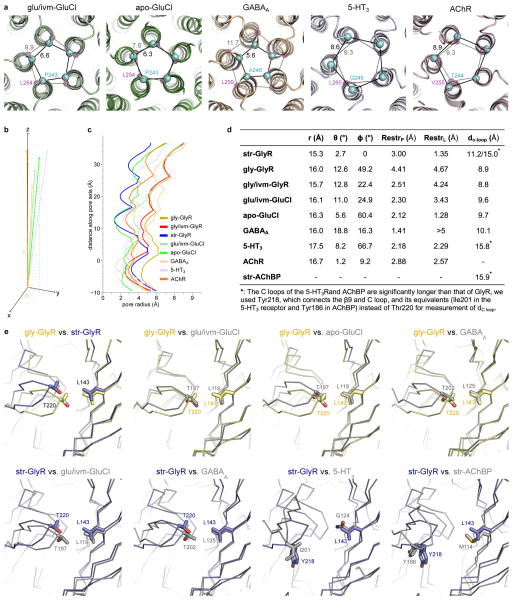
We can define a rubric for extant Cys-loop receptor structures by dividing the receptors into three groups (Extended Data Fig. 7a–d): (1) closed: str-GlyR, apo-GluCl, 5-HT3, nAChR; (2) partially open or desensitized-like: gly/ivm-GlyR, glu/ivm-GluCl, GABAA; (3) open: gly-GlyR. In group 1, the pores are restricted by 9′Leu. However, they possess distinct M2 conformations as indicated by the rotation angle (φ), perhaps reflective of their distinct states: antagonist/closed and resting/closed. In group 2, gly/ivm-GlyR and glu/ivm-GluCl share similar pore profiles and M2 conformations, suggesting they represent similar physiological states, perhaps desensitized-like or partially open, low-conductance states35. Despite a tighter restriction at the intracellular entrance, the pore properties of the GABAA receptor resemble that of gly/ivm-GlyR. This is in agreement with the suggestion that the GABAA crystal structure reflects an agonist-bound desensitized state29. In contrast, the unique large pore size and distinct M2 orientation distinguish the gly-GlyR structure and are consistent with defining a fully open, ion conducting state.
Neurotransmitter binding site
The neurotransmitter binding site of Cys-loop receptors is located at the interface of two adjacent subunits, surrounded by the C loop ‘thumb’ from the (+) side and the ‘back of the mitten’ β strands on the (−) side. Strychnine binds with a dissociation constant (Kd) of 98 nM (Fig. 3c) and, in the str-GlyR structure, we observe strong density in the neurotransmitter binding site (Fig. 3a). The almond-shaped density hews to the contours of a strychnine molecule, whose ring I forms the tip and rings III to VII form the bulbous bottom. In this pose, the relative positions of strychnine and the side chain of the conserved Tyr218 are in harmony with the crystal structure of the AChBP-strychnine complex36. A possible hydrogen bond between the carbonyl oxygen of strychnine and Arg81, proximity of the same carbonyl to Thr220 (C loop), and sandwiching of strychnine’s aromatic rings by Phe79 and Tyr218 further enhance interactions between the antagonist and receptor (Fig. 3a–b). In addition, the mutation Tyr177Ala results in strychnine insensitive GlyRs, while Tyr177Phe does not37. Despite no direct interaction to strychnine, Tyr177 sterically restricts the B loop close to the neurotransmitter binding pocket, thus explaining how a change in residue volume could indirectly perturb the binding site.
Glycine robustly activates the receptor with minimal or partial current decay upon prolonged application of agonist at intermediate or saturating concentrations, respectively (Fig. 3 f–g) and yields an EC50 of 0.26 mM. Density associated with glycine is not discernible in either the gly- or gly/ivm-bound structures due to its low molecular mass. Nevertheless, mutational analysis indicates that strychnine binding determinants such as Thr220 and Tyr218 partially overlap with those of glycine38,39,40. In particular, the Tyr218Phe mutant is reported to produce a 480-fold reduction in glycine affinity41. By comparing the ECDs of the gly- and str-bound structures, we observe that Arg81, which is involved in glutamate binding in GluCl26, exhibits substantial conformational changes, while other residues in the β-sheet forming the (−)-half of the binding pocket remain essentially unchanged (Fig. 5b–c). We propose that Arg81, Thr220 and Tyr218, together with Ser145, Phe175 and Phe223, directly participate in the binding of glycine41–43.
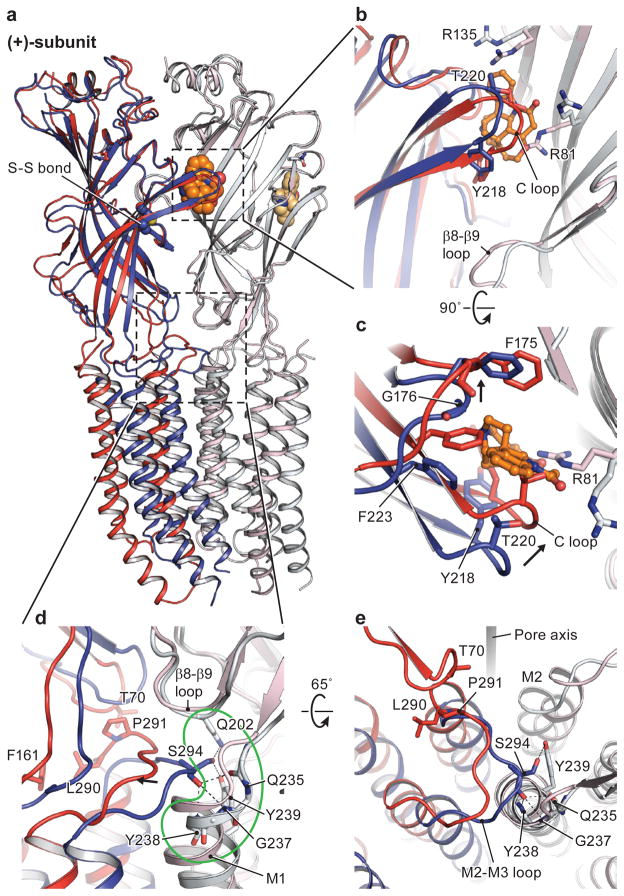
Figure 5. Conformational differences at the subunit-subunit interface between agonist- and antagonist-bound states.
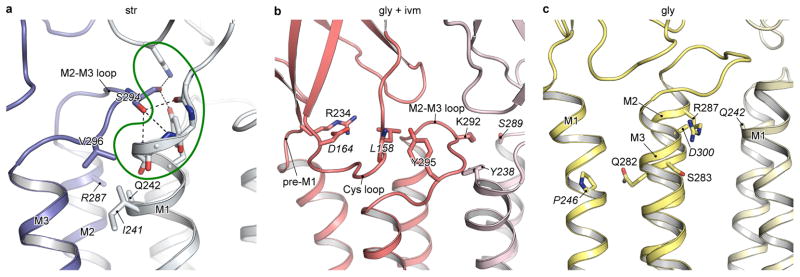
Str- and gly/ivm-bound states are in blue and red, respectively. The (−)subunits are in corresponding light colors. a, Superimposition of the ECD of the (−)-subunits showing the relative movement of the (+)-subunits. In b and c are shown conformational changes of the neurotransmitter binding pocket, viewed parallel to the membrane and from the extracellular side, respectively. The neurotransmitter binding site expands in the str-bound structure caused by repositioning of Arg135 and Arg81 in the (−)-subunit and by the opening of the C loop in the (+)subunit. Panels d and e illustrate the coupling of structural rearrangements of the ECD-TMD interface between two adjacent subunits. In the str-bound form, Ser294 in the M2-M3 loop of the (+)-subunit is inserted in the M1 N-cap in the (−)-subunit. Key residues interacting with Ser294 are highlighted in the green outline. Upon binding of glycine, the M2-M3 loop moves away from the N-cap. For clarity, the side chains of Gln235 and Tyr238 are not shown.
A central concept of the relationship between ligand binding and channel gating in Cys-loop receptors is that the conformation of the C loop ‘thumb’ is correlated to the functional state of the receptor, i.e., the C loop is “open” in the antagonist-bound, closed channel state and “closed” in the agonist-bound open channel state44–48. To examine this hypothesis, we superimposed the (−)-subunit of our GlyR models with the glu/ivm-GluCl, apo-GluCl, GABAA, 5-HT3 and str-AChBP structures and compared the size of their binding pockets by measuring the distance between the C loop and the right half of the pocket (Extended Data Fig. 7e). We observed that antagonist-bound str-GlyR and str-AChBP structures possess more open C loops and remarkably larger binding pockets than the agonist-bound structures. In addition, the gly-GlyR structure shares a more similar pocket size and shape to the glu/ivm-GluCl than the apo-GluCl structure, consistent with glycine being bound in the gly-GlyR structure.
Allosteric binding site
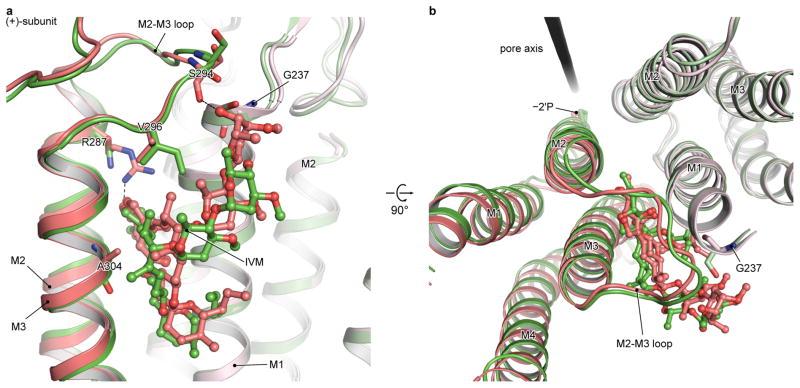
Ivermectin is a GlyR allosteric modulator that potentiates glycine-induced currents (Fig. 3h). In the gly/ivm-bound structure, the wedge-shaped ivermectin inserts into the TMD interface of two adjacent subunits and interacts with hydrophobic residues in M3 of the (+)-subunit and M1 of the (−)-subunit (Fig. 3d–e). It also likely forms hydrogen bonds with several residues, one of which is Arg287, a residue that enables a direct interaction between ivermectin and the pore-lining M2 helix. Mutations of Arg287 to Gln or Leu in the human α1 GlyR subunit reduces the sensitivity of the receptor to ivermectin by a factor of 20 and these substitutions are the most frequent mutants associated with hyperekplexia (Extended Data Fig. 9c)5,16. The potential interaction between Arg287 and ivermectin, together with the location of Arg287 on the M2 helix, explains why mutations at 287 perturb both ivermectin action and ion channel gating.
Unlike GluCl26, ivermectin is not required for GlyR activation. To probe the structural basis for this difference in function, we compared the binding pockets in both receptors (Extended Data Fig. 6). Whereas ivermectin binds similarly to both receptors, there are several notable differences. First, Arg287 in GlyR interacts with ivermectin, yet the corresponding residue in GluCl26 is an asparagine (Asn264) and the side chain is positioned further from ivermectin. Second, Val296 in the M2-M3 loop of GlyR is an isoleucine (Ile273) in GluCl, whose larger side chain prevents the O6-oxygen of ivermectin from approaching the M2-M3 loop. Third, Gly237 in the M1 and Ala304 in the M3 of GlyR are Ser217 and Gly281 in GluCl, respectively. In GlyR, the M3 residue (Ala) is larger than the M1 residue (Gly), effectively “pushing” ivermectin toward M1, creating a larger interaction surface area between ivermectin and M1 compared to M3. This situation is reversed in GluCl, however, as the M1 residue (Ser) is larger than the M3 residue (Gly). This is consistent with the experiments where the Ala304Gly substitution in the M3 dramatically enhanced the ivermectin sensitivity of human GlyR49, probably by enlarging the ivermectin-M3 interface.
Signal transduction
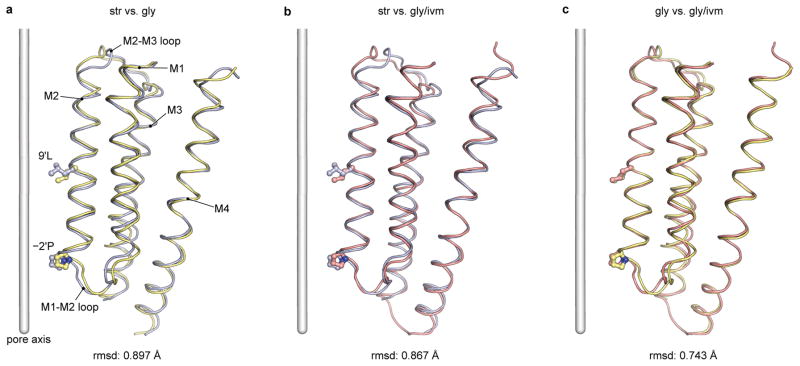
By superimposing a single subunit from the str- and gly-bound states, we see that the β-sheets and flanking loops of the ECD behave like a malleable ‘palm’, undergoing a ‘flexing-like’ conformational change that is distributed throughout the entire domain, thus defying the demarcation of a simple, local hinge (Fig. 4a). The superposition also reveals that binding of ivermectin does not cause significant conformational changes of the ECD and ECD-TMD interface (Fig. 4b–c). In the following comparison of the ECD and ECD-TMD interfaces, the gly/ivm-GlyR structure, whose reconstruction is of better quality, is used (Fig. 4d–e and Fig. 5). By contrast, the TMD behaves as an approximate rigid body upon comparison of the three structures (Extended Data Fig. 8). The M3-M4 loop is on the cytoplasmic side of the TMD, is truncated in this GlyREM construct, and may have an impact on the conformational changes of the TMD. Thus further studies are necessary in order to clarify the role of the M3-M4 loop in receptor gating. Nevertheless, when we analyze the ECD interface in the str- and gly-bound states by superposition using the ECD of the (−)-subunit (Fig. 5a), the ‘flexing’ of the β-sheets leads to prominent conformational changes in two critical regions – the interfaces between the ECDs that span the crevices between the orthosteric and allosteric agonist binding sites, and the interfaces between the ECD and the TMD.
Figure 4. Conformational changes within an individual subunit.
Str-, gly- and ivm/gly-bound states are in blue, yellow and red, respectively. a–c, Superimposition of the three GlyR structures using the ECD (residues 1-235), showing the motion of TMD. Relative rotation angles of the pore-lining M2 are indicated. Conformational changes in the ECD-TMD interface upon transition from the gly- (or gly/ivm-) to the str-bound states are shown in panels d and e, viewed parallel to the membrane and from the extracellular side, respectively. The displacement of the β8-β9 loop leads to a rotation of pre-M1/M1, pushing the lower half of M2 toward the pore axis; meanwhile, this displacement repositions the Cys loop through β10, which results in the coupling of the M2-M3 loop with the β1-β2 loop through the interaction between Pro291 and Thr70. Consequently, the upper half of M2 rotates outward.
At the neurotransmitter binding site, we observed an expansion in the str-bound structure caused by displacement of Arg81 and Arg135 on the core of the β-sheet in the (−)-subunit and by the opening of the C loop, as indicated by the outward movement Cα position of Thr220 by 4 Å (Fig. 5b–c). The motion of the C loop is mainly a consequence of the movement of the entire ECD, as also observed in GluCl27, together with a subtle ‘flexing’ at the loop tip. At the ECD-TMD interface, in the str-bound closed state, there are extensive interactions between subunits at the interface near the ECD-TMD boundary50, defined by interactions between the β1-β2 loop, Cys-loop and M2-M3 loop of the (+)-subunit with the β1-β2 loop, β8-β9 loop and pre-M1/M1 of the (−)-subunit. There are also multiple contacts between subunits within the TMD, mediated by intersubunit interactions between the M1 and M3 helices, and between M2 and M2 helices. By contrast, in the gly-bound state, the interactions between subunits at the ECD-TMD boundary have largely ruptured, simply due to an increase in the separation of subunits, and the contacts between the TMD have also diminished by more than 50% (540 A2 in str-GlyR and 240 A2 in gly-GlyR), also because the subunits have moved apart. Remarkably, the increase in the separation of the intersubunit M1-M3 interaction in the gly-bound state provides the initial ‘indentation’ of the cavity that is occupied by ivermectin in the gly/ivm complex.
To further understand how the conformational changes within the ECD are transduced to the TMD, we divide the M2 helix into upper and lower halves and follow their movements. On the one hand, the upper half of M2 couples to both the β1-β2 loop and Cys loop through the M2-M3 loop (Fig. 4d–e) in the gly-bound form. The Cys loop further interacts with pre-M1/β10, which move along with the β8-β9 loop. On the other hand, the lower half of M2 is connected to M1, which is coupled to the β8-β9 loop through the pre-M1 loop. Thus, the movement of M2 can be traced to the β8-β9 loop, a loop that, in turn, not only connects to the C loop covering the binding pocket but that also sits underneath the pocket and thus indirectly participates in the binding pocket of the adjacent subunit (Fig. 5b). In going from the str- to gly-bound states (Fig. 4d–e), the C loop ‘thumb’ switches from “open” to “closed” and the β8-β9 loop is displaced. This leads to a concomitant rotation of pre-M1 and M1, thus ‘pushing’ the lower half of M2 towards the pore axis. Meanwhile, the β8-β9 loop repositions the Cys loop through β10, which results in the interaction of the M2-M3 loop with the β1-β2 loop causing an outward rotation of the upper half of M2.
Particularly important are the interactions at the ECD-TMD interface, where the β1-β2 and Cys loop of the (+)-subunit, together with the β8-β9 loop and preM1/M1 of the (−)-subunit, interact with the crucial M2-M3 loop of the (+)-subunit (Fig. 5d–e). Here, in the str-bound form, we suggest that Ser294 in the M2-M3 loop of the (+)-subunit stabilizes the channel in a closed conformation by capping the amino terminus of the M1 helix of the (−)-subunit (N-cap), making van der Waals contacts with residues in the pre-M1/M1 region and participating in interactions with residues at the β8-β9 loop tip. Upon binding of glycine, the M2-M3 loop moves away from capping the amino terminus of the M1 helix and instead interacts with the β1-β2 loop via Pro291 and Thr70, stabilizing the channel in an open form. Several startle disease mutants in the M1 helix and the M2-M3 loop cause spontaneous activation of the receptor, perhaps by disruption of the interactions between Ser294 and the M1 N-cap (Extended Data Fig. 9a–b)10.
Comparisons of the gly- and gly/ivm-bound structures reveal that binding of ivermectin causes the most notable changes at the cytoplasmic half of the TMD by ‘tilting’ the lower half of the M2 helix toward the pore lumen by 4.5° (Fig. 4c), contracting the intercellular opening of the ion channel pore at −2′Pro (Fig. 2j–k). However, the extracellular regions of the pore in the gly- and gly/ivm-bound states are similar, possibly constrained by interactions between ivermectin and the M2 and M2-M3 loops.
Gating mechanism
The str- and gly-bound structures of the GlyR, elucidated free from constraints of a crystal lattice or Fab fragments, unambiguously define antagonist-locked/closed and agonist-activated/open states of the receptor, respectively, while the glycine and ivermectin complex is suggestive of an allosteric modulator-bound low conductance or possibly desensitized state. A mechanism of ligand-dependent gating emerges from analysis of these structures in which the ‘palm’ of the ECD and the ‘forearm’ of the TMD are coupled by a flexible joint at the ECD-TMD interface, a joint reinforced by a ‘ligament-like’ cuff composed of interacting ECD and TMD loops. Agonist binding closes the C loop ‘thumb’, promotes a concerted counterclockwise rotation around the pore axis (viewed from the extracellular side, the same below for TMD) of all 5 ‘palm’ domains about an axis formed by finger-palm joints, and thus an iris-like expansion and counterclockwise rotation of the entire TMD ‘forearms’ (Fig. 6a–c and Supplementary Movie 1). Together, these structures not only allow us to describe a molecular mechanism for ligand-dependent gating in GlyRs, but they also define a framework for interpreting the wealth of structural information on Cys loop receptors and bacterial orthologs, which in turn will ground the search for new therapeutic agents on a solid structural foundation.
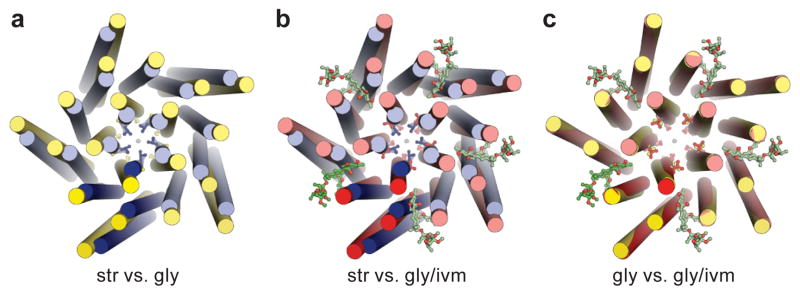
Figure 6. Overall conformational changes of the TMD.
Str-, gly- and gly/ivm-bound states are in blue, yellow and red, respectively. Comparison between str- and gly-GlyR (a), str- and gly/ivm-GlyR (b), gly- and gly/ivm-GlyR (c), viewed from the extracellular side. Side chains of −2′Pro and 9′Leu are shown in sticks to denote the change of pore sizes. In going from the str- to the gly-bound form, the TMD of each individual subunit undergoes a counter-clockwise outward rotation, enlarging the pore size by ‘pulling’ the side chains of 9′Leu and −2′Pro away from the channel axis. Binding of ivermectin to the gly-GlyR causes a clockwise inward rotation of the TMD. As a result, while the extracellular half of the pore undergoes little change, the intracellular entrance shrinks.
Methods
Receptor constructs
The gene encoding the zebra fish α1 glycine receptor (NP_571477) construct shares 92% amino acid similarity with the human α1 GlyR (Supplementary Fig. 1). The construct was chemically synthesized and further modified in accordance with the GluClcryst construct, which involved removing 8 residues (Arg26-Pro33) from the amino terminus and 10 residues (Ile435-Gln444) from the carboxyl terminus, and replacing the long, highly variable M3/M4 loop (Gln334-Arg400) with the Ala-Gly-Thr tripeptide. This construct is named GlyREM and was subcloned into the pFastBac1 vector for baculovirus expression in Sf9 insect cells. A thrombin cleavage site (Leu-Val-Pro-Arg-Ser) and an octa-histidine tag were introduced at the carboxyl terminus of GlyREM.
Expression and purification
The bacmid and baculovirus of GlyREM in pFastBac1 were generated using standard methods26. GlyREM P2 virus was used to infect Sf9 insect cells at 27°C. At 72 hours post infection, the cells were harvested and disrupted by sonication. The cell debris was removed by centrifugation at 8,000 rpm and membranes were pelleted from the supernatant by centrifugation for 1 hour at 40K (Ti45 rotor) in 150mM NaCl, 20mM Tris 8.0 (TBS buffer) in the presence of 1mM PMSF, 0.8 μM aprotinin, 2 μg/ml leupeptin, and 2 mM pepstatin A. The receptor was extracted from membranes with a buffer (affinity buffer) containing 1 mM n-dodecyl-β-D-maltopyranoside (C12M), 200mM NaCl and 20mM Tris 8.0 for 2 hours at 4°C. The solubilized materials were incubated with TALON resin overnight and washed with affinity buffer in presence of 35 mM imidazole. The receptors were eluted with 250 mM imidazole at pH 8.0 and were concentrated for overnight digestion by 1/100 (w/w) thrombin at 4°C. The receptors were further purified by size exclusion chromatography (SEC) in a buffer containing 150mM NaCl, 20mM Tris 8.0 and 1 mM C12M (SEC buffer) at 4°C. Fractions containing the receptor were pooled and concentrated to 3.3 mg/ml.
Two-electrode voltage clamp electrophysiology (TEVC)
The zebra fish α1 GlyREM was subcloned into the pGEM vector for TEVC experiments. The RNAs were then transcribed using the mMessage mMachine T7 Ultra Kit (Ambion). Xenopus laevis oocytes were injected with 30ng of mRNA and were incubated at 18°C for 2–3 days in a solution containing 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES pH 7.5, and 250 μg/ml amikacin (incubation buffer). The borosilicate pipettes were filled with 3 M KCl. The stock solutions of strychnine and glycine were prepared in H2O at concentrations of 50 mM and 2M, while ivermectin and picrotoxinin were solved in dimethyl sulfoxide (DMSO) at concentrations of 100 mM and 1 M, respectively. The ligands were solved in recording solution containing 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5 mM HEPES pH 7.5. Recordings were performed at −60mV. All recording experiments were performed three times independently. Half-maximal concentration of glycine (EC50) and Hill coefficient (nH) values were obtained using the Hill equation fitted with a non-linear least squares algorithm using GraphPad Prism. The result is averaged from three independent experiments and the error bars represent s.e.m..
Ligand binding assays
The strychnine binding constant was determined by scintillation-proximity assay (SPA). Purified GlyREM-His8 (20 nM) was incubated with 1 mg/ml copper yttrium silicate (Cu-YSi) beads (Perkin Elmer) and 3H-labelled strychnine (1:9 3H:1H) in SEC buffer with a final volume of 100μl. Non-specific binding was determined by the addition of 100 mM imidazole. Assay plates were read using a MicroBeta TriLux 1450 LSC and luminescence counter and data were fit to the Hill equation using GraphPad Prism.
Crystallization and molecular replacement
GlyREM was crystallized using the hanging-drop vapor-diffusion method in the presence of 2 mM glycine. One microliter of protein solution (1.8 mg/ml) was mixed with 1 μl reservoir solution containing 30% PEG400, 0.1 M MES 6.5, and 0.2 M CaCl2. Diffraction data was collected on the 8.2.1 beamline at the Advanced Light Source of Lawrence Berkeley Laboratories (ALS), using a wavelength of 0.9762 Å. The data were indexed, integrated and scaled using XDS51. The space group is P212121 and cell parameters are a=117.77 Å, b=121.35 Å, c=503.81 Å and α=β=γ=90°. Each asymmetric unit contains two pentameric receptors (A and B). Due to radiation damage, the overall completeness is only 62.9% (62.9%) to a resolution of 4.32 Å. Rmeas and CC1/2 are 0.101 (0.529) and 0.998 (0.309), respectively. Values in parentheses represent the highest resolution shell (4.46–4.35 Å). The structure was solved by molecular replacement using PHASER, with the crystal structure of GluCl (PDB code: 3RHW) as the initial search model. The LLG and TFZ of the solution are 615 and 20.6, respectively. The overall map quality of the pentamer A is better than that of the pentamer B. In pentamer A, the electron density is best for the transmembrane helices, where grooves and ridges can be observed. In contrast, the densities for extracellular domains are partially missing, likely caused by a lack of crystal contacts.
Sample preparation and data acquisition for cryo-EM analysis
Purified GlyREM in C12M was mixed with 10mM glycine/5μM ivermectin, with 1mM strychnine or with 10 mM glycine a few hours before grid preparation. Next, 2.5μl of protein sample at a concentration of 3.3 mg/ml was applied to a glow-discharged (10s on each side) Quantifoil holey carbon grid (copper, 1.2μm/1.3μm hole size/hole space, 200 mesh), blotted using a Vitrobot Mark III (FEI company) using 3.5s blotting time with 100% humidity, and then plunge-frozen in liquid ethane cooled by liquid nitrogen.
The data sets were collected on a Titan Krios cryo-electron microscope (FEI company) at the CryoEM Facility at the Janelia Research Campus or on a Polara microscope (FEI) at UCSF. The data for the gly/ivm-bound state was collected on Titan Krios I and the data for the str-bound state was collected on Titan Krios II at Janelia. Krios I is equipped with a CETCOR Image Corrector for spherical aberration correction and a Gatan Image Filter (GIF). A 30 eV energy slit as well as a 70 um objective aperture (corresponding to a cutoff of 2 Å) was used during data collection. The Image Corrector tuned the Cs from an original 2.7 to 0.01 mm. The data for the gly-bound form was collected on the TF30 Polara at UCSF. All the microscopes are equipped with a field emission source and operated at 300 kV. Images were recorded on the Gatan K2 Summit direct electron detector operated in super-resolution counting mode. At Janelia the dose rate was 10 e−/pixel/s, determined in ‘empty’ holes. At UCSF the dose rate was 10.9 e−/pixel/s, determined through a layer of vitreous ice. The total exposure time was either 5 s or 6 s, with an accumulation time of 0.2 s for each frame. The dose-fractionated images were recorded using the automated acquisition program SerialEM or the semi-automated acquisition program UCSFImage4 (written by X. Li) at Janelia or UCSF, respectively. Nominal defocus values ranged from −1.5 to −2.5 μm or −1.5 to −3.0 μm. (Extended Data Table 1)
Image processing
Super resolution counting images were 2×2 binned in Fourier space, resulting in a pixel size of 1.0400 Å (Krios I), 1.0100 Å (Krios II), or 1.2156 Å (Polara). Motion correction was done using MotionCorr52, with a B-factor of 250 or 1000 pixel2. All the motion-corrected frames (25 or 30) were summed to a single micrograph for subsequent processing with RELION53. Defocus values were estimated using CTFFIND354. For each dataset, approximately 1000 particles were manually picked for an initial reference-free 2D classification. Seven to eight representative 2D class averages were selected as templates for automated particle picking. The auto-picked particles were visually checked and false positives were removed. The particles were further cleaned-up by two rounds of 2D classification using RELION. An initial 3D model was generated from the GluCl crystal structure26 (PDB code: 3RHW) and low-pass filtered to 50 Å using EMAN255. No symmetry was applied during 3D classification in RELION. Classes with characteristic features of Cys loop receptors were then selected. Particles belonging to the chosen classes were used for 3D auto-refinement using RELION, followed by 3D refinement with movie particles, in which the movement of each individual particle was determined. In the subsequent ‘polishing’ step, the movement of each particle was corrected. In addition, resolution-dependent radiation damage was modeled by performing a weighted average of all aligned movie frames for each particle, using a B-factor estimated by the polishing algorithm. The resulting ‘shiny’ particles were used for a final round of 3D classification and refinement. The polishing procedure typically increased the estimated resolution by 0.2–0.3 Å. Five-fold symmetry was applied during all refinement steps. In the post-processing step in RELION, a soft mask was calculated and applied to the two half-maps before the Fourier shell coefficient (FSC) was calculated. B-factor estimation and map sharpening were also performed in the post-processing step. The resolutions reported in Extended Data Table 1 are based on the gold standard FSC 0.143 criteria56. Further details related to data processing are summarized in Extended Data Table 1.
Model building
A molecular replacement solution from a low-resolution and incomplete x-ray diffraction data, using the GluCl structure (PDB code: 3RHW) as a search probe, was used as an initial model (see Crystallization and molecular replacement). The mismatched residues are replaced with the correct ones using the SWISS-MODEL online server57. The resulting model was fit into the density map of the gly/ivm-bound state using UCSF Chimera58. Further model building and real space refinement was done using COOT59. For the other GlyR structures, the gly/ivm-bound structure was used as a starting model. Strict five-fold symmetry was used throughout model building and refinement. The final masked maps were put into P1 unit cells with cell dimensions equal to the box sizes used to extract particles (Extended Data Table 1) and structure factors were calculated using the program SFTOOLS in the CCP4 suite60. The resulting structure factors were used for maximum likelihood refinement in reciprocal space using Phenix.refine with secondary structure and NCS restraints. For cross validation61, the refined structures were randomly displaced by 0.1 Å. The displaced models were then refined against one of the half maps produced by RELION following the same procedure described above. FSC curves were calculated between the refined model and half map 1 (“work”, used for refinement), refined model and half map 2 (“free”, not used for refinement), and the refined model and summed map. No significant separation of “work” and “free” FSC curves were observed, suggesting the atomic models were not over-refined. The geometries of the atomic models were evaluated using MolProbity62. Our models have the same helical registers as the crystal structures of GluCl and the GABAA and 5-HT3receptors, but differ by one turn from that of the 4 Å cryo-EM Torpedo AChR structure. All figures were prepared using Pymol (Schrödinger, LLC)63, UCSF Chimera and Prism 5 (GraphPad Software, La Jolla). Pore radii were calculated using the program HOLE64.
Extended Data
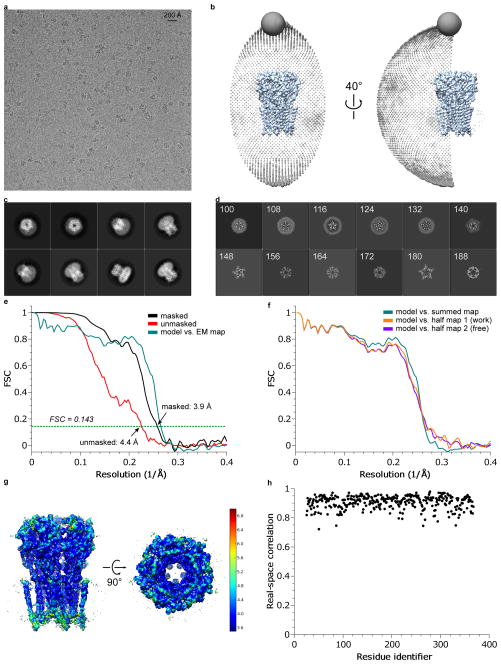
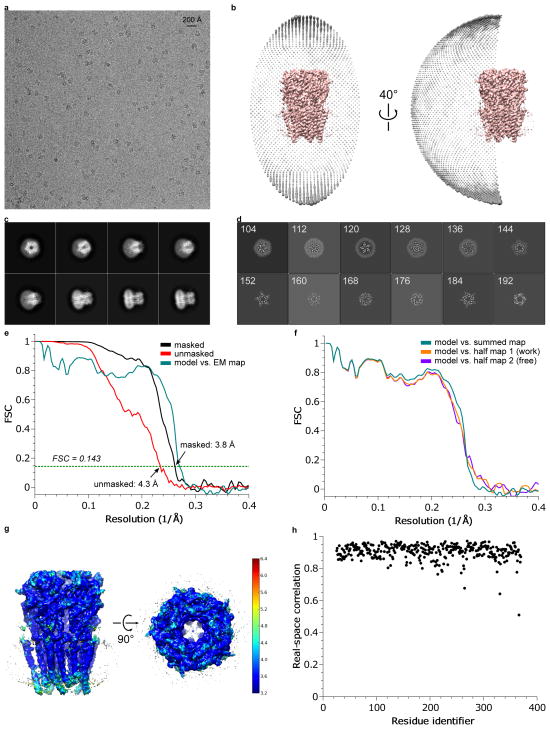
Extended Data Figure 1. 3D reconstruction of strychnine-bound GlyR.
a, A representative micrograph (out of 1829 micrographs) of str-bound GlyR in vitreous ice. b, Angular distribution of particle projections, and c, selected 2D classes are shown. In panel (c), the radius of the sphere is proportional to the number of particles assigned to it. The plot is drawn with respect to the 3D reconstruction shown in the center, taking the C5 symmetry of the receptor into account. d, Selected ‘slice’ views of the final reconstruction along the pore axis. The slice numbers are indicated, starting from the intracellular side. e, FSC curves for the density maps before (red) and after postprocessing in RELION (black). The FSC curve between the refined atomic model and the final reconstruction map is shown in green. f, FSC curves for cross-validation: model versus summed map (full data set, green), model versus half map 1 (used in test refinement, orange) and model versus half map 2 (not used in test refinement, blue). g, Unfiltered and unsharpened 3D density map colored according to local resolution estimated using RESMAP65. h, Real-space correlation between atomic model and density map calculated using PHENIX.
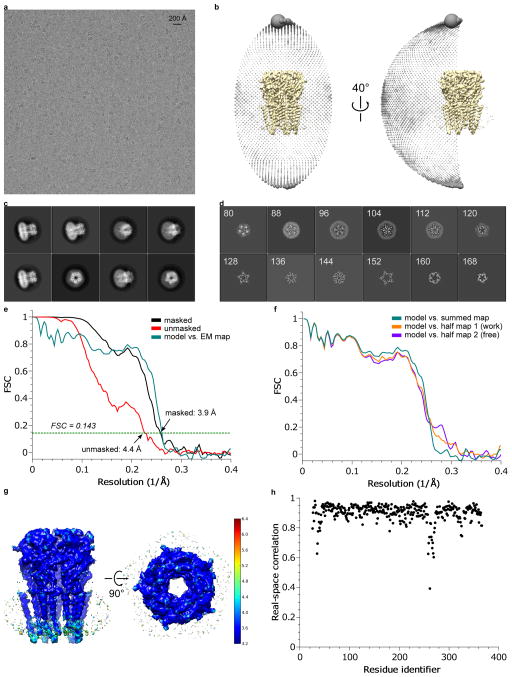
Extended Data Figure 2. 3D reconstruction of glycine-bound GlyR.
a, A representative micrograph (out of 1460 migrographs) of gly-bound GlyR in vitreous ice. b, Angular distribution of particle projections, and c, selected 2D classes are shown. In panel (c), the radius of the sphere is proportional to the number of particles assigned to it. The plot is drawn with respect to the 3D reconstruction shown in the center, taking the C5 symmetry of the receptor into account. d, Selected ‘slice’ views of the final reconstruction along the pore axis. The slice numbers are indicated, starting from the intracellular side. e, FSC curves for the density maps before (red) and after (black) post-processing in RELION. The FSC curve between the refined atomic model and the final reconstruction map is shown in green. f, FSC curves for cross-validation: model versus summed map (full data set, green), model versus half map 1 (used in test refinement, orange) and model versus half map 2 (not used in test refinement, blue). g, Unfiltered and unsharpened 3D density map colored according to local resolution estimated using RESMAP. h, Real-space correlation between atomic model and density map calculated using PHENIX.
Extended Data Figure 3. 3D reconstruction of glycine/ivermectin bound GlyR.
a, A representative micrograph (out of 2489 micrographs) of gly/ivm-bound GlyR in vitreous ice. b, Angular distribution of particle projections, and c, selected 2D classes are shown. In panel (c), the radius of the sphere is proportional to the number of particles assigned to it. The plot is drawn with respect to the 3D reconstruction shown in the center, taking the C5 symmetry of the receptor into account. d, Selected ‘slice’ views of the final reconstruction along the pore axis. The slice numbers are indicated, starting from the intracellular side. e, FSC curves for the density maps before (red) and after (black) post-processing in RELION. The FSC curve between the refined atomic model and the final reconstruction map is shown in green. f, FSC curves for cross-validation: model versus summed map (full data set, green), model versus half map 1 (used in test refinement, orange) and model versus half map 2 (not used in test refinement, blue). g, Unfiltered and unsharpened 3D density map colored according to local resolution estimated using RESMAP. h, Real-space correlation between atomic model and density map calculated using PHENIX.
Extended Data Figure 4. Representative densities of the three reconstructions of GlyR. Densities are sharpened using RELION unless indicated.
The densities in each panel are for the str-, gly/ivm-, gly-, and unsharpened gly-bound states, respectively, from left to right. a, Representative densities of the β-sheets in ECD, contoured at 8 σ. b, Densities of Cys-loop and the M2-M3 loop, contoured at 7 σ. c, Densities of helices M1 and M2, contoured at 7 σ. d, Densities of M3 and M4, contoured at 7 σ. e, Densities of −2′Pro, contoured at 7 σ except for the gly-bound state (6.5 σ). f, Densities of 9′Leu, contoured at 6.0 σ except for the gly-bound state (5.0 σ).
Extended Data Figure 5.
a–c, A single subunit of glycine/ivermectin bound GlyR, viewed in parallel to the membrane plane, with secondary structure elements labeled. b, the domain arrangement resembles an upright forearm, clad with a mitten, consisting of thumb (C loop), palm (β strands of ECD) and ligament (ECD-TMD interface).
Extended Data Figure 6. Comparison of ivermectin binding site in GlyR (red) and GluCl (green), viewed in parallel to the membrane (a) or from the extracellular side (b).
The (+)subunits are shown in darker colors. The residue corresponding to Arg287, which forms a hydrogen bond with the ivmermectin in GlyR, is an asparagine (Asn264) in GluCl. The corresponding residue of Val296 in the M2-M3 loop of GlyR is an isoleucine (Ile273) in GluCl, whose larger side chain prevents the upper tip of ivermectin from approaching and interacting with the main chain oxygen atom of Ser721 in the M2-M3 loop (Ser294 in GlyR). The Gly237 in the M1 and Ala304 in the M3 of GlyR are Ser217 and Gly281 in GluCl, respectively. Such differences on side chains weaken or strengthen the interaction of ivermectin with M3 or M1 in GlyR, respectively, in comparison to that in GluCl.
Extended Data Figure 7. Comparison of GlyR with other Cys-loop receptors.
a, The two restriction sites, viewed from the cytoplasmic side. The Cα of −2′Pro equivalents (cyan) and 9′Leu equivalents (magenta) are shown as spheres. Distances between adjacent Cα atoms are labeled. b, Plot of the vector connecting the −2′ProCα equivalent and 9′LeuCα equivalent, with −2ProCα equivalent as the origin, the tilt angle θ and the rotation angle φ relative to the pore axis. The φ of str-bound GlyR is arbitrarily set to zero. c, Pore radii as a function of distance along the pore axis, calculated using the program HOLE, where the Cα position of 0′Arg is set to zero. d, Table showing parameters of the vector connecting the −2′ProCα equivalent and 9′LeuCα equivalent, where r is the distance from −2′ProCα equivalent to 9′LeuCα equivalent, RestrP and RestrL are the pore radii at −2′Pro equivalent and 9′Leu equivalent, respectively. The dC loop is the distance between Cα of Thr220 equivalent and Leu143 equivalent, representing the opening of the C loop shown in panel (e). e, Comparison of ligand binding pockets. The side chains of marker residues are shown in sticks.
Extended Data Figure 8. Superimposition of TMD between str- and gly-GlyR (a), str- and gly/ivm-GlyR (b), gly- and gly/ivm-GlyR (c) using main chain atoms of residues Met236-Lys362.
The M2-M3 loop, residues Ser289-Ala298, is excluded from the comparison. The r.m.s.d. are 0.9, 0.9 and 0.7 A, respectively, suggesting that the movement of the TMD is rigid-body-like. Most differences are located in the termini of transmembrane helices, which are either close to the M2-M3 loop, or close to the intracellular gate −2′Pro.
Extended Data Figure 9. Positions of residues whose mutations are associated with human startle disease.
Residues that likely interact with disease-causing residues are labeled in italics. a, The str-GlyR model is used to show residues whose mutations cause spontaneous activation. The mutation of Gln242 in M1 to glutamate may enhance its electrostatic attraction to Arg287 in M2 of the adjacent subunit, tilt the upper part of M2 away from pore axis, resulting in a constitutively open channel. On the other hand, the mutation Val296Met in M2-M3 loop may cause steric collision with Ile241 in M1 of the adjacent subunit, and prevent Ser294 from interacting to the N-cap formed by pre-M1, M1 and the β8-β9 loop, thereby destabilizing the closed conformation. b, The gly/ivm-GlyR model is used to show residues in the ECD-TMD interface whose mutations reduce sensitivity to glycine and single channel conductance. The mutation of Arg234 in pre-M1 to glutamine may disturb its electrostatic interaction with Asp164 in the Cys-loop. Similarly, the mutation of Tyr295 in the M2-M3 loop to cysteine or serine may disturb its interaction with the main chain nitrogen atom of Leu158 in the Cys-loop. In both cases, the signal induced by agonist binding may be blocked. The mutation Lys292Glu in the M2-M3 loop possibly affects the cooperative interaction between two adjacent subunits by altering the van der Waals contacts between Lys292 and Tyr238. c, The gly-GlyR model is used to show residues in M2 whose mutations reduce sensitivity to glycine and diminish single channel conductance. These mutations may directly influence the pore properties by modifying the interactions with adjacent residues, for instance, between Gln282His and Pro246, and between Arg287Gln/Leu and Gln242.
Extended Data Table 1.
Statistics of 3D reconstruction and model refinement.
| strychnine | glycine | glycine/ivermectin | |
|---|---|---|---|
| Data collection/processing | |||
| Microscope | Krios | Polara | Krios |
| Voltage (kV) | 300 | 300 | 300 |
| Defocus range (μM) | −1.5–−3.0 | −1.5–−2.5 | −1.5–−3.0 |
| Exposure time (s) | 5 | 6 | 5 |
| Dose rate (e−/pixel/s) | 10.0 | 10.9 | 10.0 |
| Pixel size (Å) | 1.0100 | 1.2156 | 1.0400 |
| Particles processed | 82913 | 127276 | 160585 |
| Particles refined | 37094 | 58188 | 56957 |
| Resolution (unmasked, Å) | 4.4 | 4.4 | 4.3 |
| Resolution (masked, Å) | 3.9 | 3.9 | 3.8 |
| Map sharpening B-factor (Å2) | −146 | −151 | −156 |
|
| |||
| Refinement | |||
| Cell dimensions | |||
| a = b = c (Å) | 282.8 | 291.7 | 291.2 |
| α = β = γ (°) | 90 | 90 | 90 |
| Resolution (Å) | 3.9 | 3.9 | 3.8 |
| Number of atoms | 12705 | 12810 | 13870 |
| Protein | 12440 | 12670 | 13420 |
| Ligand | 265 | 140 | 450 |
| r.m.s. deviations | |||
| Bond length (Å) | 0.004 | 0.005 | 0.005 |
| Bond angle (°) | 0.960 | 1.115 | 1.256 |
| Ramachandran plot | |||
| Favored (%) | 97.4 | 99.1 | 99.4 |
| Allowed (%) | 2.6 | 0.9 | 0.6 |
| Disallowed (%) | 0 | 0 | 0 |
Supplementary Material
DR, Danio rerio; HS, Homo sapiens; CE, Caenorhabditis elegans; TM, Torpedo marmorata; GLOVI, Gloeobacter violaceus; ERWINIA, Erwinia chrysanthemi. Secondary structures are represented by cylinders and arrows.
Acknowledgments
We are grateful to Z.H. Yu, N. Grigorieff, J. Cruz and C. Hong (Janelia Campus), C. Arthur (FEI/OHSU) and M. Braunfeld (UCSF) for assistance with microscope operation, data collection and for constructive comments and to R. Stites, M. Hakanson and A. Trzynka (OHSU) for computational support. We acknowledge the support of R. Goodman, J. Gray, the OHSU Center for Spatial Systems Biomedicine and the Multiscale Microscopy Core. We thank L. Vaskalis for help with illustrations and H. Owen for proofreading. R. Hibbs is gratefully acknowledged for pre-screening the GlyR constructs and D. P. Claxton for optimizing the constructs. We are thankful to Gouaux and Baconguis lab members for discussions. This work was supported by the National Institute of Health (E.G). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contribution
J.D., W.L. and E.G. designed the project, J.D. and W.L. performed sample preparation, cryo-EM data collection and data analysis, J.D., W.L. and E.G. wrote the manuscript, S.W. and Y.C. assisted in cryo-EM experiments at UCSF and participated in discussion and editing of the manuscript.
Author information
Three 3D cryo-EM density maps and coordinates of α1 glycine receptors in str-bound, gly-bound and gly/ivm-bound forms have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-6344, EMD-6345 and EMD-6346 and deposited in the RCSB Protein Data Bank under the accession codes 3JAD, 3JAE and 3JAF, respectively.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Literature
- 1.Katz B, Thesleff S. A study of the ‘desensitization’ produced by acetylcholine at the motor end-plate. J Physiol. 1957;138:63–80. doi: 10.1113/jphysiol.1957.sp005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aprison MH, Werman R. The distribution of glycine in cat spinal cord and roots. Life Sci. 1965;4:2075–83. doi: 10.1016/0024-3205(65)90325-5. [DOI] [PubMed] [Google Scholar]
- 3.Curtis DR, Hosli L, Johnston GA. Inhibition of spinal neurons by glycine. Nature. 1967;215:1502–3. doi: 10.1038/2151502a0. [DOI] [PubMed] [Google Scholar]
- 4.Werman R, Davidoff RA, Aprison MH. Inhibition of motoneurones by iontophoresis of glycine. Nature. 1967;214:681–3. doi: 10.1038/214681a0. [DOI] [PubMed] [Google Scholar]
- 5.Curtis DR, Hosli L, Johnston GA, Johnston IH. The hyperpolarization of spinal motoneurones by glycine and related amino acids. Exp Brain Res. 1968;5:235–58. doi: 10.1007/BF00238666. [DOI] [PubMed] [Google Scholar]
- 6.Lynch JW. Molecular structure and function of the glycine receptor chloride channel. Physiol Rev. 2004;84:1051–95. doi: 10.1152/physrev.00042.2003. [DOI] [PubMed] [Google Scholar]
- 7.Galzi JL, Edelstein SJ, Changeux J. The multiple phenotypes of allosteric receptor mutants. Proc Natl Acad Sci U S A. 1996;93:1853–8. doi: 10.1073/pnas.93.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legendre P. The glycinergic inhibitory synapse. Cell Mol Life Sci. 2001;58:760–93. doi: 10.1007/PL00000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch JW, Callister RJ. Glycine receptors: a new therapeutic target in pain pathways. Curr Opin Investig Drugs. 2006;7:48–53. [PubMed] [Google Scholar]
- 10.Bode A, Lynch JW. The impact of human hyperekplexia mutations on glycine receptor structure and function. Mol Brain. 2014;7:2. doi: 10.1186/1756-6606-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young AB, Snyder SH. Strychnine binding associated with glycine receptors of the central nervous system. Proc Natl Acad Sci USA. 1973;70:2832–2836. doi: 10.1073/pnas.70.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeiffer F, Graham D, Betz H. Purification by affinity chromatography of the glycine receptor of rat spinal cord. J Biol Chem. 1982;257:9389–93. [PubMed] [Google Scholar]
- 13.Graham D, Pfeiffer F, Betz H. Photoaffinity-labelling of the glycine receptor of rat spinal cord. Eur J Biochem. 1983;131:519–25. doi: 10.1111/j.1432-1033.1983.tb07292.x. [DOI] [PubMed] [Google Scholar]
- 14.Bormann J, Hamill OP, Sakmann B. Mechanism of anion permeation through channels gated by glycine and gamma-aminobutyric acid in mouse cultured spinal neurones. J Physiol. 1987;385:243–86. doi: 10.1113/jphysiol.1987.sp016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fatima-Shad K, Barry PH. Anion permeation in GABA- and glycine-gated channels of mammalian cultured hippocampal neurons. Proc Biol Sci. 1993;253:69–75. doi: 10.1098/rspb.1993.0083. [DOI] [PubMed] [Google Scholar]
- 16.Shan Q, Haddrill JL, Lynch JW. Ivermectin, an unconventional agonist of the glycine receptor chloride channel. J Biol Chem. 2001;276:12556–64. doi: 10.1074/jbc.M011264200. [DOI] [PubMed] [Google Scholar]
- 17.Changeux JP, Edelstein SJ. Allosteric receptors after 30 years. Neuron. 1998;21:959–80. doi: 10.1016/s0896-6273(00)80616-9. [DOI] [PubMed] [Google Scholar]
- 18.Miller PS, Smart TG. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol Sci. 2010;31:161–74. doi: 10.1016/j.tips.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 20.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–8. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 21.Spurny R, et al. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc Natl Acad Sci U S A. 2012;109:E3028–34. doi: 10.1073/pnas.1208208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauguet L, et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci U S A. 2014;111:966–71. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moraga-Cid G, et al. Allosteric and hyperekplexic mutant phenotypes investigated on an alpha1 glycine receptor transmembrane structure. Proc Natl Acad Sci U S A. 2015;112:2865–70. doi: 10.1073/pnas.1417864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–55. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 25.Unwin N. Refined structure of the nicotinic acetylcholine receptor. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 26.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassaine G, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–81. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- 29.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–5. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.daCosta CJ, Baenziger JE. Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure. 2013;21:1271–83. doi: 10.1016/j.str.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Cecchini M, Changeux JP. The nicotinic acetylcholine receptor and its prokaryotic homologues: Structure, conformational transitions & allosteric modulation. Neuropharmacology. 2014 doi: 10.1016/j.neuropharm.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Hille B. Ion channels of excitable membranes. Sinauer Associates, Inc; Sunderland, MA: 2001. [Google Scholar]
- 33.Kinde MN, et al. Conformational Changes Underlying Desensitization of the Pentameric Ligand-Gated Ion Channel ELIC. Structure. 2015 doi: 10.1016/j.str.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gielen M, Thomas P, Smart TG. The desensitization gate of inhibitory Cys-loop receptors. Nat Commun. 2015;6:6829. doi: 10.1038/ncomms7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akabas MH. Using molecular dynamics to elucidate the structural basis for function in pLGICs. Proc Natl Acad Sci U S A. 2013;110:16700–1. doi: 10.1073/pnas.1316157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brams M, et al. A structural and mutagenic blueprint for molecular recognition of strychnine and d-tubocurarine by different cys-loop receptors. PLoS Biol. 2011;9:e1001034. doi: 10.1371/journal.pbio.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandenberg RJ, Handford CA, Schofield PR. Distinct agonist- and antagonist-binding sites on the glycine receptor. Neuron. 1992;9:491–6. doi: 10.1016/0896-6273(92)90186-h. [DOI] [PubMed] [Google Scholar]
- 38.Marvizon JC, et al. The glycine receptor: pharmacological studies and mathematical modeling of the allosteric interaction between the glycine- and strychnine-binding sites. Mol Pharmacol. 1986;30:590–7. [PubMed] [Google Scholar]
- 39.Ruiz-Gomez A, Morato E, Garcia-Calvo M, Valdivieso F, Mayor F., Jr Localization of the strychnine binding site on the 48-kilodalton subunit of the glycine receptor. Biochemistry. 1990;29:7033–40. doi: 10.1021/bi00482a012. [DOI] [PubMed] [Google Scholar]
- 40.Rajendra S, Schofield PR. Molecular mechanisms of inherited startle syndromes. Trends Neurosci. 1995;18:80–2. [PubMed] [Google Scholar]
- 41.Rajendra S, et al. The unique extracellular disulfide loop of the glycine receptor is a principal ligand binding element. EMBO J. 1995;14:2987–98. doi: 10.1002/j.1460-2075.1995.tb07301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu R, et al. Agonist and antagonist binding in human glycine receptors. Biochemistry. 2014;53:6041–51. doi: 10.1021/bi500815f. [DOI] [PubMed] [Google Scholar]
- 43.Grudzinska J, et al. The beta subunit determines the ligand binding properties of synaptic glycine receptors. Neuron. 2005;45:727–39. doi: 10.1016/j.neuron.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 44.Mukhtasimova N, Free C, Sine SM. Initial coupling of binding to gating mediated by conserved residues in the muscle nicotinic receptor. J Gen Physiol. 2005;126:23–39. doi: 10.1085/jgp.200509283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen SB, et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–46. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purohit P, Auerbach A. Loop C and the mechanism of acetylcholine receptor-channel gating. J Gen Physiol. 2013;141:467–78. doi: 10.1085/jgp.201210946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–14. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 48.Huang S, et al. Complex between alpha-bungarotoxin and an alpha7 nicotinic receptor ligand-binding domain chimaera. Biochem J. 2013;454:303–10. doi: 10.1042/BJ20130636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Lynch JW. A comparison of glycine- and ivermectin-mediated conformational changes in the glycine receptor ligand-binding domain. Int J Biochem Cell Biol. 2012;44:335–40. doi: 10.1016/j.biocel.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Purohit P, Gupta S, Jadey S, Auerbach A. Functional anatomy of an allosteric protein. Nat Commun. 2013;4:2984. doi: 10.1038/ncomms3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 55.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–4. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 64.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–60. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 65.Swint-Kruse L, Brown CS. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics. 2005;21:3327–8. doi: 10.1093/bioinformatics/bti511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DR, Danio rerio; HS, Homo sapiens; CE, Caenorhabditis elegans; TM, Torpedo marmorata; GLOVI, Gloeobacter violaceus; ERWINIA, Erwinia chrysanthemi. Secondary structures are represented by cylinders and arrows.