Abstract
In this review we briefly describe the development of CRISPR tools for genome editing and control of transcription in bacteria. We focus on the Type II CRISPR/Cas9 system, provide specific examples for use of the system, and highlight the advantages and disadvantages of CRISPR versus other techniques. We suggest potential strategies for combining CRISPR tools with high-throughput approaches to elucidate gene function in bacteria.
Introduction
Bacteria exist in a sea of foreign DNA that is internalized via phage infection or various DNA transfer and uptake systems. About 40% of bacterial species use CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) as a genetic adaptive immune system to defend against invading DNA [1][2]. CRISPR consists of CRISPR (cr) RNAs that target specific foreign DNA sequences, primarily via RNA-DNA binding, and their associated Cas (CRISPR-associated) proteins that then cleave the targeted DNA [3][4]. The sequences of crRNAs that dictate DNA targeting by CRISPR are a direct result of acquisition of foreign DNA into the CRISPR array during phage or plasmid exposure; in this way CRISPR records previous encounters and allows specific restriction of remembered “attackers.” Furthermore, there is increasing evidence that CRISPR arrays and Cas proteins often carry additional roles in endogenous gene regulation and genome rearrangements; sometimes by distinct mechanisms than those employed in immunity (reviewed in [5], and [6]). Recent discoveries of diverse CRISPR systems that function in immunity by alternative mechanisms as well as anti-CRISPR systems deployed by bacteriophage as part of an evolutionary arms race (reviewed in [5] and [7]) suggest that we have only begun to appreciate the fascinating biology and technological potential of CRISPR.
There are three major types of CRISPRs, some of which use multiple Cas proteins to target and degrade DNA, but the Type II system was the starting point for genome engineering due to its simplicity [8][9]. Most work has focused on the Streptococcus pyogenes Type II CRISPR/Cas9 system in which the natural two RNA duplex has been further simplified to a chimeric single guide RNA (sgRNA) which targets Cas9 to specified DNA sequences. The endonucleolytic activity of Cas9 then causes a double-strand (ds) break in the target DNA [8][9]. Cas9 has HNH-and RuvC-like nuclease domains, both of which are required for cutting dsDNA [9][10], and an alpha-helical lobe that makes contacts with the sgRNA (other Cas9 structural features are reviewed in [11]). In addition to the 20 nt sequence specified in the sgRNA, Cas9 requires a short protospacer adjacent motif (PAM; NGG in S. pyogenes) to recognize DNA. Once bound to DNA, the Cas9-sgRNA complex is extremely stable; in vitro binding kinetics indicate nearly irreversible binding over long timescales [12]. The most important feature of CRISPR is its programmability: Cas9 can be directed to any PAM-adjacent sequence in the genome by modifying the basepairing region of the sgRNA. A second important feature is its versatility; whereas Cas9 is used for precise genome editing, a catalytically dead Cas9 (dCas9) unable to cleave DNA is used to precisely activate and repress gene expression [13]**[14]*. CRISPR/Cas9 is revolutionizing genome engineering, and many reviews have detailed its use in eukaryotes [15][16][17][18][19]*. Here, we briefly review CRISPR/Cas9 systems for editing and gene regulation in bacteria.
CRISPR editing
CRISPR/Cas9 genome editing, used as a tool for site-directed mutagenesis in vivo, has radically increased the ease and throughput of genetic studies in eukaryotic and especially mammalian systems, but has been applied only sporadically to bacteria so far (reviewed in [20]). CRISPR/Cas9 editing studies in γ-proteobacteria [21]**[22][23] represent improvements to existing genome editing technology, while those in less studied Firmicutes [24][25], and Actinobacteria [26] [27] mark the transfer of a highly portable technology into bacterial species that previously lacked sufficient genetic tools.
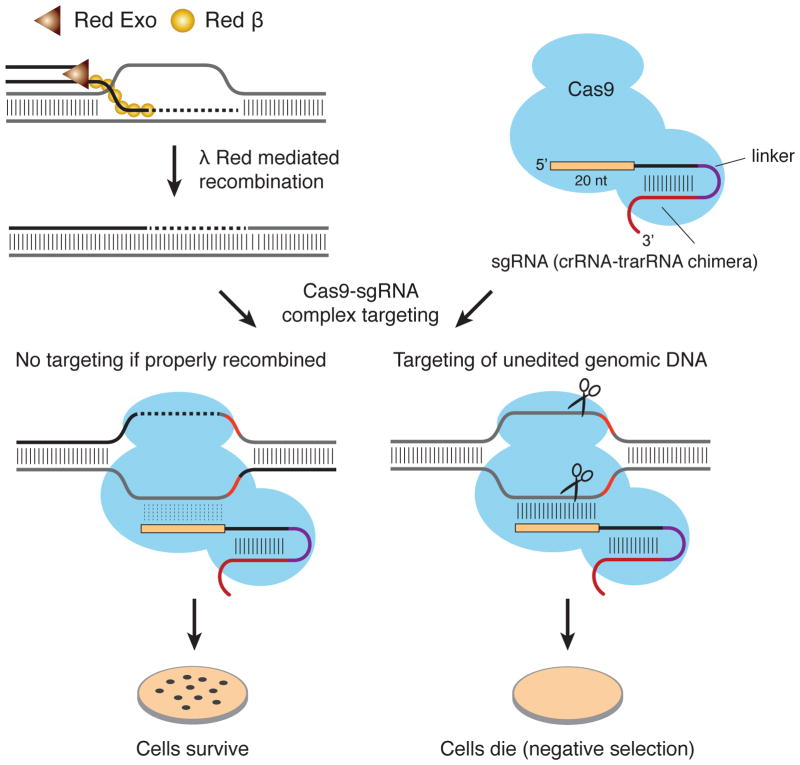
The basic protocol for editing relies on the fact that double-strand DNA breaks caused by Cas9 are fatal events in most bacterial genomes. Cas9 is targeted in a sequence-specific manner to un-edited genomes, thereby selecting for recombinants that are designed to lack the targeting or nearby PAM sequence (Figure 1). This CRISPR-based negative selection in combination with traditional recombineering is the primary methodology for Cas9 genome engineering in bacteria [20]*. In bacteria with a low intrinsic frequency of recombination, expression of recombinases—such as the lambda-red recombinase [28]—in addition to the active CRISPR editing system enhances recovery of bacteria that have undergone the desired recombination event [21]**.
Figure 1.
CRISPR genome editing in bacteria. sgRNAs are designed to target the “unedited” genome. The Cas9-sgRNA complex cuts unedited target DNA, resulting in a lethal double-strand break; this is a negative selection. DNA edited with the assistance of recombinases (e.g., λ Red in E. coli) is immune to cleavage by the Cas9-sgRNA complex that targets unedited DNA. Exo is an exonuclease that processes double stranded DNA to single stranded ends, while β drives recombination of single stranded DNA.
Thus far, bacterial CRISPR editing is plasmid-based and introduced to recipients via direct transformation (e.g., electroporation) [25][23][24], or conjugation [27][26]. Plasmid-based approaches allow high-throughput assembly of large numbers of gene-specific sgRNAs by pooled cloning. Bacteriophage may offer another route of delivering the system for editing, as they have been shown to efficiently deliver CRISPR/Cas9 as a sequence-specific antimicrobial [29].
Existing systems for expression of the active CRISPR system in recipient bacteria may require optimization. Cas9 toxicity makes it important to limit exposure of cells to the editing system by placing Cas9 under tight control of an inducible promoter and employing a curable plasmid (e.g., with a temperature-sensitive origin of replication). An inducible cas9 gene has two main benefits: it reduces the opportunity for potential off target effects and it lessens the likelihood of suppressor mutations that inactivate Cas9 or sgRNA. These suppressor mutations may result in “escaper” colonies that are initially indistinguishable from correct recombinants [21]**[20]*. Codon optimization of Cas9 may also be necessary. S. pyogenes cas9 has low GC content (35% GC), and optimizing Cas9 codons may be necessary in high GC organisms, as has been found in Streptomyces [27][26]. Problems with optimizing and regulating Cas9 expression have been circumvented in eukaryotic systems by electroporation of active Cas9-sgRNA complexes directly into eukaryotic cells [30][31]; this possibility remains unexplored in bacteria.
CRISPR interference (CRISPRi)
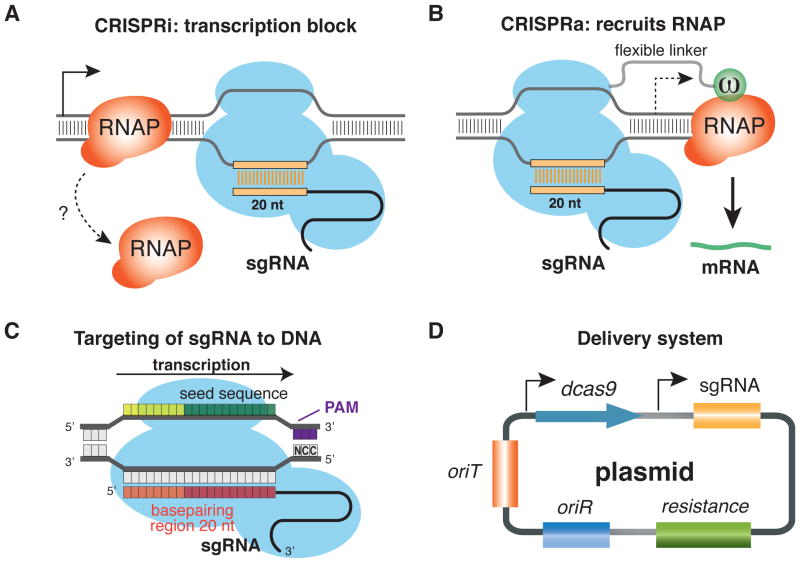
Bacterial CRISPRi has been extensively characterized in E. coli [13] **[14]*[32]. In this organism, the dCas9-sgRNA complex represses transcription either by occluding RNA polymerase (RNAP) binding to promoter DNA [13]**[14]* or by causing a steric block to transcription elongation, as demonstrated by deep sequencing of nascent transcripts (Figure 2A) [13]**[33]. An elongation block targeting the non-template strand is most effective (up to 1000-fold repression). The repression is highly specific, as determined by RNA-seq [13]**. CRISPRi has been used to predictably control genetic circuits in model bacteria [13]**, modulate essential gene expression in pathogenic bacteria [34], and alter flux through metabolic pathways [32]. Upcoming studies have established a chromosomal CRISPRi system in the model Gram-positive bacterium Bacillus subtilis (Peters JM et al., unpublished) and in the important human microbiome commensal Bacterioides thetaiotaomicron (Mimee et al., in press).
Figure 2.
Bacterial CRISPRi and CRISPRa: mechanisms, delivery, and screening. (A) Stable binding of the dCas9-sgRNA complex to DNA sterically blocks the progression of elongating RNA polymerase (RNAP), repressing transcription. (B) dCas9::ω-sgRNA bound upstream of promoter DNA recruits RNAP via direct interactions between ω and RNAP, activating transcription. (C) Design of genome-wide CRISPRi/a libraries in bacteria requires computational tools to predict sgRNA efficacy. S. pyogenes dCas9 induces the strongest transcriptional repression when the target DNA contains both the PAM (NGG recognized by dCas9) and a perfect 20-nt match between the sgRNA basepairing region and the target DNA. Mismatches in the PAM-distal region (orange) are tolerated and result in reduced efficacy of repression, but mismatches in the seed sequence (green) or the PAM (purple) severely attenuate repression. (D) Replicative plasmids containing a complete CRISPRi system (i.e., dcas9 and sgRNA), a plasmid origin of replication (oriR), and an origin of transfer (oriT) can be mated from E. coli into other bacteria.
CRISPRi has important advantages compared to traditional techniques for regulating gene expression. First, it is easy to repress new targets simply by inserting a new 20 nt base-pairing region into the sgRNA (methods for sgRNA construction are described in [35][36]). Second, CRISPRi is scalable; thousands of sgRNAs can be constructed using pooled cloning strategies with complex oligo libraries [37][38][39]*[40]. Third, CRISPRi is inducible, which enables manipulation of essential genes by partial repression [34]. Further, inducible CRISPRi libraries of non-essential genes are likely more genetically stable (i.e., less likely to accumulate suppressor mutations) during passaging and amplification than transposon or gene deletion libraries in which gene products are constitutively inactivated. Finally, CRISPRi can be multiplexed; i.e., several genes can be simultaneously repressed in the same cell using multiple sgRNAs [13]**[32]. Multiplexing is important for genetic and synthetic biology applications that require modulating the expression of many genes; this is particularly useful for bacteria with extremely long doubling times (e.g., 24 hours for Mycobacterium tuberculosis [34]), as making multiple deletion mutations by standard techniques in such strains is very time consuming. The major disadvantage of CRISPRi is that it reduces expression of downstream genes in operons (i.e., polarity) due to the dCas9 block to elongating RNAP. In some cases, we have also observed reduced expression of upstream genes in operons (i.e., “reverse polarity”); these effects may be transcript or organism-specific (Peters, JM et al., unpublished). The fact that operons often contain genes of related function somewhat mitigates this issue. As is the case with CRISPR editing, the strong selective pressure associated with CRISPRi knockdown of genes required for growth (i.e., essential genes) may result in mutations that inactivate the CRISPR system.
CRISPR activation (CRISPRa)
CRISPRa enhances transcription of target genes by using a modified dCas9-sgRNA complex containing activator domains to recruit RNA polymerase to promoter DNA. In the existing E. coli CRISPRa system, dCas9 is fused to the ω subunit of RNAP (dCas9::ω). In the presence of an appropriate sgRNA, RNAP is recruited to a position upstream of the promoter, thereby activating transcription (Figure 2B) [14]*. Initial characterization based on four promoters indicates that there may be a narrow window for efficient activation: repression occurs if dCas9::ω is too close, and activation fails when dCas9::ω is too far upstream. Activation works best for weak promoters (the weakest promoter tested was upregulated by ~28-fold [14]*). This system requires further characterization and optimization to be broadly applicable.
An optimized bacterial CRISPRa system could be used to perform genome-scale gain-of-function screens (without the need to clone thousands of genes), to mitigate polar effects of CRISPRi (by targeting the many weak promoter-like sequences in intregenic regions), or to create an “allelic series” of gene overexpression and depletion strains using CRISPRa and CRISPRi, respectively (as demonstrated in mammalian cells [39]*). The last application is a powerful way to interrogate both gene function and mechanisms of antibiotic action. To realize this potential, we may need alternative strategies for bacterial CRISPRa that are less dependent on precise spacing from the promoter. The dCas9 variants that recruit activators via long, flexible linkers, analogous to those used in mammalian CRISPRa systems [41][39]* could reduce the spacing dependency. Additionally, the published E. coli CRISPRa system requires a strain that lacks the native copy of ω [14]*, which may alter complex regulatory pathways such as the stringent response [42].
sgRNA design
One of the most attractive features of CRISPR is the ability to flexibly and precisely target Cas9 to essentially any genomic location. DNA targeting requires recognition of a short PAM by Cas9 [8][12], and base-paring of the 20-nt target sequence with the spacer region of the sgRNA (Figure 2C). Thus, sgRNA designs must meet two criteria: specific targeting to a region with a PAM (e.g., 3 nt PAM +20 nt sgRNA for S. pyogenes) and minimized off-target binding. The strongest sgRNAs (i.e., resulting in highest fold repression by dCas9) have a perfect match for a PAM-adjacent target, but imperfect matches may still bind the dCas9-sgRNA complex. A single mismatch in the 12 nucleotides at the 3′ end of the sgRNA or in the mandatory GG bases of the PAM sequence will greatly curtail the repressive effect of dCas9-sgRNA [13]**[14]*. Multiple mismatches between the sgRNA and a potential target will further reduce the repressive effect [13]**[14]*[39]*. Repression can also be tuned by targeting different positions within the gene: locating the sgRNA target in the 3′ end or template strand of a gene reduces repression [13]**[34].
Although imperfectly matched sgRNA may have lower efficacy, they may still result in off-target cutting by Cas9 or mitigate CRISPRi repression by titrating dCas9. Computational tools like Bowtie [43] can be used to compare candidate sgRNA sequences to all other PAM-adjacent off-target sites, using a weighted threshold function to discard sgRNA designs with potential off target effects [39]*. More elaborate algorithms incorporating data from assays such as ChIP-Seq have also been developed [44][45], although transient off-target association of dCas9-sgRNA with DNA detected by ChIP-seq may not be functionally relevant, especially in the case of repression.
Future Directions
There are no published high-throughput CRISPR screens in bacteria, likely because there are outstanding issues to be resolved before such screens can be performed. Eukaryotic CRISPR editing screens rely on non-homologous end joining (NHEJ) pathways to mitigate Cas9 killing by repairing double strand DNA breaks (DSBs) in DNA [16]; this mutagenic process leads to loss of function insertion/deletion mutations. Many bacteria lack a NHEJ pathway ([46]; e.g., E. coli K-12), or express it conditionally (e.g., B. subtilis [47]). The simplest bacterial NHEJ systems consist of just two proteins, Ku and LigD [48], suggesting that CRISPR editing screens may be possible by inducing NHEJ concomitant with Cas9 cutting. However, even in bacterial strains with active NHEJ (e.g., Mycobacterium smegmatis), DSBs cause significant killing [49].
Genome-scale CRISPRi screens also require optimization. Mammalian CRISPRi utilizes a chromatin silencer (KRAB) fused to dCas9 [39]*[50], whereas dCas9 alone is highly effective in bacteria [13]**. Therefore, rules for CRISPRi sgRNA function in bacteria will likely differ from those established in mammalian cells; such rules can be determined by empirically testing sgRNA efficacy in large-scale pooled experiments using deep sequencing of sgRNAs. Polarity effects may mean that CRISPRi screens report on operon rather than individual gene knockdowns, especially for organisms in which CRISPRi knockdowns cause reverse polarity.
We will also have to solve the question of CRISPRi delivery to a broad range of bacteria. Ji et al., took an initial step using a matable plasmid to transfer a CRISPRi system between two E. coli strains [51] (Figure 2E). However, it is likely that there will be no “one size fits all” CRISPRi system that functions in all bacteria; instead, a modular approach in which components (e.g., regulatable promoters) can be swapped in and out of the matable system has more versatility, and, thus, more potential to be robust to species specific issues.
Numerous powerful tools originating from bacteria and phages have transformed the landscape of biological research. The CRISPR-Cas9 system may revolutionize modern genetics in ways comparable in magnitude to the advent of recombinant DNA technologies. Bacterial CRISPR-Cas9 holds enormous promise to accelerate the pace of gene function discovery by radically increasing the scale of genetic screens and by providing novel genetic tools for previously intractable organisms.
Highlights.
Cas9 provides a powerful counter-selection tool for genome editing in bacteria.
CRISPRi is a versatile method for repressing gene expression.
With further optimization, CRISPR tools will enable genome-scale screens in microbes.
CRISPR tools present new opportunities for genetic engineering in non-tractable bacterial species
Acknowledgments
We would like to thank members of the Gross and Qi labs for critically reading the manuscript. L.S.Q. was supported by an NIH Director’s Early Independence Award (OD017887) and NIH R01 (DA036858); J.M.P. is a Ruth L. Kirchstein Fellow (F32 GM108222).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R. The roles of CRISPR–Cas systems in adaptive immunity and beyond. Curr Opin Immunol. 2015;32:36–41. doi: 10.1016/j.coi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Bondy-Denomy J, Davidson AR. To acquire or resist: the complex biological effects of CRISPR-Cas systems. Trends Microbiol. 2014;22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. The authors demonstrate the efficacy of CRISPR-based gene silencing in bacteria using only dCas9 and a chimeric sgRNA. They show that transcriptional repression by dCas9-sgRNA can result from a steric block to the progress of RNA polymerase during elongation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. The authors demonstrate CRISPR activation in bacteria by directing a dCas9::ω translational fusion upstream of weak promoters that control expression of reporter genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 16.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 18.Harrison MM, Jenkins BV, O’Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes Dev. 2014;28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. This review places CRISPR in the context of other genome engineering tools, provides an excellent description of the history of CRISPR function and mode of action and briefly discusses its applications for editing and gene regulation in eukaryotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Selle K, Barrangou R. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015;23:225–232. doi: 10.1016/j.tim.2015.01.008. An excellent, detailed review devoted entirely to bacterial genome editing using CRISPR-Cas9. [DOI] [PubMed] [Google Scholar]
- 21**.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. The first report of marker-free mutations in bacteria assisted by Cas9-sgRNA sequence-specific counter-selection in E. coli and S. pneumoniae. In combination with recombineering, the authors demonstrate that the majority of cells surviving Cas9 cutting have edited genomes, as well as simultaneous editing of multiple loci. They also establish guidelines for abolishing Cas9-sgRNA recognition in the desired editing template, including mutations in the PAM and seed sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh J-H, van Pijkeren J-P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang Z-T, Seo S-O, Choi K, Lu T, Jin Y-S, Blaschek HP. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol. 2015;200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Cobb RE, Wang Y, Zhao H. High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ACS Synth Biol. 2015;4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Zheng G, Jiang W, Hu H, Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin. 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 28.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho SW, Lee J, Carroll D, Kim J-S, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013;195:1177–1180. doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung YH, Kim JM, Kim H-T, Lee J, Jeon J, Jin Y, Choi J-H, Ban YH, Ha S-J, Kim C-H, et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24:125–131. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv L, Ren Y-L, Chen J-C, Wu Q, Chen G-Q. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: Controllable P(3HB-co-4HB) biosynthesis. Metab Eng. 2015;29:160–168. doi: 10.1016/j.ymben.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Qi LS, Arkin AP. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat Rev Microbiol. 2014;12:341–354. doi: 10.1038/nrmicro3244. [DOI] [PubMed] [Google Scholar]
- 34.Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 35.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawkins JS, Wong S, Peters JM, Almeida R, Qi LS. Targeted Transcriptional Repression in Bacteria Using CRISPR Interference (CRISPRi) Methods Mol Biol Clifton NJ. 2015;1311:349–362. doi: 10.1007/978-1-4939-2687-9_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 38.LeProust EM, Peck BJ, Spirin K, McCuen HB, Moore B, Namsaraev E, Caruthers MH. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. In a tour de force, Gilbert et al. use genome-scale CRISPRi and CRISPRa in mammalian cells to screen for genes involved in toxin resistance or sensitivity. The authors demonstrate the incredible power of genome-scale CRISPR screens, providing a framework for such studies that could be optimized for functional genomics in bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–2387. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 45.Singh R, Kuscu C, Quinlan A, Qi Y, Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 48.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 49.Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji W, Lee D, Wong E, Dadlani P, Dinh D, Huang V, Kearns K, Teng S, Chen S, Haliburton J, et al. Specific gene repression by CRISPRi system transferred through bacterial conjugation. ACS Synth Biol. 2014;3:929–931. doi: 10.1021/sb500036q. [DOI] [PMC free article] [PubMed] [Google Scholar]