Abstract
High-throughput functional genomic technologies are accelerating progress in understanding the diversity of bacterial life and in developing a systems-level understanding of model bacterial organisms. Here we highlight progress in deep-sequencing-based functional genomics, show how phenotyping based on whole genome sequencing is enabling phenotyping in organisms recalcitrant to genetic approaches, and recount the rapid proliferation of functional genomic approaches to non-growth phenotypes, and discuss how advances are enabling genome-scale resource libraries for many different bacteria.
Introduction
There is an increasingly urgent need for the broad application of high-throughput, genome-scale genetic approaches in bacteria. First, there are large gaps in our knowledge of gene function and pathway connections even in well-studied model bacterial organisms. Yet, functional annotation of these bacteria to understand gene function and pathway connections is foundational knowledge that is broadly used in understanding both prokaryotic and eukaryotic organisms. Second, the advent of rapid, inexpensive DNA sequencing has fuelled large-scale genomics and metagenomics projects, providing blueprints for a wide range of bacterial species and insights into complex communities such as the gut microbiome. This has catalyzed the study of many new organisms. Increasing emphasis on these new species, including both commensal and pathogenic microbiome organisms, environmental organisms, and a variety of organisms important for specialized studies and applications, requires us to rapidly characterize the gene functions and circuits of diverse new organisms, including those with limited or no genetic tools. Finally, unforeseen effects of cellular context often undermine the promise of a knowledge-based bio-economy based on synthetic biology, because it remains so challenging to control or predict how engineered pathways will interact with the host cell [1]. This underlines the importance of probing in vivo relationships between pathways, an important outcome of unbiased genome-scale screens.
Efforts in many laboratories, carried out in diverse organisms, are rising to the challenge of addressing this need. Brochado and Typas [2]** provided an excellent review of this area in 2013, with an emphasis on reverse genetic approaches. Recent advances are dramatically increasing the power of deep-sequencing-based functional genomic experiments. This includes the use of whole-genome sequencing for forward genetic approaches in organisms recalcitrant to genetic tools. In addition, many non-growth and other phenotypes can now be investigated at the genomic level. Finally, recent innovations are facilitating the construction of arrayed libraries for diverse species that represent important community resources. In this review, we focus on advances in these areas. For reference, key terms and concepts underlying these approaches are discussed in Box 1.
Box 1. Glossary of key terms and concepts in high-throughput genetics.
Metagenome
Genetic material from multiple organisms obtained directly from an environmental sample. Metagenomics allows for genetic characterization of non-culturable bacteria.
Library
A large (usually genome-scale) collection of strains or plasmids. Here, used primarily to describe a collection of strains with mutant alleles covering a significant fraction of the genome.
Arrayed library
A library in which individual strains are grown and stored in pure culture. Arrayed libraries are typically stored in a format that allows manipulation with an automated liquid handling system. Because strains are grown individually in a defined array, phenotypes can be measured without needing to simultaneously track genotype.
Pooled library
A library in which strains are grown and stored together in a single mixed culture.
Fitness
Fitness is a general term for phenotypes that are related to strain growth. Colony size is the fitness parameter used in arrayed approaches while digital counting is the parameter most used in pooled screens.
Multiplexing
Coverage depth of NGS can be sacrificed to sequence multiple samples simultaneously. During library preparation, short indexing barcodes present in amplification primers are added to individual samples, allowing them to be identified and parsed in the data processing step.
Next Generation Sequencing (NGS)
Refers to the current collection of sequencing technologies that provide large numbers of sequencing reads. A variety of technologies (reviewed in [80]) can be applied to the approaches described in this review.
Forward genetic screen
A library of unknown composition is screened to identify strains with a particular phenotype. The genotypes of these strains are then characterized to understand the genetic basis of the phenotype.
Reverse genetic screen
A library of known composition is assayed, and the phenotypes of each individual strain are measured.
Chemical phenotypic profiling
A library of mutants is assayed for fitness across a large set of chemical stresses. The resultant phenotypic signatures can be subjected to hierarchical clustering to group genes with similar phenotypes, suggesting shared or related functions.
Phenotypic signature
A set of quantitative phenotypes for an individual mutant strain.
Hierarchical clustering
A computational process for grouping objects based on similarity in a hierarchical manner, generally represented as a dendrogram or similar tree structure. Hierarchical clustering is used to mine profiling data for new insights by associating strains with similar phenotypic signatures.
Loss-of-function allele
A mutant allele of a gene that results in either a complete loss of gene function or reduced functionality. Common types include full gene knockouts and gene disruptions such as transposons (provided that the disruption results in loss of function). Gene knockdown (e.g., CRISPRi) can also be used to induce loss-of-function by blocking gene expression.
Gain-of-function allele
A mutant allele of a gene that results in increased expression, activity or gain of a novel function. The most common type of allele in large-scale libraries is a constitutive overexpression mutant that results in increased activity.
DNA barcode
A short DNA sequence that uniquely identifies a specific strain in a library. Barcodes can be generated randomly and then associated with a mutation via sequencing (e.g., RB-TnSeq or barcoded deletion libraries), derived from flanking genomic sequence (e.g., Tn-seq) or rationally designed (e.g., CRISPRi).
Deep sequencing
A next-generation sequencing application where particular regions are sequenced many times (high coverage). Adequate depth of sequencing is critical for robust digital counting.
Digital counting
Uses the number of sequencing reads associated with a particular mutant strain as an estimate of cell count. Digital counting can be used to quantify fitness of strains in a pooled library under various growth/stress conditions.
Type IIS restriction enzyme
A restriction endonuclease that cleaves DNA downstream of its recognition sequence. For example, MmeI cleaves 20 bp downstream from last nucleotide in its recognition site.
Synthetic lethal
A combination of two or more mutations that results in cell death whereas each of these mutation does not.
Deep-sequencing-based functional genomics
DNA sequence-based assays for determining the fitness of individual strains in a pooled library have been exceedingly valuable since their introduction two decades ago [3,4]**. Now, advances in methods for generating libraries, coupled with continued improvements in throughput and multiplexing for next generation sequencing (NGS), have increased the power and decreased the cost of these approaches. In addition to forward and reverse genetic screening to identify genes involved in particular microbial cell processes, functional genomic profiling across many conditions (chemical phenotypic profiling) using deep sequencing is now feasible [5,6]. Importantly, such studies enable hierarchical clustering of the resultant phenotypic signatures to reveal functional associations between genes as well as higher order characterization of interaction networks.
The evolution of transposon methodologies has played a pivotal role in enabling deep-sequencing-based functional genomics. Since their introduction as a genetic tool in the 1970s, transposons have been a driving force for analysis of mutant phenotypes. They were first used extensively in forward genetic selections, and later adapted for use in reverse genetic screens as well. Now, transposons are also likely to play a central role in the expanding functional genomics to new organisms, given the diversity of organisms amenable to random transposon insertion mutagenesis [3,7]. Further, developments in transposon engineering and delivery methods continue to expand both the repertoire of available applications and range of targetable organisms for transposon mutagenesis [4]**[8]. Transposon insertions generate loss-of-function (LOF) alleles via gene disruption. However, since transposons often carry outward-facing promoters to alleviate polar effects on gene expression, they can also be used to generate gain-of-function (GOF) alleles via overexpression of downstream genes in an operon [9].
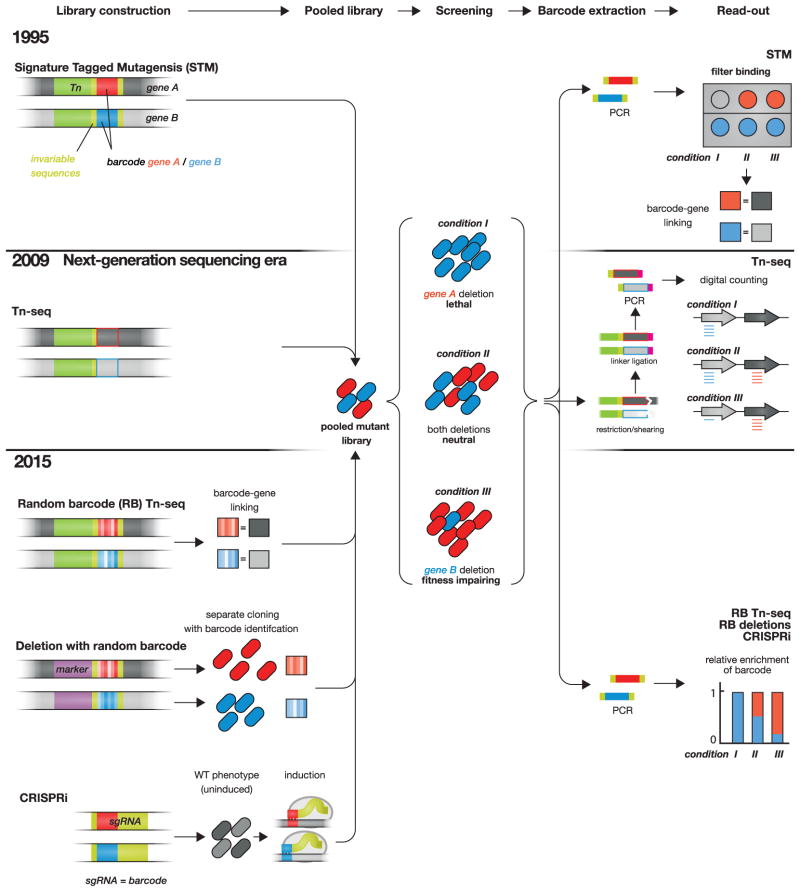
Historically, signature-tagged mutagenesis (STM) was the first transposon-based technology to systematically query gene function [10], and was especially important in identifying genes required for pathogen virulence [11]. Here, each transposon carried a DNA barcode flanked by common priming sites, serving as a unique identifier for each insertion mutant (Figure 1). Individual insertion mutants were arrayed, pooled in small groups (50–100), and used in various infection models. Virulence mutants were identified as those present in the input pool but not in the output pool, determined by hybridization to barcode arrays [12,13]. This first iteration of STM required use of an arrayed transposon library to generate corresponding barcode arrays. Later adaptations such as transposon site hybridization (TraSH) [14,15] and microarray tracking of transposon mutants (MATT) [16] enabled amplification of probes from flanking genomic DNA and hybridization to genomic oligonucleotide microarrays, obviating the need for arrayed libraries. However, both the difficulty of sample preparation and high levels of background hybridization in microarrays limited the utility of this approach.
Figure 1. Timeline and comparison of screening technologies used in functional genomics.
All technologies result in a library of pooled mutant strains (middle), which are identified by different barcodes (e.g., red and blue for gene A and gene B). Libraries can then be screened under different test conditions. Differences in technologies lie in the construction of the strains (left) and steps during analysis after screening (right). In Signature Tagged Mutagenesis [10], transposon mutants with barcodes of known sequence are pooled. After screening, barcodes are amplified using invariant priming sequences (yellow), labeled and detected via filter binding. Tn-seq and related methodologies (including INSeq, HITS, TraDIS [4]) use flanking DNA-sequences as barcodes. Sequencing library preparation requires restriction cleavage or DNA shearing, followed by linker ligation and amplification using priming sequences in the transposon (yellow) and linker (color). Relative enrichment of mutants is then determined via deep sequencing and genome alignment (middle right). Random barcode Tn-seq [36]** incorporates random unique barcodes within the transposon, which are linked with a corresponding site of transposon insertion via Tn-seq before screening. As in Signature Tagged Mutagenensis, these barcodes are flanked by invariant priming sequences for barcode amplification. The relative enrichment of mutants can then be determined by barcode amplification and sequencing. Instead of relying on random insertion of transposons into the genome, Random barcode deletion libraries consist of arrayed and pooled deletion strains, which allows more control over the composition of the initial mutant library. CRISPRi uses small guide RNAs (sgRNA) to repress a specific target gene when induced. The 20 bp variable portion of the sgRNA defines the target and also serves as a unique barcode.
The advent of next generation sequencing and its coupling to transposon mutagenesis revolutionized and revitalized transposon-based genetic approaches. Four contemporaneously published methods, reviewed extensively in [4]**, enabled NGS-based deep sequencing and digital counting of individual transposon mutants in a pooled population; in this review we refer to these methods collectively as transposon sequencing (Tn-seq). Tn-seq uses genomic DNA flanking the site of transposon insertion both to identify the site of mutation and as a barcode for quantification (Figure 1). Flanking DNA is obtained either using Type IIS restriction enzyme sites at the transposon ends, oriented such that cutting occurs about 20 bp downstream of the transposon [17,18], or DNA shearing followed by size selection [19,20]; preparation of the DNA for sequencing requires additional steps, including adapter oligonucleotide ligation and PCR amplification.
Deep sequencing provides a highly accurate method for quantifying all strains in a genome-scale pooled library, a vast improvement on the previous methodology. A typical NGS run (e.g., HiSeq 2500, single flow cell) produces up to 300 million sequence reads. Since accurate quantification requires e.g. somewhat over 100 reads/strain, fitness can be calculated at genome scale, even if some strains are underrepresented in the starting population. With this high depth of coverage, initial library sequencing can itself be highly informative, as genes that are missing (or severely underrepresented with statistical significance) in transposon libraries are candidates for essential genes. Indeed, the broad applicability of transposon mutagenesis has shed light on the essential gene sets in several bacterial species [21]. Hyper-saturated libraries can even yield information on protein domains, promoters, non-coding RNA, and intergenic regions [20,22,23].
The genome-scale evaluation of mutant fitness afforded by Tn-seq has illuminated biology in many organisms. Identifying S. aureus transposon insertion mutants sensitive to the wall teichoic acid (WTA) biosynthesis inhibitor tunicamycin revealed an interaction network connecting the WTA pathway with other cell envelope related genes [24]. Screening the S. pneumonia transposon library in multiple conditions both in vivo and in vitro resulted in a genotype-phenotype virulence map [25]. Similarly, assessing Vibrio cholerae transposon mutant fitness in a both a model host and pond water, representing different stages of its life cycle, identified genes important for host infection and dissemination [26]*. Tn-seq has also revealed unique requirements for host colonization in vivo, including biofilm development, particular metabolic requirements during nutrient restriction, and other novel factors [27–34]. Finally, Tn-seq has also been useful in a limited sense for genetic interaction analysis: Tn-seq mutagenesis of B. subtilis strain lacking 4 multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily genes identified a novel lipid II flippase gene, amj, via a synthetic lethal interaction with the known lipid II flippase gene, murJ [35].
While enormously useful for screens involving single or small sets of conditions, the multi-step process involved in Tn-seq sequencing library preparation is laborious enough to be prohibitive for larger-scale profiling across many conditions. A recent modification of Tn-seq, however, has greatly increased the scalability of the approach, and promises to overcome this limitation. Termed random bar code transposon-site sequencing (RB-Tn-seq) [36]**, this method comes full-circle to the principle of STM, incorporating random unique DNA barcodes into the transposon itself (Figure 1). Barcodes are then associated with disrupted genes using an initial round of Tn-seq. From there, mutants can be quantified simply via barcode amplification and sequencing. The authors show that as many as 48 to 96 samples can be multiplexed and sequenced in a single Illumina HiSeq lane, while still yielding adequate coverage for reproducible quantification of mutant fitness. This is enormously important for cost-effective profiling across large sets of diverse conditions, which until now has been the primary limiting factor for deep-sequencing-based functional genomics approaches. As proof-of-principle, in this study, the authors conduct fitness profiling for five organisms across a wide range of carbon sources, providing insights into metabolic gene functions and networks, including the identification of a novel mannitol utilization pathway in Phaeobacter inhibens.
Tn-seq and RB-Tn-seq are powerful methodologies for exploring gene function, but have drawbacks. Transposon library composition can be biased against low fitness mutants after outgrowth [37], there is a constant selective pressure for compromised strains to accumulate second site suppressors, and essential genes cannot be studied. Additionally, an advantage of transposon mutagenesis–dense random coverage of the genome–itself introduces certain issues; there is no simple way to target a subset of genes, and full genome coverage results in multiple insertions in most genes, decreasing the throughput of deep sequencing. Some of these problems can be alleviated by instead pooling clones from an arrayed barcoded deletion library; this approach also allows all strains to be initially present in similar amounts.
A promising new technology, CRISPRi (Clustered Regularly Interspaced Short Palindromic Repeats interference), addresses many of these issues and promises to offer a new platform for functional genomics in bacteria. CRISPRi uses computationally designed single guide RNAs (sgRNAs) to direct a catalytically inactivate variant of the Cas9 endonuclease (dCas9) to target genes, repressing transcription [38], (reviewed in this issue [Peters et al., 2015]). The system is inherently well-suited to deep sequencing applications, because the portion of the sgRNA that mediates gene targeting also serves as a unique DNA barcode. Components of the CRISPRi system can be expressed under the control of inducible promoters. Because repression is inducible and tunable, essential genes can be targeted [39], and the library can be maintained in the non-perturbed (uninduced) state, reducing pressure for accumulating suppressor mutations. Because sgRNA libraries are rationally designed, they can target subsets of genes for in-depth genetic exploration of particular pathways [40]*. Finally, CRISPRi in bacteria may require only a single sgRNA per gene for efficient knockdown leading to a null phenotype [Peters JM et al., unpublished], which would increase the throughput of sequencing based assays. Efficacy may first require optimization in other species [39], however, and has yet to be tested across diverse bacteria. A further potential weakness of CRISPRi is that the dCas9-sgRNA complex sterically blocks transcription elongation [38], causing a polar effect on downstream gene expression in an operon. Fortunately, this polar effect is quantitatively predictable for operons without internal promoters [Peters, JM, unpublished], and genes in the same operon are often functionally related, minimizing pleiotropy.
Phenotyping approaches based on whole-genome sequencing
Though Tn-seq is a powerful technology for functional genomics, some important organisms are refractory to transposon mutagenesis, and indeed to introduction of any foreign genetic elements. In these organisms, chemical or UV mutagenesis followed by selection and then whole gene sequencing (WGS) is proving an effective alternative for genetic interrogation, albeit at lower throughput than transposon analysis. Recent advances in sequencing technology and tools for data analysis allow the rapid identification of mutations with single nucleotide resolution. For example, chemical mutagenesis coupled with WGS was used to dissect genetic changes underlying the plaque phenotypes of Chlamydia trachomatis plated on a macrophage lawn [41]** and to identify novel magnetosome genes in Desulfovibrio magneticus RS-1 [42]*.
For centuries, genetic crosses were the gold standard for determining the association of phenotype with genotype. However, with the advent of WGS, Genome wide association studies (GWAS), which report on the correlation of single nucleotide polymorphisms (SNPs) to diseases states or other phenotypes, have been successfully used as a basis to narrow in on genes of interest for further functional studies [43]. In bacteria, these methodologies are now being applied to understand drug resistance mechanisms [44]* and compensatory mutations that improve the competitive fitness of drug-resistant strains [45,46]. For example, sequencing of over 100 drug-resistant Mycobacteria tuberculosis strains revealed genomic and intragenic SNPs that were highly associated with resistance [44]*. Of the top ~20 hits, 10 were already known to be associated with drug resistance, suggesting that these SNPs were high confidence leads for further study. Large-scale phenotyping of natural isolates is also being used to identify naturally-occurring determinants of fitness. For example, WGS in conjunction with phenotyping of Pseudomonas aeruginosa clinical isolates has been used to characterize evolution and diversification of the pathogen during chronic infection [47].
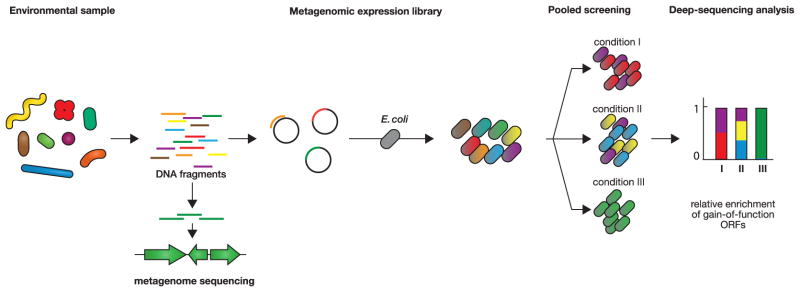
The rapid expansion of next generation sequencing has enabled culture-independent characterization of microbial communities such as the human microbiome, in particular via 16S rRNA gene deep sequencing and metagenome assembly. To improve our understanding of how microbial communities in different environments function and to identify useful proteins for biotechnology, gain-of-function approaches to discover the functions of genes from the metagenome, called functional metagenomics, have been developed [48]. Metagenomic libraries are expressed in host strains with high transformation efficiency (e.g., E. coli) and then screened for functions of interest such as antibiotic resistance, enzymatic degradation of dietary fiber, or aspects of bacteria-host interactions [49–52]. One of the major challenges in functional metagenomics is efficient expression and maintaining functionality of foreign genes in the host strain. Expression of libraries in other highly transformable bacteria, as well as shuttle vectors that allow screening of libraries in a range of transformable bacteria, are now being used to improve the recovery of gene functions [53,54]. A related approach termed “Temporal Functional Metagenomics sequencing (TFUMseq) identified genes improving fitness of a gut commensal bacterium, Bacteroides thetaiotaomicron, by transforming a B. thetaiotaomicron genome fragment library into E. coli, and determining the ability of the transformants to survive in germ-free mice by tracking the population over time using deep sequencing [55]*. Similar approaches will be useful for mining metagenomic genes that contribute to fitness of bacteria both in vitro and in vivo. Finally, microfluidic platforms have been developed for cultivating and screening rare species isolated directly from the environment [56–58]* (see also [Morten Sommer review reference] in this issue).
Phenotypes that are not measurable by growth differences
Bacteria carry out many activities that cannot be assayed via growth fitness, and genome-wide screening methods have been developed for many of them. Visible phenotypes of developmental processes such as biofilms, motility and sporulation can be directly monitored using arrayed ordered deletion libraries [[59,60], Koo BM et al., unpublished], or from pooled transposon libraries [61–63]*. Cell morphology phenotypes can also be screened with increasing throughput; for example, cell shape mutants in Helicobacter pylori were identified by sorting pooled mutants with differing cell morphology by flow cytometry followed by deep sequencing [64]*. This approach can also be used in conjunction with fluorescent dyes or reporters for phenotyping based on pathway activation, cell damage or metabolic state [65].
Assays have also been developed to identify players in phenotypes that lack obvious visual or growth-related correlates. For example, genes important for acquisition of the B. subtilis integrative and conjugative element ICEBs1 were identified by mating a donor element with a kanamycin resistance marker into an existing transposon library marked with spectinomycin resistance, followed by deep sequencing of transconjugants [66]*. Campylobacter jejuni genes required for motility and host invasion were identified by comparing strains present inside vs. outside of host cells after infection in cell culture [67]. Paradis-Bleau et al. identified mutants defective in the permeability barrier and envelope integrity using a colorimetric assay of the arrayed E. coli single gene deletion library [68]*. In this study, many of the 100 genes identified as important for envelope biogenesis were of unknown function, and had no corresponding fitness defect in the chemical genomics screen of the same library. The analytical tools developed to measure this phenotype has immediate application for ordered libraries and plate-based colorimetric assays, including Gram staining, pH indicators and biofilm indicators. Incorporation of colorimetric reporters for other phenotypes enables readout of other specific responses. High-throughput image analysis may allow for characterization of colony morphology and single cell imaging can generate multiple phenotypic indicators as shown in yeast [69]. High-throughput and high-content screening promises to increase the sensitivity and power of phenotypic analyses.
Is it feasible to have community resource libraries for many bacteria?
E. coli was the first bacterium with a dedicated single gene deletion library. Since its publication in 2006 [70], this library has had an enormous impact on research in the community. To date, Baba et al. has been cited >1400 times, and has spawned global chemical-genomic, gene-gene, and drug-drug interaction analyses providing leads for numerous mechanistic studies, most recently several related to the E. coli envelope [71–77].
There are many advantages of an arrayed and barcoded single gene deletion/antibiotic replacement library as a starting point for genome scale studies [2]**. The same library is available to all researchers, enabling cross comparison of results. Slow growing strains are retained in the library. Relevant strains are immediately available for directed follow-up experiments. Many single-cell phenotyping assays are currently feasible only with arrayed libraries. Additionally, since strains are physically isolated from one another, they cannot be complemented by other strains in the population, which is especially important when assaying secreted factors. Finally, when desirable, arrayed libraries can be used as pooled libraries, but input ratios of the initial library can be adjusted as necessary for equal representation of strains with different fitness. However, arrayed gene deletion libraries are expensive to construct, and many bacteria lack the genetic tools necessary to make construction feasible.
As an alternative, rapid methods are being developed to array transposon libraries. Pioneered in B. thetaiotaomicron [18], combinatorial pooled sequencing strategies can be used to uniquely identify individual archived clones at fractional cost, and increasingly sophisticated pooling strategies [78]*[79] have made this cost-effective even with large initial libraries. As these technologies continue to be adapted, ordered libraries–either transposon or CRISPRi-based–will be feasible in a much wider set of bacteria.
Summary and Prospects
Sequencing based approaches are being carried out to perform functional genomic analyses of genetic circuits, phenotypes, and physiological responses. This advance relies on easier methods of preparation of pooled samples and increases in the power of deep sequencing--in addition to longer reads, newer machines can achieve greater depth of reads in a shorter time. Multiplexing samples— up to ~100/sequencing lane for RB-Tn-seq [36]**, decreases cost and enables sampling multiple conditions, moving beyond the analysis of a single phenotype to query functional associations among genes and gene networks. At the same time, CRISPRi technology is providing a new, streamlined method for creating a genome-scale knockdown library based on small guide RNAs that target the dCas9 repression to each ORF. Because CRISPRi works in trans, it may be possible to deliver multiple guide RNAs to cells in a configuration where they can be detected by deep sequencing, which will enable us to develop the one method currently missing from our arsenal-- genetic interaction analysis of a pooled library at a genome or subgenome scale.
Bacterial life is incredibly diverse, having adapted to some of the harshest and most varied environments on earth. How do they thrive in so many niches? What biological functions underpin the solutions they have found, what adaptations to their profound lineage of ever-changing evolutionary pressures? If we can make sense of these blueprints in a systematic way, we stand to revolutionize our understanding of the diversity of bacterial life, at both a systems and mechanistic level. Genomic resources for high throughput phenotyping are constantly improving, and being applied to an increasingly large number of bacteria, as well as to an increasingly large number of phenotypes. For some bacteria, these efforts are at the beginning; for others, like E. coli, we are steadily moving towards unraveling the circuitry and functions that enable its success. It is exciting to be at the beginning of this vista.
Figure 2. Deep-sequencing dependent identification of protein functions in environmental samples.
Metagenomic libraries are constructed with DNA from an environmental sample (left), and introduced into a transformable model organism, here E. coli, as host (middle). Pooled screening of the library followed by deep sequencing analysis then allows identification of ORFs that confer gain-of-function, increasing viability of the host strain under the conditions tested (right).
Highlights.
High-throughput phenotyping accelerates understanding of gene function and network.
Tn-seq is enabling functional genomics in a diverse set of bacteria.
Whole genome sequencing is accelerating forward genetic screens.
New approaches are expanding the types of phenotypes assayed on a global scale.
Acknowledgments
We would like to thank members of the Gross lab and Jeffery Cox for critically reading the manuscript. C.A.G. is supported by National Institutes of Health grants (R01GM102790 and R01GM036278).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended reading
- 1.Cardinale S, Arkin AP. Contextualizing context for synthetic biology--identifying causes of failure of synthetic biological systems. Biotechnol J. 2012;7:856–866. doi: 10.1002/biot.201200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Brochado AR, Typas A. High-throughput approaches to understanding gene function and mapping network architecture in bacteria. Curr Opin Microbiol. 2013;16:199–206. doi: 10.1016/j.mib.2013.01.008. An excellent review that described different types of high-throughput reverse genomics approaches and their application to the probing gene-gene, gene-drug, and drug-drug interactions. [DOI] [PubMed] [Google Scholar]
- 3.Barquist L, Boinett CJ, Cain AK. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 2013;10:1161–1169. doi: 10.4161/rna.24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11:435–442. doi: 10.1038/nrmicro3033. An excellent review of Tn-seq and related methodologies, describing their similarities and differences and applications in investigating bacterial gene function and gentic networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutschbauer A, Price MN, Wetmore KM, Tarjan DR, Xu Z, Shao W, Leon D, Arkin AP, Skerker JM. Towards an informative mutant phenotype for every bacterial gene. J Bacteriol. 2014;196:3643–3655. doi: 10.1128/JB.01836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin T, Troy EB, Hu LT, Gao L, Norris SJ. Transposon mutagenesis as an approach to improved understanding of Borrelia pathogenesis and biology. Front Cell Infect Microbiol. 2014;4:63. doi: 10.3389/fcimb.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reznikoff WS, Winterberg KM. Transposon-based strategies for the identification of essential bacterial genes. Methods Mol Biol. 2008;416:13–26. doi: 10.1007/978-1-59745-321-9_2. [DOI] [PubMed] [Google Scholar]
- 9.Pozsgai ER, Blair KM, Kearns DB. Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Appl Environ Microbiol. 2012;78:778–785. doi: 10.1128/AEM.07098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 11.Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat Rev Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- 12.Mecsas J. Use of signature-tagged mutagenesis in pathogenesis studies. Curr Opin Microbiol. 2002;5:33–37. doi: 10.1016/s1369-5274(02)00282-5. [DOI] [PubMed] [Google Scholar]
- 13.Autret N, Charbit A. Lessons from signature-tagged mutagenesis on the infectious mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 2005;29:703–717. doi: 10.1016/j.femsre.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Badarinarayana V, Estep PW, 3rd, Shendure J, Edwards J, Tavazoie S, Lam F, Church GM. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat Biotechnol. 2001;19:1060–1065. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- 15.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42:D574–580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2015;112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santa Maria JP, Jr, Sadaka A, Moussa SH, Brown S, Zhang YJ, Rubin EJ, Gilmore MS, Walker S. Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc Natl Acad Sci U S A. 2014;111:12510–12515. doi: 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 2013;9:e1003800. doi: 10.1371/journal.ppat.1003800. A clear demonstration of the utility of Tn-seq, this study compared the growth requirements of V. cholera both during infection and in an environmental reservoir. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks JF, 2nd, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A. 2014;111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio. 2014;5:e01163–01114. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhagen LM, de Jonge MI, Burghout P, Schraa K, Spagnuolo L, Mennens S, Eleveld MJ, van der Gaast-de Jongh CE, Zomer A, Hermans PW, et al. Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLoS One. 2014;9:e89541. doi: 10.1371/journal.pone.0089541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA, Santa Maria J, Jr, Lazinski DW, Camilli A, Walker S, Hooper DC, et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. MBio. 2014;5:e01729–01714. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson JG, Livny J, Dirita VJ. High-throughput sequencing of Campylobacter jejuni insertion mutant libraries reveals mapA as a fitness factor for chicken colonization. J Bacteriol. 2014;196:1958–1967. doi: 10.1128/JB.01395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palace SG, Proulx MK, Lu S, Baker RE, Goguen JD. Genome-wide mutant fitness profiling identifies nutritional requirements for optimal growth of Yersinia pestis in deep tissue. MBio. 2014:5. doi: 10.1128/mBio.01385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio. 2015;6:e00306–00315. doi: 10.1128/mBio.00306-15. This was the first demonstration of RB-TnSeq, a technology that promises to enable scalable phenotype profiling via transposon library deep-sequencing, accelerating discovery via functional genomics in diverse bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Opijnen T, Camilli A. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol. 2010;Chapter 1(Unit1E 3) doi: 10.1002/9780471729259.mc01e03s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 40*.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. The authors demonstrated the power of genome-scale CRISPRi screens in mammalian cells, providing a framework for adaptating such studies in bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17:716–725. doi: 10.1016/j.chom.2015.03.014. The authors generated an ordered mutant library of C. trachomatis using chemical mutagenesis and whole genome sequencing. C. trachomatis is refractory to genetic manipulation, and this study provides a blueprint for functional genomics in other refractory bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Rahn-Lee L, Byrne ME, Zhang M, Le Sage D, Glenn DR, Milbourne T, Walsworth RL, Vali H, Komeili A. A genetic strategy for probing the functional diversity of magnetosome formation. PLoS Genet. 2015;11:e1004811. doi: 10.1371/journal.pgen.1004811. Working with the genetically intransigent bacterium D. magneticus, the authors used an ingenious phenotype enrichment step with chemical mutagenesis and WGS to quickly identify novel genes involved in magnetosome biogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, Liu Z, Li C, Galwey N, Deng J, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45:1255–1260. doi: 10.1038/ng.2735. Whole genome sequencing of 161 drug resistant M. tuberculosis identified a complex genetic basis for drug resistance and demonstrated the utility of WGS of natural isolates for functional genomics. [DOI] [PubMed] [Google Scholar]
- 45.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markussen T, Marvig RL, Gomez-Lozano M, Aanaes K, Burleigh AE, Hoiby N, Johansen HK, Molin S, Jelsbak L. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. MBio. 2014;5:e01592–01514. doi: 10.1128/mBio.01592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ufarte L, Potocki-Veronese G, Laville E. Discovery of new protein families and functions: new challenges in functional metagenomics for biotechnologies and microbial ecology. Front Microbiol. 2015;6:563. doi: 10.3389/fmicb.2015.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tasse L, Bercovici J, Pizzut-Serin S, Robe P, Tap J, Klopp C, Cantarel BL, Coutinho PM, Henrissat B, Leclerc M, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakhdari O, Cultrone A, Tap J, Gloux K, Bernard F, Ehrlich SD, Lefevre F, Dore J, Blottiere HM. Functional metagenomics: a high throughput screening method to decipher microbiota-driven NF-kappaB modulation in the human gut. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon SS, Kim EK, Lee WJ. Functional genomic and metagenomic approaches to understanding gut microbiota-animal mutualism. Curr Opin Microbiol. 2015;24:38–46. doi: 10.1016/j.mib.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Wexler M, Richardson DJ, Bond PL, Johnston AW. Screening a wide host-range, waste-water metagenomic library in tryptophan auxotrophs of Rhizobium leguminosarum and of Escherichia coli reveals different classes of cloned trp genes. Environ Microbiol. 2005;7:1927–1936. doi: 10.1111/j.1462-2920.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- 54.Taupp M, Mewis K, Hallam SJ. The art and design of functional metagenomic screens. Curr Opin Biotechnol. 2011;22:465–472. doi: 10.1016/j.copbio.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 55*.Yaung SJ, Deng L, Li N, Braff JL, Church GM, Bry L, Wang HH, Gerber GK. Improving microbial fitness in the mammalian gut by in vivo temporal functional metagenomics. Mol Syst Biol. 2015;11:788. doi: 10.15252/msb.20145866. This study provided a new framework for using metagenomic DNA to construct large-scale heterologous expression libraries that are tracked in vivo over time by deep sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma L, Kim J, Hatzenpichler R, Karymov MA, Hubert N, Hanan IM, Chang EB, Ismagilov RF. Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project’s Most Wanted taxa. Proc Natl Acad Sci U S A. 2014;111:9768–9773. doi: 10.1073/pnas.1404753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Najah M, Calbrix R, Mahendra-Wijaya IP, Beneyton T, Griffiths AD, Drevelle A. Droplet-based microfluidics platform for ultra-high-throughput bioprospecting of cellulolytic microorganisms. Chem Biol. 2014;21:1722–1732. doi: 10.1016/j.chembiol.2014.10.020. Using a microfluidics-based high-throughput screening system, uncultured bacteria from environment were screened directly for cellobiohydrolase activity. [DOI] [PubMed] [Google Scholar]
- 59.Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, Andrews-Polymenis HL. Identification of novel factors involved in modulating motility of Salmonella enterica serotype typhimurium. PLoS One. 2014;9:e111513. doi: 10.1371/journal.pone.0111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niba ET, Naka Y, Nagase M, Mori H, Kitakawa M. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 2007;14:237–246. doi: 10.1093/dnares/dsm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giles SK, Stroeher UH, Eijkelkamp BA, Brown MH. Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol. 2015;15:116. doi: 10.1186/s12866-015-0440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alonso AN, Perry KJ, Regeimbal JM, Regan PM, Higgins DE. Identification of Listeria monocytogenes determinants required for biofilm formation. PLoS One. 2014;9:e113696. doi: 10.1371/journal.pone.0113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. MBio. 2015;6:e02383. doi: 10.1128/mBio.02383-14. Using Tn-seq, the authors describe screening for genes required for sporulation in Clostridium difficile. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Sycuro LK, Rule CS, Petersen TW, Wyckoff TJ, Sessler T, Nagarkar DB, Khalid F, Pincus Z, Biboy J, Vollmer W, et al. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol. 2013;90:869–883. doi: 10.1111/mmi.12405. To identify genes affecting cell morphology, transposon insertion mutants were sorted by flow cytometry according to scattering properties, allowing identification of sorted mutants with altered cell shape via deep sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson CM, Grossman AD. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol. 2014;93:1284–1301. doi: 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67*.Gao B, Lara-Tejero M, Lefebre M, Goodman AL, Galan JE. Novel components of the flagellar system in epsilonproteobacteria. MBio. 2014;5:e01349–01314. doi: 10.1128/mBio.01349-14. The authors used Tn-seq to identify genes affecting conjugation in B. subtilis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Paradis-Bleau C, Kritikos G, Orlova K, Typas A, Bernhardt TG. A genome-wide screen for bacterial envelope biogenesis mutants identifies a novel factor involved in cell wall precursor metabolism. PLoS Genet. 2014;10:e1004056. doi: 10.1371/journal.pgen.1004056. The authors developed a high-throughput colorimetric assay as an alternative readout for phenotyping. An arrayed E. coli deletion library was screened for identification of mutants defective in cell envelope biogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin K, Li J, Vizeacoumar FS, Li Z, Min R, Zamparo L, Vizeacoumar FJ, Datti A, Andrews B, Boone C, et al. PhenoM: a database of morphological phenotypes caused by mutation of essential genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:D687–694. doi: 10.1093/nar/gkr827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Babu M, Diaz-Mejia JJ, Vlasblom J, Gagarinova A, Phanse S, Graham C, Yousif F, Ding H, Xiong X, Nazarians-Armavil A, et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 2011;7:e1002377. doi: 10.1371/journal.pgen.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science. 2013;340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Babu M, Arnold R, Bundalovic-Torma C, Gagarinova A, Wong KS, Kumar A, Stewart G, Samanfar B, Aoki H, Wagih O, et al. Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli. PLoS Genet. 2014;10:e1004120. doi: 10.1371/journal.pgen.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chevereau G, Bollenbach T. Systematic discovery of drug interaction mechanisms. Mol Syst Biol. 2015;11:807. doi: 10.15252/msb.20156098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gray AN, Egan AJ, Van’t Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AF, Damen MJ, Huang KC, et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife. 2015:4. doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuehl JV, Price MN, Ray J, Wetmore KM, Esquivel Z, Kazakov AE, Nguyen M, Kuehn R, Davis RW, Hazen TC, et al. Functional genomics with a comprehensive library of transposon mutants for the sulfate-reducing bacterium Desulfovibrio alaskensis G20. MBio. 2014;5:e01041–01014. doi: 10.1128/mBio.01041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79*.Vandewalle K, Festjens N, Plets E, Vuylsteke M, Saeys Y, Callewaert N. Characterization of genome-wide ordered sequence-tagged Mycobacterium mutant libraries by Cartesian Pooling-Coordinate Sequencing. Nat Commun. 2015;6:7106. doi: 10.1038/ncomms8106. The authors report a direct, cost-effective protocol to generate an ordered mutant library using transposon mutagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]