Abstract
According to a Pew Research study published in February 2015, there are 37 antibacterial programs currently in clinical trials in the United States. Protein structure-based methods for guiding small molecule design were used in at least 34 of these programs. Typically, this occurred at an early stage (drug discovery and/or lead optimization) prior to an Investigational New Drug (IND) application, although sometimes in retrospective studies to rationalize biological activity. Recognizing that structure-based methods are resource-intensive and often require specialized equipment and training, the NIAID has funded two Structural Genomics Centers to determine structures of infectious disease species proteins with the aim of supporting individual investigators’ research programs with structural biology methods.
Introduction
The primary use of protein structure for the development of drug compounds is to determine the structure of a protein in complex with a tool compound (a known ligand or lead inhibitor) for the purpose of suggesting a new chemical hypothesis in order to improve inhibitor affinity by suggesting new chemical modifications. These are usually guided by the three dimensional scaffold of the protein surrounding the ligand, including hydrogen bond donors or acceptors, hydrophobic patches, and neighboring pockets near the compound binding site. Medicinal chemists use this information to design and synthesize variants of the tool compound, which are then tested for inhibitory activity. This approach, known as Structure-Based Drug Design (SBDD), is the traditional and most well-known use of protein structure and often occurs in an iterative cycle where new molecules are synthesized, tested and crystallized with the target protein. In addition to traditional SBDD there are numerous other methods and variations that utilize protein structure in the discovery and development of new drug entities, including X-ray crystallography- and NMR-based fragment screening, and virtual (in silico) screening [1, 2]. Several previous reviews have discussed the techniques and technology of SBDD, as well as the application of SBDD methods towards the development of new drug molecules [3, 4]. Here we will summarize recent applications of structure-based methods for the development of antibacterial agents.
A recent Pew Research study [5] identified 37 antibacterial molecules currently in active clinical trials. Analysis of the PDB identified 34 of these compounds as having protein complex structural data available for the compound or a similar compound derivative. The three compounds (Brilacidin, Surotomycin, and SMT19969) without direct structural data have unknown mechanisms of action or act on the cell membrane and thus no target structure is available. Brilacidin is a defensin-mimetic that is proposed to act through depolarization of the membrane [6]. Surotomycin is a lipopeptide derivative of daptomycin that also acts through a membrane depolarization mechanism [7]. The mechanism of action of SMT19969 is unknown, but it has been suggested to inhibit DNA synthesis and is structurally similar to Hoechst dyes which bind in the minor groove of double-stranded DNA [8]. The remaining 34 compounds in clinical trials can be grouped into several broad classes with different mechanisms of action, and include fluoroquinolones, oxazolidinones, and β-lactams. Published structural data is available for some specific compounds directly, but usually structural information is available indirectly through a published protein structure bound to a close chemical derivative of the specific clinical trial compound. Understanding the true impact structural data has during the development cycle can be difficult to determine from published literature because the work is often done in commercial laboratories that don’t always publish structural coordinate files [9]. However, published retrospective studies, academic investigations and the Protein Data Bank (PDB) provide a wealth of structural information.
Bacterial protein structures in the Protein Data Bank
The Protein Data Bank (PDB) is the primary worldwide location where structural data is deposited [10], and many scientific journals require authors to submit structural coordinates as a condition of publication. In addition, US government-funded structural genomics centers are required to deposit structural coordinates regardless of publication status or intent [11]. In 2013, 10,566 structures were deposited in the PDB, while in 2014, 10,367 structures were deposited. As of the end of the third quarter of 2015, PDB depositions are on track to reach a similar 10,000 per year rate with 7381 deposited as of September 8th, 2015 (http://www.wwpdb.org/stats/deposition). Of 25,196 structural coordinates released between January 1st, 2013 and August 31st, 2015, 9387 were from a bacterial sourceiwith 3497 bacterial structures containing ligands larger than 300 Daltons, including drug-like molecules and cofactors such as ATP, NADP, etcii. A total of 884 bacterial structures can be identified by searching with the keyword “inhibitor”iii. These data suggest that about a third of all structure determination is focused on bacterial proteins, of which about 10 to 20% of bacterial structure determination is directly related to structure-based small molecule development. A few examples of how structure-based drug design has been used recently are explored in more detail below.
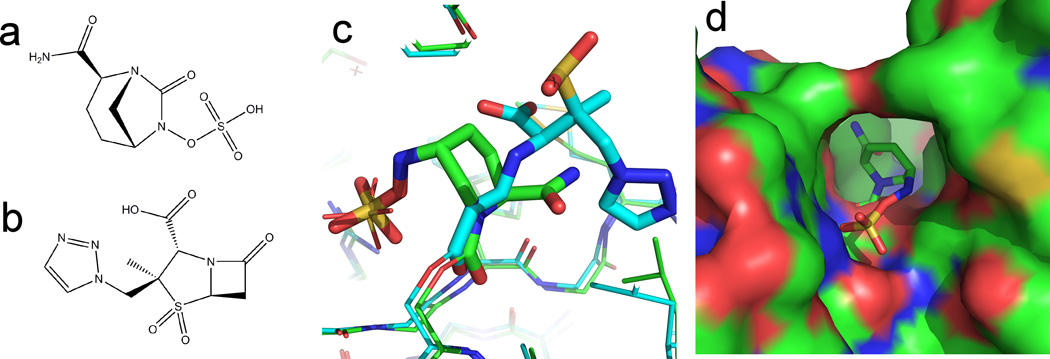
Avibactam, a new β–lactamase inhibitor
The most widely used antibiotics are β-lactam containing compounds, which inhibit bacterial cell wall synthesis and include penicillin derivatives (penams), cephalosporins (cephems), monobactams, and carbapenems. The development of antimicrobial resistance (AMR) has initiated a search for new molecules that overcome resistance. A primary mechanism of β- lactam resistance is over-expression of β-lactamases that degrade the pharmaceutical compounds. One strategy to overcome resistance caused by β-lactamases is to co-administer a β-lactamase inhibitor along with traditional β-lactam-containing antibiotic in order to prevent degradation of the drug [12]. New combinations of β-lactam/β-lactamase inhibitor are under development and six of the 37 new drugs reported by the Pew Foundation contain β-lactamase inhibitors. Prominent among these is Avibactam, a bicyclic diazobicylcooctane (Figure 1a), with a mechanism of action that was recently shown in a series of papers describing the X-ray structures of Avibactam bound to Class A [13], C [14], and D [15] β–lactamases. Avibactam binds the highly conserved active site of β–lactamases with a conformation in which the bicyclic ring mimics the β–lactam ring (see Figure 1c). In the high resolution Class C structure the sulfamite moiety of Avibactam is seen to displace a water molecule responsible for β-lactam hydrolysis. Structures in the ring-opened and ring-closed conformation of Avibactam show that the open ring maintains close positioning to the reactive center allowing re-cyclization and release of re-activated drug from the enzyme. Hydrolysis of β–lactam inhibitors are deactivated upon hydrolysis. However, re-cyclization of Avibactam releases an active drug molecule that can return and inactivate the enzyme. This mechanism accounts for the observed efficiency and long half-life of Avibactam [16, 17].
Figure 1. New β-lactamase inhibitors.
(a) Chemical diagram of Avibactam. (b) Chemical diagram of Tazobactam. (c) Overlay of the protein structure OXA-24 from Acinetobacter baumannii bound to Avibactam (PDB: 4WM9) and Tazobactam (PDB: 3ZNT). Avibactam is a non β-lactam containing compound which binds OXA- 24 in similar ring-open conformation to the β-lactam containing compound Tazobactam. Avibactam structures shown with green carbons. Tazobactam structures shown with cyan carbons. (d) Surface of OXA-24 from A. baumannii bound to Avibactam. A hydrophobic bridge in Class-D β -lactamases covers the active site thus restricting access. Surface colored by atom (blue=nitrogen, red=oxygen, green=carbon).
Avibactam has broad activity against Class A and Class C β–lactamases, as well as activity against some Class D β–lactamases. The structure of Avibactam with Oxa-24 and Oxa-48 Class D β–lactamases allowed the identification of the structural features responsible for this selectivity. A hydrophobic bridge at the entrance of the Class D enzymes was identified that restricts entry into the active site (Figure 1d). A series of structure-based sequence alignments of 310 known Class D β–lactamases found the residues that form the hydrophobic bridge can rationalize and predict the activity of Avibactam against Class D enzymes. Larger residues in this conserved region block entry into the active site acting as a thermodynamic barrier to entry and reduced inhibitory activity.
Fragment-based discovery of new gyrase inhibitors
Fragment-based drug discovery is an alternative to high throughput screening for the identification of new compounds active against a target protein. Fragment screening uses biophysical methods, such as Surface Plasmon Resonance (SPR), Nuclear Magnetic Resonance (NMR), or mass spectrometry (MS), to detect binding of small (<300 Da) compounds to a protein. Once a small molecule is identified, a 3-dimensional structure of the molecule in complex with the target protein is used to visualize the precise binding mode. The small molecules identified by these binding studies may not show inhibitory activity in enzymatic or phenotypic assays due to low affinity. The “fragment” provides a starting point for development of a new chemical series by subsequent chemical modification and expansion of the molecule to increase affinity, phenotypic activity, and drug-like characteristics.
Fluoroquinolones have been a mainstay of antibacterial treatment for over 40 years by targeting the bacterial DNA gyrase. However, the emergence of antimicrobial resistance has prompted renewed efforts to identify non-quinolone containing compounds, and 5 of the 37 compounds in current clinical trials target this enzyme. Fragment-based discovery efforts have been conducted to “scaffold-hop” away from the quinolone core or to target different parts of the enzyme, for example the ATPase domain. AstraZeneca[18] recently used structure-based development of a lead fragment with an initial IC50 of 32 µM to develop a lead compound, which has a final IC50 of 10 nm and activity in mouse models. The new compound overcomes resistance mutations in GyrA and ParC enzymes by binding in the Streptococcus pneumoniae ParE ATPase domain. Previous work at AstraZeneca also published the development of additional scaffolds through an NMR-based screen and subsequent X-ray structure of fragments bound to the Staphylococcus aureus GyrB ATPase domain[19]. Brvar [20] used computational methods to identify fragment molecules based on the structure of Escherichia coli GyrB ATPase domain in complex with the natural product clorobiocin. Structural methods were then used to elucidate the mechanism of binding and validate the hypothesized binding mode of lead compounds.
Nuclear Magnetic Resonance (NMR) spectroscopy in antibacterial drug development
The resonance frequencies of certain nuclei naturally present (1H) or easily incorporated (13C,15N) into proteins are extremely sensitive to their local inter- and intramolecular chemical environment [21]. This fundamental property is harnessed in various NMR experiments to provide detailed information on the molecular structure and interactions of proteins [22]. Consequently, NMR is extensively used to assist antimicrobial drug development in various stages of the SBDD process including hit identification, hit validation, and lead optimization [23–25]. Numerous NMR experiments, based on differences in the NMR properties between big (target) and small (fragment) molecules, have been designed to identify and validate lead compounds [24]. These experiments, some of which can be used in high-throughput mode, include WaterLOGSY [26], SLAPSTIC [27], TINS [28], transferred NOEs [29] DOSY-NMR [30] and saturation transfer difference (STD)-NMR [31, 32] For example, STD-NMR was used to identify a series of compounds with low-micromolar affinity for the macrophage infectivity potentiator BpML1 from Burkholderia pseudomallei, the etiological agent for melioidosis [33]. In terms of lead optimization, the use of NMR to determine structures for protein less than ~25 kDa in size is well established[21, 34]. However, because X-ray crystallography can often determine protein structures at higher resolution much more rapidly, NMR is typically employed to determine structures of proteins that are recalcitrant to forming well-diffracting crystals [34, 35] and is particularly well suited to studying intrinsically disordered proteins, an underexplored area of great interest for new drug design strategies [36]. For proteins under ~30 kDa with a solved structure (via XRD or NMR) and an assigned 1H-15N or 1H-13C HSQC spectrum, NMR can be used to obtain structure-activity relationships (SARs) on identified fragment hits to assist the early stages of lead optimization [37] via chemical shift perturbation experiments [38].
In eukaryotic cells, targeting the proteins involved in cell division (cytokinesis) has proven highly successful in the discovery and development of anticancer drugs such as the vinca alkaloids (vinblastine and vincristine) and taxanes (docetaxel, paclitaxel, and cabazitaxel), which act by destabilizing and stabilizing, respectively, microtubules [39] [40]. Prokaryotic cells contain a tubulin homologue, FtsZ, which is the most abundant of at least eight proteins involved in prokaryotic cell division [41] and is highly conserved in both Gram+ and Gram- bacteria [42]. Upon binding GTP it polymerizes to form a ring-like structure at the site of cell division and queues the recruitment and assembly of the cell division machinery. Consequently, agents that block Z-ring formation or protein recruitment may represent a powerful new class of antimicrobial drugs [43]. In the quest for such agents, NMR-based methods have been used at a number of stages in the SBDD process. For example, NMR methods were used to determine the solution structure for the C-terminal domain of ZipA [44], an essential component of the cell division complex that forms around FtsZ [45]. Using a library of 850 compounds and the 15N-labelled C-terminal domain of ZipA, two-dimensional 1H-15N HSQC chemical shift perturbation experiments were collected on sets of these compounds to identify seven hits for further SBDD [46]. In another example with cinnamaldehyde, a plant-based small molecule inhibitor of FtsZ, STD-NMR was used to determine the pharmacophoric groups responsible for binding to FtsZ [47]. Traditional small molecule NMR experiments were used to assist the characterization of novel natural antimicrobial products named chrysophaentins, with STD-NMR and NMR competition experiments showing these compounds bind FtsZ at the GTP-binding site [48]. A combination of STD-NMR epitope mapping and transfer NOE (trNOESY) experiments were used to deduce differences in the recognition mode of Methanococcus jannaschii and Bacilus subtilis FtsZ for C8-substituted guanine nucleotides [49], information that may facilitate the design of FtsZ inhibitors based on GTP analogs.
Structures from orthologous species can be used as surrogate for SBDD
The discovery of new drugs and novel chemical scaffolds can be assisted by structure-guided efforts; however, the corresponding structures are often not available in the PDB. If structure determination of the target of interest fails, it is possible to use the structure of the target from an orthologous species as a surrogate model. For example, a structure of Mycobacterium smegmatis served as a surrogate for the target from M. tuberculosis. Recently, Baugh et al [50] conducted an extensive comparison of structures from M. tuberculosis to other non-TB mycobacterial (NTM) species by comparing 106 pairs of Mtb and NTM structures. NTM structures with >55% sequence identity were shown to share similar active site conformation and >85% identity within the active site of the enzyme. Kling et al. [51] solved ligand-bound structures of both M. tuberculosis and M. smegmatis showing that the griselimycins bound to DnaN in identical conformation. Shirude et al. [52] utilized the structure of GyrB from M. smegmatis to validate hits from a high-throughput screen (HTS) and develop lead inhibitor molecules with activity against M. tuberculosis.
NIAID Structural Genomics Centers provide access to structural biology methods
The NIAID has funded two structural genomics centers, which offer structure determination services to the infectious disease research community and deposit all structures to the PDB. The Seattle Center for Structural Genomics (SSGCID; www.ssgcid.org) and the Center for Structural Genomics for Infectious Disease (CSGID; www.csgid.org) have together determined and deposited >1500 structures to the PDB from Category A, B, and C Biodefense organism list [11, 53]. In addition to structure determination services for researchers, the NIAID structural genomics centers determine structures of potential drug targets, essential genes, as well as proteins of unknown function to assist in greater understanding of the proteome of infectious disease organisms. Baugh et al. [54] reported the structures of 88 proteins from Burkholderia species derived from a genomic essentiality screen. Forty-nine gene families were covered by at least one structure from an initial pool off 406 putative essential gene families providing a resource for future drug development
Conclusion
Protein structure-based methods will continue to contribute to the discovery and development of anti-infective compounds and will remain an important tool in the arsenal of drug discovery scientists. Advancements in structural biology techniques have reduced the primary bottleneck of structure determination to be protein expression and solubility, rather than technical methodology. The remaining challenge of structural biology is to address the significant challenge of integral membrane proteins, which include many important drug targets.
Highlights.
New beta-lactamase inhibitor with novel mechanism of action is reviewed.
Methods of targeting bacterial gyrase compounds are discussed.
Structures of proteins from orthologous species can be used for drug design and discovery.
Applications of Nuclear Magnetic Resonance to drug discovery are discussed.
Structural genomics programs funded by NIAID provide support to independent investigators.
Acknowledgements
SSGCID is funded by Federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, under Contract No.: HHSN272201200025C from September 1, 2012. SSGCID was funded under NIAID Contract No.: HHSN272200700057C from September 28, 2007 through September 27, 2012.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DepositDateQuery: database_PDB_rev.date_original.comparator=between database_PDB_rev.date_original.min=2013-01-01 database_PDB_rev.date_original.max=2015-08-31 database_PDB_rev.mod_type.comparator=< database_PDB_rev.mod_type.value=1 and TAXONOMY is Bacteria (eubacteria)
DepositDateQuery: database_PDB_rev.date_original.comparator=between database_PDB_rev.date_original.min=2013-01-01 database_PDB_rev.date_original.max=2015-08-31 database_PDB_rev.mod_type.comparator=< database_PDB_rev.mod_type.value=1 and TAXONOMY is Bacteria (eubacteria) and Ligand Search : Has free ligands=yes
DepositDateQuery: database_PDB_rev.date_original.comparator=between database_PDB_rev.date_original.min=2013-01-01 database_PDB_rev.date_original.max=2015-08-31 database_PDB_rev.mod_type.comparator=< database_PDB_rev.mod_type.value=1 and Text Search for: inhibitor and TAXONOMY is Bacteria (eubacteria)
References
- 1.Begley DW, et al. Fragment screening of infectious disease targets in a structural genomics environment. Methods Enzymol. 2011;493:533–556. doi: 10.1016/B978-0-12-381274-2.00021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grinter SZ, Zou X. Challenges, applications, and recent advances of protein-ligand docking in structure-based drug design. Molecules. 2014;19(7):10150–10176. doi: 10.3390/molecules190710150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuenemann MA, et al. In silico design of low molecular weight protein-protein interaction inhibitors: Overall concept and recent advances. Prog Biophys Mol Biol. 2015 doi: 10.1016/j.pbiomolbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Kellici TF, Tzakos AG, Mavromoustakos T. Rational drug design and synthesis of molecules targeting the angiotensin II type 1 and type 2 receptors. Molecules. 2015;20(3):3868–3897. doi: 10.3390/molecules20033868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antibiotics Currently in Clinical Development. Washington, D. C.: Pew Research Center; 2014. [Google Scholar]
- 6.Mensa B, et al. Comparative mechanistic studies of brilacidin, daptomycin, and the antimicrobial peptide LL16. Antimicrob Agents Chemother. 2014;58(9):5136–5145. doi: 10.1128/AAC.02955-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider T, et al. Cyclic lipopeptides as antibacterial agents - potent antibiotic activity mediated by intriguing mode of actions. Int J Med Microbiol. 2014;304(1):37–43. doi: 10.1016/j.ijmm.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Weiss W, Pulse M, Vickers R. In vivo assessment of SMT19969 in a hamster model of clostridium difficile infection. Antimicrob Agents Chemother. 2014;58(10):5714–5718. doi: 10.1128/AAC.02903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seddon G, et al. Drug design for ever, from hype to hope. J Comput Aided Mol Des. 2012;26(1):137–150. doi: 10.1007/s10822-011-9519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stacy R, et al. Structural genomics of infectious disease drug targets: the SSGCID. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 9):979–984. doi: 10.1107/S1744309111029204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris PN, Tambyah PA, Paterson DL. beta-lactam and beta-lactamase inhibitor combinations in the treatment of extended-spectrum beta-lactamase producing Enterobacteriaceae: time for a reappraisal in the era of few antibiotic options? Lancet Infect Dis. 2015;15(4):475–485. doi: 10.1016/S1473-3099(14)70950-8. [DOI] [PubMed] [Google Scholar]
- 13. Lahiri SD, et al. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-lactamases. Antimicrob Agents Chemother. 2013;57(6):2496–2505. doi: 10.1128/AAC.02247-12. ** The authors describe the structures of Avibactam bound to both a Class A and Class C β-lactamase enzyme. A detailed description of the reversible mechanism of action of Avibactam is proposed which provides a rationale for the broad spectrum activity of Avibactam.
- 14. Lahiri SD, et al. Avibactam and class C beta-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother. 2014;58(10):5704–5713. doi: 10.1128/AAC.03057-14. * The authors present the structures of a class C β-lactamase bound with Avibactam in both a ring-open and ring-closed conformation.
- 15. Lahiri SD, et al. Molecular basis of selective inhibition and slow reversibility of avibactam against class D carbapenemases: a structure-guided study of OXA-24 and OXA-48. ACS Chem Biol. 2015;10(2):591–600. doi: 10.1021/cb500703p. * The structures of two Class D β-lactamases are described bound to Avibactam. The authors use the structures to rationalize the spectrum of activity of Avibactam against the broader family of Class D enzymes and provide a hypothesis for predicting activity of Avibactam based on sequence features of Class D family members.
- 16.Karpiuk I, Tyski S. Looking for the new preparations for antibacterial therapy. II. Clinical trials; new beta-lactam antibiotics and beta-lactamase inhibitors. Przegl Epidemiol. 2013;67(1):51–56. 135–140. [PubMed] [Google Scholar]
- 17.Ehmann DE, et al. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci U S A. 2012;109(29):11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Basarab GS, et al. Fragment-to-hit-to-lead discovery of a novel pyridylurea scaffold of ATP competitive dual targeting type II topoisomerase inhibiting antibacterial agents. J Med Chem. 2013;56(21):8712–8735. doi: 10.1021/jm401208b. ** The process of optimizing a fragment hit into a potent inhitior through medicinal chemistry is described. The discovery process utilizes a blend of technologies including X-ray crystallography, computational modelling and enzymatic assays.
- 19.Eakin AE, et al. Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents. Antimicrob Agents Chemother. 2012;56(3):1240–1246. doi: 10.1128/AAC.05485-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brvar M, et al. Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors. J Med Chem. 2012;55(14):6413–6426. doi: 10.1021/jm300395d. [DOI] [PubMed] [Google Scholar]
- 21.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 22.Barrett PJ, et al. The quiet renaissance of protein nuclear magnetic resonance. Biochemistry. 2013;52(8):1303–1320. doi: 10.1021/bi4000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellecchia M, Sem DS, Wuthrich K. NMR in drug discovery. Nat Rev Drug Discov. 2002;1(3):211–219. doi: 10.1038/nrd748. [DOI] [PubMed] [Google Scholar]
- 24.Pellecchia M, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat Rev Drug Discov. 2008;7(9):738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klages J, Coles M, Kessler H. NMR-based screening: a powerful tool in fragment-based drug discovery. Analyst. 2007;132(7):692–705. doi: 10.1039/b709658p. [DOI] [PubMed] [Google Scholar]
- 26.Dalvit DA, Dziarmaga J, Zurek WH. Unconditional pointer states from conditional master equations. Phys Rev Lett. 2001;86(3):373–376. doi: 10.1103/PhysRevLett.86.373. [DOI] [PubMed] [Google Scholar]
- 27.Jahnke W, Rudisser S, Zurini M. Spin label enhanced NMR screening. J Am Chem Soc. 2001;123(13):3149–3150. doi: 10.1021/ja005836g. [DOI] [PubMed] [Google Scholar]
- 28.Vanwetswinkel S, et al. TINS, target immobilized NMR screening: an efficient and sensitive method for ligand discovery. Chem Biol. 2005;12(2):207–216. doi: 10.1016/j.chembiol.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Fejzo J, et al. The SHAPES strategy: an NMR-based approach for lead generation in drug discovery. Chem Biol. 1999;6(10):755–769. doi: 10.1016/s1074-5521(00)80022-8. [DOI] [PubMed] [Google Scholar]
- 30.Hajduk PJ, Olejniczak ET, Fesik SW. One-Dimensional Relaxation- and Diffusion-Edited NMR Methods for Screening Compounds That Bind to Macromolecules. Journal of the American Chemical Society. 1997;119(50):12257–12261. [Google Scholar]
- 31.Mayer M, Meyer B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angewandte Chemie International Edition. 1999;38(12):1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 32.Wagstaff JL, Taylor SL, Howard MJ. Recent developments and applications of saturation transfer difference nuclear magnetic resonance (STD NMR) spectroscopy. Mol Biosyst. 2013;9(4):571–577. doi: 10.1039/c2mb25395j. [DOI] [PubMed] [Google Scholar]
- 33.Begley DW, et al. A structural biology approach enables the development of antimicrobials targeting bacterial immunophilins. Antimicrob Agents Chemother. 2014;58(3):1458–1467. doi: 10.1128/AAC.01875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yee A, et al. An NMR approach to structural proteomics. Proc Natl Acad Sci U S A. 2002;99(4):1825–1830. doi: 10.1073/pnas.042684599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myler PJ, et al. The Seattle Structural Genomics Center for Infectious Disease (SSGCID) Infect Disord Drug Targets. 2009;9(5):493–506. doi: 10.2174/187152609789105687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habchi J, et al. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- 37.Shuker SB, et al. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274(5292):1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 38.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41(1):1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 39.Altmann KH. Microtubule-stabilizing agents: a growing class of important anticancer drugs. Curr Opin Chem Biol. 2001;5(4):424–431. doi: 10.1016/s1367-5931(00)00225-8. [DOI] [PubMed] [Google Scholar]
- 40.Ojima I, et al. Drug discovery targeting cell division proteins, microtubules and FtsZ. Bioorg Med Chem. 2014;22(18):5060–5077. doi: 10.1016/j.bmc.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutkenhaus J, Addinall SG. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 42.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67(1):52–65. doi: 10.1128/MMBR.67.1.52-65.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Respicio L, et al. Characterizing septum inhibition in Mycobacterium tuberculosis for novel drug discovery. Tuberculosis (Edinb) 2008;88(5):420–429. doi: 10.1016/j.tube.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Moy FJ, et al. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry. 2000;39(31):9146–9156. doi: 10.1021/bi0009690. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Molecular Microbiology. 1999;31(6):1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 46.Tsao DH, et al. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorg Med Chem. 2006;14(23):7953–7961. doi: 10.1016/j.bmc.2006.07.050. [DOI] [PubMed] [Google Scholar]
- 47.Domadia P, et al. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem Pharmacol. 2007;74(6):831–840. doi: 10.1016/j.bcp.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 48.Plaza A, et al. Chrysophaentins A-H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J Am Chem Soc. 2010;132(26):9069–9077. doi: 10.1021/ja102100h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcelo F, et al. Interactions of bacterial cell division protein FtsZ with C8-substituted guanine nucleotide inhibitors. A combined NMR, biochemical and molecular modeling perspective. J Am Chem Soc. 2013;135(44):16418–16428. doi: 10.1021/ja405515r. [DOI] [PubMed] [Google Scholar]
- 50. Baugh L, et al. Increasing the structural coverage of tuberculosis drug targets. Tuberculosis (Edinb) 2015;95(2):142–148. doi: 10.1016/j.tube.2014.12.003. ** The authors make pairwise structural comparisons of 106 pairs of Mtb and non-TB mycobacterial enzymes and identify quantitative criteria for using homolog structures as surrogates for structure based drug design.
- 51.Kling A, et al. Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science. 2015;348(6239):1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- 52.Shirude PS, et al. Aminopyrazinamides: novel and specific GyrB inhibitors that kill replicating and nonreplicating Mycobacterium tuberculosis. ACS Chem Biol. 2013;8(3):519–523. doi: 10.1021/cb300510w. [DOI] [PubMed] [Google Scholar]
- 53.Stacy R, Anderson WF, Myler PJ. Structural Genomics Support for Infectious Disease Drug Design. ACS Infect Dis. 2015;1(3):127–129. doi: 10.1021/id500048p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baugh L, et al. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS One. 2013;8(1):e53851. doi: 10.1371/journal.pone.0053851. [DOI] [PMC free article] [PubMed] [Google Scholar]