Abstract
We report a simple, robust, and general strategy for protein detection based on supramolecular dissociation. The simplicity of the design is exemplified by the fact that the host assemblies can be widely varied and that these assemblies can be achieved from commercially available surfactants. An operating mechanism that is consistent with all the data has been proposed.
Development of new biosensors for recognizing biological analytes and reporting their presence is important due to the implications in proteomics, medical diagnostics, and pathogen detection. Approaches to developing new sensors can be broadly classified into two categories, both of which are inspired by nature: (i) array-based sensing, where several less-specific receptors are used to develop a response pattern for each analyte;1 and (ii) sensing based on ‘lock-and-key’ design, where specific receptors are needed to selectively bind the analytes of interest.2 The former method has the advantage of being simple as the receptor design is less intense and provides convenient opportunities to transduce the analyte recognition. In contrast, the latter approach has the promise of being specific to the target analyte even when the analyte mixture becomes complex. An approach that captures the simplicity of the former and the specificity of the latter would certainly be desirable for ultimately implementing these strategies in practical systems. Here, we disclose a simple supramolecular dissociation approach to protein sensing based on specific ligand-analyte interactions.
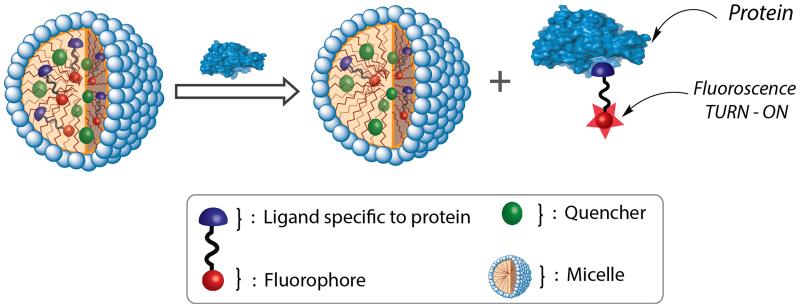
A sensor system requires two important elements, viz. specific recognition of the target analyte and generation of a signal that transduces the recognition event. Enabled by the efforts in drug discovery to impact pharma, specific ligands have been developed for many important proteins.3 However, methods that utilize these ligand discoveries to develop protein sensors are scarce, as these are hampered by strategies that transduce these binding events. Prior approaches that utilize these ligands require a supramolecular disassembly event to occur in response to a specific ligand-protein binding event.4 This strategy therefore requires a design strategy that requires the ligand-containing molecule to assemble in the absence of the protein, but disassemble in its presence. We envisaged a simple supramolecular dissociation strategy that obviates this rigorous design requirement (Figure 1). Briefly, in this approach, a probe is generated by simply tethering the protein-specific ligand moiety to a hydrophobic fluorophore, which is non-covalently incorporated into a micelle along with a corresponding hydrophobic fluorescence quencher. The micelle itself is chosen such that it is known to exhibit good guest exchange dynamics with the bulk solvent, i.e. the aqueous phase.5 Here, since the probe molecule is hydrophobic, its thermodynamic distribution coefficient will dictate that most of it is present inside the micelle, where the fluorescence is quenched due to its co-confinement with the quencher. We hypothesized that in the presence of the analyte protein, the binding event between the ligand moiety in the probe and the protein should cause the complex to decidedly favor the bulk solvent. This anticipation is because, in contrast to the probe by itself, the probe-protein complex should not have the hydrophobic driving force to re-bind to the interior of the micellar host. This specific protein-driven dissociation of the probe from the micelle also drastically decreases the proximity of the fluorophore and the quencher, which can be conveniently read as fluorescence increase, as schematically illustrated in Figure 1. The extent of this dissociation should also be dependent on the concentration of analyte protein.
Figure 1.
Schematic illustration of the supramolecular dissociation based protein sensor. The difference in the equilibrium concentrations (between the interior of the micelle and the bulk aqueous phase) of the pristine fluorescent probe and the probe-protein complex affords a simple, turn-on fluorescent sensor.
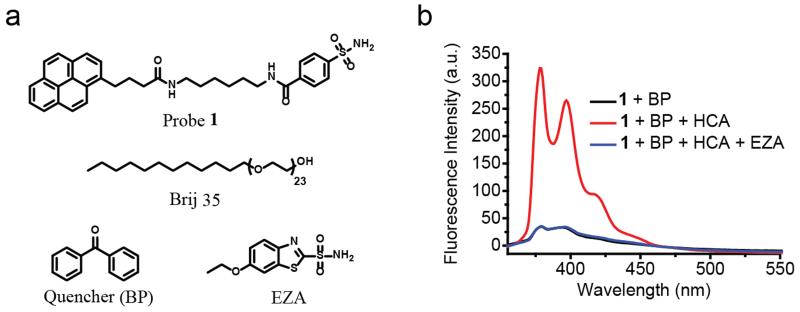
To test our design hypothesis, we chose human carbonic anhydrase I (HCA) as the target analyte because of its disease relevance.6 This protein also has several well-established ligand moieties.6,7 Brij 35, a commercially available surfactant, is used as the micelle-forming macromolecule because its non-ionic head group, composed of polyethylene glycol unit, helps reduce nonspecific interactions. Benzenesulfonamide, which is used as a specific ligand to bind HCA,6,7 is tethered to pyrene as the fluorophore using a simple hydrophobic linker to obtain the fluorescent probe 1 (Figure 2a).
Figure 2.
(a) Chemical structures of probe 1, quencher and micelle. (b) Fluorescence spectra of probe 1 (1 μM) and benzophenone (5 mM) in Brij 35 (1 mM) Tris buffer solution (25 mM, pH 7.4) treated without or with HCA (5 μM) or with HCA (5 μM) and EZA (100 μM). All spectra were taken after 3-hour HCA incubation at room temperature.
For the design to be functional, it’s pivotal that probe 1 is incorporated into the micellar assemblies. To test this, probe 1 was dissolved in a buffer solution (pH 7.4) in the presence and absence of Brij 35 in the solution. Absorption spectra of the solutions indicated that while the solution without Brij 35 has negligible absorbance at 340 nm, the solution with Brij 35 exhibits strong absorption peaks that correspond to pyrene (Figure S1a). This indicates micellar assemblies help solubilize the hydrophobic probe 1, which otherwise has limited solubility in aqueous solution. Thus, this observation also provides the support that the majority of the probe in the solution will be present inside the Brij 35 micelle, compared to the bulk solvent. To achieve a fluorescence “OFF” state, a known photoinduced electron transfer quencher (benzophenone (BP))8 was co-encapsulated in the micelle. This co-encapsulation indeed causes the pyrene fluorescence to be predominantly quenched (Figure S2a).
Next, we examined the possibility of utilizing this assembly to sense the presence of the analyte protein. Accordingly, HCA was added to the Brij 35 micellar assembly containing probe 1 and BP. Indeed, upon addition of the protein, the fluorescence of pyrene increased dramatically, as shown in Figure 2b. To check if this fluorescence enhancement is indeed due to the ligand-protein binding mechanism illustrated in Figure 1, we added a competitive ligand to HCA. The presence of the competitive ligand should displace the fluorescent probe from the protein. Since the probe is hydrophobic, it should be re-encapsulated into the micelle upon release from the protein to cause the fluorescence to be quenched again. As shown in Figure 2b, addition of 6-Ethoxy-2-benzothiazolesulfonamide (EZA), a strong competitive inhibitor of HCA,9 caused the fluorescence to go back to the original fluorescence level, i.e. significantly quenched. In addition to supporting the design hypothesis outlined in Figure 1, this observation also suggests that the micellar assembly itself is intact and that the BP quencher stays encapsulated in this assembly to cause the re-encapsulated fluorophore to be quenched.
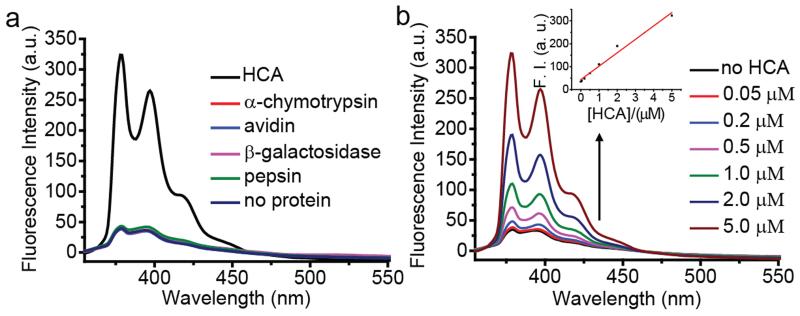
To further confirm that the fluorescence enhancement was indeed caused by the specific protein-ligand interaction, an unmodified pyrene fluorophore, which lacks the sulfonamide ligand moiety, was encapsulated along with benzophenone in Brij 35 micelle assemblies. Upon exposure to HCA, no fluorescence enhancement was observed (Figure S3). Similarly, to test whether the probe design is specific to HCA, solutions of BP quenched probe 1 in Brij 35 micelles were exposed to four different proteins with different isoelectric points (α-chymotrypsin, avidin, β-galactosidase, and pepsin) that have no known binding affinity toward benzenesulfonamide. As shown in Figure 3a, none of these proteins caused a significant change in pyrene fluorescence. In addition, we also found that the HCA-induced fluorescence enhancement occurred in a 50% fetal bovine serum solution (Figure S4). Even though the background fluorescence signal was much higher compared to that in buffer solution, the fluorescence enhancement after adding HCA was also higher in 50% serum solution. This is possibly due to the decreased stability of micelles in serum and nonspecific binding of probe to serum proteins. Further serum optimization experiments should be done in the future to minimize this background fluorescence. Nevertheless, the initial serum study suggests that this sensing strategy has the potential to be translated to a complex milieu containing many biological macromolecules.
Figure 3.
Fluorescence spectra of probe 1 (1 μM) and benzophenone (5 mM) in Brij 35 (1 mM) Tris buffer solution (25 mM, pH 7.4) treated with (a) 5 μM of proteins; (b) different concentrations of HCA. The inset shows fluorescence intensity at λem = 379 nm. All spectra were taken after 3-hour protein incubation at room temperature.
We were also interested in investigating the sensitivity of this sensing strategy. If our mechanistic hypothesis is correct, then we should have a linear increase in fluorescence with increase in concentration of the protein. This is because, the inherent preference for the probe is to be within the micellar assembly and therefore exhibit fluorescence quenching. However, the bound probes have the opposite preference, i.e. to stay in the bulk aqueous phase where there is no fluorescent quenching. Since increasing concentrations of protein should afford higher amounts of bound probes, the fluorescence increase should be linear. Indeed, we found that the fluorescence increased linearly with increasing HCA concentrations (Figure 3b). We found that a reproducible change in the fluorescence can be achieved even at 50 nM concentration of the protein and an appreciable change starts occurring in the protein above 200 nM HCA. Overall, the sensitivity of this surfactant-probe-quencher combination is in the high nM range.
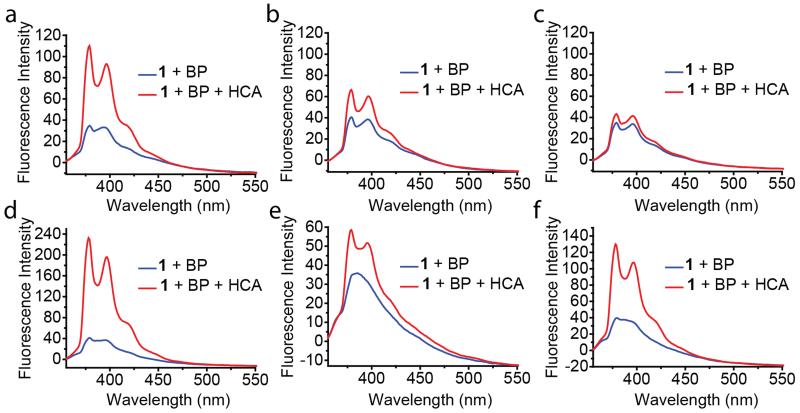
While investigating the factors that could affect the sensitivity of this method, we found that the sensitivity of the probe was affected by the concentration of the surfactant. As indicated in Figure 4a-c, fluorescence enhancement becomes less obvious as the concentration of Brij 35 increases. In these cases, the probe and quencher concentrations were kept constant. These observations seem to be consistent with our mechanistic hypothesis that the probe binds to the protein in the bulk solvent, as it has some propensity to partition to the bulk solvent. It is reasonable to suggest that the probe partitions between the micellar core and bulk solvent with the equilibrium largely favoring the micelle core. Increasing the surfactant concentration increases the effective micelle concentration, which should shift the equilibrium even further toward the micellar core. This should render the availability of the probe for protein binding even lesser and therefore the sensitivity of the probe becomes lesser.
Figure 4.
Fluorescence spectra of probe 1 (1 μM) and benzophenone (5 mM) treated without or with 1 μM HCA in different surfactant solutions: (a) 1 mM Brij 35; (b) 5 mM Brij 35; (c) 20 mM Brij 35; (d) 4 mM Triton X-100; (e) 1 mM Tween 20; (f) 1 mg/mL PEG-dodecyl random copolymer. All experiments were carried out in Tris buffer solution (25 mM, pH 7.4). All spectra were taken after 3-hour protein incubation at room temperature.
Next, we were interested in testing the broad utility of this approach. From the perspective of varying the fluorophore and the quencher, it is perhaps obvious that other combinations can be utilized for this purpose. However, we were interested in testing the versatility of this approach by investigating whether it will accommodate variations in the micellar assembly and the target protein. Instead of Brij 35, we investigated other charge neutral micelles generated from Triton X-100, Tween 20 and an amphiphilic random copolymer based on PEG-acrylate and dodecyl-acrylate. First of all, we were gratified to find that in all these cases the protein-induced fluorescence change can be observed (Figure 4d-f). Next, we also tested whether the sensitivity to the concentration of the surfactants would be different in each of these cases. All surfactants exhibited the same trend in that the sensitivity decreased with increasing surfactant concentration (Figure S6), suggesting similar operating mechanism. It is however interesting that the relative sensitivity itself was different at concentrations above their respective critical micelle concentration. Triton X-100 was found to provide the most sensitive sensing system and Tween 20 was found to be the least sensitive system. Although we do not understand the reasons for these differences at this time, this observation suggests that a systematic structure-property relationship study in the future could provide an even more sensitive system.
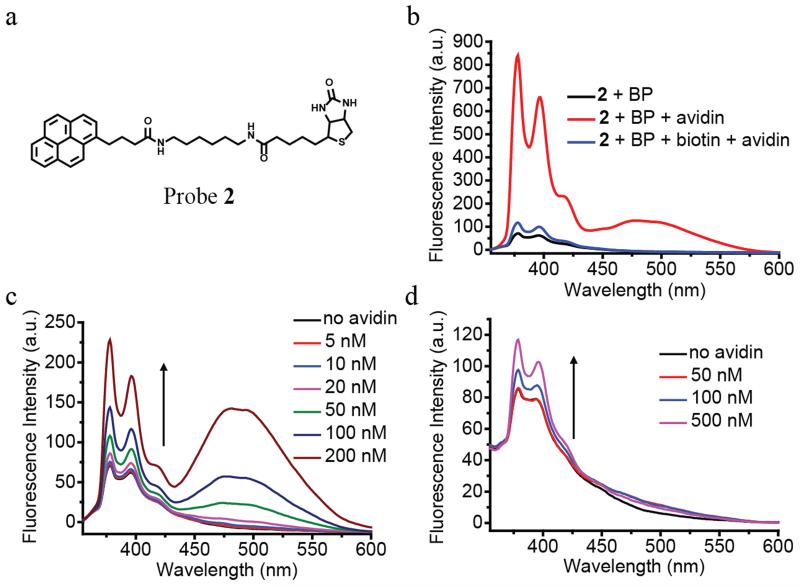
Finally, to test whether this design principle can be applied to detect other proteins, we synthesized a structurally similar probe to target avidin (probe 2, Figure 5a). Biotin, which has extraordinary binding affinity toward avidin, was linked to the pyrene fluorophore in the place of benzenesulfonamide. Similar to that observed with probe 1 and HCA, the fluorescence intensity was very weak when probe 2 and benzophenone were co-encapsulated in Brij 35. On the other hand, the fluorescence was dramatically enhanced upon the addition of avidin (Figure 5b). As biotin binds strongly and irreversibly to avidin, the displacement method using a competitive inhibitor for probe 1 would not be viable for probe 2. Therefore, to test the recognition-driven dissociation mechanism, a solution of BP-quenched probe 2 in Brij 35 micelles was pre-incubated with 20-fold excess biotin, which can compete with probe 2 to bind to the protein. As expected, upon exposure to avidin, very little fluorescence enhancement was observed for above solution compared to the one without the free biotin ligand (Figure 5b). Interestingly, in addition to the increase in the monomeric pyrene emission peak, a new peak appears around 485 nm, which correspond to the pyrene excimer emission. Since high fluorophore concentration is required for excimer formation, we attribute this to the tetravalent binding sites of avidin and possible hydrophobic association. Probe 2 is capable of detecting avidin concentration even at 5 nM concentrations (Figure 5c), likely due to the high binding affinity of biotin avidin interaction. To further demonstrate the generality of this sensing strategy, we also showed that probe 2 can also be used to detect avidin using the random copolymer micelles (Figure 5d).
Figure 5.
(a) Chemical structure of probe 2. (b) Fluorescence spectra of probe 2 (1 μM) and benzophenone (5 mM) in Brij 35 (1 mM) solution treated without or with avidin (1 μM) or with avidin (1 μM) and biotin (20 μM). Fluorescence spectra of probe 2 (1 μM) and benzophenone (5 mM) treated with different concentrations of avidin in (c) Brij 35 (1 mM) solution; (d) PEG-dodecyl random copolymer (1 mg/mL) solution. All experiments were done in Tris buffer solution (25 mM, pH 7.4). All spectra were taken after 3-hour protein incubation at room temperature.
In summary, we have developed a new supramolecular dissociation based sensing strategy to detect specific proteins with turn-on fluorescence signals. The sensing approach is based on non-covalent encapsulation of ligand-tethered fluorophore/ quencher pair in the micellar assemblies, where the fluorescence is in the ‘off’ state. Protein-binding induced dissociation of the ligand-fluorophore combination away from micelle turns the fluorescence to the ‘on’ state, since its proximity with the quencher is compromised. The versatility of this approach lies in its simplicity: (i) well-established and commercially available surfactants can be used; (ii) other than being hydrophobic, the fluorophore-ligand combination does not have to exhibit inherent self-assembly features and therefore does not require extensive molecular design; (iii) the strategy is conveniently extendable to any fluorophore-quencher combinations to modulate the colour of detection; and (iv) the approach is potentially extendable to most target protein analytes. Overall, we anticipate that the design principle has the potential to open up fundamentally new avenues in supramolecular chemistry for generation of fluorescent signal or even other spectroscopic signals in response to specific protein-ligand recognition events.
Supplementary Material
Acknowledgments
Support from Army Research Office (63889-CH) and the National Institutes of Health (GM-065255) are acknowledged.
Footnotes
Electronic Supplementary Information (ESI) available: [synthetic procedures and characterizations of the probes]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.(a) Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR. Chem. Rev. 2000;100:2595. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]; (b) Adams M, Joyce L, Anslyn EV. In: Supramolecular Chemistry: From Molecules to Nanomaterials. Gale PA, Steed JW, editors. Vol. 2. Wiley; Chichester: 2012. pp. 709–732. [Google Scholar]; (c) Gutiérrez J, Horrillo MC. Talanta. 2014;124:95. doi: 10.1016/j.talanta.2014.02.016. [DOI] [PubMed] [Google Scholar]; (d) You L, Zha D, Anslyn EV. Chem. Rev. 2015;115 doi: 10.1021/cr5005524. ASAP. [DOI] [PubMed] [Google Scholar]; (e) You C-C, Miranda OR, Gider B, Ghosh PS, Kim I-B, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat. Nanotechnol. 2007;2:318. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]; (f) Sandanaraj BS, Demont R, Thayumanavan S. J. Am. Chem. Soc. 2007;129:3506. doi: 10.1021/ja070229f. [DOI] [PubMed] [Google Scholar]; (g) Goodwin S, Gade AM, Byrom M, Herrera B, Spears C, Anslyn EV, Ellington AD. Angew. Chem. Int. Ed. 2015;54:6339. doi: 10.1002/anie.201501822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Behr J-P. The Lock-and-Key Principle, The State of the Art – 100 Years On. John Wiley & Sons; Chichester, U.K.: 2008. [Google Scholar]; (b) Ma X, Zhao Y. Chem. Rev. 2015;115 doi: 10.1021/cr500392w. ASAP. [DOI] [PubMed] [Google Scholar]; (c) Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]; (d) Oh KJ, Cash KJ, Plaxco KW. J. Am. Chem. Soc. 2006;128:14018. doi: 10.1021/ja0651310. [DOI] [PubMed] [Google Scholar]; (e) Thurley S, Röglin L, Seitz O. J. Am. Chem. Soc. 2007;129:12693. doi: 10.1021/ja075487r. [DOI] [PubMed] [Google Scholar]
- 3.(a) Stockwell BR. Nature. 2004;432:846. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kodadek T, Reddy MM, Olivos HJ, Bachhawat-Sikder K, Alluri PG. Acc. Chem. Res. 2004;37:711. doi: 10.1021/ar030145l. [DOI] [PubMed] [Google Scholar]
- 4.(a) Takaoka Y, Sakamoto T, Tsukiji S, Narazaki M, Matsuda T, Tochio H, Shirakawa M, Hamachi I. Nat. Chem. 2009;1:557. doi: 10.1038/nchem.365. [DOI] [PubMed] [Google Scholar]; (b) Yoshii T, Mizusawa K, Takaoka Y, Hamachi I. J. Am. Chem. Soc. 2014;136:16635. doi: 10.1021/ja508955y. [DOI] [PubMed] [Google Scholar]; (c) Azagarsamy M, Yesilyurt V, Thayumanavan S. J. Am. Chem. Soc. 2010;132:4550. doi: 10.1021/ja100746d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Amado Torres D, Garzoni M, Subrahmanyam A, Pavan GM, Thayumanavan S. J. Am. Chem. Soc. 2014;136:5385. doi: 10.1021/ja500634u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiwpanich S, Ryu JH, Bickerton S, Thayumanavan S. J. Am. Chem. Soc. 2010;132:10683. doi: 10.1021/ja105059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Chem. Rev. 2008;108:946. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Alterio V, Di Fiore A, D’Ambrosio K, Supuran CT, De Simone G. Chem. Rev. 2012;112:4421. doi: 10.1021/cr200176r. [DOI] [PubMed] [Google Scholar]
- 7.(a) Taylor PW, King RW, Burgen ASV. Biochemistry. 1970;9:2638. doi: 10.1021/bi00815a012. [DOI] [PubMed] [Google Scholar]; (b) Matulis D, Kranz JK, Salemme FR, Todd MJ. Biochemistry. 2005;44:5258. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 8.Bandyopadhyay P, Ghosh AK. J. Phys. Chem. B. 2010;114:11462. doi: 10.1021/jp104463u. [DOI] [PubMed] [Google Scholar]
- 9.Krishnamurthy VM, Semetey V, Bracher PJ, Shen N, Whitesides GM. J. Am. Chem. Soc. 2007;129:1312. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.