Abstract
In the past decade, the development of next-generation sequencing (NGS) has paved the way for whole-genome analysis in individuals. Research on the human leukocyte antigen (HLA), an extensively studied molecule involved in immunity, has benefitted from NGS technologies. The HLA region, a 3.6-Mb segment of the human genome at 6p21, has been associated with more than 100 different diseases, primarily autoimmune diseases. Recently, the HLA region has received much attention because severe adverse effects of various drugs are associated with particular HLA alleles. Owing to the complex nature of the HLA genes, classical direct sequencing methods cannot comprehensively elucidate the genomic makeup of HLA genes. Thus far, several high-throughput HLA-typing methods using NGS have been developed. In HLA research, NGS facilitates complete HLA sequencing and is expected to improve our understanding of the mechanisms through which HLA genes are modulated, including transcription, regulation of gene expression and epigenetics. Most importantly, NGS may also permit the analysis of HLA-omics. In this review, we summarize the impact of NGS on HLA research, with a focus on the potential for clinical applications.
Introduction
The sequencing of the entire human genome in 2007, after a 4-year process, provided important insights into our complete genomic makeup.1 Subsequently, the genomic sequences of J. Watson, African, Chinese, Korean and Japanese genomes were reported.2, 3, 4, 5, 6 Analyses of the personal genomes of individuals have provided information on human genetic variation and complexity. Additionally, rapid progress in next-generation sequencing (NGS) technology has led to revolutionary changes in medical genomics, supplying massive sequencing data for human samples. Indeed, the 1000 Genome Project has already reported novel variants, both rare and common, from population-scale sequencing.7
Various study designs have been applied to NGS, including DNA target resequencing, RNA sequencing for transcriptome analysis, chromatin immunoprecipitation sequencing, bisulfite sequencing for methylome analysis and others. The Encyclopedia of DNA Elements (ENCODE) project has examined the role of 99% of non-protein-coding DNA,8 revealing substantial interactions between proteins and DNA and the transcription of functional elements other than mRNA encoding proteins. Moreover, various types of NGS technologies have been developed, including smaller-scale benchtop and long-read NGS systems. Benchtop NGS systems, such as GS Jr, ionPGM and MiSeq, allow researchers to make fine adjustments for various smaller-scale studies. For example, some panels of focused target genes, such as genes related to cancer and inherited diseases, are now available for sequencing.
Human leukocyte antigen (HLA) genes have a long research history as important targets in biomedical science and treatment. The HLA region on chromosome 6p21 comprises six classical HLA genes and at least 132 protein-coding genes. This region has important roles in regulation of the immune system as well as fundamental molecular and cellular processes.9 The sequencing of a continuous 3.6-Mb HLA genomic region with annotation of 224 genes was reported by the MHC Sequencing Consortium in 1999.10 In addition, the MHC Haplotype Project was carried out between 2000 and 2006 by the Sanger Institute, providing genomic sequences and gene annotations of eight different HLA-homozygous haplotypes to build a framework and resource for association studies of all HLA-linked diseases; these haplotypes were registered as UCSC hg19 or NCBI GRCh37 reference assemblies.11, 12, 13 This small segment of 3.6 Mb occupies only 0.13% of the human genome but is associated with more than 100 diseases, mostly autoimmune diseases such as diabetes, rheumatoid arthritis, psoriasis and asthma. Furthermore, specific alleles of the HLA genes are strongly associated with hypersensitivities to specific drugs. For example, strong associations between carbamazepine-induced Stevens-Johnson syndrome or toxic epidermal necrolysis and HLA-B*15:02,14, 15 abacavir-induced liver injury and HLA-B*57:01,16, 17, 18, 19 and allopurinol-induced Stevens-Johnson syndrome or toxic epidermal necrolysis and HLA-B*58:0120 have been reported in various populations. For a better understanding of the disease causality and adverse effects of drugs, the haplotype structure of the HLA region should be extensively and unambiguously determined. Therefore, a specific analytical procedure should be developed for completion of HLA sequencing and haplotype determination. NGS technologies have potential advantages over the Sanger method in the sequencing of HLA genes, that is, sequences of haplotype structure can be obtained at high throughput.
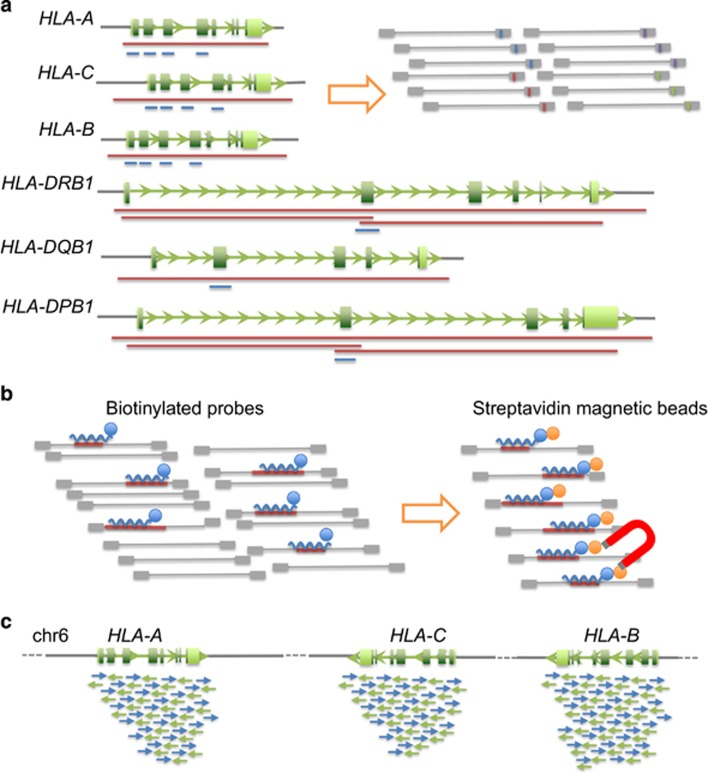
To date, several high-throughput HLA-typing methods using NGS have been developed.21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 Importantly, HLA typing using NGS provides both high-throughput and high-resolution capabilities (Figure 1). Additionally, as reported by the ENCODE Project, HLA gene sequencing alone is not sufficient for developing a complete understanding of the genetic makeup of the HLA locus. The expression levels of HLA genes can have crucial roles in the pathogenesis of diseases; thus, detection of regulatory single-nucleotide variants (SNVs) and insertions and deletions (Indels) located outside of exons is necessary. If phase-defined complete sequencing of HLA genes, including functional regulatory regions, is performed, novel alleles associated with disease risks and adverse effects of drugs could be obtained, and the expression levels of genes that affect biological processes could be clarified.
Figure 1.
HLA typing to provide sequencing data for the HLA gene(s) and regions. The HLA sequencing data of NGS could be analyzed from various points of view. The minimum scope of polymorphisms is the genotype of an SNV, and the maximum scope is the HLA haplotype sequence as a set of alleles from each HLA gene. The phase-determined sequence of the HLA allele can be applied for HLA typing as a reference. The resolution of HLA typing is classified into the following four categories: two-digit for alleles, four-digit for specific HLA proteins, six-digit for specific HLA coding sequence (CDS) and eight-digit as specific HLA genome sequences including untranslated regions and introns.
PCR-based HLA sequencing using NGS
PCR-based methods, involving an amplicon-sequence step and a sequence capture step, are commonly used for library preparation. Most of the NGS-based HLA-typing methods have been developed using such techniques. In 2009, two HLA-typing methods using a Roche GS FLX system were reported (Table 1).21, 22 The first NGS-based HLA-typing method focused only on key exons, which have commonly been analyzed using sequence-specific oligonucleotide-primed PCR (PCR-SSO) with fluorescent beads and sequencing-based typing (PCR-sequencing-based typing) using direct sequencing. Additionally, various PCR designs, such as long PCR and reverse transcription-PCR, have been applied for NGS HLA typing.21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 These PCR-based HLA-typing methods are primarily different based on primer design and the type of sequencer (Figure 2a). In particular, the long PCR method enables sequencing of the entire HLA gene, including the intron, untranslated region, and upstream and downstream regions, thus realizing high-resolution and high-throughput HLA typing. Importantly, HLA typing should be carried out by determining complete HLA gene sequences based on the physical determination of DNA sequences, but not HLA-type imputation or estimation based on the IMGT/HLA database. Indeed, the phase-defined sequencing method includes an HLA-typing method as a part of the pipeline for determination of complete HLA gene sequences.33 Moreover, some studies have shown that PCR dropout or allelic imbalance may occur during the PCR step; these issues are unpredictable and tedious to resolve. Several companies have recently released NGS HLA-typing kits based on long PCR products for library preparation; these kits include Illumina TruSight HLA, One Lambda NXType, GenDX NGS-go AmpX and Omixon Holotype HLA. Using these kits, 11 (HLA-A, -C, -B, -DRB3, -DRB4, -DRB5, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1), 8 (HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1), 5 (HLA-A, -C, -B, -DRB1 and -DQB1) and 5 (HLA-A, -C, -B, -DRB1 and -DQB1) genes have been amplified, respectively.
Table 1. PCR-based HLA typing using NGS.
| Year | Author | Journal | Locus | PCR method | Target region | Sequencer | Data analysis |
|---|---|---|---|---|---|---|---|
| 2009 | Gabriel C et al.21 | Human Immunology | HLA-A, -B | PCR for each exon | Exons 1, 2, 3 and 4 | GS FLX (Roche) | AVA (Roche) Assign SBT(Conexio Genomics) |
| 2009 | Bentley G et al.22 | Tissue Antigens | HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, DPB1 | PCR for each exon | Exons 2, 3 and 4 of A, B and C; exon 2 of DRB1, DPB1, DQA1; exons 2 and 3 of DQB1 | GS FLX (Roche) | HLA typing software (Conexio Genomics) |
| 2010 | Lind C et al.23 | Human Immunology | HLA-A, -C, -B, -DRB1, -DQB1 | Long PCR | Entire gene of A, B and C exons 2–3 of DRB1 and DQB1 | GS FLX (Roche) | Assign MPS software (Conexio Genomics) |
| 2010 | Lank SM et al.24 | Human Immunology | HLA-A, -C, -B | RT-PCR | Exons 2, 3 and 4 of A, B and C | GS FLX (Roche) | BLAT |
| 2011 | Erlich et al.25 | BMC Genomics | HLA-A, -C, -B | PCR | Exons 2, 3 and 4 of A, B and C | GS FLX (Roche) | GATK |
| 2011 | Holcomb CL et al.26 | Tissue Antigens | HLA-A, -C, -B, -DPB1, -DQA1, -DQB1, -DRB1, -DRB3/4/5 | PCR for each exon | Exons 2, 3 and 4 of A, B and C; exon 2 of DRB1, DRB3/4/5, DPB1, DQA1; exons 2 and 3 of DQB1 | GS FLX (Roche) | Assign ATF (Conexio Genomics) |
| 2012 | Wang C et al.27 | Proc Natl Acad Sci U S A. | HLA-A, -C, -B, -DRB1 | Long PCR | Exons 1 -7 of A, B and C exons 2 -5 of DRB1 | GAIIx (Illumina) HiSeq2000 (Illumina) MiSeq (Illumina) | BLASTN |
| 2012 | Shiina T et al.28 | Tissue Antigens | HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1, -DPB1 | Long PCR | Entire gene (2 amplicons for DRB1and DPB1) | GS Junior (Roche) ionPGM (Thermo) | BLAT Sequencher (GeneCodes) |
| 2012 | Lank SM et al.29 | BMC Genomics | HLA class I, all HLA class II loci | RT-PCR | Exons 1 -7 of HLA class I (two amplicons) exons 1 -4 of HLA class II | GS Junior (Roche) | BLAT |
| 2013 | Moonsamy PV et al.30 | Tissue Antigens | HLA-A, -C, -B, -DRB1, -DRB3/4/5 | PCR | Exons 2 and 3 of A, B and C exon 2 of DRB1, DRB3/4/5, DQB1 | GS FLX (Roche) | Assign ATF 454 (Conexio Genomics) |
| 2013 | Ringquist S et al.31 | PLoS One | HLA-DRB1 | PCR | Exon 2 | GS FLX (Roche) | CAPSeq (Original) |
| 2013 | Danzer M et al.32 | BMC Genomics | HLA-A, -C, -B, -DRB1, -DRB3/4/5,-DQB1, -DPB1 | PCR for each exon | Exons 2, 3 and 4 of A exons 1, 2, 3 and 4 of B; exons 1, 2, 3, 4 and 7 of C; exon 2 and 3 of DRB1, DRB3/4/5, DQB1; exon 2 of DPB1 | GS Junior (Roche) | Assign ATF (Conexio Genomics) |
| 2013 | Hosomichi K et al.33 | BMC Genomics | HLA-A, -C, -B, -DRB1,-DQB, -DPB1 | Long PCR | Entire gene | MiSeq (Illumina) | Phase-defined sequencing (Original) |
| 2013 | Trachtenberg EA et al.34 | Methods Mol Biol | HLA-A, -C, -B, -DRB1, -DRB3/4/5,- DQA1, -DQB1, -DPB1 | PCR for each exon | Exons 2, 3 and 4 of A, B and C; exon 2 of DRB1, DRB3/4/5, DPB1, DQA1; exons 2 and 3 of DQB1 | GS FLX (Roche) | Assign ATF (Conexio Genomics) |
| 2014 | Ozaki Y et al.35 | Tissue Antigens | HLA- DRB1, -DRB3/4/5 | Long PCR | Exons 2–6 of DRB1, DRB3/4/5 | GS Junior (Roche) | BLAT Sequencher (GeneCodes) |
| 2014 | Hajeer AH et al.36 | Tissue Antigens | HLA-A, -C, -B, -DRB1,-DQB1 | PCR | Exons 2 and 3 of A, B and C exon 2 of DRB1 exons 2 and 3 of DQB1 | GS FLX (Roche) GS Junior (Roche) | Assign ATF 454 (Conexio Genomics) |
| 2014 | Hosomichi K et al.37 | BMC Genomics | HLA-B | Long PCR | entire gene | MiSeq (Illumina) | Phase-defined sequencing (Original) |
| 2014 | Smith AG et al.38 | Human Immunology | HLA-DRB1, -DRB3/4/5, -DQA1, -DQB1, -DPB, -DPA1 | PCR for each exon | Exons 2 and 3 of DQA1, DQB1, DRB1, DRB3/4/5 exon 2 of DPA1 and DPB1 | MiSeq (Illumina) | GeMS (Scisco Genetics) |
| 2014 | Ehrenberg PK et al.39 | BMC Genomics | HLA-A, -C, -B, -DRB1 | Long PCR | Entire gene | MiSeq (Illumina) | Omixon target (Omixon) |
| 2014 | Zhou M et al.40 | Tissue Antigens | HLA-A, -C, -B, -DRB1, -DQB1 | PCR | Exons 1–7 of A, B and C (4 amplicons); exon 2 of DRB1; exons 2 and 3 of DQB1 | HiSeq2000 (Illumina) | BGI computing procedure (Original) |
| 2015 | Lan JH et al.41 | Human Immunology | HLA-A, -C, -B, - DRB1, -DQB1 | Long PCR | Entire gene (2 amplicons for DRB1) | MiSeq (Illumina) | NGSengine (Gen Dx) |
| 2015 | Ozaki Y et al.42 | BMC Genomics | HLA-A, -C, -B, -DRB1, -DRB3/4/5,- DQA1, -DQB1, -DPB1 | Long PCR | Entire genes of A, B and C exons 2–4 of DRB1, DRB3/4/5, DQA1, DQB1 exons 2–6 of DPB1 | ionPGM (Thermo) | SeaBass (Original, In-house) |
Figure 2.
Preparation of HLA gene fragments for the DNA library. DNA fragments of the HLA genes are prepared by PCR-based (a) or hybridization-based (b) methods. (a) Many publications describing PCR-based methods have used different PCR designs such as short PCR for target exons (blue bar) or long PCR for entire genes (red bar). After PCR amplification, each of the pooled PCR products is applied for library preparation with/without fragmentation to add adapters with/without indexes for each sequencer. In the PCR-based method, the first step is PCR for HLA genes and the second step is library preparation. (b) The sequence capture method based on hybridization is also commonly used to enrich HLA gene fragments. DNA/RNA probes with the HLA gene sequence are hybridized to the DNA library, which includes the HLA gene sequence. The biotinylated probes-bound DNA libraries are collected using a magnet and streptavidin magnetic beads. In the sequence capture method, the first step is library preparation and the second step is enrichment for HLA genes. (c) After sequencing, HLA gene sequences of each individual are reconstructed by alignment to reference HLA gene sequences. The consensus sequences constructed by the aligned reads are searched for specific HLA alleles in the IMGT/HLA database. In the NGS-based HLA-typing method, the basic data analysis approach is similar between PCR-based and sequence capture methods.
The capture method for HLA sequencing
Target resequencing of the HLA genes using the sequence capture method has not been well developed compared with PCR-based HLA typing. The sequence capture method is based on hybridization between DNA of an adapter-ligated library and a biotinylated DNA/RNA probe designed based on target sequences of genes or the genomic region (Figure 2b).43 Hybridized DNA fragments are enriched for the target region using streptavidin magnetic beads. Wittig and colleagues44 reported the first automated HLA-typing method based on the sequence capture technology. This method uses targeted capturing of the classical class I (HLA-A, HLA-C and HLA-B) and class II (HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1) HLA genes. The DNA fragments from these eight HLA genes can be simultaneously enriched by a hybridization reaction in a single tube, without allele dropout, which is frequently observed in PCR-based methods. The results showed high accuracy for allele call (99%) and identified errors in the IMGT/HLA reference database. It is also notable that the sequence capture method is generally applicable for NGS-based target resequencing of larger genomic regions and a larger number of genes than the PCR-based methods. On the basis of these features and the findings from the automated NGS-typing method, the sequence capture method has major advantages over PCR-based methods and is a promising method for HLA sequencing.
Sequencers for HLA gene sequencing
Various sequencing machines have been developed for NGS HLA typing. The majority of published methods have been established using Roche sequencers. However, for the last few years, the Illumina MiSeq instrument has also been used for HLA typing (Table 1). The types of NGSs used for HLA typing may often change along with improvements in NGS technologies. The Pacific Biosciences PacBio RS II sequencer, which is capable of generating enormously long reads in a single molecule using real-time sequencing, was recently developed for HLA typing. Single molecule using real-time sequencing is highly effective in generating accurate, phased sequences of full-length alleles of HLA genes. Complete phasing of the HLA genes from single molecule using real-time sequencing may resolve phase ambiguity, which is a fundamental problem of conventional HLA-typing methods.
HLA typing from WGS and WES data
During the past few years, whole exome sequencing (WES) has identified the causalities of a large number of Mendelian diseases by analyzing familial samples and/or sporadic patients.45 In addition, WES has facilitated the acquisition of massive amounts of data in various genome sequencing projects, such as the 1000 Genomes Project,7 NHLBI GO Exome Sequencing Project (https://esp.gs.washington.edu/drupal/) and UK10K project (http://www.uk10k.org), which are expected to improve our understanding of variations in the human genome. The sequence capture method has also been applied for WES using various kits, such as the Agilent Human All Exon kit, Roche SeqCap EZ Human Exome kit and Illumina TruSeq Exome Enrichment kit. The respective libraries of capture oligo-probes, which cover all human exons, are designed to target all exons of all HLA genes. For example, 820 exons of 182 genes in the HLA region are found in the Agilent Human All Exon kit design. Because DNA sequence reads for HLA genes are included in the whole genome sequencing (WGS) and WES datasets, HLA typing could be carried out using both the datasets. Within WGS and WES datasets, HLA gene sequences represent only a small portion of the data, but these sequences are phased HLA gene sequences as HLA alleles. Therefore, HLA typing from WGS or WES datasets should be the key analysis method used to promote higher accuracy rates compared with those of the existing PCR-SSO or PCR-sequencing-based typing results.
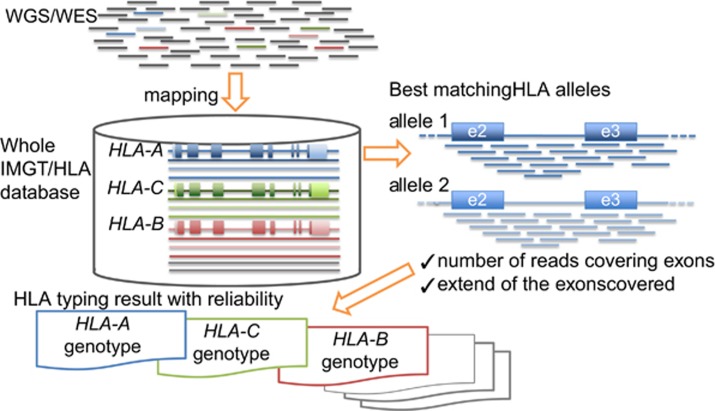
NGS HLA-typing software
As described in Table 2, various HLA-typing software programs, including the aforementioned Omixon Target HLA and HLAminer, as well as academic software and commercial software packages, have been developed for HLA typing from various types of data, including WGS, WES, RNA sequencing and amplicon datasets.46, 47, 48, 49, 50, 51, 52, 53, 54 An example of HLA-typing software for WGS or WES, including a brief overview of the Omixon Target HLA typing system, is described in Figure 3. For statistical methods, sequence reads are first aligned to the whole IMGT/HLA database (all known HLA alleles). Then, the best matching alleles are selected based on various alignment statistics, such as the number of reads covering exons and the extent of exons covered. During statistical analysis, only reads that are mappable as homologous to any allele in the IMGT/HLA database with a low number of mismatches should be stored. In the Omixon publication, which used data from the 1000 Genomes Project, the concordance rate between the NGS-based method and PCR-SSO was around 90%, which was not considered high.54 For the analysis, sequence reads from all exons of HLA genes were applied. At least 10 reads are required to counterbalance random noise. However, the sequence reads were not evenly distributed for each gene region, and the average depth implied that there may be holes in coverage. Another publication in which the authors utilized the HLAminer software also mentioned 92.8% concordance rate between these two methods for allele group prediction.46 NGS HLA typing can call for all the HLA genes recorded in the IMGT/HLA database and for novel HLA alleles. On the other hand, it is not currently possible to detect rare HLA alleles by PCR-SSO. Therefore, if sequence reads of rare HLA alleles are in WGS or WES, the HLA-typing results from NGS would be expected to be discordant with those from PCR-SSO. In the near future, the reliability of these HLA-typing methods from WGS and WES data may be improved. These programs are the next step in developing methods with greater specificity and sensitivity of HLA-typing results. In particular, the specificity is dependent on the HLA-typing resolution, for example, two-, four-, six- and eight-digit, each of which is based on the composition of the allele group, the specific allele protein, the specific DNA sequence with synonymous substitutions in the coding region and the specific DNA sequence of the entire gene, respectively. The high-resolution HLA typing of NGS is advantageous compared with the existing PCR-SSO and PCR-sequencing-based typing methods. In practice, it is not possible to execute complete eight-digit HLA typing because of limitations in the number of known HLA allelic sequences deposited in the IMGT/HLA database, where most HLA allelic sequences have been recorded as coding sequences or partial exons. Only HLA alleles recorded as full-length HLA gene sequences can be used for allele-call with eight-digit resolution. To put eight-digit resolution typing into practice, the NGS-based phase-defined complete sequencing methods for the HLA genes will be applicable as a high-resolution tool for the detection of novel alleles, and will facilitate the development of expanded databases with full-length HLA allelic sequences for eight-digit HLA typing.33 The success of complete HLA gene sequencing with high accuracy should be determined based on high sequence read depth. In the case of HLA-B sequencing, the minimum depth for complete phasing was approximately 800 folds the average depth.37
Table 2. HLA-typing software and category of acceptable reads.
Figure 3.
Overview of data analysis for HLA typing using WGS/WES. Massive sequence reads from WGS/WES are aligned to the whole IMGT/HLA database (all known HLA alleles) to search for best matching alleles based on alignment statistics, number of reads covering exons and the extent of exon coverage. The HLA allele can be identified by only storing reads that are mappable as homologous to any allele in the IMGT/HLA database with a low number of mismatches by statistical analysis.
HLA in the ENCODE project data
In HLA research, NGS technologies have influenced HLA typing as well as our understanding of the functional regulatory regions within the HLA region, which could affect the expression of HLA genes. Thus far, HLA-associated diseases have been understood on the hypothesis that antigen presentation of HLA molecule affects the immune system. Four-digit HLA typing is sufficient to explain the importance of antigen presentation in disease causality. On the other hand, the HLA-associated phenotypes could also be affected by the expression levels of HLA genes or by allelic imbalance. In 2012, the ENCODE project succeeded in the systematic arrangement of transcript regions and transcription factor (TF)-binding sites in the genome, and showed the genomic patterns of chromatin structure and histone modifications.8 The achievements of the project also include the discovery of putative functional elements and domains within the HLA region. The knowledge obtained in the ENCODE project could be extended to understand HLA-associated diseases and phenotypes. Certain diseases are associated with specific HLA alleles, and many variants within the HLA genes are also associated with HLA alleles in linkage disequilibrium; therefore, it is quite difficult to genetically determine which variant is associated with the disease because the disease is associated with a haplotype carrying the HLA alleles and many variants. Furthermore, there are limitations to genetic analyses with limited numbers of samples and minor genetic effects; however, the ENCODE project highlights the functional regions of the entire human genome including the HLA region.
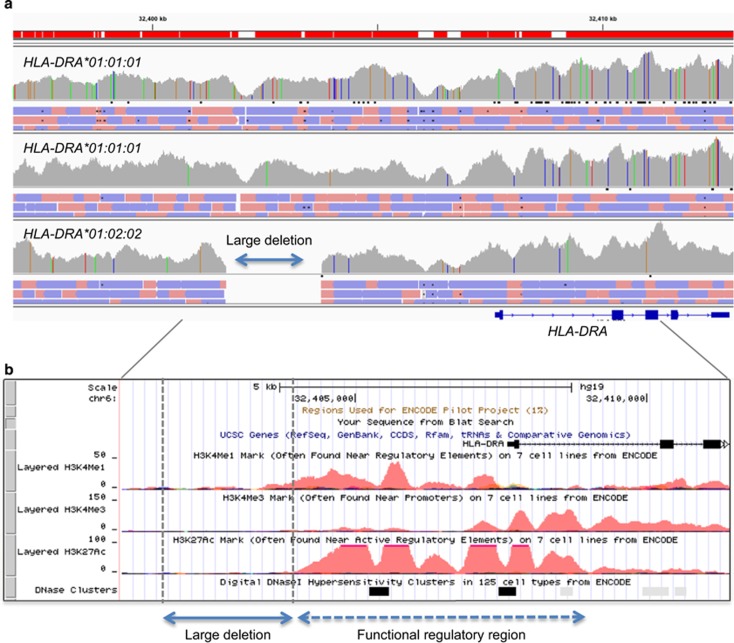
Two examples of HLA genes and the associated diseases are described in Figure 4, Table 3 and Table 4. HLA-DRA is less polymorphic than HLA-A, -C, -B or -DRB1. However, many variants have been observed in the upstream region among HLA-DRA alleles, particularly between the same six-digit HLA-DRA alleles, HLA-DRA*01:01:01 (Figure 4, unpublished data). A deletion of about 2 kb was also detected in the upstream region of HLA-DRA*01:02:02 by HLA target resequencing data. Before the completion of the ENCODE project, it was difficult to understand the effects of deletions. Now, we can see the possibility of a functional regulatory region around the deletion. Interestingly, two haploid genome sequences of HLA-DRA*01:01:01 had different sequences within the intron and upstream regions. Some of the variants also may affect the expression levels of HLA-DRA by mediating TF binding to the variants. The haplotype of the variants in the upstream region could be significantly different, even though the HLA allele was found to be the same as the HLA-DRA*01:01:01 sequence with six-digit resolution. The ENCODE project stressed the importance of complete HLA gene sequencing, including the upstream regulatory region, to determine the haplotype. In total, 3619 SNVs in the HLA region were selected as expression Quantitative Trait Loci (eQTL) SNVs for HLA gene expression (Table 3).55 These eQTL SNVs were identified in the RegulomeDB database (http://www.regulomedb.org), which have provided annotations of SNPs with known and predicted regulatory elements in the intergenic regions of the human genome. The database includes public datasets from the ENCODE project, in addition to GEO and publications. Known and predicted regulatory DNA elements from DNAase hypersensitivity, TF-binding sites and promoter regions that have been biochemically characterized to regulate transcription are also included. Recorded variants have been classified into various categories according to TF binding and target gene expression. The 3619 HLA eQTL SNVs are likely to affect the binding of TFs to mediate expression of the HLA gene. For variants and deletions near HLA-DRA, new hypotheses concerning the biological functions of the gene could be generated to improve our understanding of HLA-DRA-associated phenotypes.
Figure 4.
Example of target resequencing to detect variants and functional prediction of the regulatory region. Target resequencing of the HLA region clarifies all variants in the target region. For example, several variants and approximately 2-kb deletions have been detected in the upstream region of HLA-DRA. (a) Alignment view of mapped reads (pink: forward strand read, purple: reverse strand read) in the alignment track for detection of SNVs (A: green, C: blue, G: yellow, T: red) and the deletion as displayed in the coverage track. (b) The region was located in cis-regulatory elements as active (H3K27ac-marked) enhancers and a DNase I-hypersensitive site defined by ENCODE chromatin immunoprecipitation sequencing and DNaseI-seq peaks. The deletion and SNVs may affect the expression level of HLA-DRA by influencing the binding of TFs.
Table 3. Number of eQTL SNVs linking expression level of HLA genes.
| HLA gene | Number of eQTL SNVs |
|---|---|
| HLA-A | 821 |
| HLA-C | 12 |
| HLA-B | 773 |
| HLA-DRA | 288 |
| HLA-DRB1 | 580 |
| HLA-DQA1 | 544 |
| HLA-DQB1 | 473 |
| HLA-DPA1 | 2 |
| HLA-DPB1 | 126 |
| Total | 3619 |
Abbreviations: eQTL, expression Quantitative Trait Loci; HLA, human leukocyte antigen; SNV, single-nucleotide variant.
Table 4. Lead SNVs linking rheumatoid arthritis association with regulatory information in the human genome.
| Chr | Position | Lead SNP | Distance to nearest TSS | GENCODE v7 location | RegulomeDB Score | |
|---|---|---|---|---|---|---|
| 6 | 29 789 171 | rs1610677 | 23 582 bp | Intergenic region | No regulatory annotation | |
| 6 | 31 379 931 | rs1063635 | 9903 bp | Coding region | 6 | Motif |
| 6 | 32 218 989 | rs9296015 | 27 283 bp | Intergenic region | 6 | Motif |
| 6 | 32 282 854 | rs6910071 | 49 238 bp | Intron | 6 | Motif |
| 6 | 32 429 643 | rs9268853 | 68 359 bp | Intergenic region | 6 | Motif |
| 6 | 32 574 171 | rs615672 | 35 847 bp | Intergenic region | 6 | Motif |
| 6 | 32 577 380 | rs660895 | 32 638 bp | Intergenic region | 4 | ChIP-seq peak + DNaseI-seq peak |
| 6 | 32 602 269 | rs9272219 | 44 749 bp | Intergenic region | 6 | Motif |
| 6 | 32 663 851 | rs6457617 | 27 409 bp | Intergenic region | 6 | Motif |
| 6 | 32 663 999 | rs6457620 | 27 557 bp | Intergenic region | 6 | Motif |
| 6 | 32 671 103 | rs13192471 | 34 661 bp | Intergenic region | No regulatory annotation | |
| 6 | 32 680 928 | rs7765379 | 44 486 bp | Intergenic region | No regulatory annotation |
Abbreviations: SNP, single nucleotide polymorphism; SNV, single-nucleotide variant; TSS, transcription start site.
In another example, 12 SNVs with regulatory functions have been shown to be associated with rheumatoid arthritis (Table 4). Of the lead SNVs, rs660895 is located 32.6 kb upstream of HLA-DRB1 (from the nearest transcription start site) and has been described as a tag SNP for the HLA-DRB1*04:01 allele. The HLA-DRB1*04:01 allele has been shown to be associated with a higher risk of rheumatoid arthritis (OR: 6.2).56 From chromatin immunoprecipitation sequencing and DNaseI-seq peak data from the ENCODE project, it was found that the SNP is located within regulatory regions. Other eight SNVs are shown to be located within TF-binding sites predicted by in silico motif discovery. The information from ENCODE data will help decision-making for additional and follow-up experiments to obtain reliable evidence for the mechanism through which SNVs in the HLA region contribute to the development of RA.
Future directions
In 2005, Roche launched the first NGS instrument, the Genome Sequencer 20. The Genome Sequencer 20 was able to achieve a read length of about 100 bp and could sequence 20 Mbp per run. Within the last decade, rapid progress in NGS technology has resulted in revolutionary changes in medical genomics for applications in genetic diagnosis, called clinical sequencing or medical exome. However, the two commonly utilized methods for HLA typing, PCR-SSO and PCR-sequencing-based typing, are still the first-line methods in HLA research and diagnosis for more than 10 years. Recently, several manufacturers have begun to develop HLA-typing kits for NGS; thus, elucidation of the complete HLA gene sequence will soon provide new knowledge that will be useful for medical science. However, gene sequence of the HLA region alone will be insufficient for a complete understanding of HLA and all of the HLA-associated phenomena. For this purpose, phase-defined sequencing and haplotype determination of all regions including the HLA genes and regulatory sequences in the HLA region are essential. Further analyses will be required to determine the transcription of the fundamental ‘HLA' unit, including the HLA genes and all associated targets involved in the HLA functional pathway, along with physically interacting targets and regulatory regions containing TF-binding sites. These must all be considered carefully to develop a complete understanding of ‘HLA', that is, HLA-omics analysis. Finally, the goal of HLA typing as complete gene sequencing should be clinical applications that will benefit patients. Future HLA-typing methods will help realize the goal of ‘precision medicine' by determining biologically distinct subgroups for precisely targeted treatments.57
The authors declare no conflict of interest.
References
- 1Levy, S., Sutton, G., Ng, P. C., Feuk, L., Halpern, A. L., Walenz, B. P. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Wheeler, D. A., Srinivasan, M., Egholm, M., Shen, Y., Chen, L., McGuire, A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008). [DOI] [PubMed] [Google Scholar]
- 3Bentley, D. R., Balasubramanian, S., Swerdlow, H. P., Smith, G. P., Milton, J., Brown, C. G. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Wang, J., Wang, W., Li, R., Li, Y., Tian, G., Goodman, L. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Kim, J. I., Ju, Y. S., Park, H., Kim, S., Lee, S., Yi, J. H. et al. A highly annotated whole-genome sequence of a Korean individual. Nature 460, 1011–1015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Fujimoto, A., Nakagawa, H., Hosono, N., Nakano, K., Abe, T., Boroevich, K. A. et al. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat. Genet. 42, 931–936 (2010). [DOI] [PubMed] [Google Scholar]
- 71000 Genomes Project Consortium, Abecasis, G. R., Auton, A., Brooks, L. D., DePristo, M. A., Durbin, R. M. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Pennisi, E. ENCODE project writes eulogy for junk DNA. Science 337, 1159–1161 (2012). [DOI] [PubMed] [Google Scholar]
- 9Shiina, T., Hosomichi, K., Inoko, H. & Kulski, J. K. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 54, 15–39 (2009). [DOI] [PubMed] [Google Scholar]
- 10The MHC sequencing consortium Complete sequence and gene map of a human major histocompatibility complex. Nature 401, 921–923 (1999). [DOI] [PubMed] [Google Scholar]
- 11Stewart, C. A., Horton, R., Allcock, R. J., Ashurst, J. L., Atrazhev, A. M., Coggill, P. et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 14, 1176–1187 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Horton, R., Gibson, R., Coggill, P., Miretti, M., Allcock, R. J., Almeida, J. et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 60, 1–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Traherne, J. A., Horton, R., Roberts, A. N., Miretti, M. M., Hurles, M. E., Stewart, C. A. et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2, e9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Alfirevic, A., Jorgensen, A. L., Williamson, P. R., Chadwick, D. W., Park, B. K. & Pirmohamed, M. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics 7, 813–818 (2006). [DOI] [PubMed] [Google Scholar]
- 15Hung, S. I., Chung, W. H., Jee, S. H., Chen, W. C., Chang, Y. T., Lee, W. R. et al. Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmacogenet. Genomics 16, 297–306 (2006). [DOI] [PubMed] [Google Scholar]
- 16Hetherington, S., Hughes, A. R., Mosteller, M., Shortino, D., Baker, K. L., Spreen, W. et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 359, 1121–1122 (2002). [DOI] [PubMed] [Google Scholar]
- 17Mallal, S., Nolan, D., Witt, C., Masel, G., Martin, A. M., Moore, C. et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 359, 727–732 (2002). [DOI] [PubMed] [Google Scholar]
- 18Martin, A. M., Nolan, D., Gaudieri, S., Almeida, C. A., Nolan, R., James, I. et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc. Natl Acad. Sci. USA 101, 4180–4185 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Saag, M., Balu, R., Phillips, E., Brachman, P., Martorell, C., Burman, W. et al. High sensitivity of human leukocyte antigen-b*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin. Infect. Dis. 46, 1111–1118 (2008). [DOI] [PubMed] [Google Scholar]
- 20Dainichi, T., Uchi, H., Moroi, Y. & Furue, M. Stevens-Johnson syndrome, drug-induced hypersensitivity syndrome and toxic epidermal necrolysis caused by allopurinol in patients with a common HLA allele: what causes the diversity? Dermatology 215, 86–88 (2007). [DOI] [PubMed] [Google Scholar]
- 21Gabriel, C., Danzer, M., Hackl, C., Kopal, G., Hufnagl, P., Hofer, K. et al. Rapid high-throughput human leukocyte antigen typing by massively parallel pyrosequencing for high-resolution allele identification. Hum. Immunol. 70, 960–964 (2009). [DOI] [PubMed] [Google Scholar]
- 22Bentley, G., Higuchi, R., Hoglund, B., Goodridge, D., Sayer, D., Trachtenberg, E. A. et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens 74, 393–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Lind, C., Ferriola, D., Mackiewicz, K., Heron, S., Rogers, M., Slavich, L. et al. Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Hum. Immunol. 71, 1033–1042 (2010). [DOI] [PubMed] [Google Scholar]
- 24Lank, S. M., Wiseman, R. W., Dudley, D. M. & O'Connor, D. H. A novel single cDNA amplicon pyrosequencing method for high-throughput, cost-effective sequence-based HLA class I genotyping. Hum. Immunol. 71, 1011–1017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Erlich, R. L., Jia, X., Anderson, S., Banks, E., Gao, X., Carrington, M. et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics 12, 42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Holcomb, C. L., Höglund, B., Anderson, M. W., Blake, L. A., Böhme, I., Egholm, M. et al. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens 77, 206–217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Wang, C., Krishnakumar, S., Wilhelmy, J., Babrzadeh, F., Stepanyan, L., Su, L. F. et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc. Natl Acad. Sci. USA 109, 8676–8681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Shiina, T., Suzuki, S., Ozaki, Y., Taira, H., Kikkawa, E., Shigenari, A. et al. Super high resolution for single molecule-sequence-based typing of classical HLA loci at the 8-digit level using next generation sequencers. Tissue Antigens 80, 305–316 (2012). [DOI] [PubMed] [Google Scholar]
- 29Lank, S. M., Golbach, B. A., Creager, H. M., Wiseman, R. W., Keskin, D. B., Reinherz, E. L. et al. Ultra-high resolution HLA genotyping and allele discovery by highly multiplexed cDNA amplicon pyrosequencing. BMC Genomics 13, 378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Moonsamy, P. V., Williams, T., Bonella, P., Holcomb, C. L., Höglund, B. N., Hillman, G. et al. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array™ system for simplified amplicon library preparation. Tissue Antigens 81, 141–149 (2013). [DOI] [PubMed] [Google Scholar]
- 31Ringquist, S., Bellone, G., Lu, Y., Roeder, K. & Trucco, M. Clustering and alignment of polymorphic sequences for HLA-DRB1 genotyping. PLoS ONE 8, e59835–e59837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Danzer, M., Niklas, N., Stabentheiner, S., Hofer, K., Pröll, J., Stückler, C. et al. Rapid, scalable and highly automated HLA genotyping using next-generation sequencing: a transition from research to diagnostics. BMC Genomics 14, 221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Hosomichi, K., Jinam, T. A., Mitsunaga, S., Nakaoka, H. & Inoue, I. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics 14, 355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Trachtenberg, E. A. & Holcomb, C. L. Next-generation HLA sequencing using the 454 GS FLX system. Methods Mol. Biol. 1034, 197–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Ozaki, Y., Suzuki, S., Shigenari, A., Okudaira, Y., Kikkawa, E., Oka, A. et al. HLA-DRB1, -DRB3, -DRB4 and -DRB5 genotyping at a super-high resolution level by long range PCR and high-throughput sequencing. Tissue Antigens 83, 10–16 (2013). [DOI] [PubMed] [Google Scholar]
- 36Hajeer, A. H., Al Balwi, M. A., Aytül Uyar, F., Alhaidan, Y., Alabdulrahman, A., Al Abdulkareem, I. et al. HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in Saudis using next generation sequencing technique. Tissue Antigens 82, 252–258 (2013). [DOI] [PubMed] [Google Scholar]
- 37Hosomichi, K., Mitsunaga, S., Nagasaki, H. & Inoue, I. A Bead-based Normalization for Uniform Sequencing depth (BeNUS) protocol for multi-samples sequencing exemplified by HLA-B. BMC Genomics 15, 645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Smith, A. G., Pyo, C. W., Nelson, W., Gow, E., Wang, R., Shen, S. et al. Next generation sequencing to determine HLA class II genotypes in a cohort of hematopoietic cell transplant patients and donors. Hum. Immunol. 75, 1040–1046 (2014). [DOI] [PubMed] [Google Scholar]
- 39Ehrenberg, P. K., Geretz, A., Baldwin, K. M., Apps, R., Polonis, V. R., Robb, M. L. et al. High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics 15, 864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Zhou, M., Gao, D., Chai, X., Liu, J., Lan, Z., Liu, Q. et al. Application of high-throughput, high-resolution and cost-effective next generation sequencing-based large-scale HLA typing in donor registry. Tissue Antigens 85, 20–28 (2014). [DOI] [PubMed] [Google Scholar]
- 41Lan, J. H., Yin, Y., Reed, E. F., Moua, K., Thomas, K. & Zhang, Q. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum. Immunol. 76, 166–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Ozaki, Y., Suzuki, S., Kashiwase, K., Shigenari, A., Okudaira, Y., Ito, S. et al. Cost-efficient multiplex PCR for routine genotyping of up to nine classical HLA loci in a single analytical run of multiple samples by next generation sequencing. BMC Genomics 16, 318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Gnirke, A., Melnikov, A., Maguire, J., Rogov, P., LeProust, E. M., Brockman, W. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Wittig, M., Anmarkrud, J. A., Kässens, J. C., Koch, S., Forster, M., Ellinghaus, E. et al. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucleic Acids Res. 43, e70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Rabbani, B., Mahdieh, N., Hosomichi, K., Nakaoka, H. & Inoue, I. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J. Hum. Genet. 57, 621–632 (2012). [DOI] [PubMed] [Google Scholar]
- 46Warren, R. L., Choe, G., Freeman, D. J., Castellarin, M., Munro, S., Moore, R. et al. Derivation of HLA types from shotgun sequence datasets. Genome Medicine 4, 95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Boegel, S., Löwer, M., Schäfer, M., Bukur, T., de Graaf, J., Boisguérin, V. et al. HLA typing from RNA-Seq sequence reads. Genome Med 4, 102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Liu, C., Yang, X., Duffy, B., Mohanakumar, T., Mitra, R. D., Zody, M. C. et al. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 41, e142–e142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Szolek, A., Schubert, B., Mohr, C., Sturm, M., Feldhahn, M. & Kohlbacher, O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Kim, H. J. & Pourmand, N. HLA haplotyping from RNA-seq data using hierarchical read weighting. PLoS ONE 8, e67885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Bai, Y., Ni, M., Cooper, B., Wei, Y. & Fury, W. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics 15, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Huang, Y., Yang, J., Ying, D., Zhang, Y., Shotelersuk, V., Hirankarn, N. et al. HLAreporter: a tool for HLA typing from next generation sequencing data. Genome Med. 7, 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Nariai, N., Kojima, K., Saito, S., Mimori, T., Sato, Y., Kawai, Y. et al. HLA-VBSeq: accurate HLA typing at full resolution from whole-genome sequencing data. BMC Genomics 16, S7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Major, E., Rigó, K., Hague, T., Bérces, A. & Juhos, S. HLA typing from 1000 genomes whole genome and whole exome Illumina data. PLoS ONE 8, e78410–e78419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Boyle, A. P., Hong, E. L., Hariharan, M., Cheng, Y., Schaub, M. A., Kasowski, M. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Gorman, J. D., David-Vaudey, E., Pai, M., Lum, R. F. & Criswell, L. A. Particular HLA-DRB1 shared epitope genotypes are strongly associated with rheumatoid vasculitis. Arthritis Rheum. 50, 3476–3484 (2004). [DOI] [PubMed] [Google Scholar]
- 57Katsnelson, A. Momentum grows to make 'personalized' medicine more 'precise'. Nat. Med. 19, 249 (2013). [DOI] [PubMed] [Google Scholar]