SUMMARY
The human TEAD family of transcription factors (TEAD1-4) is required for YAP-mediated transcription in the Hippo pathway. Hyperactivation of TEAD’s co-activator YAP contributes to tissue overgrowth and human cancers, suggesting that pharmacological interference of TEAD-YAP activity may be an effective strategy for anticancer therapy. Here we report the discovery of a central pocket in the YAP-binding domain (YBD) of TEAD that is targetable by small molecule inhibitors. Our X-ray crystallography studies reveal that flufenamic acid, a non-steroidal anti-inflammatory drug (NSAID), binds to the central pocket of TEAD2 YBD. Our biochemical and functional analyses further demonstrate that binding of NSAIDs to TEAD inhibits TEAD-YAP-dependent transcription, cell migration and proliferation, indicating that the central pocket is important for TEAD function. Therefore, our studies discover a novel way of targeting TEAD transcription factors and set the stage for therapeutic development of specific TEAD-YAP inhibitors against human cancers.
Keywords: TEAD, flufenamates, cancer therapy, Hippo pathway
INTRODUCTION
The Hippo pathway controls organ size by blocking cell proliferation and promoting apoptosis (Hansen et al., 2015; Wackerhage et al., 2014). Two homologous oncoproteins YAP and TAZ are the major downstream targets that are inhibited by the Hippo signaling network. YAP/TAZ interact with the TEA domain transcription factors (TEADs) to promote cell growth and proliferation and inhibit apoptosis. In humans, there are four closely related TEAD proteins, TEAD1-4. All four TEADs contain an N-terminal TEA domain that binds DNA, and a C-terminal YAP-binding domain (YBD) that binds to YAP/TAZ (Figure 1A). Because TEADs do not contain an activation domain and YAP/TAZ do not contain a DNA-binding domain, TEADs and YAP/TAZ together form a functional heterodimeric transcription complex that activates the expression of Hippo-responsive genes critical for tissue homeostasis, stem cell regeneration and maintenance, and tumorigenesis (Pobbati and Hong, 2013).
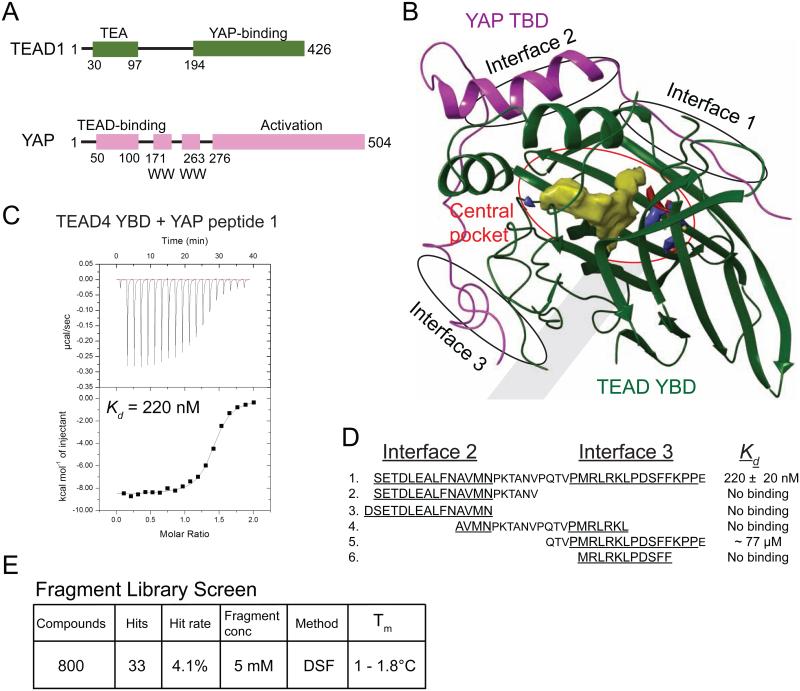
Figure 1. The YAP-binding Domain of TEADs Has a Central Pocket.
(A) Domain architecture of TEAD1 and YAP. All the four TEAD genes have a N-terminal DNA-binding TEA domain and a C-terminal YAP-binding domain (YBD). YAP has an N-terminal TEAD-binding domain (TBD) followed by one or two WW domains and a C-terminal activation domain.
(B) The crystal structure of the TEAD-YAP complex (PDB code 3KYS) reveals three interfaces between TEAD and YAP. TEAD YBD is colored green and YAP TBD is colored magenta. TEAD YBD has a large pocket in the center (red circle); the hydrophobic volume of this pocket is shown in yellow. On both ends of this pocket are hydrophilic areas, shown in red and blue that could be used to improve the specificity of TEAD-binding drugs. All structural figures were generated with Schrödinger software suite or Chimera (UCSF).
(C) Isothermal titration calorimetry (ITC) showing heat response during TEAD-YAP interaction. The binding affinity (Kd) between the YAP peptide 1 and TEAD was indicated.
(D) Sequences and binding affinities of the YAP peptides used in this study. Residues from Interface 2 and 3 are underlined.
(E) Summary of the results from fragment library screen.
Genetic studies have shown that TEAD-YAP promotes cell proliferation and inhibit cell death (Hong and Guan, 2012; Sawada et al., 2008). Hyperactivation of TEAD-YAP contributes to various human cancers, such as prostate cancer (Knight et al., 2008) and pancreatic cancer (Hucl et al., 2007; Kapoor et al., 2014). TEADs also bind to the Vgll family of proteins that modulates YAP oncogenic function (Jiao et al., 2014; Pobbati et al., 2012; Pobbati and Hong, 2013). For cancers to progress, cells need to possess certain attributes (Hanahan and Weinberg, 2011). Remarkably, YAP and TAZ, with the help of TEADs, endow cells with many such attributes, such as resistance to contact inhibition (Zhao et al., 2007), ability to sustain anchorage independent growth (Overholtzer et al., 2006), transition to a mesenchymal state for migration, invasion and maintenance of stemness (Chan et al., 2008; Diepenbruck et al., 2014; Lei et al., 2008), and ability to survive in suspension or resistance to anoikis for metastasis (Lamar et al., 2012; Zhao et al., 2012). To achieve this effect, the TEAD-YAP/TAZ complexes upregulate the expression of a myriad of target genes involved in anti-apoptosis and proliferation, such as CTGF (Zhao et al., 2008), Cyr61 (Zhao et al., 2008), c-Myc (Dong et al., 2007), Axl (Xu et al., 2010), and Jagged-1 (Tschaharganeh et al., 2013). There is also a good correlation between YAP/TAZ overexpression or nuclear localization and poor prognosis in various cancers (Moroishi et al., 2015). Taken together, YAP and TAZ are potential targets for cancer therapy.
Targeting an intracellular protein is conventionally achieved through binding of small molecule drugs to inhibit the activity of the target protein. For a small molecule to bind, the protein should possess a pocket with certain electrostatic and geometric properties. The available structures of YAP and TAZ indicate that they do not have such a pocket and are therefore not deemed targetable by small molecules. This is generally the case for more than ninety percent of the proteins. However, we noticed that the YAP-binding domain (YBD) of TEADs have a pocket in the center of the protein that appears to be druggable. TEAD YBD has been shown to bind YAP/TAZ through a defined surface (Chen et al., 2010; Li et al., 2010; Tian et al., 2010). Because YAP/TAZ relies on TEADs for activating gene expression, inhibiting the oncogenic activities of YAP/TAZ can be achieved by direct targeting TEAD-YAP protein-protein interactions or by inhibiting the activity of TEADs. Another impetus for focusing on TEADs is that TEADs appear to be largely dispensable for tissue homeostasis in adults (Liu-Chittenden et al., 2012), therefore, inhibiting TEADs should not perturb normal tissue growth or result in major adverse toxicity effects.
The majority of drugs or drug candidates that potentially target the Hippo pathway affect the nucleocytoplasmic transport of YAP/TAZ (Johnson and Halder, 2014; Park and Guan, 2013). These include: GPCR agonists, such as dobutamine (Bao et al., 2011); inhibitors of ILK (Serrano et al., 2013), PI3K (Fan et al., 2013), and ROCK (Dupont et al., 2011) kinases; statins that inhibit HMG-CoA reductase (Sorrentino et al., 2014); and compounds, such as forskolin and rolipram (Yu et al., 2013), that increase cAMP levels. As diverse cellular signals regulate YAP/TAZ nucleocytoplasmic transport, and as these regulator proteins are also involved in other important cellular signaling networks, it is unclear if these compounds could be effective to treat cancers caused by a defective Hippo pathway. There are also reported molecules that could disrupt TEAD-YAP interactions, such as verteporfin (Liu-Chittenden et al., 2012) and cyclic YAP peptides (Zhang et al., 2014), but these molecules suffer from having a low plasma half-life or cell penetrating ability. Therefore, there is an urgent need to find new strategies for developing drugs against human cancers caused by a dysregulated Hippo pathway.
In this study, using both computational and experimental approaches, we have evaluated the targetability of the YAP-binding domain (YBD) of TEADs. We show that TEAD YBD binds to flufenamates (nonsteroidal anti-inflammatory drugs) with appreciable affinity. We have determined the crystal structure of TEAD2 YBD bound to flufenamic acid, which reveals that flufenamates bind to the central pocket of TEAD. We also show that flufenamates, such as flufenamic acid and niflumic acid, can inhibit both TEAD function and TEAD-YAP-dependent processes, such as cell migration and proliferation. Taken together, our studies bring to light the critical function of the central pocket in TEADs that is generally overlooked in the field and suggest that it is biologically relevant to target the central pocket of TEADs for anticancer therapy. Our work has therefore established the basis for future development of more potent and specific TEAD-YAP inhibitors to treat human cancers.
RESULTS and DISCUSSION
The YAP-binding Domain of TEAD Has a Central Pocket
TEADs (TEAD1-4) all have an N-terminal TEA domain and a C-terminal YAP-binding domain (YBD) (Figure 1A). TEADs bind to transcription co-activator YAP through the YBD. YAP binds to TEADs through the N-terminal TEAD-binding domain (TBD). YAP TBD is followed by one or two WW domains and an activation domain (Figure 1A). The TEAD-YAP complexes regulate the transcription of Hippo-responsive genes to promote cell proliferation and inhibit apoptosis. Hyperactivation of TEAD-YAP causes tumors; therefore, the TEAD-YAP complexes are promising targets for cancer treatment.
For drug development, TEAD appears to be a suitable target. TEAD YBD has a central pocket that is very hydrophobic and seems druggable (Figure 1B). The amino acid residues lining the central pocket are also conserved (Figure S1). The volume of the central pocket is large enough to bind and fully envelope a small molecule. The central pocket also has an excellent druggability score, Dscore = 1.39 (Halgren, 2009), meaning that it has the right electrostatics and enclosure for binding drug-like molecules with high affinity. Therefore, TEADs belong to the small group of proteins that have an internal druggable pocket.
YAP binding requires a surface pocket on TEAD YBD that is located at the back of the seven-stranded b-sheet (Figure 1B) (Tian et al., 2010). However, this surface pocket does not have a good druggability score. Careful inspection of the crystal structures of TEAD1-YAP (PDB code 3KYS) and TEAD4-YAP (PDB code 3JUA) reveals that the YAP-binding site is formed by a shallow surface that cannot provide enclosure for a small molecule. Although in theory, the surface of TEADs could be potentially targeted by small molecules to prevent its interaction with YAP, the absence of deep pockets with good druggability scores will likely make it very challenging to develop potent inhibitors to disrupt the TEAD-YAP protein-protein interactions.
Fragment Screen for TEAD4 YBD
We have used human TEAD4 YBD to biochemically analyze the druggability of the TEAD proteins. TEAD4 YBD is well folded, and is both necessary and sufficient to interact with YAP/TAZ (Chen et al., 2010; Li et al., 2010; Tian et al., 2010). It also has the hydrophobic central pocket. Unlike the full-length TEAD4 protein, sufficient quantities of purified recombinant TEAD4 YBD could be obtained.
We next evaluated whether the TEAD4-YAP interaction is conducive for disruption by small molecule drugs. Because TEADs and YAP form a functional transcriptional heterodimer, disruption of TEAD-YAP interactions will hamper cell proliferation. However, the interactions between TEADs and YAP are quite extensive involving three interfaces with YAP encircling roughly three-quarters of TEAD YBD (Figure 1B). For drug development, it would be beneficial to identify the hot spots that contribute mainly to the binding affinity between TEADs and YAP. In order to do this, we used several YAP peptides spanning various boundaries at interface 2 and 3 and measured their affinity towards TEAD4 YBD using isothermal titration calorimetry (ITC). We did not use the YAP peptides containing residues in interface 1 because it has been shown that interface 1 is dispensable for YAP binding (Chen et al., 2010; Tian et al., 2010).
The YAP peptide that contains interface 2 and 3 residues (peptide 1) interacts with TEAD4 YBD with a nanomolar affinity (Figure 1C), which is in line with our previous study (Tian et al., 2010). Therefore, it seems plausible to predict that a small molecule that binds to TEADs with lower nanomolar affinity should be able to disrupt the TEAD-YAP interactions. We noticed that the YAP peptides containing only interface 2 residues did not bind to TEAD4 YBD (Figure 1D). The YAP peptide containing only interface 3 residues (peptide 5) had very weak binding to TEAD4 YBD with a dissociation constant (Kd) about 77 μM (Figure 1D), which is about 350-fold weaker than that of the YAP peptide containing interface 2 and 3 (peptide 1). This suggests that interface 3 is the major YAP-binding site but interface 2 is also required for more efficient binding. However, the distance between interface 2 and 3 imposes a lower size limit on a dual-site ligand that is larger than what is generally deemed acceptable for small drug-like molecules (Veber et al., 2002). Furthermore, interface 3 has a poor predicted druggability score. Due to the above reasons, we decided to focus our effort to target the hydrophobic central pocket of TEADs.
The central pocket of TEADs has an excellent druggability score mainly due to its enclosure and high hydrophobicity. This led us to believe that a fragment-based screen would be a viable approach for testing the druggability of the central pocket of TEADs. Interestingly, the crystal structure of TEAD2 YBD (Tian et al., 2010) shows that there is an opening at the surface of TEAD2 YBD that is capable of allowing small molecules to access the central pocket. In fact, some small molecules used in the crystallization buffers were found to occupy the central pocket of TEADs.
To experimentally verify the druggability of TEADs, we screened our in-house fragment library. These compounds are small molecules about 150 – 300 daltons in mass and are used to test the druggability of proteins (Hung et al., 2009). Qualitative binding of fragments to TEAD4 YBD was evaluated using differential scanning fluorimetry (DSF). Change in the melting temperature (Tm) of TEAD4 YBD in the presence of fragments is indicative of binding (Lo et al., 2004). Among the 800 compounds screened, 33 were found to increase the Tm of TEAD4 YBD, giving a hit rate of about 4% (Figure 1E). We obtained similar hit rates in the fragment-based screens of more conventional drug targets, such as kinases (unpublished data). Therefore, our data strongly suggest that the YAP-binding domain (YBD) of TEADs is druggable by small molecules. The high hit rate from the fragment screen raises the possibility that small molecules bind to the central pocket as opposed to the surface of TEAD YBD.
Identification of Flufenamic Acid as a TEAD-binding Drug
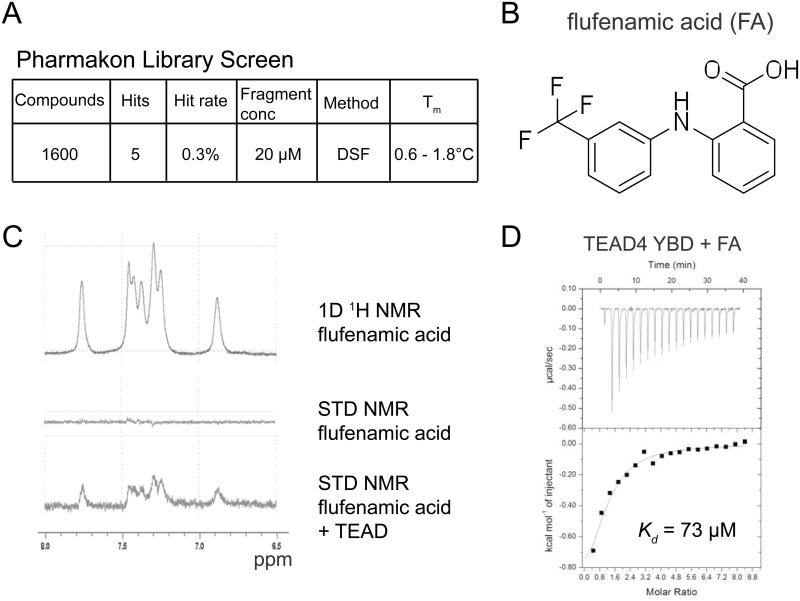
Encouraged by the results from the initial fragment screen, we proceeded to screen a Pharmakon library that contained a collection of FDA-approved drugs to identify more potent drugs that bind to TEAD4 YBD. We found that a small portion of these drugs increased the Tm of TEAD4 YBD (Figure 2A). The hit rate was found to be much lower than that of the fragment screen, as the Pharmakon library contained larger and more complex molecules. We identified flufenamic acid (FA) as the best hit in the Pharmakon screen (Figure 2B). Flufenamic acid is used to treat inflammation and belongs to a class of flufenamates called non-steroidal anti-inflammatory drugs (NSAIDs).
Figure 2. Identification of Flufenamic Acid as a TEAD-binding Drug.
(A) Summary of the results from Pharmakon library screen.
(B) Structure of flufenamic acid (FA).
(C) Saturation-Transfer Difference (STD) NMR spectra of FA showing a conspicuous change after the addition of TEAD. Upper panel shows the 1D 1H NMR spectrum of free FA. Middle and lower panels are STD spectra of FA in the absence and presence of TEAD.
(D) ITC measurement of the affinity (Kd) between TEAD and FA.
We also confirmed the binding of FA to TEAD4 YBD qualitatively through Saturation-Transfer Difference (STD) NMR (Begley et al., 2013). Addition of TEAD4 YBD triggered a positive signal in the STD spectrum of FA, which was indicative of binding (Figure 2C). Using ITC, we were able to measure the dissociation constant (Kd) between FA and TEAD4 YBD, which is about 73 μM (Figure 2D). Thus, the TEAD protein is directly targetable by small molecule drugs.
Structure of the TEAD2–flufenamic acid Complex
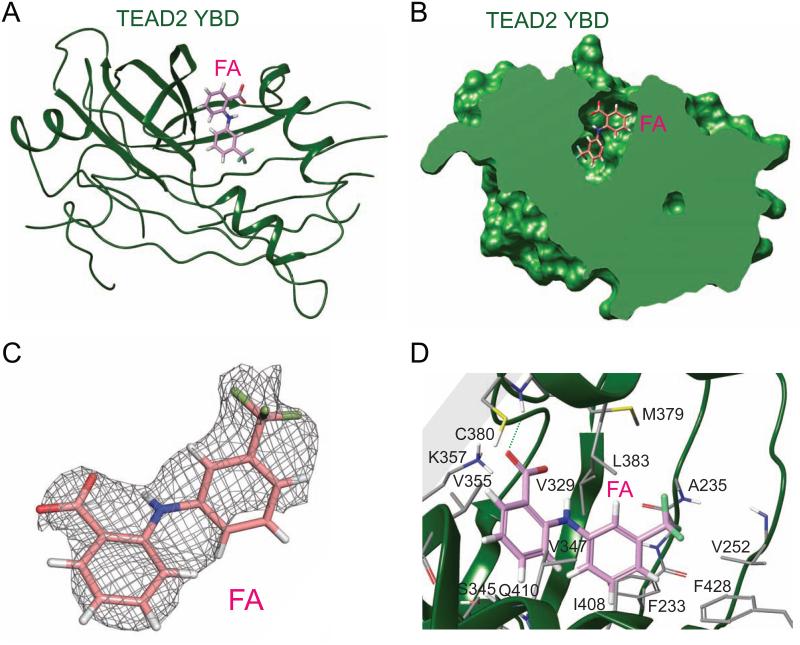
Having confirmed flufenamic acid (FA) binding to TEAD4 YBD qualitatively and quantitatively (Figure 2), we were interested to locate the FA binding site of TEADs. Based on our druggability assessment of TEADs, we postulated that FA very likely binds to the hydrophobic central pocket. To test our hypothesis, we determined the crystal structure of FA-bound human TEAD2 YBD (Figure 3A). There are two molecules of TEAD2 YBD per asymmetric unit in the crystal and reassuringly, FA is seen in the central pocket of both TEAD2 molecules. A cross section of TEAD2 YBD with FA occupying the central pocket is shown in Figure 3B. FA could be nicely fitted to the omit electron density map that is observed in the TEAD2 central pocket (Figure 3C).
Figure 3. Structure of the TEAD2-flufenamic acid Complex.
(A) Crystal structure of the TEAD2-FA complex reveals that FA binds to the central pocket in TEAD. TEAD2 YBD is colored green. FA is shown as pink sticks.
(B) A cross-section of the surface drawing of TEAD2 YBD shows FA occupying the central pocket.
(C) Simulated annealing omit map contoured at 1 σ for FA.
(D) Detailed view of the TEAD-FA interaction. FA and FA-contacting residues in the central pocket are shown as pink and grey sticks, respectively. The hydrogen bond between FA and C380 is shown as a green dash line.
A closer look at the TEAD2-FA structure revealed that the hydrophobic portions of FA make contacts with hydrophobic residues lining the central pocket (Figure 3D). The carboxylate group of FA binds to an area where TEAD2 would interact favorably with an electronegative group. The steric and electrostatic complementarities between FA and the TEAD2 central pocket support the validity of the TEAD2-FA structure.
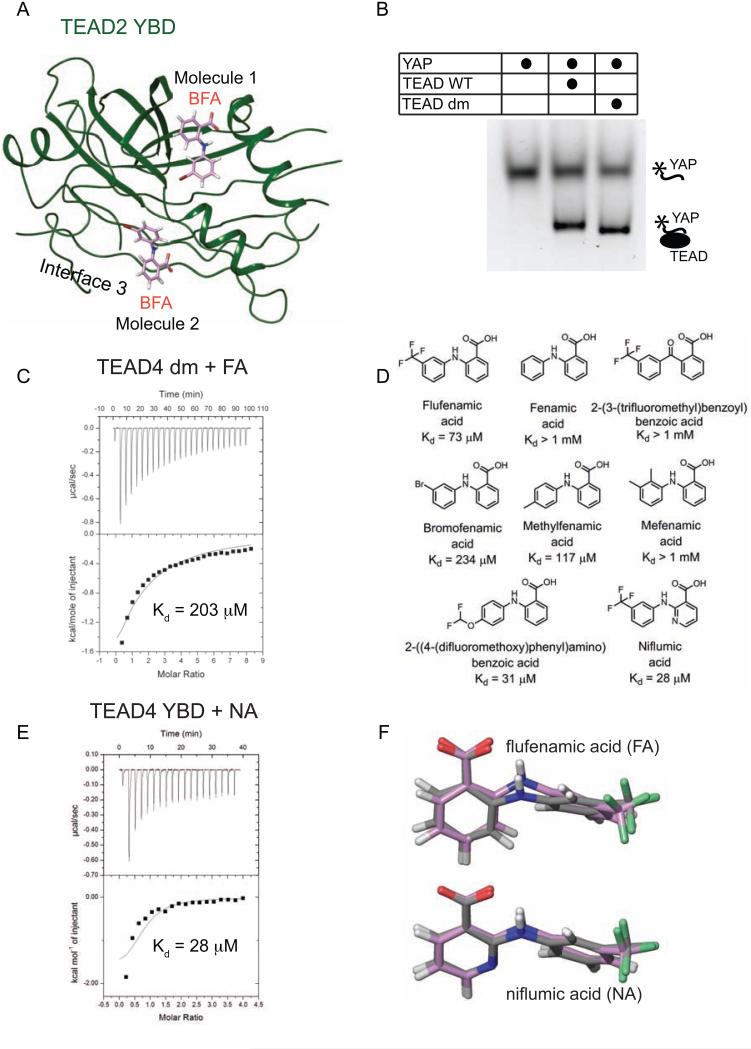
To further confirm that the density observed in the central pocket indeed corresponded to the FA molecule, we also crystallized TEAD2 YBD in the presence of bromo-fenamic acid (BFA), an FA derivative where the fluorines are replaced by bromine. Bromine scatters X-rays anomalously and this property is commonly used in X-ray crystallography to locate the position of bromine atoms and thereby the bromoderivatives in the crystal. As bromine is a much larger atom than fluorine, in our bromo-fenamic acid (BFA), we replaced the trifluoromethyl group with one bromine atom. We then solved the crystal structure of BFA-bound TEAD2 YBD (Figure 4A). As expected, BFA (molecule 1) binds to the central pocket of TEAD2 YBD in the same way as FA does. This result confirms that FA indeed binds to the central pocket of TEAD2 YBD. The data collection and refinement statistics for the TEAD-FA and TEAD-BFA structures are shown in Table 1.
Figure 4. Specificity of the TEAD-fenamate Interactions.
(A) Crystal structure of the TEAD2-BFA complex. Molecule 1 of BFA binds to the central pocket. Molecule 2 of BFA binds to the YAP-binding Interface 3 region of TEAD. TEAD YBD is colored green. BFA are shown as pink sticks.
(B) Monitoring TEAD-YAP interaction using TEAD4 YBD and a fluorescent-labeled YAP peptide. The native gel shows that TEAD wild-type (WT) and TEAD double mutant (dm) interact with YAP with similar efficiency.
(C) The affinity (Kd) between flufenamic acid and TEAD dm was measured by ITC.
(D) Structures of FA analogs and their binding affinities towards TEAD.
(E) ITC measurement of the affinity (Kd) between TEAD and niflumic acid (NA).
(F) The minimal energy conformations of FA and NA are shown as grey sticks. The conformation of FA in the crystal structure of TEAD2-FA and the docked conformation of NA are shown as pink sticks.
Table 1.
Data collection and refinement statistics for TEAD2-FA and TEAD2-BFA structures
| Data collection | ||
|---|---|---|
| Crystal | TEAD2 – FA | TEAD2 – BFA |
| Space group | C2 | C2 |
| Wavelength (Å) | 0.97918 | 0.91925 |
| Unit cell | ||
| a, b, c (Å) | 120.82, 61.45, 80.42 | 121.47, 61.58, 80.42 |
| b (°) | 117.55 | 117.70 |
| Resolution range (Å) | 50.0 – 2.30 (2.34 – 2.30) | 40.0 – 2.18 (2.22 – 2.18) |
| Unique reflections | 20,054 (1,222) | 24,263 (1,480) |
| Multiplicity | 5.1 (4.5) | 4.0 (3.1) |
| Data completeness (%) | 99.7 (99.7) | 98.4 (86.6) |
| Rmerge (%)a | 11.9 (>100) | 5.4 (43.9) |
| I/σ (I) | 17.2 (3.39) | 24.0 (3.14) |
|
| ||
| Refinement statistics | ||
|
| ||
| Resolution range (Å) | 33.7 – 2.30 (2.43 – 2.30) | 38.7 – 2.18 (2.27 – 2.18) |
| No. of reflections Rwork/Rfree | 20,053/1,002 (1,222/60) | 24,260/1,213 (1,480/74) |
| Data completeness (%) | 86.2 (55.0) | 88.1 (55.0) |
| Atoms (non-H protein/solvent/inhibitor) |
3,304/85/60 | 3,304/109/51 |
| Rwork (%) | 17.1 (22.4) | 17.9 (21.2) |
| Rfree (%) | 23.2 (27.4) | 22.7 (26.8) |
| R.m.s.d. bond length (Å) | 0.015 | 0.015 |
| R.m.s.d. bond angle (°) | 1.55 | 1.50 |
| Mean B-value (Å2) (protein/solvent/inhibitor) |
44.7/40.3/60.2 | 45.0/44.8/63.5 |
| Ramachandran plot (%) (favored/additional/disallowed)b |
97.9/2.1/0.0 | 97.2/2.8/0.0 |
Statistics for the highest-resolution shell are shown in parentheses.
, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
As defined by the validation suite MolProbity.
Surprisingly, on the surface of one of the two TEAD2 molecules in the asymmetric unit of the TEAD2-BFA structure, we found a second molecule of BFA (molecule 2) (Figure 4A). We noticed that the second BFA molecule is located close to interface 3 of the TEAD-YAP complex. However, we only observed partial density for this second BFA molecule, suggesting that BFA might not fully occupy this site and bind to interface 3 with much weaker affinity. Furthermore, a careful inspection of the TEAD2-FA structure suggests that a second FA molecule also binds to the interface 3 region of TEAD2 YBD. This indicates that small molecule inhibitors can also bind to the surface pocket of TEAD YBD but might be with weak affinity.
Specificity of the TEAD-fenamate Interactions
After confirming that the molecule in the central pocket is indeed FA, we examined in detail the interactions between FA and TEAD2 YBD. There are 14 residues of TEAD2 YBD, mostly hydrophobic, within 4 Å distance to FA (Figure 3D). FA mostly forms hydrophobic interactions with these residues. However, there is also a hydrogen bond between the carboxylate group in FA and the main chain amide nitrogen of C380, as well as a putative salt bridge between the carboxylate group in FA and K357 in TEAD2 YBD (Figure 3D). As expected, the trifluoromethyl group interacts with the neighboring hydrophobic residues. Five residues in TEAD2 YBD contact the fluorines in FA, including A235, V252, F428, F233 and L383. Residues I408, L383, F233 and Q410 contact the rest of the trifluromethyl-benzene ring. The benzoic acid part of FA makes contact with residues S345, V355, V329, V347, C380, M379 and K357 in TEAD2 YBD.
To further evaluate the specificity of the TEAD-FA interactions, we decided to mutate the FA-interacting residues in TEAD4 YBD. Based on the TEAD2-FA structure, an A235I and C380P double mutation (dm) of TEAD2 is expected to sterically interfere with FA binding (Figure 3D). Therefore, we made a corresponding dm mutant in TEAD4 YBD (TEAD dm, A231I and C367P). We first tested if the TEAD dm could fold properly. Proper folding is needed to maintain the YAP-binding interface on TEAD4 YBD. The binding between TEAD4 YBD and YAP was assayed by native gel using TEAD4 YBD and a fluorescently labeled YAP peptide (peptide 1). The mobility of the YAP peptide is shifted due to its interaction with TEAD4 YBD (TEAD WT) (Figure 4B). Indeed, TEAD dm interacts with YAP with similar efficiency as TEAD WT does (Figure 4B), indicating that TEAD dm is properly folded. We then measured the affinity between TEAD dm and FA by ITC (Figure 4C). As predicted, TEAD dm binds weaker to FA than TEAD WT does, validating the observed interactions.
TEAD Binds to Niflumic Acid
We next set out to test commercially available FA analogues aiming to find compounds with higher affinity for TEAD. We measured affinities of various FA analogs to TEAD4 YBD by ITC (Figure 4D). The FA analog that binds to TEAD4 YBD with the highest affinity is Niflumic acid (NA) (Figure 4E). NA is structurally similar to FA, except that it has nicotinic acid in place of the benzoic acid. Because we could not obtain the crystal structure of NA-bound TEAD, we docked NA onto the FA molecule in the TEAD2-FA structure and observed that the heterocyclic nitrogen does not contact any residues. To understand the basis for improved affinity observed for NA, we did a conformational analysis on FA and NA. We observed that the conformation of docked NA in the NA-TEAD complex is closer to its minimal conformational energy, as compared to the conformation of TEAD-bound FA (Figure 4F). The decreased conformational energy might be the reason for the improved affinity of NA towards TEAD. The strain energies of different ligands of TEAD are tabulated in Table 2.
Table 2.
Strain energies of cocrystalized ligands in kJ/mol
| TITLE | OPLS-2005 | B3LYP LAV3P** |
B3LYP LACV3P** |
B3LYP LAC3P**++ |
|---|---|---|---|---|
| Niflumic acid | 1.1 | 10.0 | 8.2 | 6.1 |
| Flufenamic acid | 4.6 | 19.0 | 17 | 15.8 |
| Bromo–fenamic acid | 3.4 | 7.7 | 6.2 | 5.2 |
Biological Consequence of Fenamate Binding to TEADs
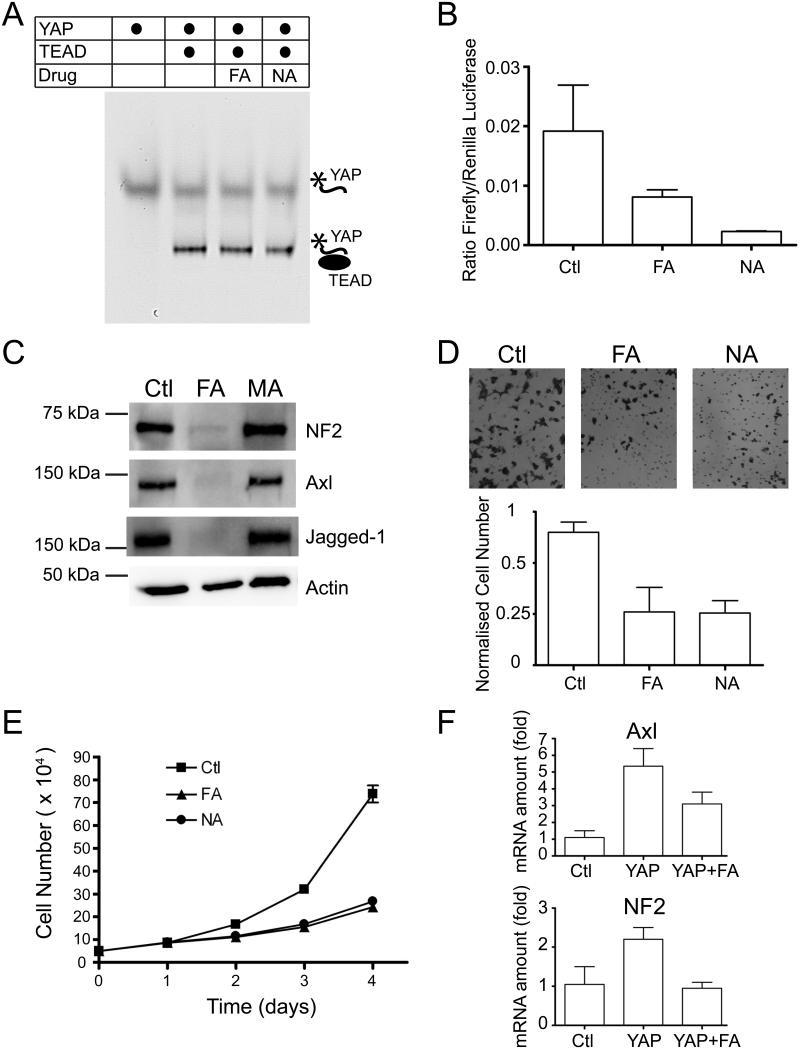
We next tested the biological consequence of flufenamate binding to TEADs. The effect of flufenamate binding on TEAD-YAP interaction was first assayed by native gel using TEAD4 YBD and a fluorescently labeled YAP peptide. The YAP peptide interacted with TEAD4 YBD with similar efficiency in the presence of the flufenamate drugs (FA or NA), indicating that TEAD-YAP interaction is unaffected by flufenamates binding in vitro (Figure 5A). Our data also suggested that binding of FA or NA does not cause any conformational change at the YAP-binding surface of TEAD.
Figure 5. Biological Consequence of Fenamate Binding to TEADs.
(A) Native gel showing that the binding of TEAD to fluorescently labeled YAP peptide is unaffected in the presence of the flufenamate drugs.
(B) Significant reduction in the TEAD reporter activity was observed after the treatment of cells with flufenamic acid (FA) and niflumic acid (NA).
(C) The expression of Hippo-responsive genes, such as NF2, Axl and Jagged-1, were greatly reduced after flufenamic acid (FA) treatment. However, no significant decrease was observed when the cells were treated with mefanamic acid (MA).
(D) Migration of HEK293 cells was measured after FA and NA treatments using a transwell assay. The cells that migrate across the membrane were visualized using crystal violet staining. The quantification is shown below.
(E) Proliferation of HEK293 cells was measured after treatment with FA and NA.
(F) The qPCR data shows that flufenamic acid (FA) reduces the expression of genes, such as Axl and NF2 that are stimulated upon YAP overexpression. All the error bars in the figure represent SD.
The central pocket of TEAD is seen in all available TEAD YBD structures, including TEAD1 (PDB code 3KYS), TEAD2 (PDB code 3L15), and TEAD4 (PDB code 4EAZ). The residues lining the central pocket are well conserved among TEAD genes from various species (Figure S1). Thus, it is plausible to assume that the conserved central pocket is important for the biological function of TEADs. We hypothesized that flufenamates, through binding to the central pocket, might disrupt the biological activity of TEADs. To test this hypothesis, we measured the TEAD transcriptional activity in the presence of FA or NA using a TEAD reporter construct (Dupont et al., 2011). The TEAD binding sites are placed upstream of a luciferase reporter, therefore, the expression level of luciferase correlates with TEAD transcriptional activity. We observed significant decrease in luciferase expression level in the presence of flufenamates, FA and NA (Figure 5B). This suggests that FA and NA indeed compromise TEAD function in vivo. We next examined TEAD-YAP mediated expression of Hippo-responsive genes after FA treatment. Expression levels of Hippo-responsive genes, such as NF2, Axl and Jagged-1, were greatly reduced after FA treatment in MCF10A cells (Figure 5C). To ascertain that the observed reduction in gene expressions was due to TEAD binding and not mediated by other targets of NSAID, we tested the expression levels of these genes after treating the cells with mefanamic acid (MA), another NSAID that does not bind to TEAD (Figure 4D). Reassuringly, the Hippo-responsive gene expression was reduced only in FA-treated cells, but not in MA-treated cells (Figure 5C). Because we did not observe the disruption of TEAD-YAP interactions by FA or NA in vitro, it is not exactly clear how flufenamates, such as FA, inhibit the TEAD-YAP mediated gene expression. Future experiments are needed to address this important question.
Since the Hippo-responsive genes promote cell migration and proliferation, we next tested whether cell migration and proliferation were affected in the presence of FA and NA. Interestingly, we observed significant reduction in cell migration and proliferation after treating cells with these flufenamate drugs (Figure 5D and E). These results suggest that cellular processes dependent on gene expression driven by TEAD-YAP are also affected by flufenamate treatment.
We next evaluated whether flufenamates, such as FA, could also inhibit the expression of genes that were induced by YAP overexpression. We generated the stable MCF10A cells that express YAP S127A, the Hippo refractory mutant of YAP. The expression levels of genes, such as Axl and NF2, were measured using qPCR. Our data indicated that the expression of these genes did indeed increase in YAP S127A overexpressing cells (Figure 5F). Again, we observed that the FA treatment in these cells reduced the expression levels of both Axl and NF2 (Figure 5F).
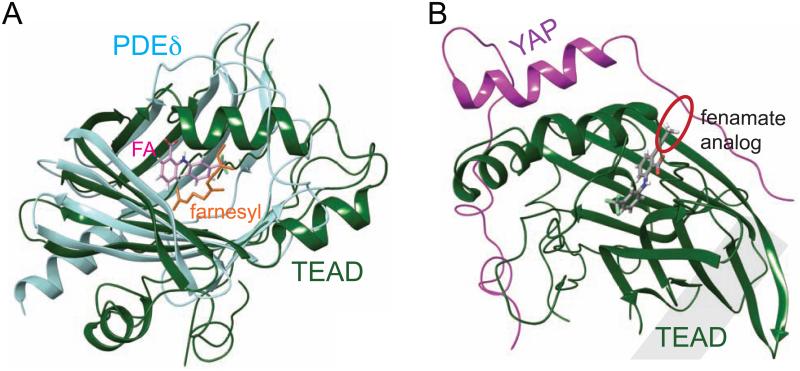
The YAP-binding domain (YBD) of TEADs has a fold similar to that of PDEδ and RhoGDI, which can solubilize the lipid moieties of GTPases, such as Ras and Rho (Figure 6A). In these proteins, farnesyl or geranylgeranyl lipid groups from Ras or Rho also occupy a hydrophobic central pocket similar to that of TEADs. We hypothesize that a fully functional TEAD-YAP transcriptional complex requires the association of TEAD with an unknown lipidated regulator where the lipid group of this protein occupies the central pocket of TEADs. This interaction is disrupted when small molecule inhibitors, such as flufenamates, occupy the central pocket (Figure 6A), which, in turn, compromises the TEAD-YAP function. We propose that the inhibition of TEAD activity observed in our cellular assays is largely due to the binding of flufenamates to the central pocket of TEADs. However, we cannot completely rule out the possibility that the second flufenamate-binding site, which is close to the interface 3 region of TEADs, could also play a synergistic role in inhibition. Fenamate analogs could also be potentially used as a tool to disrupt TEAD-YAP interactions. The entrance of the central pocket in TEADs is close to the surface where YAP interacts with TEADs. Fenamate analog, such as the one shown in Figure 6B, spans the central pocket in TEADs and also extends to the surface at the TEAD-YAP interface. It would be of great interest to test if any of these analogs could disrupt TEAD-YAP interactions.
Figure 6. TEAD and PDEδ Have a Similar Hydrophobic Central Pocket.
(A) Superposition of TEAD-FA and PDEδ-farnesyl (PDB code 3T5I) complexes. Farnesyl (shown as orange sticks) and FA (shown as pink sticks) occupy the central pocket of PDEδ and TEAD, respectively.
(B) A structural model to show that a fenamate analog (grey sticks) extends from the central pocket to interfere with TEAD-YAP interaction (red circle). A propyne substituent was added ortho to the acid of FA and the compound was docked onto the TEAD1-YAP complex (PDB code 3KYS). Bulky substituents in this position may disrupt YAP binding.
In summary, we have shown that specific targeting of the hydrophobic central pocket of TEADs is conceptually and biochemically feasible. Similar approach has been successfully used to disrupt the interaction between PDEδ and KRAS. Compounds that can disrupt the PDEδ-KRAS interaction have been shown to have a great potential to treat pancreatic cancers (Zimmermann et al., 2013). This provides a strong incentive for the development of potent small molecule inhibitors that specifically target the central pocket of TEADs. These inhibitors can potentially be used to suppress the oncogenicity of YAP by disrupting the TEAD activity.
EXPERIMENTAL PROCEDURES
Purification of TEAD4 YBD
The YBD domain of TEAD4, residues 217-434, was expressed as a His-tagged protein in BL21 (DE3) cells. The protein was purified using a two-step procedure, using IMAC and size exclusion chromatography. TEAD dm and TEAD2 YBD was also purified using the same procedure. With the exception of crystallography, the YBD domain of TEAD4 was used in all our assays. TEAD4 did not crystallize either alone or in the presence of drugs. For this purpose the YBD domain of TEAD2, residues 215-445, was used.
Isothermal Titration Calorimetry
ITC experiments were performed on an Auto-iTC200 instrument from Microcal Inc. at 25°C. TEAD in 20 mM HEPES, 150 mM NaCl, 2 mM 2-mercaptoethanol at pH 7.5 was loaded into the ITC cell at concentrations of 100-120 μM with 5 % v/v DMSO solution. Ligands in concentrations of 2.5-5 mM were dissolved in the same buffer/DMSO solution and auto-loaded into the syringe. Each titration was carried out with 18 injections of 2.4 μl were performed over a period of 30 min with stirring at 1000 rpm. Commercial ligands were purchased from Sigma Aldrich, VitasM laboratories, Ark Pharm Inc and Enamine.
Differential Scanning Fluorimetry
Thermal shift experiments were performed on a Roche LC480 PCR machine in a 384-well plate format. Fragments were tested at concentrations of 5 mM and Pharmakon library compounds were tested at concentrations of 20 μM. Each well consisted of 4 μM TEAD and spyro orange.
STD NMR
STD NMR experiments were performed on a Bruker 600 MHz NMR machine equipped with cyroprobe. FA at 2.5 mM was prepared in a buffer containing 20 mM HEPES, 150 mM NaCl and 10% D2O. Proton NMR experiment with water suppression was collected and processed using Topspin (2.1). STD experiments were conducted in the absence and presence of 30 μM TEAD4.
Crystallization and Data collection
Purification of recombinant human TEAD2 YBD was carried out as previously described (Tian et al., 2010). Purified TEAD2 YBD was concentrated to 4 mg/ml in a buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, 2 mM MgCl2, 1 mM TCEP and 5% glycerol (v/v). The TEAD2 YBD domain was crystallized at 20°C using the hanging-drop vapor-diffusion method with a reservoir solution containing 0.1 M HEPES (pH 7.2) and 2.4 M sodium formate. The crystals were soaked and cryo-protected with reservoir solution supplemented with 3 mM inhibitors (FA or BFA) and 25% glycerol (v/v) for one hour and then flash-cooled in liquid nitrogen. Crystals exhibited the symmetry of space group C2 and contained two molecules per asymmetric unit. Diffraction data were collected at beamline 19-ID (SBC-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois, USA) and processed with HKL3000. Phases were obtained by molecular replacement with Phaser (McCoy et al., 2007) using the crystal structure of human TEAD2 YBD domain (PDB code: 3L15) as the search model. Iterative model building and refinements were carried out with Coot and PHENIX, respectively (Adams et al., 2010; Emsley et al., 2010). The dataset for TEAD2-BFA was collected at a wavelength near the absorption edge of bromine. Bromide atoms in the bound BFA molecules were confirmed from peaks in the anomalous difference map created with data truncated to 4.5 Å resolution without refining the structure. The final model for TEAD2-FA (Rwork = 17.1%, Rfree = 22.4%) contains 401 residues, 85 water molecules and three flufenamic acid (FA) molecules. The final model for TEAD2-BFA (Rwork = 17.9%, Rfree = 22.7%) contains 401 residues, 109 water molecules and three BFA molecules. MolProbity was used for structure validation to show that all models have good geometry (Davis et al., 2007). Data collection and structure refinement statistics are summarized in Table 1. All structural figures were generated with Schrodinger software suite or Chimera (UCSF)(Pettersen et al., 2004).
Molecular Modeling
The protein-ligand complexes were prepared using the protein preparation wizard in Maestro (Schrödinger, LLC). This included adding hydrogens, removing water molecules beyond 5Å from the ligand and optimizing the hydrogen-bonding network. Finally, the complex was relaxed using the OPLS-2005 force field and the restrained minimization procedure of the protein preparation wizard. The resulting conformation of the ligand was taken as the bioactive conformation. The global energy conformation was found by conformational search using MacroModel (Schrödinger, LLC), the OPLS-2005 force field, GB/SA salvation model and torsional sampling. The search was continued until all low energy conformations were found at least 10 times. The global energy minima was found to be within 2 kJ/mol of the conformation obtained by minimizing the bioactive conformation. 2kJ/mol is within the uncertainty of the method. DFT calculations were done using Jaguar (Schrödinger, LLC) the PBF solvation model with water as solvent and the basis sets are listed in Table 2. Grid density was set to fine and Accuracy level to Accurate. For each bioactive conformation, two optimizations were done, one full optimization and one with all torsions constrained. The strain energy was found by subtracting the energy of the fully optimized bioactive conformation from that of the bioactive conformation optimized with constraints.
Luciferase Reporter Assay
HEK293 cells were grown at 37°C in the presence of 5% CO2 in DMEM supplemented with 10% FBS and antibiotic-antimycotic (Life Technologies). TEAD activity reporter developed by (Dupont et al., 2011) was used in the assay. Cells in 6-cm dish were transfected with 150 ng TEAD firefly luciferase and 25 ng CMV Renilla luciferase constructs. The cells were incubated with FA (150 μM) or NA (150 μM) overnight. At 150 μM concentration the TEAD activity was inhibited by fifty percent. Luciferase assay was performed using Promega Dual luciferase assay kit as per manufacturer guidelines.
Western Blot
The antibodies used in the Western blot in this study includes NF2 - Cell signaling (D1D8); Axl - Cell signaling (C2B12); Jagged1 - Cell signaling (28H8) and Actin - Santa Cruz (C4).
Migration Assay
HEK293 cells were treated with 150 μM FA or NA, and then cells were serum-starved overnight. Around 18,000 cells were seeded into the transwell inserts (BD BioCoat, control inserts) in triplicates. The inserts were immersed in DMEM supplemented with 10% FBS and the cells were allowed to migrate overnight. The membranes were then washed in PBS and fixed in 4% PFA. The migrated cells were visualized through staining with 1% crystal violet.
qPCR
The following TaqMan probes were used, Axl - Hs01064444_m1, NF2 - Hs00966302_m1, 18S - Hs03003631_g1. The qPCR assay was performed as per manufacturer’s guidelines.
Supplementary Material
TEAD transcription factors have a large hydrophobic central pocket
This central pocket is targetable by small molecule inhibitors
Crystal structure of small molecule drug flufenamic acid bound to TEAD
Flufenamates such as flufenamic acid inhibit TEAD activity
Acknowledgments
We thank Xiangyu Ouyang for performing preliminary experiments for the manuscript, Diana R. Tomchick for structure refinement, and Liu Chen Ying for the stable MCF10A cells. This work is financially supported by the Agency for Science, Technology, and Research (A*STAR), Singapore, and NIH/NIGMS R01GM107415 (to X. L.). We thank A*STAR JCO grant No. 1231B015 for G.Y.C’s postdoctoral fellowship support. The US DOE under contract DE-AC02-06CH11357 supported use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A.P., A.V.P. and W.H. conceived the project. A.P. and A.W.H. designed experiments related to drug screening and binding assays. Screening experiments were performed by A.W.H., S.W., N.H., and G.Y.C. C.S.B.C synthesized the YAP peptide. Protein purification and cell biology assays were performed by A.V.P. X-ray crystallography was done by X.H. and X.L. A.P. did molecular modeling. A.V.P. and X.L. wrote the paper.
ACCESSION NUMBERS
The atomic coordinates and structure factors for human TEAD2-FA and TEAD2-BFA have been deposited in the Protein Data Bank with the PDB ID codes 5DQ8 and 5DQE, respectively.
The authors declare no conflicts of interest.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Nakagawa K, Yang Z, Ikeda M, Withanage K, Ishigami-Yuasa M, Okuno Y, Hata S, Nishina H, Hata Y. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J Biochem. 2011;150:199–208. doi: 10.1093/jb/mvr063. [DOI] [PubMed] [Google Scholar]
- Begley DW, Moen SO, Pierce PG, Zartler ER. Saturation transfer difference NMR for fragment screening. Curr Protoc Chem Biol. 2013;5:251–268. doi: 10.1002/9780470559277.ch130118. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, Song H. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepenbruck M, Waldmeier L, Ivanek R, Berninger P, Arnold P, van Nimwegen E, Christofori G. Tead2 expression levels control the subcellular distribution of Yap and Taz, zyxin expression and epithelial-mesenchymal transition. J Cell Sci. 2014;127:1523–1536. doi: 10.1242/jcs.139865. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Kim NG, Gumbiner BM. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc Natl Acad Sci U S A. 2013;110:2569–2574. doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hansen CG, Moroishi T, Guan KL. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucl T, Brody JR, Gallmeier E, Iacobuzio-Donahue CA, Farrance IK, Kern SE. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 2007;67:9055–9065. doi: 10.1158/0008-5472.CAN-07-0474. [DOI] [PubMed] [Google Scholar]
- Hung AW, Silvestre HL, Wen S, Ciulli A, Blundell TL, Abell C. Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase. Angew Chem Int Ed Engl. 2009;48:8452–8456. doi: 10.1002/anie.200903821. [DOI] [PubMed] [Google Scholar]
- Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JF, Shepherd CJ, Rizzo S, Brewer D, Jhavar S, Dodson AR, Cooper CS, Eeles R, Falconer A, Kovacs G, et al. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 2008;99:1849–1858. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109:E2441–2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012 doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Guan KL. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol Sci. 2013;34:581–589. doi: 10.1016/j.tips.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Chan SW, Lee I, Song H, Hong W. Structural and Functional Similarity between the Vgll1-TEAD and the YAP-TEAD Complexes. Structure. 2012;20:1135–1140. doi: 10.1016/j.str.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14 doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano I, McDonald PC, Lock F, Muller WJ, Dedhar S. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- Tian W, Yu J, Tomchick DR, Pan D, Luo X. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc Natl Acad Sci U S A. 2010;107:7293–7298. doi: 10.1073/pnas.1000293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530–1542. doi: 10.1053/j.gastro.2013.02.009. e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal. 2014;7:re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- Xu MZ, Chan SW, Liu AM, Wong KF, Fan ST, Chen J, Poon RT, Zender L, Lowe SW, Hong W, et al. AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2010 doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhang Y, Park HW, Jewell JL, Chen Q, Deng Y, Pan D, Taylor SS, Lai ZC, Guan KL. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27:1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lin Z, Zhou Z, Shen HC, Yan SF, Mayweg AV, Xu Z, Qin N, Wong JC, Rong Y, et al. Structure-Based Design and Synthesis of Potent Cyclic Peptides Inhibiting the YAP-TEAD Protein-Protein Interaction. ACS Med Chem Lett. 2014;5:993–998. doi: 10.1021/ml500160m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.