SUMMARY
Leishmania infantum causes visceral leishmaniasis (VL) in the New World. The diagnosis of VL is confirmed by parasitological and serological tests, which are not always sensitive or specific. Our aim was to design new primers to perform a Polymerase Chain Reaction (PCR) for detecting L. infantum. Sequences of the minicircle kinetoplast DNA (kDNA) were obtained from GenBank, and the FLC2/RLC2 primers were designed. Samples of DNA from L. infantum, Leishmania amazonensis,Leishmania braziliensis, Leishmania guyanensis, Leishmania naiffi, Leishmania lainsoni, Leishmania panamensis,Leishmania major and Trypanosoma cruzi were used to standardize the PCR. PCR with FLC2/RLC2 primers amplified a fragment of 230 bp and the detection limit was 0.2 fg of L. infantum DNA. Of the parasite species assayed, only L. infantum DNA was amplified. After sequencing, the fragment was aligned to GenBank sequences, and showed (99%) homology with L. infantum. In the analysis of blood samples and lesion biopsy from a dog clinically suspected to have VL, the PCR detected DNA from L. infantum. In biopsy lesions from humans and dogs with cutaneous leishmaniasis, the PCR was negative. The PCR with FLC2/RLC2 primers showed high sensitivity and specificity, and constitutes a promising technique for the diagnosis of VL.
Keywords: Visceral leishmaniasis, Polymerase Chain Reaction, Leishmania infantum, Diagnosis
RESUMO
Leishmania infantum causa leishmaniose visceral (LV) no Novo Mundo. O diagnóstico de LV é confirmado por testes parasitológicos e sorológicos, os quais nem sempre são sensíveis ou específicos. Nosso objetivo foi desenhar novos iniciadores para realizar uma Reação em Cadeia da Polimerase (PCR) para detecção de L. infantum. Sequências do DNA do minicírculo do cinetoplasto (kDNA) foram obtidos do GenBank, e os iniciadores FLC2/RLC2 foram desenhados. Amostras de DNA de L. infantum,Leishmania amazonensis, Leishmania braziliensis, Leishmania guyanensis,Leishmania naiffi, Leishmania lainsoni,Leishmania panamensis, Leishmania major e Trypanosoma cruzi foram usados para padronizar a PCR. PCR com iniciadores FLC2/RLC2 amplificou um fragmento de 230 pb e detectou 0,2 fg de DNA de L. infantum. Das espécies de parasitos analisadas, somente DNA de L. infantum foi amplificado. Após sequenciamento, o fragmento foi analisado no GenBank, que mostrou homologia com L. infantum. Em análises de amostras de sangue e lesão de cão com suspeita clínica de LV, a PCR detectou DNA deL. infantum. Em amostras de lesão de humanos e cães com leishmaniose cutânea, a PCR foi negativa. A PCR padronizada com os iniciadores FLC2/RLC2 mostrou alta sensibilidade e especificidade, sendo técnica promissora para o diagnóstico de LV.
INTRODUCTION
Visceral leishmaniasis (VL) is an infectious, parasitic disease with a worldwide distribution; India, Sudan, Nepal and Brazil account for 90% of cases35. In the New World, VL is caused byLeishmania infantum (syn. Leishmania chagasi), and in Brazil there were 74,980 cases of VL from 1990 to 20137. VL is a severe and eventually lethal disease, and produces signs and symptoms common to other diseases, making its diagnosis complex and time-consuming19.
Confirmation of clinical suspicion of VL is based on clinical, epidemiological and laboratory tests. Among the conventional diagnostic methods for confirmation of VL are parasitological and serological tests8. Parasitological examination can directly demonstrate the presence of L. infantum in aspirates from the liver, spleen, lymph nodes and bone marrow. However, due to the low sensitivity of direct microscopic examination many patients do not receive confirmation of the diagnosis2. The serological tests include the indirect immunofluorescence assay (IFA) and enzyme-linked immunosorbent assay (ELISA)3 , 14. However, these techniques may produce false negative results and cross-reactions
Molecular biology techniques have been proposed as an alternative in the diagnosis of VL. The Polymerase Chain Reaction (PCR) enables rapid diagnosis of disease, is more sensitive than conventional methods, and can help to identify the parasite species27 , 30. Another advantage is that PCR can be performed in different biological samples, such as human and canine blood, lymph nodes, buffy coat, canine conjunctival scrapings, urine and lesions10,12,16,17,21,24.
In view of the limitations of conventional methods, it is necessary to standardize new methodologies for the diagnosis of VL. PCR has proven to be a valuable, rapid and sensitive tool, and can identify the parasite species in different clinical samples from humans, reservoirs and vectors. The aim of this study was to design new primers to perform a PCR for detecting L. infantum.
MATERIAL AND METHODS
Design of oligonucleotides: The specific forward and reverse PCR primers FLC2 (5'-GTCAGTGTCGGAAACTAATCCGC-3') and RLC2 (5'-GGGAAATTGGCCTCCCTGAG-3') were designed to amplify a 230 bp segment of the conserved region of the DNA minicircle kinetoplast (kDNA) of L. chagasi [GenBank:AF308682 LOCUS] with 716 bp (Fig. 1). The used sequence to design the primers was analyzed by CLUSTAL W (www.ebi.ac.uk) and primer-BLAST 2.0 server ("Basic Alignment Search Tool") of the National Center for Biotechnology Information (NCBI) to detect their specificity to the proposed PCR template.
Fig. 1. - Leishmania chagasi kinetoplast minicircle DNA, complete sequence (GenBank: AF308682 LOCUS) with 716 bp and the primers localization. The underlined bold sequences represent the FLC2/RLC2 primers.

Clinical samples: The sensitivity and specificity of primers FLC2/ RLC2 were analyzed in clinical samples: lesion biopsy and blood from a dog with clinical suspicion of VL, lesions biopsies from three dogs with cutaneous leishmaniasis, and five samples obtained by scraping the edge of lesions from humans with cutaneous leishmaniasis. The samples were placed in eppendorf vials with 50 μL STE buffer (10 mM TRIS, 1 mM EDTA, 0.1 M NaCl, pH 8.0) and stored at -20 ºC for later DNA extraction. The present study received approval from the Permanent Committee for Ethics in Research involving Humans (Process No. 533/2009) of theUniversidade Estadual de Maringá. All participants were informed about the importance and objectives of the study, and were assured of both anonymity and confidentiality. We obtained written informed consent from patients who agreed to participate. All procedures involving humans were conducted according to protocols approved by the National Health Council of the Brazilian Ministry of Health (Resolution No. 196/1996).
An aliquot of 2 mL of dog blood was added to an equal volume of ACD solution (25 mM citric acid, 50 mM sodium citrate, 81 mM glucose), and centrifuged at 3,500 g for 10 min. The buffy coat was then removed and added to a tube containing ACD and stored at -20 ºC for later DNA extraction.
Two hamsters were inoculated in the footpad with 50 μL of Leishmaniaculture from a lesion and venous blood of a dog with clinical suspicion of VL. After seven months, the hamsters were euthanized, and material from their regional lymph node was placed in culture medium for the isolation of the parasite and later DNA extraction. All procedures involving animals followed the Ethical Principles of Animal Experimentation established by the Brazilian College of Animal Experimentation (COBEA) and were performed according to protocols approved by the Committee for Ethical Conduct in Animal Experimentation of the Universidade Estadual de Maringá (report 015/2007 on 04/03/2007).
Parasites: L. infantum, Leishmania amazonensis, Leishmania braziliensis, Leishmania guyanensis, Leishmania naiffi, Leishmania lainsoni, Leishmania panamensis, Leishmania major and Trypanosoma cruzi (Y strain) were employed for assessing the specificity and sensitivity of PCR. Samples of L. braziliensis from different serodemes obtained from dogs and humans in the northwestern of the Parana State were also analyzed (Table 1).
Table. 1 Parasite species used in the study 1: Isolates from humans with cutaneous leishmaniasis attended at Laboratório de Ensino e Pesquisa em Análises Clínicas, Universidade Estadual de Maringá, Maringá, Paraná and identified atInstituto Evandro Chagas, Belém, Pará. 2: Provided by Dr. Jeffrey J. Shaw, Instituto Evandro Chagas. 3: Provided by Dra. Vanete T. Soccol, Universidade Federal do Paraná, Curitiba, Paraná. 4: Provided by Dr. Carlos H. N. Costa, Instituto de Doenças Tropicais Natan Portella, Universidade Federal do Piauí, Teresina, Piauí. 5: Provided by Dr. Mônica L. Gomes, Departamento de Ciências Básicas da Saúde, Universidade Estadual de Maringá, Maringá, Paraná.6: Provided by Collection of Leishmania(CLIOC), Instituto Oswaldo Cruz. 7:Isolate from dog with cutaneous leishmaniasis attended at Laboratório de Ensino e Pesquisa em Análises Clínicas da Universidade Estadual de Maringá, Maringá, Paraná and identified at Instituto Evandro Chagas, Belém, Pará.
| Species | Isolates | International Code | Sample Source |
|---|---|---|---|
| Leishmania (Viannia) braziliensis - serodeme I | M14405 | MHOM/BR/1993/M14405 | 1 |
| Leishmania (Viannia) braziliensis - serodeme II | M15490 | MHOM/BR/1995/1077 | 1 |
| Leishmania (Viannia) braziliensis - serodeme III | M14046 | MHOM/BR/1992/M14046 | 1 |
| Leishmania (Viannia) braziliensis - serodeme VII | M20932 | MHOM/BR/2002/M20932 | 1 |
| Leishmania (Viannia) braziliensis - serodeme I | M11272 | MHOM/BR/1987/M11272 | 1 |
| Leishmania (Viannia) braziliensis - serodeme I | M20171 | CAN/BR/2001/M20171 | 7 |
| Leishmania (Viannia) braziliensis | M2903 | MHOM/BR/1975/M2903 | 2 |
| Leishmania (Leishmania) infantum | CUR392 | MCAN/BR/2010/CUR392 | 3 |
| Leishmania (Leishmania) infantum | PP75 | MHOM/BR/1974/PP75 | 4 |
| Leishmania (Leishmania) infantum | L2906 | MHOM/BR/2002/LPC-RPV | 6 |
| Leishmania (Leishmania) amazonensis | M2269 | MHOM/BR/1973/M2269 | 2 |
| Leishmania (Viannia) lainsoni | M6426 | MHOM/BR/1981/M6426 | 2 |
| Leishmania (Viannia) panamensis | M4037 | MHOM/PA/1967/BOYTON | 2 |
| Leishmania (Viannia) naiffi | M5533 | MDAS/BR/1979/M5533 | 2 |
| Leishmania (Leishmania) major | LV39 | MRHO/SU/1959/P | 2 |
| Leishmania (Viannia) guyanensis | M4147 | MHOM/BR/1975/M4147 | 2 |
| Trypanosoma cruzi | Y strain | 5 |
Promastigotes of Leishmania spp. were grown in Medium 199 (Invitrogen, Paisley, Scotland, UK) supplemented with 1% female human urine, 10% fetal-calf serum (Invitrogen, Paisley, Scotland, UK) and 2 mM L-glutamine (Gibco-BR, Paisley, Scotland, UK), at 25 °C in a B.O.D. incubator (Logen Scientific, Brazil). Epimastigotes of T. cruzi were grown in LIT (Liver Infusion Tryptose) at 25 °C. The parasites were washed by centrifugation at 1,600 g for 10 min with phosphate-buffered saline (PBS) pH 7.2. Parasites were counted in a Neubauer chamber and stored at -18 °C until DNA extraction.
DNA preparation: The DNA from scrapings of skin lesions was extracted according to VENAZZI et al. 33 and from the canine lesion biopsy was extracted with a Puregene kit (Gentra, USA). The DNA from blood was extracted with guanidine isothiocyanate and phenol34 and the DNA from Leishmania isolated from hamsters was obtained by heating22. The DNA ofLeishmania spp. and T. cruzi was extracted with guanidine isothiocyanate and phenol34. The DNA was re-suspended in 50 μL of TE buffer (1 M Tris, 0.5 M Na2EDTA, pH 8.0) and stored at -18 °C until use.
The DNA samples were quantified by fluorometry using a Quant-iT(tm) dsDNA BR Assay Kit (Invitrogen, Eugene, OR, USA) and a Qubit(tm) Fluorometer Kit (Invitrogen, USA).
Polymerase Chain Reaction: For PCR standardization, some conditions were tested. The reaction mixture contained 0.5 μM of each primer (Invitrogen Life Technologies, Sao Paulo, Brazil), 0.2 mM dNTP (Invitrogen, Carlsbad, CA, USA), 1 U Taq DNA polymerase (Invitrogen Life Technologies, Sao Paulo, Brazil), 1.5 or 2.0 mM MgCl2, 1X enzyme buffer and 2 μL of the DNA sample in a final volume of 25 μL. Ultrapure water was used as a negative control. Amplification was performed in a thermocycler (Biometra Personal Cycler, Germany) under the following conditions: initial denaturation at 95 °C for five min, followed by 30 or 35 cycles of one min at 95 °C, one min at 56 °C or 58 oC and one min at 72 °C. After 10 min at 72 °C, the amplified material was maintained at 4 °C until analysis. The amplified product was analyzed by electrophoresis in 2% agarose gel (Invitrogen, Paisley, Scotland, UK) at 10-15 V/cm. The gel was stained with ethidium bromide and analyzed under UV light in a transilluminator (MacroVue(tm) UV-20, Hoefer). A 100 bp DNA ladder (Invitrogen Life Technologies, São Paulo, Brazil) was used as a molecular marker.
Analysis of the sensitivity and specificity of FLC2/RLC2 primers in parasites:Assessment of PCR sensitivity (FLC2/RLC2 primers) was performed using DNA extracted from L. infantum (MHOM/ BR/1974/PP75) at concentrations of 200 ng to 0.002 fg. To determine the specificity, DNA (100 pg) from different species of Leishmania, two strains of L. infantum,and T. cruzi were analyzed (Table 1). DNA (100 pg) from L. braziliensis (serodemes I, II, III and VII) obtained from patients in northwestern Paraná and identified by reactivity with monoclonal antibodies at the Instituto Evandro Chagas (Belém, Pará, Brazil) was also analyzed.
Analysis of the sensitivity and specificity of primers FLC2/RLC2 in clinical samples: DNA samples from the blood and lesion of a dog with clinical suspicion of VL, living in a VL-endemic area, and DNA extracted fromLeishmania isolated from hamsters that were inoculated with lesions and blood from a dog with clinical suspicion of VL were analyzed to assess the sensitivity of the primers.
DNA samples from lesions of five humans and three dogs, all with a confirmed diagnosis of cutaneous leishmaniasis [direct parasite search, IFA and PCRLeishmania (Viannia)] were also analyzed to verify the specificity of the primers. The PCR Leishmania (Viannia) was carried out using the MP3H and MP1L primers that amplify a fragment of 70 bp of the conserved region of DNA from the minicircle of the kinetoplast (kDNA) of the subgenus Leishmania (Viannia) 18.
Sequencing reaction: For the sequencing reaction, PCR products from two strains of L. infantum (MHOM/BR/1974/PP75 and MHOM/ BR/2002/LPC-RPV) were purified (Axygen Kit) and quantified using the Nanodrop Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA). Samples were diluted to a concentration of 4 ng/μL. For sequencing, 1 μL of Big Dye Terminator v3.1, 0.4x dilution buffer, 0.4 mM of each primer (forward and reverse), and 2 μL of sample, totaling 10 μL of reaction volume were used. The reaction was carried out in an Applied Biosystems Thermal Cycler, with 1 cycle at 96° C for one min, followed by 30 cycles at 96° C for 15 sec, 56 °C for 15 sec and 60 °C for four min, and stored at 4 °C overnight. The sequencing product was precipitated and purified with ethanol/EDTA according to the protocol. Next, it was re-suspended in 10 μL of formamide, and sequenced in an ABI 3500XL automated sequencer (Applied Biosystems).
Alignment of sequences: Sense and anti-sense strand sequences of the kDNA obtained were examined in the DNAstar Lasergene SeqMan v.7.00 package, which produced a consensus strand for each sample. The sequences were aligned using the software MEGA 532 for each sample that produced a single sequence kDNA. The single sequence was aligned using the primer-BLAST 2.0 server of the NCBI.
RESULTS
Initially, the conditions for the PCR, including the MgCl2 concentration, the number of cycles, and the annealing temperature for the primers were optimized. The annealing temperatures tested were based on the GC content and length of the FLC2/RLC2 primers. The condition in which only one fragment was amplified was chosen. The reaction mixture contained 0.5 μM of each primer, 1 U Taq DNA polymerase, 2.0 mM MgCl2, 1X enzyme buffer and 2 μL of the DNA sample in a final volume of 25 μL. Amplification was performed at 95 °C for five min, 35 cycles of one min at 95 °C, one min at 56 °C and one min at 72 °C, followed by 10 min at 72 °C.
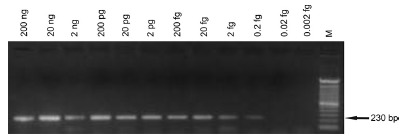
With regard to the sensitivity of the PCR, the FLC2/RLC2 primers amplified a DNA segment of 230 bp in the presence of quantities of DNA template equal to or greater than 0.2 fg (Fig. 2).
Fig. 2. - Gel showing the analytical sensitivity of the 230 bp fragment of the kDNA of Leishmania infantum. The PCR with FLC2/RLC2 primers was carried out using as template the serially diluted DNA extracted from promastigotes of L. infantum(MHOM/BR/1974/ PP75). M, 100 bp molecular marker.
Analysis of DNA from different species of Leishmania (100 pg) andT. cruzi (100 pg) showed that only a 230 bp fragment fromL. infantum was amplified. No fragment was observed with DNA ofL. braziliensis isolated from dogs and humans (100 pg). In all PCR analyses performed in this study, only L. infantum samples showed the 230 bp expected amplification product indicating that there was no
contamination or reaction inhibition (Fig. 3). Analysis of DNA samples from different human patients with a confirmed diagnosis (Fig. 4A) of cutaneous leishmaniasis indicated that the primers showed specificity for L. infantum.Figure 4B shows the specificity of the primers in DNA samples from lesions of dogs diagnosed with cutaneous leishmaniasis (parasitological, serological and molecular diagnoses) and a DNA sample from the blood and lesion of a dog with clinically suspected VL
Fig. 3. - Gel showing specificity of the 230 bp fragment of the kDNA of Leishmania infantum. The PCR with FLC2/RLC2 primers was carried out using as template the DNA extracted from promastigotes of Leishmania species (100 pg), epimastigotes of T. cruzi (100 pg) and L. braziliensis isolates from humans and dogs (100 pg). kDNA, kinetoplast DNA; M, 100 bp molecular marker. Panel A. Lane 1: L. amazonensis; Lane 2: L. lainsoni;Lane 3: L. panamensis; Lane 4: L. guyanensis; Lane 5: L. braziliensis(MHOM/BR/1987/M11272); Lane 6: L. infantum(MHOM/BR/1974/PP75); Lane 7: negative control (water).Panel B. Lane 1: L. naiffi; Lane 2: L. major; Lane 3: T. cruzi; Lane 4: L. infantum (MHOM/BR/1974/PP75); Lane 5: negative control (water). Panel C. Lane 1: L. braziliensis (CAN/BR/2001/M20171) isolated from dog (serodeme I); Lane 2: L. braziliensis(MHOM/BR/1993/M14405) isolated from human (serodeme I); Lane 3:L. braziliensis isolated from human (serodeme II); Lane 4: L. braziliensis isolated from human (serodeme III); Lane 5: L. braziliensis isolated from human (serodeme VII); Lane 6: L. braziliensis(MHOM/BR/1987/M11272) isolated from human (serodeme I); Lane 7: L. infantum(MHOM/BR/1974/PP75).
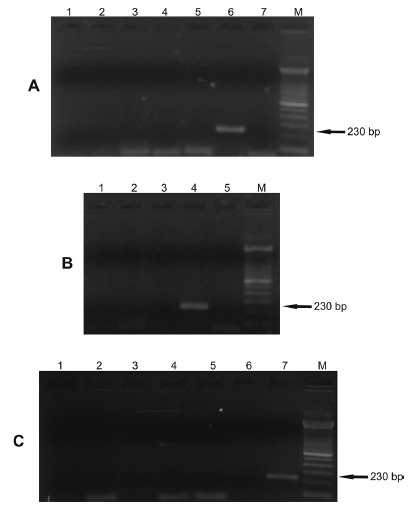
Fig. 4. - Gel showing the specificity of the 230 bp fragment of the kDNA of Leishmania infantum in clinical samples. The PCR with FLC2/RLC2 primers was carried out using as template the DNA extracted from clinical samples. kDNA, kinetoplast DNA; M, 100 bp molecular marker. Panel A. Lanes 1, 2, 3, 4 and 5: DNA from lesions in different humans with confirmed diagnosis of cutaneous leishmaniasis (parasitological, serological and molecular diagnoses); Lane 6: L. braziliensis (MHOM/BR/1987/M11272); Lane 7:L. infantum (MHOM/BR/1974/PP75). Panel B. Lane 1: DNA from the lesion of a dog with clinical suspicion of VL; Lane 2: DNA from the blood sample of a dog clinically suspected to have VL; Lanes 3, 4 and 5: DNA from lesions of different dogs with confirmed diagnosis of cutaneous leishmaniasis (parasitological, serological and molecular diagnoses); Lane 6:L. braziliensis (MHOM/BR/1987/M11272); Lane 7:L. infantum (MHOM/BR/1974/PP75). Panel C.Lane 1: DNA from promastigotes isolated from a hamster inoculated with tissues from the lesion of a dog with clinical suspicion of VL (Isolate I); Lane 2: DNA from promastigotes isolated from a hamster inoculated with the blood sample of a dog with clinical suspicion of VL (Isolate II); Lane 3: L. infantum (MCAN/ BR/2010/CUR392); Lane 4: L. infantum(MHOM/BR/1974/PP75).
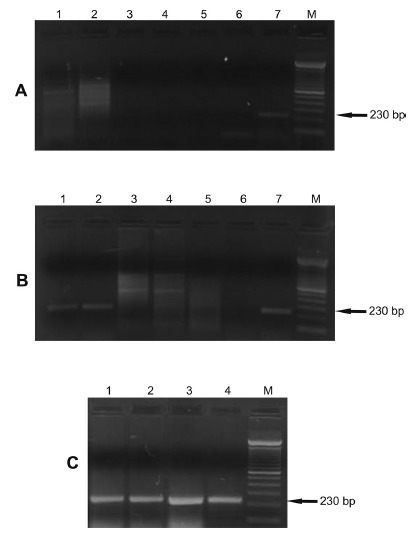
The specificity of the FLC2/RLC2 primers for different isolates of L. infantum was also analyzed (Fig. 4C). DNA of promastigotes isolated from two hamsters inoculated with the blood and lesion of a dog with clinical suspicion of VL showed an amplified fragment of 230 bp. The DNA from another strain of L. infantum also showed this fragment.
No false positive result or inhibition of the reaction was observed in the controls.
After the two samples were sequenced, the consensus strands containing 230 bp were identical, and the sequence alignment to GenBank sequences showed homology withL. chagasi (99%), L. infantum, and L. donovani (89%) (Fig. 5).
Fig. 5. - Alignment of PCR product obtained showing homology with DNA minicircle kinetoplast (kDNA) of isolates from GenBank. IncludingL. chagasi (GenBank accession No. AF308682.1|AF308682), L. infantum strain IranJWinf (GenBank accession No. AB678348.1), L. infantum strain MCAN/ES/98/10445 (GenBank accession No. EU437407.1) and L. donovani (GenBank accession No. L19877.1), respectively. Underlined nucleotides represent additional bases and bolded positions represent different bases.

DISCUSSION
In endemic areas where it is possible to find patients with different forms of leishmaniasis, detecting the correct parasite species of the genusLeishmania in a clinical sample is very important to confirm a suspected case of visceral or cutaneous leishmaniasis. Although no autochthonous cases of VL have been found in Parana, the neighboring states of Sao Paulo and Mato Grosso do Sul are endemic areas for VL. Therefore, it is necessary to standardize methodologies that allow rapid and safe diagnosis of VL, as well as for disease control.
A number of molecular techniques for the identification of Leishmaniahave been reported, such as RFLP-PCR (restriction fragment length polymorphism-PCR)6 , 29, RAPD (random amplified polymorphic DNA)5 , 26 and multiplex-PCR. Although these methods require technical skills of professional and equipped laboratories in their implementation, the continuing search for specific targets, safer and faster molecular diagnosis seems to be more advantageous than using conventional techniques.
The application of PCR in the identification of L. infantum has great potential as a valuable tool in the detection of parasite DNA. PCR can identify the species of parasite involved, and is more sensitive than microscopy and serology1 , 28. Under the conditions used here, the
standardization of PCR with FLC2/RLC2 primers showed high specificity and sensitivity, confirming the results obtained in the primer-BLAST 2.0 software of the NCBI.
The sensitivity obtained (0.2 fg) was similar to the related by OLIVEIRA et al. 25 (2 fg) and DE BRUJIN & BARKER11 (1 fg) but it was higher than the one related by SILVEIRA NETO et al. 31 (1 pg/μL) in a PCR for L. infantum. The high sensitivity was checked by the positive results obtained with low concentrations of DNA from L. infantum, demonstrating the ability to detect parasite DNA in peripheral blood and lesions, even in situations where the number of parasites is low. The primers were specific for L. infantum, since they amplify a 230 bp fragment of DNA only from L. infantum. The several different species
of Leishmania and serodemes of L. braziliensistested did not show any amplification. The sequenced fragment showed homology with the L. chagasi (99%), L. infantum and L. donovani (89%) available in the GenBank. The high specificity of the FLC2/RLC2 primers could also be seen in the clinical samples from dogs and humans diagnosed with cutaneous leishmaniasis.
MOREIRA et al. 23 compared the effectiveness of parasitological, immunological and molecular methods for diagnosing dogs with different clinical signs of VL. In samples of lymph-node aspirates from dogs, PCR showed 100% sensitivity in symptomatic, 96% in mildly symptomatic, and 95.5% in asymptomatic dogs. According to QUARESMA et al. 29, in canine visceral leishmaniasis (CVL), PCR can detect infection even when conventional tests fail to diagnose the disease. QUARESMA et al. 29 suggested PCR as a method for detection of CVL since it is highly reliable, sensitive and reproducible, requires a relatively short time for assay, and can turn into a valuable tool for epidemiological surveillance in endemic areas.
ANTINORI et al. 4 compared conventional tests with PCR on peripheral blood samples and bone-marrow aspirates from patients with VL who were infected, or not, with HIV, and reported that PCR showed 98.5% sensitivity in peripheral blood and 95.7% in bone-marrow aspirates. ALAM et al. 1 performed PCR on DNA extracted from blood collected on filter paper, and detected parasites in 38 of 39 samples tested, in contrast to direct microscopy, which gave positive results for only 66.7% of the samples. BRUSTOLONI et al. 9 extracted DNA from histological sections of patients with positive parasitological diagnoses for VL, and the PCR showed 94.8% sensitivity.
In this study, PCR with FLC2/RLC2 primers proved to be a reliable tool for the detection of DNA from L. infantum. Showing high sensitivity and specificity, PCR with FLC2/RLC2 primers detected the parasite in clinical samples of peripheral blood and lesions. In addition, this method does not involve laborious, invasive or complex procedures.
ACKNOWLEDGMENTS
Kézia Peres Gualda performed the experiments, participated in the sequence alignment, analyzed the data, and drafted the manuscript. Lílian Mathias Marcussi performed the experiments. Herintha Coeto NeitzkeAbreu performed the experiments, analyzed the data, participated in the sequence alignment, and drafted the manuscript. Sandra Mara Alessi
Aristides participated in the design of the study. Maria Valdrinez Campana Lonardoni participated in the design of the study. Rosilene Fressatti Cardoso participated in the design of the study, in the sequence alignment, and analyzed the data. Thaís Gomes Verzignassi Silveira participated in the design and coordination of the study, analyzed the data, and drafted the manuscript. All authors read and approved the final manuscript.
Funding Statement
The authors would like to thank the Complexo de Centrais de Apoio à Pesquisa (COMCAP-UEM) for their collaboration in the use of equipments. The study received financial support from the Fundação Araucária and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - award number 410550/2006-0).
Footnotes
The authors would like to thank the Complexo de Centrais de Apoio à Pesquisa (COMCAP-UEM) for their collaboration in the use of equipments. The study received financial support from the Fundação Araucária and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - award number 410550/2006-0).
REFERENCES
- 1.Alam MZ, Shamsuzzaman AK, Kuhls K, Schõnian G. PCR diagnosis of visceral leishmaniasis in an endemic region, Mymensingh district, Bangladesh. Trop Med Int Health. 2009;14:499–503. doi: 10.1111/j.1365-3156.2009.02254.x. [DOI] [PubMed] [Google Scholar]
- 2.Al-Jawabreh A, Schoenian G, Hamarsheh O, Presber W. Clinical diagnosis of cutaneous leishmaniasis: a comparison study between standardized graded direct microscopy and ITS1- PCR of Giemsa-stained smears. Acta Trop. 2006;99:55–61. doi: 10.1016/j.actatropica.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Alvar J, Cañavate C, Molina R, Moreno J, Nieto J. Canine leishmaniasis. Adv Parasitol. 2004;57:1–88. doi: 10.1016/S0065-308X(04)57001-X. [DOI] [PubMed] [Google Scholar]
- 4.Antinori S, Calattini S, Longhi E, Bestetti G, Piolini R, Magni C. Clinical use of polymerase chain reaction performed on peripheral blood and bone marrow samples for the diagnosis and monitoring of visceral leishmaniasis in HIV-infected and HIV uninfected patients: a single-center, 8-year experience in Italy and review of the literature. Clin Infect Dis. 2007;44:1602–1610. doi: 10.1086/518167. [DOI] [PubMed] [Google Scholar]
- 5.Baptista C, Schubach AO, Madeira MF, Leal CA, Pires MQ, Oliveira FS. Leishmania (Viannia)braziliensisgenotypes identified in lesions of patients with atypical or typical manifestations of tegumentary leishmaniasis: evaluation by two molecular markers. Exp Parasitol. 2009;121:317–322. doi: 10.1016/j.exppara.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Botilde Y, Laurent T, Tintaya WQ, Chicharro C, Cañavate C, Cruz I. Comparison of molecular markers for strain typing ofLeishmania infantum . Infect Genet Evol. 2006;6:440–446. doi: 10.1016/j.meegid.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Brasil. Ministério da Saúde . Casos confirmados de leishmaniose visceral, Brasil, grandes regiões e unidades federadas, 1990 a 2013. 2014. http://portalsaude.saude.gov.br/images/pdf/2014/setembro/09/LV-Casos.pdf [Google Scholar]
- 8.Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde . Leishmaniose visceral grave: normas e condutas. Brasília: Ministério da Saúde; 2006. [Google Scholar]
- 9.Brustoloni YM, Lima BR, da Cunha VR, Dorval ME, Oshiro ET, de Oliveira ALL. Sensitivity and specificity of polymerase chain reaction in Giemsa-stained slides for diagnosis of visceral leishmaniasis in children. Mem Inst Oswaldo Cruz. 2007;102:497–500. doi: 10.1590/s0074-02762007005000036. [DOI] [PubMed] [Google Scholar]
- 10.Cascio A, Calattini S, Colomba C, Scalamogna C, Galazzi M, Pizzuto M. Polymerase chain reaction in the diagnosis and prognosis of Mediterranean visceral leishmaniasis in immunocompetent children. Pediatrics. 2002;109: doi: 10.1542/peds.109.2.e27. [DOI] [PubMed] [Google Scholar]
- 11.De Brujin MHL, Barker DC. Diagnosis of New World leishmaniasis: specific detection of species of theL braziliensis complex by amplification of kinetoplast DNA. . Acta Trop. 1992;52:45–58. doi: 10.1016/0001-706x(92)90006-j. [DOI] [PubMed] [Google Scholar]
- 12.de Queiroz NMGP, da Silveira RCV, de Noronha ACF Jr, Oliveira TMFS, Machado RZ, Starke-Buzetti WA. Detection ofLeishmania (L.) chagasiin canine skin. Vet Parasitol. 2011;178:1–8. doi: 10.1016/j.vetpar.2010.12.033. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira EC, de Lana M, Carneiro M, Reis AB, Paes DV, Silva ES. Comparison of serological assays for the diagnosis of canine visceral leishmaniasis in animals presenting different clinical manifestations. Vet Parasitol. 2007;146:235–241. doi: 10.1016/j.vetpar.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Gradoni L. The diagnosis of canine leishmaniasis; Canine leishmaniasis: moving towards a solution; Proceedings of the Second International Canine Leishmaniasis Forum ; 6-9 February 2002; Sevilla: Intervet International; 2002. pp. 7–14. [Google Scholar]
- 15.Harris E, Kropp G, Belli A, Rodriguez B, Agabian N. Single-step multiplex PCR assay for characterization of New WorldLeishmania complexes. J Clin Microbiol. 1998;36:1989–1995. doi: 10.1128/jcm.36.7.1989-1995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachaud L, Chabbert E, Dubessay P, Reynes J, Lamothe J, Bastien P. Comparison of various sample preparation methods for PCR diagnosis of visceral leishmaniasis using peripheral blood. J Clin Microbiol. 2001;39:613–617. doi: 10.1128/JCM.39.2.613-617.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachaud L, Marchergui-Hammani S, Chabbert E, Dereure J, Dedet JP, Bastien P. Comparison of six PCR methods using peripheral blood for detection of canine visceral leishmaniasis. J Clin Microbiol. 2002;40:210–215. doi: 10.1128/JCM.40.1.210-215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez M, Ingá R, Cangalaya M, Echevarria J, Llanos-Cuentas A, Orrego C. Diagnosis ofLeishmaniausing the polymerase chain reaction: a simplified procedure for field work. Am J Trop Med Hyg. 1993;49:348–356. doi: 10.4269/ajtmh.1993.49.348. [DOI] [PubMed] [Google Scholar]
- 19.Luz ZMP, Carneiro M, Schall V, Rabello A. The organization of health services and visceral leishmaniasis: an integrated intervention to improve diagnosis and treatment. Cad Saude Publica. 2009;25:1177–1184. doi: 10.1590/s0102-311x2009000500024. [DOI] [PubMed] [Google Scholar]
- 20.Maia C, Campino L. Methods for diagnosis of canine leishmaniasis and imune response to infection. Vet Parasitol. 2008;158:274–287. doi: 10.1016/j.vetpar.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 21.Manna L, Vitale F, Reale S, Caracappa S, Pavone LM, Morte RD. Comparison of different tissue sampling for PCR based diagnosis and follow-up of canine visceral leishmaniosis. Vet Parasitol. 2004;125:251–262. doi: 10.1016/j.vetpar.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Marcussi VM, Marcussi LM, Barbosa-Tessmann IP, Lonardoni MVC, Silveira TGV. Leishmania (Viannia) braziliensis: new primers for identification using polymerase chain reaction. Exp Parasitol. 2008;120:300–305. doi: 10.1016/j.exppara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Moreira MAB, Luvizotto MCR, Garcia JF, Corbett CEP, Laurenti MD. Comparison of parasitological, immunological and molecular methods for the diagnosis of leishmaniasis in dogs with different clinical signs. Vet Parasitol. 2007;145:245–252. doi: 10.1016/j.vetpar.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Motahareh M, Fakhar M, Motazedian MH, Hatam G, Mikaeili F. A urine-based polymerase chain reaction method for the diagnosis of visceral leishmaniasis in immunocompetent patients. Diagn Microbiol Infect Dis. 2008;60:151–154. doi: 10.1016/j.diagmicrobio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira DM, Lonardoni MVC, Teodoro U, Silveira TGV. Comparison of different primes for PCR-based diagnosis of cutaneous leishmaniasis. Braz J Infect Dis. 2011;15:20410–20410. doi: 10.1016/s1413-8670(11)70176-3. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira JP, Fernandes F, Cruz AK, Trombela V, Monteiro E, Camargo AA. Genetic diversity ofLeishmania amazonensisstrains isolated in northeastern Brazil as revealed by DNA sequencing, PCR-based analyses and molecular karyotyping. Kinetoplastid Biol Dis. 2007;6:5–5. doi: 10.1186/1475-9292-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshaghi MA, Ravasan MH, Hide M, Javadian E-A, Rassi Y, Sedaghat MM. Development of species-specific PCR and PCR-restriction fragment length polymorphism assays forL. infantum/L. donovanidiscrimination. Exp Parasitol. 2009;122:61–65. doi: 10.1016/j.exppara.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Ozerdem D, Eroglu F, Genc A, Demirkazik M, Koltas IS. Comparison of microscopic examination, rK39 and PCR for visceral leishmaniasis diagnosis in Turkey. Parasitol Res. 2009;106:197–200. doi: 10.1007/s00436-009-1650-3. [DOI] [PubMed] [Google Scholar]
- 29.Quaresma PF, Murta SMF, Ferreira EC, da Rocha-Lima ACVM, Xavier AAP, Gontijo CMF. Molecular diagnosis of canine visceral leishmaniasis: identification ofLeishmaniaspecies by PCR-RFLP and quantification of parasite DNA by real-time PCR. Acta Trop. 2009;111:289–294. doi: 10.1016/j.actatropica.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Reithinger R, Dujardin JC. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol. 2007;45:21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silveira OJ, Neto, Duarte SC, Costa HX, Linhares GFC. Design of primer pairs for species-specific diagnosis ofLeishmania (Leishmania) infantum chagasi using PCR. Rev Bras Parasitol Vet. 2012;21:304–307. doi: 10.1590/s1984-29612012000300024. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, volutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venazzi EAS, Roberto ACBS, Barbosa-Tessmann IP, Zanzarini PD, Lonardoni MVC, Silveira TGV. Polymerase chain reaction with lesion scrapping for the diagnosis of human American tegumentary leishmaniasis. Mem Inst Oswaldo Cruz. 2006;101:42730–42730. doi: 10.1590/s0074-02762006000400014. [DOI] [PubMed] [Google Scholar]
- 34.Venazzi EAS, Roberto ACBS, Barbosa-Tessmann IP, Zanzarini PD, Lonardoni MVC, Silveira TGV. Detection ofLeishmania (Viannia)DNA in blood from patients with American cutaneous leishmaniasis. Exp Parasitol. 2007;115:399–402. doi: 10.1016/j.exppara.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization Leishmaniasis. 2014 http://goo.gl/PiM9n


