Abstract
Aggregatibacter actinomycetemcomitans QseBC regulates its own expression and is essential for biofilm growth and virulence. However, the signal that activates the QseC sensor has not been identified and the qseBC regulon has not been defined. In this study, we show that QseC is activated by catecholamine hormones and iron but not by either component alone. Activation of QseC requires an EYRDD motif in the periplasmic domain of the sensor and site-specific mutations in EYRDD or the deletion of the periplasmic domain inhibits catecholamine/iron-dependent induction of the ygiW-qseBC operon. Catecholamine/iron-dependent induction of transcription also requires interaction of the QseB response regulator with its binding site in the ygiW-qseBC promoter. Whole genome microarrays were used to compare gene expression profiles of A. actinomycetemcomitans grown in a chemically defined medium with and without catecholamine and iron supplementation. Approximately 11.5% of the A. actinomycetemcomitans genome was differentially expressed by at least two-fold upon exposure to catecholamines and iron. The expression of ferritin was strongly induced, suggesting that intracellular iron storage capacity is increased upon QseBC activation. Consistent with this, genes encoding iron binding and transport proteins were down-regulated by QseBC. Strikingly, 57% of the QseBC up-regulated genes (56/99) encode proteins associated with anaerobic metabolism and respiration. Most of these up-regulated genes were recently reported to be induced during in vivo growth of A. actinomycetemcomitans. These results suggest that detection of catecholamines and iron by QseBC may alter the cellular metabolism of A. actinomycetemcomitans for increased fitness and growth in an anaerobic host environment.
Keywords: Aggregatibacter actinomycetemcomitans, anaerobic respiration, catecholamine, iron, QseBC, two component system
Introduction
Aggregatibacter actinomycetemcomitans is a Gram-negative opportunistic oral pathogen that is strongly associated with aggressive forms of periodontitis and other systemic diseases and infections such as cardiovascular diseases (Yew et al., 2014), atherosclerosis (Zhang et al., 2010), urinary tract infections (Townsend & Gillenwater, 1969) and brain abscesses (Rahamat-Langendoen et al., 2011). The organism resists killing mediated by neutrophils (Permpanich et al., 2006; Ji et al., 2007) and produces a variety of potential virulence factors including a cytolethal distending toxin (Cdt), a leukotoxin (LtxA) of the RTX family of bacterial toxins, and a collagenase (Robertson et al., 1982; Kachlany et al., 2010; Jinadasa et al., 2011). Aggregatibacter actinomycetemcomitans also produces autoinducer-2 (AI-2) and this quorum-sensing mechanism has been shown to be essential for virulence and growth under iron-limiting conditions (Fong et al., 2001, 2003; Shao et al., 2007). Novak et al. (2010) have also shown that AI-2 regulates qseBC and that this two-component system is essential for A. actinomycetemcomitans biofilm growth and virulence. However, the mechanism that links the detection of AI-2 to the activation of QseBC, or the signal(s) that activate the QseC sensor have not yet been determined.
In Eschericha coli and Salmonella enterica, QseBC is activated by catecholamine hormones such as epinephrine (Ep) and norepinephrine (Ne) and these hormones regulate the expression of the genes involved in the formation of the attaching and effacing lesion, the Type Three Secretion System contained on the locus of enterocyte effacement, and flagella and motility genes (Sperandio et al., 2003). Catecholamine hormones have also been suggested to control virulence-associated genes in Actinobacillus pleuropneumoniae (Li et al., 2012). However, the QseBC paralogue of Haemophilus influenza (designated FirRS) is not activated by catecholamines, but instead responds to ferrous iron (Steele et al., 2012). Interestingly, Ep and Ne have been shown to increase virulence and stimulate the growth of Bordetella, E. coli and S. enterica by functioning as pseudo-siderophores (Freestone et al., 2000, 2008). Ep and Ne are capable of extracting iron from host proteins such as transferrin and lactoferrin (Freestone et al., 2000) which is subsequently imported into the cell by the enterobactin transporter complex (Burton et al., 2002; Freestone et al., 2003; Anderson & Armstrong, 2006). Catecholamine hormones have also been suggested to stimulate the growth of some periodontal pathogens (Jentsch et al., 2013) but their mechanism of action is not known. In addition, recent studies show that activated phagocytic cells (e.g. neutrophils, polymorphonuclear cells and macrophages) release catecholamine hormones and lactoferrin in response to inflammatory stimuli (Brown et al., 2003; Flierl et al., 2007, 2008, 2009) and can produce local concentrations of catecholamines in the millimolar range (Brown et al., 2003). This suggests that the inflamed gingival pocket may be an environment that is rich in both catecholamines and lactoferrin.
In A. actinomycetemcomitans, qseBC is co-expressed with ygiW, which encodes a periplasmic solute binding protein in the bacterial OB-fold family (Juárez-Rodríguez et al., 2013b). The functional outcomes of QseBC activation have only been broadly defined as influencing biofilm growth and virulence. Little is known about how QseBC activation influences these complex phenotypes, in part because the qseBC regulon has not been defined and the signals (ligands) detected by QseC have not been identified. In this report, we show that qseBC expression is maximally induced in the presence of both iron and catecholamine hormones, and that activation requires the periplasmic domain of QseC. This suggests that iron and catecholamines may be signals that activate the QseC sensor. In addition, using genome microarrays, we show that exposure of A. actinomycetemcomitans to catecholamines and iron significantly induces genes that are involved in anaerobic respiration, anaerobic metabolism and iron storage, and downregulates genes involved in iron uptake. Together, these results suggest that the detection of catecholamines and iron by the QseBC two-component system may play an important role in the adaptation of A. actinomycetemcomitans to the host cell environment.
Methods
Bacterial strains, plasmid and media
The bacterial strains and plasmids used in this study are listed in the Supplementary material (Table S1). Luria–Bertani (LB) broth and LB agar (LB broth plus 1.5% agar), or brain–heart infusion (BHI) broth and BHI agar (all from Difco, BD Biosciences, Franklin Lakes, NJ) were routinely used for the propagation and plating of E. coli or A. actinomycetemcomitans strains, respectively. The A. actinomycetemcomitans (afimbriated, smooth-colony-morphotype strain 652, serotype c) was grown at 37°C under microaerophilic conditions in a candle jar. For some experiments, A. actinomycetemcomitans strains were grown in a chemically defined medium (CDM) essentially as described by Socransky et al. (1985) or in CDM supplemented with 100 μm FeCl2 or FeCl3 and/or 50 μm Ep or Ne. The composition of CDM is shown in detail in the Supplementary material (Table S2) and after combining the individual components, CDM was adjusted to pH 7.3 with HCl, supplemented with 0.2% glucose and sterilized by filtration through a 0.22-μm filter before use. If necessary for plasmid maintenance, medium was also supplemented with 25 μg ml−1 kanamycin or 50 μg ml−1 spectinomycin.
Aggregatibacter actinomycetemcomitans growth kinetics
To determine the effect of catecholamines and iron on cell growth, a frozen stock of A. actinomycetemcomitans 652 was inoculated into BHI and was grown to an optical density at 600 nm (OD600) of 0.3–0.4. This culture was then inoculated into fresh BHI at a 1 : 30 dilution. At an OD600 of 0.5–0.6, cells were washed with CDM and inoculated into freshly prepared CDM at a 1 : 30 dilution. At various time-points, aliquots were removed from each of triplicate cultures and the OD600 was measured using a BioRad SmartSpec Plus uV-vis spectrophotometer (BioRad, Hercules, CA). Growth kinetics were determined for two independently grown cultures for each condition tested.
DNA procedures
DNA manipulations were carried out as described by Sambrook & Russell (2001). Transformation of E. coli and A. actinomycetemcomitans was carried out by electroporation as previously described (Juárez-Rodríguez et al., 2013a). Transformants containing plasmids were selected on LB agar plates supplemented with the appropriate antibiotics and plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). Restriction digestions were carried out following the protocols as recommended by the manufacturer (New England Biolabs, Ipswich, MA). All primers used in this study were synthesized by Integrated DNA Technology, Inc. (Coralville, IA) and are shown in Table S3. Restriction enzyme sites to facilitate cloning of the resulting polymerase chain reaction (PCR) products are underlined in the primer sequences. All primer sequences were designed based on the genome sequence of A. actinomycetemcomitans D11S-1, serotype c, which is available from the Pathosystems Resource Integration Center (http://www.genome.jp/kegg-bin/show_organism?org=aat). All constructs were verified by DNA sequencing (University of Louisville Core Sequencing Facilities).
Site-specific mutagenesis of the putative EYRDD iron-binding motif of QseC
The conserved amino acid residues tyrosine (Y) and arginine (R) of the putative iron-binding motif of QseC at positions 155 and 156, respectively, were individually substituted by alanine (A). The qseC codon substitutions for Y155A (TAT to GCT) and R156A (CGC to GCC) were achieved by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using plasmid pDJR28 as the template and primer set MDJR-163F/MDJR-164R. The successful incorporation of the desired mutations was verified by sequencing the resulting plasmid construct, pDJR28-M1-2.
Integration of QseC mutant alleles in single-copy into A. actinomycetemcomitans qseBC
To integrate a single copy of the qseC mutant allele encoding QseCY155A, R156A into the A. actinomycetemcomitans chromosome, an approach that was used previously to integrate a single copy of a ygiW promoter-lacZ fusion into the A. actinomycetemcomitans genome was used (Juarez-Rodriguez et al., 2014). Briefly, the qseC wild-type or mutant alleles along with the upstream ygiW and qseB genes were amplified by PCR from the chromosome of A. actinomycetemcomitans 652 and pDJR28-M1-2, respectively, using the primer sets MDJR-124F/MDJR-125R. The resulting 3135-base-pair PCR fragments were digested with NotI–PstI and cloned into the NotI–PstI-digested suicide vector pJT1. The recombinant suicide plasmids (20 μg) were then introduced individually into A. actinomycetemcomitans ΔqseBC by electroporation. Subsequently, 10 Spr colonies containing a single-copy of the suicide vector inserted into the chromosome were selected and verified for the appropriate insertion event by PCR using primer sets MDJR-63F/MDJR-61R and MDJR-54F/MDJR-77R. One of the PCR-verified Spr colonies was designated A. actinomycetemcomitans ΔqseBC::61 and contained the mutated qseC allele (ygiW-qseB-qseCY155A, R156A). A second colony containing the wild-type qseC allele was also selected for further study and was designated ΔqseBC::46. The pDJR29 lacZ reporter plasmid was subsequently introduced into each strain by electroporation.
β-Galactosidase assays
β-Galactosidase (β-gal) activity was qualitatively assessed on BHI agar plates that were supplemented with 50 μg ml−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). Quantitative evaluation of β-gal activity was carried out using permeabilized cells incubated with o-nitrophenyl-β-d-galactopyranoside (ONPG) substrate (Sigma, St Louis, MO) as previously described (Miller, 1972). Briefly, a primary culture of the desired strain (OD600 of 0.3–0.4) was diluted 1 : 30 into 1.5 ml of BHI in a 1.7-ml propylene centrifuge tube and grown standing for 24 h at 37°C. Subsequently, an aliquot of the secondary overnight culture was diluted 1 : 30 into 1.5 ml of CDM in 1.7-ml propylene centrifuge tubes and grown standing for 24 h at 37°C. An aliquot of 0.1 ml was then used to determine the OD600 of the culture and triplicate aliquots of 0.1 ml were used to measure β-galactosidase activity. Average values (± the standard deviations) for activity units were routinely calculated from three independent experiments using GraphPad Prism v5 software (GraphPad, San Diego, CA).
Microarray analysis of the A. actinomycetemcomitans transcriptome
A custom A. actinomycetemcomitans gene expression microarray was printed by Agilent Technologies (Santa Clara, CA; GE 8 × 15,000 grids per slide, 60-mer oligonucleotide probes were used). Oligonucleotide probe sequences were obtained from the University of Michigan OligoArray Database that is available for A. actinomycetemcomitans D11S-1 (http://berry.engin.umich.edu/oligoarraydb/organismPage.php?ORG=Aggregatibacter&actinomycetemcomitans&D11S-1). This database comprises 5638 oligonucleotide probes representing 2062 transcripts encoded by the A. actinomycetemcomitans D11S-1 genome. Of the 2062 transcripts, 1552 are represented by three independent oligonucleotide probes, 215 are represented by two probes and 221 by a single specific oligonucleotide. Each oligonucleotide was printed twice on each 15,000-spot grid; hence the majority of genes are represented by six separate spots on the array and no transcript is represented by fewer than two spots. Seventy-two putative transcripts of A. actinomycetemcomitans D11S-1 are not represented in the oligonucleotide database; these mostly comprise hypothetical proteins (n = 37) and transposases/integrases of mobile genetic elements (n = 11).
Aggregatibacter actinomycetemcomitans 652 was cultured in CDM or in CDM supplemented with 50 μm Ne and 100 μm FeCl2 for 14 h as described previously. The late exponential phase cultures were then adjusted to equivalent OD600 and labeling of RNA, microarray hybridization, array scanning and data analysis were carried out by the MicroArray Core Facility at the University of Louisville. Briefly, RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) following the protocols supplied by the manufacturer. The quality of the isolated RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). RNA samples were considered to have little or no degradation if they exhibited an RNA integrity number >9 (Schroeder et al., 2006). Total RNA (100 ng) was labeled with Cyanine 3-CTP using the Low Input Quick Amplification WT Labeling Kit for one color (Agilent Technologies). This kit uses random nucleotide-based T7 promoter primers to first synthesize cDNA, which is then reacted with T7 RNA polymerase in the presence of NTPs and Cyanine 3-CTP to generate labeled cRNA. The labeled cRNA was purified using the RNeasy Mini Elute kit (Qiagen) and total yield and Cy3 incorporation efficiency was determined using a NanoDrop Technologies spectrophotometer.
Array hybridizations were carried out using the Agilent Gene Expression Hybridization kit as described by the manufacturer. Briefly, 600 ng of each labeled cRNA sample was incubated at 60°C for 30 min and then hybridized to the custom A. actinomycetemcomitans gene expression array at 65°C for 17 h. After hybridization, the microarray slides were washed with Agilent gene expression wash buffer 1 at 37°C for 5 min followed by wash buffer 2 at 37°C for 1 min. After hybridization and washing, the slides were scanned using an Agilent microarray scanner (Model G2505C) set for one-color using the green channel and 5-μm resolution. The one-color microarray images (.tif) were extracted with the aid of Feature Extraction software (v 9.5.1; Agilent Technologies). The raw data files (.txt) were imported into GeneSpring (GX 11.1) and the data were transformed to bring any negative value or value <0.01–0.01. Normalization was then performed using a per-chip 75 centile method that normalizes each chip on its 75th centile to facilitate comparison among chips. A per-gene on median normalization was then performed to normalize the expression of every gene on its median among samples. An expression threshold of at least two-fold was applied and the differentially expressed genes were evaluated using a pair-wise t-test and genes with P-values < 0.05 were considered to be significantly upregulated or downregulated. The array data have been submitted to the Gene Expression Omnibus Database with the accession number GSE68749.
Determination of formate concentration
Extracellular formate concentration was determined using the formate assay kit supplied by r-BioPharm (Darmstadt, Germany). This kit measures formate concentration by the conversion of formate and NAD+ to bicarbonate and NADH through the action of formate dehydrogenase. The stoichiometric production of NADH is measured by absorbance at 365 nm. Culture supernatants from mid-exponential phase cultures (6–8 h post inoculation) were collected and treated with 30 mm trichloroacetic acid (Sigma-Aldrich) at a ratio of 1 part supernatant to 2 parts trichloroacetic acid to precipitate proteins. After neutralizing with 1 m KOH, the samples were filtered (0.22 μm) to remove insoluble proteins and 100 μl of the filtered product was used in a formic acid assay following the protocol supplied by the manufacturer. Formate concentration was calculated from the levels of NADH produced as determined by measuring the OD365 and the data were normalized for cell density in the CDM culture. Assays were performed in triplicate and results are presented from two independent experiments.
Results
Catecholamines and iron activate the A. actinomycetemcomitans QseBC two component system
We previously showed that the ygiW-qseBC operon of A. actinomycetemcomitans is auto-regulated by QseB (Juárez-Rodríguez et al., 2014) and is essential for biofilm growth and virulence (Novak et al., 2010). However, the signal that activates the A. actinomycetemcomitans QseBC two-component system was not previously identified. In E. coli and S. enterica, qseBC is activated by catecholamine hormones. In contrast, the qseBC paralog of H. influenzae is activated by ferrous iron and does not appear to respond to catecholamines or ferric iron (Steele et al., 2012). To determine if the ygiW-qseBC operon is induced by catecholamines and/or iron, A. actinomycetemcomitans 652 was transformed with pDJR29 containing lacZ fused to the ygiW-qseBC promoter (Juárez-Rodríguez et al., 2013b) and cultures were grown in CDM supplemented with catecholamines (50 μm Ep or Ne) and/or 100 μm FeCl2 or FeCl3. As shown in Fig. 1, the presence of Ep, Ne, Fe2+ or Fe3+ alone did not significantly induce lacZ expression. However, the addition of Ep or Ne together with either Fe2+ or Fe3+ induced lacZ expression by approximately five-fold over the control (CDM alone), suggesting that QseBC activation in A. actinomycetemcomitans requires both components.
Figure 1.
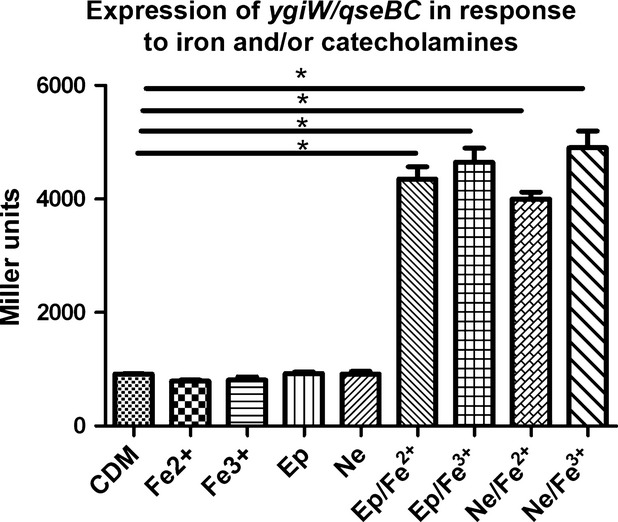
Expression of the ygiW-qseBC operon in Aggregatibacter actinomycetemcomitans cultures exposed to catecholamines and iron. Aggregatibacter actinomycetemcomitans 652 harboring the ygiW-qseBC promoter-lacZ reporter plasmid pDJR29 was grown in chemically defined medium (CDM) broth or in CDM supplemented with either ferrous or ferric chloride (Fe2+ or Fe3+; 100 μm), epinephrine (Ep; 50 μm), norepinephrine (Ne; 50 μm, or a combination of both catecholamine (50 μm) and iron (100 μm). β-galactosidase activity was determined after 24 h of growth. Significant differences (P < 0.05) are indicated by asterisks.
To determine if catecholamines and iron function as signals that directly activate the QseC sensor, pJDR29 was transformed into non-polar gene deletion mutants of A. actinomycetemcomitans that lacked qseC or qseB (ΔqseC and ΔqseB, respectively), or into a strain in which qseC was replaced by a single copy of the qseC gene that did not encode the periplasmic region of the sensor kinase (qseCΔp, see Juárez-Rodríguez et al., 2013b). As shown in Fig. 2, lacZ expression in CDM alone was reduced by approximately four-fold in the ΔqseC and ΔqseB strains and no induction of lacZ expression occurred in the presence of Ep/FeCl2. These results are consistent with our previous finding that ygiW-qseBC is auto-regulated by QseC-mediated activation of QseB (Juárez-Rodríguez et al., 2014). Importantly, no Ep/FeCl2-dependent induction of lacZ occurred in A. actinomycetemcomitans that expressed a QseC protein that lacked the periplasmic domain. These results suggest that Ep and Fe2+ function as signals that activate the QseC sensor and that activation requires the periplasmic domain.
Figure 2.
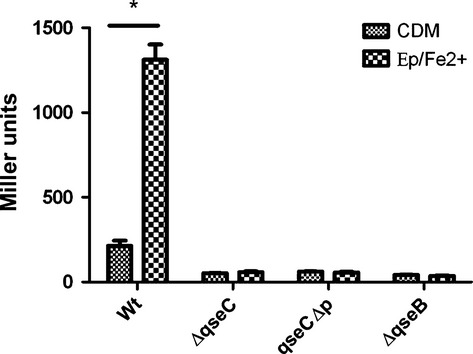
Catecholamine/iron-dependent induction of ygiW-qseBC requires the periplasmic domain of QseC and the QseB response regulator. Aggregatibacter actinomycetemcomitans 652 (WT), isogenic non-polar gene deletion strains lacking qseC (ΔqseC) or qseB (ΔqseB), or a strain expressing the QseC sensor without the periplasmic domain (qseCΔp; see Methods) each harboring plasmid pDJR29 were grown in chemically defined medium (CDM) broth alone or in CDM supplemented with both Ep and FeCl2 (Ep/Fe2+; 50 μm and 100 μm, respectively). β-galactosidase activity was determined after 24 h of growth. Significant differences (P < 0.05) are indicated by asterisks.
To confirm that Ep/FeCl2-dependent induction of lacZ was mediated by QseB, a family of reporter plasmids containing a nested series of ygiW-qseBC promoter deletions was tested (Fig. 3A). These constructs have been previously described and were used to map the –10, –35 and QseB binding sites in the ygiW-qseBC promoter (Juárez-Rodríguez et al., 2014). As shown in Fig. 3(B), ygiW-qseBC promoter activity was reduced by approximately 35% when nucleotides –94 to –138 were deleted (compare pDJR63, pDJR57 and pDJR56), consistent with our previous results (Juárez-Rodríguez et al., 2014). However, Ep/FeCl2-dependent induction of lacZ expression still occurred in each of these constructs. Ep/FeCl2-dependent induction of lacZ was significantly reduced only after deletion (pDJR58) or site-specific mutation (pDJR86) of the QseB binding site. This indicates that the interaction of QseB with its binding site is essential for Ep/FeCl2-dependent induction of the operon and is consistent with a model where catecholamines and iron represent signals that are recognized by and activate QseC, which in turn activates the QseB response regulator.
Figure 3.
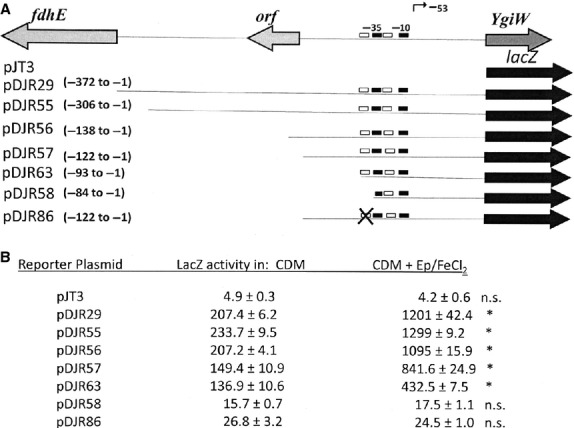
Catecholamine/iron-dependent induction of ygiW-qseBC requires a functional QseB binding site. (A) Schematic diagram of the ygiW-qseBC promoter region and transcriptional fusion constructs pDJR29, pDJR55, pDJR56, pDJR57, pDJR63, pDJR58, pDJR85 and pDJR86 (Juárez-Rodríguez et al., 2013b, 2014) showing the binding regions for QseB (white boxes), the –10 and –35 promoter elements (black boxes) and the primary transcriptional start site (bent arrow). Site-specific mutations in the QseB binding site are indicated with the × symbol. The numbering of the nucleotides is relative to the ygiW translational start codon. (B) β-galactosidase activity in A. actinomycetemcomitans 652 transformed individually with each reporter plasmid. Cultures were grown in chemically defined medium (CDM) or CDM supplemented with epinephrine (Ep; 50 μm) and FeCl2 (100 μm) and β-galactosidase activity was determined after 24 h of growth. Values are means of results from three independent experiments ± standard deviations. Statistical analysis was performed by using one-way analysis of variance followed by Tukey's multiple-comparison test. Significant differences (P < 0.05) are indicated by asterisks; n.s., not significant.
A putative iron responsive motif regulates the activation of QseC
The periplasmic region of A. actinomycetemcomitans QseC contains the sequence EYRDD (residues 154–157) that resembles the DYRED motif previously shown to be important for the sensing of iron by the QseC paralogs of H. influenzae (Steele et al., 2012) and Salmonella typhimurium (Merighi et al., 2009). To determine if this motif in A. actinomycetemcomitans is important for Ep/FeCl2-dependent activation of QseC, EYRDD was altered to EAADD by site-directed mutagenesis and the mutant (encoding QseCY155A, R156A) or wild-type qseC alleles together with ygiW and qseB were integrated by homologous recombination into the chromosome of A. actinomycetemcomitans ΔqseBC to generate strains ΔqseBC::61 and ΔqseBC::46, respectively. As shown in Table 1, lacZ expression from pDJR29 in ΔqseBC::61 was not significantly induced when cells were cultured in CDM supplemented with Ep and FeCl2. In contrast, ΔqseBC::46 exhibited an approximately five-fold increase in lacZ activity, similar to the wild-type strain. However, lacZ expression in ΔqseBC::61 was approximately 90-fold higher than in the wild type even when cultured in CDM alone. These results suggest that QseCY155A, R156A cannot sense Ep/FeCl2 and that mutations in EYRDD may lock QseC in an activated conformation that results in constitutive activation of QseB and high expression of the ygiW-qseBC operon.
Table 1.
The EYRDD motif regulates the QseC response to Ep/FeCl2
| LacZ activity in |
|||
|---|---|---|---|
| Strain | CDM | CDM + Ep/FeCl2 | Fold induction |
| WT | 280.6 ± 44.0 | 1240.0 ± 207.6 | 4.4 (P < 0.05) |
| ΔqseBC::46 | 273.2 ± 26.7 | 1422.0 ± 234.2 | 5.2 (P < 0.05) |
| ΔqseBC::61 | 25,066 ± 11,041 | 17,246 ± 10,247 | n.s.1 |
n.s., No significant difference; CDM, chemically defined medium; Ep, epinephrine; WT, wild-type.
Catecholamines and iron stimulate A. actinomycetemcomitans growth
Exposure to catecholamine stress hormones has recently been reported to stimulate the growth of several potential periodontal pathogens such as Fusobacterium nucleatum and Tannerella forsythia but had little effect on the growth of Porphyromonas gingivalis (Jentsch et al., 2013). To determine if exposure to catecholamines and iron influence A. actinomycetemcomitans growth, cells were cultured in CDM alone, CDM supplemented with 50 μm Ne or 100 μm FeCl2, or in CDM supplemented with both Ne and FeCl2. As shown in Fig. 4, there was no significant difference in the growth of A. actinomycetemcomitans in CDM or medium supplemented with Ne. However, addition of FeCl2 resulted in a significant increase in growth over CDM alone and a further significant stimulation of A. actinomycetemcomitans growth was observed when CDM was supplemented with both Ne and FeCl2. Similar results were observed when medium was supplemented with Ep or Ep/FeCl2 (not shown).
Figure 4.
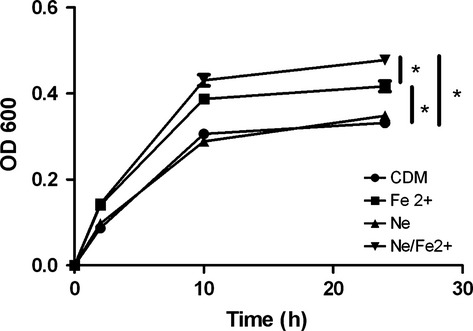
Catecholamine/iron-mediated induction of Aggregatibacter actinomycetemcomitans growth. The A. actinomycetemcomitans 652 was grown in chemically defined medium (CDM) alone or in CDM supplemented either with FeCl2 (Fe2+), norepinephrine (Ne), or both norepinephrine and iron (Ne/Fe2+). At the designated times, growth was measured by determining the optical density at 600 nm (OD600). Significant differences (P < 0.05) are indicated by asterisks.
Exposure to catecholamines and FeCl2 induces genes involved in anaerobic respiration/metabolism and decreases expression of genes involved in iron acquisition
The role of the QseBC two component system in A. actinomycetemcomitans has only been broadly described as regulating complex phenotypes such as biofilm formation and virulence. To better understand how this regulation occurs, we generated custom microarrays based on the genome sequence of A. actinomycetemcomitans D11S-1 (see Methods) and used these arrays to compare the transcriptomes of cultures grown in CDM and CDM supplemented with Ne and FeCl2. As shown in Supplementary material (Table S4), 235 genes (approximately 11.5% of the A. actinomycetemcomitans genome) were differentially expressed at greater than or equal to two-fold in cells cultured in Ne/Fe2+-supplemented medium relative to cells grown in CDM alone. Ninety-nine genes (∼4.9% of the genome) were upregulated 2- to 18.5-fold and 135 genes (∼6.7% of the genome) were down-regulated 2- to 9.5-fold in cells exposed to Ne/FeCl2. Consistent with the results presented in Fig. 1, ygiW-qseBC was one of the most strongly induced operons detected in cells grown in supplemented medium and this was further confirmed by reverse transcription PCR (see Table S4). Strikingly, 56 of the 99 upregulated genes encoded proteins associated with anaerobic metabolism and respiration, as shown in Table 2. Six operons that were induced encode components associated with anaerobic electron transport, e.g. the nap operon (D11S_205–210), an operon involved in the biogenesis of c-type cytochromes (D11S_2000–1990), the hydrogenase-4 complex and hydN (D11S1735–1747 and D11S_1092/1093) involved in electron transport from formate to hydrogen and the oxidation of hydrogen, D11S_1412/1413 involved in trimethylamine N-oxide anaerobic respiration, and D11S_493/494 encoding a dimethylsulfoxide terminal reductase. At least four additional induced operons and several other single genes encode proteins that may be associated with anaerobic metabolism. D11S_303 and D11S_1771 each encode a C4 dicarboxylate transporter involved in the utilization of aspartate and fumarate. Consistent with this, D11S_597 encodes aspartate ammonia lyase, which converts aspartate to fumarate, D11S_810–812 encodes a fumarate reductase, and D11S_1061 encodes fumarate hydratase. Other upregulated operons code for oxaloacetate decarboxylase (D11S_1379-1381) which converts oxaloacetate to pyruvate, and D11S_1749/1748 and D11S_1989-1986, which encode formate dehydrogenase and catalyze the formate-dependent reduction of nitrite to ammonia, respectively. To confirm the microarray results, 15 genes representative of many of the operons listed above were selected for verification of induction using reverse transcription PCR. The results obtained, shown in the Supplementary material (Table S4) were consistent with the array data. As formate dehydrogenase was induced 10-fold and several of the metabolic pathways that were predicted to be induced from the array data use formate, the array results were further confirmed by determining the levels of formate in cultures grown in CDM alone or in supplemented media. As shown in Fig. 5, formate levels were significantly increased when cells were cultured in medium containing FeCl2 and a further significant increase occurred when the medium was supplemented with Ne/Fe2 +. Together, these data suggest that the activation of QseBC may function to prime A. actinomycetemcomitans for growth in an anaerobic niche by upregulating genes involved in anaerobic metabolism and electron transport.
Table 2.
Differentially expressed genes associated with anaerobic respiration and metabolism
| ID tag | Product | Fold change | Direction | Putative function |
|---|---|---|---|---|
| D11S_0205 | Cytochrome c-type protein TorC | 3.05 | Up | Trimethylamine N-oxide anaerobic respiration |
| D11S_0206 | Periplasmic nitrate reductase, diheme cytochrome | 2.94 | Up | Diheme cytochrome c subunit |
| D11S_0207 | Ferredoxin-type protein NapH | 3.36 | Up | Quinol dehydrogenase membrane component |
| D11S_0208 | Quinol dehydrogenase periplasmic component | 2.74 | Up | Nitrate respiration electron transport |
| D11S_0209 | Periplasmic nitrate reductase, large subunit | 4.92 | Up | Nitrate redutase periplasmic component |
| D11S_0210 | NapD protein | 6.69 | Up | Assembly of D11S_208 and D11S_209 |
| D11S_0303 | Anaerobic C4-dicarboxylate membrane transporter | 2.41 | Up | Anaerobic utilization of aspartate/fumarate |
| D11S_0383 | Alkylhydroperoxidase AhpD core | 4.03 | Up | Anti-oxidant protein |
| D11S_0493 | Anaerobic dimethyl sulfoxide reductase chain A | 2.26 | Up | Anaerobic terminal reductase |
| D11S_0494 | Anaerobic dimethyl sulfoxide reductase chain B | 2.23 | Up | Anaerobic terminal reductase |
| D11S_0597 | Aspartate ammonia-lyase | 6.86 | Up | Converts aspartate to fumarate |
| D11S_0810 | Fumarate reductase subunit C (Fumarate reductase) | 2.39 | Up | Anaerobic conversion of fumarate to succinate |
| D11S_0811 | Fumarate reductase iron-sulfur subunit | 2.39 | Up | Anaerobic conversion of fumarate to succinate |
| D11S_0812 | Fumarate reductase flavoprotein subunit | 3.86 | Up | Anaerobic conversion of fumarate to succinate |
| D11S_1061 | Fumarate hydratase, class II | 2.43 | Up | Reversible hydration/dehydration of fumarate to malate |
| D11S_1092 | Hydrogenase accessory protein HypB | 1.81 | Up | GTP hydrolase; assembly of hydrogenase |
| D11S_1093 | Hydrogenase expression/formation protein HypD | 1.57 | Up | Hydrogenase maturation protein |
| D11S_1376 | Hydrogenase assembly chaperone HypC/HupF | 1.88 | Up | Hydrogenase maturation protein |
| D11S_1379 | Oxaloacetate decarboxylase γ chain 3 | 4.74 | Up | Conversion of OAA to pyruvate |
| D11S_1380 | Oxaloacetate decarboxylase α subunit | 4.44 | Up | Conversion of OAA to pyruvate |
| D11S_1381 | Oxaloacetate decarboxylase β chain | 4.31 | Up | Conversion of OAA to pyruvate |
| D11S_1412 | Cytochrome c-type protein TorY | 7.66 | Up | Trimethylamine N-oxide anaerobic respiration |
| D11S_1413 | Trimethylamine-n-oxide reductase 2 | 4.67 | Up | Trimethylamine N-oxide anaerobic respiration |
| D11S_1676 | Cytochrome c peroxidase | 5.05 | Up | Metabolism of reducing equivalents from cytochrome c |
| D11S_1735 | [NiFe] hydrogenase maturation protein HypF | 2.58 | Up | Hydrogenase maturation protein |
| D11S_1736 | Electron transport protein HydN | 18.44 | Up | Electron transport from formate to hydrogen |
| D11S_1737 | Hydrogenase-4 component B | 14.93 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1738 | Hydrogenase-4 component B | 15.02 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1739 | Hydrogenase-4 component C | 10.68 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1740 | Hydrogenase-4 component D | 10.59 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1741 | Hydrogenase-4 component E | 7.95 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1742 | Hydrogenase-4 component F | 7.76 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1743 | Hydrogenase-4 component G | 5.89 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1744 | Hydrogenase-4 component H | 5.87 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1745 | Hydrogenase-4 component I | 5.4 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1746 | Hydrogenase-4 component J | 4.62 | Up | Oxidation of hydrogen; coupled to electron transport |
| D11S_1747 | Hydrogenase maturation peptidase Hyc | 4.79 | Up | C-terminal processing of hydrogenase |
| D11S_1748 | Formate dehydrogenase H | 10.42 | Up | Anaerobic metabolism of formate to H2 and CO2 |
| D11S_1749 | Formate dehydrogenase, α subunit | 10.0 | Up | Oxidation of formate to CO2 |
| D11S_1771 | C4-dicarboxylate membrane transporter | 1.73 | Up | Anaerobic utilization of aspartate/fumarate |
| D11S_1811 | Bifunctional acetaldehydeCoA/alcohol dehydrogenase | 2.72 | Up | Oxidation aldehydes and alcohols |
| D11S_1888 | TorCAD operon transcriptional regulatory protein | 1.86 | Up | Regulator of trimethylamine N-oxide respiration genes |
| D11S_1984 | Cytochrome c-type biogenesis protein CcmF | 3.87 | Up | Assembly of c-type cytochromes |
| D11S_1986 | NrfD protein | 11.59 | Up | Electron transfer from quinone to type c cytochromes |
| D11S_1987 | Cytochrome c nitrite reductase, Fe-S protein | 13.63 | Up | Reduction of nitrite to ammonia |
| D11S_1988 | Cytochrome c nitrite reductase, pentaheme | 17.08 | Up | Formate dependent reduction of nitrite to ammonia |
| D11S_1989 | Nitrite reductase (cytochrome; ammonia-forming) | 18.43 | Up | Reduction of nitrite to ammonia |
| D11S_1993 | Cytochrome c-type biogenesis protein CcmH | 1.57 | Up | Biogenesis of cytochrome c |
| D11S_1994 | Thiol-disulfide interchange protein | 2.04 | Up | Biogenesis of cytochrome c |
| D11S_1995 | Cytochrome c-type biogenesis protein CcmF | 2.39 | Up | Assembly of c-type cytochromes |
| D11S_1996 | Cytochrome c-type biogenesis protein CcmE | 2.44 | Up | Heme chaperone; assembly of c-type cytochromes |
| D11S_1997 | Heme exporter protein D (CcmD) | 2.65 | Up | Heme transporter |
| D11S_1998 | CcmC | 3.15 | Up | Cytochrome c biosynthesis |
| D11S_1999 | Heme exporter protein CcmB | 2.69 | Up | Heme exporter; cytochrome c biosynthesis |
| D11S_2000 | Heme ABC exporter, ATP-binding protein CcmA | 3.3 | Up | Heme exporter; cytochrome c biosynthesis |
| D11S_2128 | Anaerobic ribonucleoside-triphosphate reductase | 2.7 | Up | Reduction of CTP to dCTP |
Figure 5.
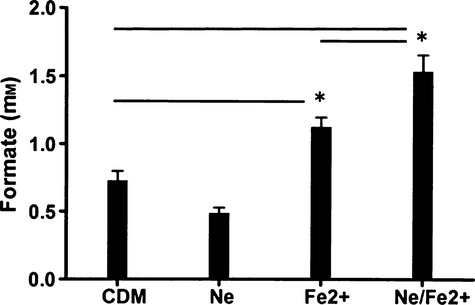
Formate concentration in spent media from Aggregatibacter actinomycetemcomitans 652 cultures. Cells were grown in chemically defined medium (CDM) alone or CDM supplemented with iron (Fe2+), norepinephrine (Ne) or both norepinephrine and iron (Ne/Fe2+). Formate concentration was determined by measuring the production of NADH after the enzymatic conversion of formate to bicarbonate by formate dehydrogenase. Significant differences (P < 0.05) are indicated by asterisks.
The genes that were downregulated were more diverse in their putative functions. However, 20 of the 135 downregulated genes (∼15%) encode proteins associated with iron transport (see Table 3). Within this group, four operons encode putative ABC-type iron transporters (e.g. D11S_621/622, D11S_815-818, D11S_1128-1131 and D11S_1557/1558). Other downregulated genes encode putative outer membrane iron receptors (D11S_1630 and D11S_1864) and consistent with this, the expression of tonB components (D11S_487–489) is also reduced by approximately five-fold. In contrast to the downregulation of iron acquisition genes, the expression of ferritin (D11S_1330/1331) was significantly increased (∼10-fold), suggesting that intracellular iron storage is increased when QseBC is activated.
Table 3.
Differentially expressed genes associated with iron/heme acquisition
| ID Tag | Product | Fold change | Direction |
|---|---|---|---|
| D11S_0487 | TonB-system energizer ExbB | 0.208 | Down |
| D11S_0488 | TonB system transport protein ExbD | 0.223 | Down |
| D11S_0489 | Protein TonB | 0.229 | Down |
| D11S_0621 | Iron(III)-transport system permease protein FbpB | 0.182 | Down |
| D11S_0622 | Ferric iron binding protein | 0.195 | Down |
| D11S_0815 | Iron(III) dicitrate transport ATP-binding | 0.123 | Down |
| D11S_0816 | ABC transporter, iron chelate uptake transporter | 0.108 | Down |
| D11S_0817 | Iron(III) dicitrate transport system permease | 0.166 | Down |
| D11S_0818 | Iron(III) dicitrate-binding periplasmic protein | 0.157 | Down |
| D11S_1128 | Iron(III) dicitrate transport ATP-binding | 0.215 | Down |
| D11S_1129 | Iron(III) transport system permease protein | 0.331 | Down |
| D11S_1130 | Putative iron/heme permease | 0.23 | Down |
| D11S_1131 | Putative periplasmic siderophore binding protein | 0.143 | Down |
| D11S_1330 | Nonheme iron-containing ferritin | 9.87 | Up |
| D11S_1331 | Ferritin | 10.59 | Up |
| D11S_1557 | High-affinity Fe2+/Pb2+ permease | 0.245 | Down |
| D11S_1558 | High affinity Fe2+ transporter | 0.122 | Down |
| D11S_1559 | Putative Fe2+ permease | 0.138 | Down |
| D11S_1630 | Putative TonB-dependent iron receptor | 0.48 | Down |
| D11S_1643 | AfeC periplasmic iron binding protein | 0.403 | Down |
| D11S_1809 | Heme acquisition system receptor | 0.463 | Down |
| D11S_1864 | OMP 64 (heme/hemogobin receptor) | 0.106 | Down |
Discussion
In A. actinomycetemcomitans, QseBC regulates biofilm growth and virulence (Shao et al., 2007; Novak et al., 2010) but little is known about how this two-component system controls these complex phenotypes, in large part because the signal that activates the QseC sensor has not been identified and the qseBC regulon has not been defined. A variety of stimuli have been shown to activate the QseBC two-component system in other organisms. In E. coli and S. enterica, the QseC sensor is activated by catecholamine hormones and/or AI-3, an autoinducer of unknown structure (Clarke et al., 2006; Bearson et al., 2008;). In contrast, the qseBC paralog in H. influenzae (firRS) is activated by cold shock or by Fe2+ (but not Fe3+) and does not respond to catecholamines (Steele et al., 2012). Using the AI-3 purification scheme described by Sperandio et al. (2003), we were unable to detect AI-3 produced by A. actinomycetemcomitans and furthermore, cold shock at either 25 or 4°C did not induce qseBC expression (data not shown). Our results indicate that A. actinomycetemcomitans QseC is activated by catecholamine hormones, but only in the presence of iron. Furthermore, activation of QseC occurs in the presence of either Fe2+ or Fe3+ and so appears to be distinct from FirS of H. influenzae, which is not activated by Fe3+ (Steele et al., 2013). This suggests that a complex of catecholamines and iron may function as the signal to activate QseC and consistent with this, catecholamines have been reported to function as pseudosiderophores (Freestone et al., 2000; Anderson & Armstrong, 2008; Bearson et al., 2008; Sandrini et al., 2010). However, we cannot exclude the possibility that catecholamines and iron interact individually with the sensor and that activation of QseC only occurs when both are bound.
Interestingly, qseBC expression in E. coli is also induced by elevated levels of Fe3+ but the QseC sensor itself is not directly activated by iron. Instead, iron activates the PmrB sensor, which in turn phosphorylates the PmrA response regulator. Activated PmrA binds to the qseBC promoter and induces expression of the operon. However, PmrB also phosphorylates the non-cognate QseB response regulator, which autoregulates the qseBC operon (Guckes et al., 2013). The QseB binding site in the ygiW-qseBC promoter of A. actinomycetemcomitans matches the consensus PmrA binding sequence in the pmrAB operon of E. coli (Juárez-Rodríguez et al., 2014), but the pmrAB genes are not present in the A. actinomycetemcomitans genome. Hence, it is possible that the A. actinomycetemcomitans QseC sensor integrates the iron and catecholamine sensory functions of the PmrAB and QseBC two component systems of E. coli. Finally, although QseC is activated by catecholamines in both A. actinomycetemcomitans and enteric organisms, the virulence strategies employed by pathogenic strains of E. coli differ significantly from A. actinomycetemcomitans. Many of the virulence genes that are induced by QseBC in E. coli (Clarke et al., 2006; Hughes et al., 2009) are not present in the A. actinomycetemcomitans genome. This suggests that the regulons controlled by the QseBC two-component system may have evolved in a species-specific manner.
The periplasmic domain of A. actinomycetemcomitans QseC is required for interacting with catecholamines and iron as deleting this region of the protein prevented the induction of ygiW-qseBC in cells grown in the presence of catecholamine and iron. Deleting the periplasmic domain also significantly reduced the overall expression of ygiW-qseBC in cells that were cultured in CDM alone, consistent with our previous results showing that QseBC regulates its own expression (Juárez-Rodríguez et al., 2013b). Aggregatibacter actinomycetemcomitans qseCΔp was also previously shown to exhibit defective biofilm growth (Juárez-Rodríguez et al., 2013b). The periplasmic region of QseC contains the EYRDD motif that is related to the iron-responsive elements that are present in the periplasmic domains of the E. coli PmrB (Wosten et al., 2000) and H. influenza FirS (Steele et al., 2013) sensors. Site-specific mutation of this motif significantly reduced iron-dependent expression of firRS in H. influenzae (Steele et al., 2013) and reduced lacZ expression of a pmrAB-regulated promoter–reporter construct in E. coli (Wosten et al., 2000). In A. actinomycetemcomitans, mutations in EYRDD not only prevented catecholamine/Fe2+-dependent induction of qseBC, but also resulted in constitutively high expression of qseBC (approximately 90-fold greater than the wild-type grown in CDM and ∼12-fold higher than wild-type grown in CDM supplemented with catecholamines and iron). Hence, although EYRDD is important for catecholamine/Fe2+-dependent induction of qseBC, mutations in this motif may also induce a conformational change in QseC that locks the protein in an active state which constitutively phosphorylates QseB, resulting in high-level constitutive expression of the ygiW-qseBC operon. In contrast, deletion of the entire periplasmic domain completely uncouples the sensory and kinase regions of QseC, preventing activation of QseB and reducing transcription of the ygiW-qseBC operon.
A variety of virulence-associated genes are differentially expressed after activation of the E. coli QseBC system (Clarke et al., 2006; Hughes et al., 2009) but although qseBC has been clearly shown to be required for virulence of A. actinomycetemcomitans in an animal model of periodontitis (Novak et al., 2010), the specific genes that potentially contribute to this process have not been identified. Our results indicate that a significant portion of the A. actinomycetemcomitans genome (>11%) is differentially expressed when QseBC is activated by catecholamines and iron. Sixty-three percent of the differentially expressed genes (147/235) were organized in operons where at least one other gene in the operon was also identified from the array data as being differentially expressed. Interestingly, none of the genes encoding the well characterized virulence factors of A. actinomycetemcomitans, e.g. the leukotoxin, cytolethal distending toxin, tad fimbriae, autotransporter epithelial cell adhesins, EmaA or the pga matrix biogenesis components, were upregulated by QseBC when a threshold of at least two-fold difference in expression was applied. Indeed, the leukotoxin (ltxC and ltxA) and pgaB were downregulated by ∼2.1-fold upon activation of QseBC. The striking result was that 57% of the upregulated genes (56/99) that were identified encode proteins associated with anaerobic metabolism or respiration. This group includes electron transport components such as a hydrogenase complex and proteins involved in the reduction of nitrate, dimethyl sulfoxide, trimethylamine-N-oxide, fumarate and formate. In addition, enzymes associated with the metabolism of aspartate, fumarate, malate, oxaloacetate, pyruvate and formate were significantly induced upon activation of QseBC. Also striking is that many of the genes induced by catecholamine/iron activation of QseBC were identified by Jorth et al. (2013) as being upregulated during in vivo growth in a study that used RNAseq to compare A. actinomycetemcomitans gene expression during subcutaneous growth in vivo (mouse abscess model) and biofilm growth in vitro. Similar to the results reported here, Jorth et al. (2013) showed that ∼14% of the A. actinomycetemcomitans genome was differentially expressed during in vivo growth and a preponderance of these genes encoded proteins involved in anaerobic respiration and metabolism. In light of these data, our results suggest that the detection of catecholamines and iron by the QseBC two component system may alter A. actinomycetemcomitans gene expression to prime cellular metabolism for growth in an anaerobic environment, such as that which occurs in the subgingival pocket in vivo. Interestingly, recent studies suggest that activated phagocytic cells (e.g. neutrophils, polymorphonuclear cells and macrophages) release catecholamines in response to inflammatory stimuli (Brown et al., 2003; Flierl et al., 2007, 2008, 2009) and can produce local concentrations of catecholamines in the millimolar range (Brown et al., 2003). Hence, the inflamed gingival pocket may be an environment that is rich in catecholamines, which A. actinomycetemcomitans may detect via QseBC as a stimulus to increase its fitness to survive in this niche. Consistent with this, our results also showed that catecholamines and iron stimulate A. actinomycetemcomitans growth. It is also possible that the pattern of gene expression observed by Jorth et al. (2013) in the mouse abscess arose from the response of QseBC to catecholamines produced by phagocytic cells responding to A. actinomycetemcomitans infection. A model that illustrates this potential QseBC signaling cascade is shown in Fig. 6. Finally, we can speculate that the shift in cellular metabolism and energy production may be the primary factor that links the QseBC two-component system with A. actinomycetemcomitans virulence as QseBC does not appear to be a major regulator of virulence gene expression but is essential for A. actinomycetemcomitans virulence (Novak et al., 2010).
Figure 6.
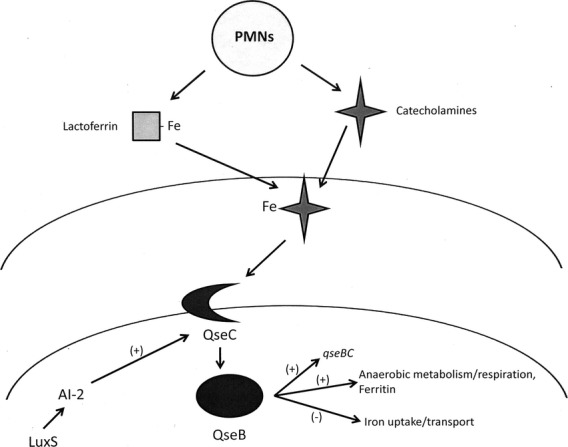
Schematic model of the QseBC signaling cascade of Aggregatibacter actinomycetemcomitans. Catecholamines released by phagocytic cells responding to A. actinomycetemcomitans infection may function as pseudosiderophores and extract iron from host proteins such as lactoferrin or transferrin. Iron and catecholamines, either in complex or individually function to activate QseC by interacting with the periplasmic domain of the sensor. Activated QseC then phosphorylates QseB, which induces the expression of the qseBC operon and numerous genes encoding proteins involved in anaerobic metabolism, electron transport and intracellular iron storage. Activated QseB also downregulates several operons encoding putative inner and outer membrane iron acquisition and transport proteins. The QseBC two-component system is also influenced by autoinducer-2-dependent quorum sensing but the mechanism that links the detection of the autoinducer with induction of qseBC expression has not yet been determined.
The genes that are down-regulated when QseBC is activated are associated with a variety of metabolic processes. However, 15% of the downregulated genes code for iron binding and transport proteins, including the AfuABC ferric iron transporter (D11S_0622-0620) and the FecBCDE operon (D11S_0818-0815). In addition, the expression of several putative outer membrane iron/heme-binding proteins was reduced and consistent with this, genes encoding TonB and its accessory energizer proteins ExbB and ExbD were also downregulated. These results are not surprising because CDM supplemented with catecholamines and FeCl2 represents an iron-rich medium and under these growth conditions, high-level expression of high-affinity iron binding and transport proteins is not likely to be required. Consistent with this, the expression of ferritin was induced by ∼10-fold, suggesting that intracellular iron storage capacity is being increased when QseBC is activated. As lactoferrin is also released by neutrophils, the inflamed gingival pocket may represent an iron replete environment for A. actinomycetemcomitans as catecholamines are known to function as pseudosiderophores that can extract iron from transferrin and lactoferrin (Freestone et al., 2000; Anderson & Armstrong, 2008; Bearson et al., 2008; Sandrini et al., 2010). Indeed, host catecholamines are used by Bordetella and other micro-organisms to acquire iron (Freestone et al., 2000; Burton et al., 2002; Anderson & Armstrong, 2008; Bearson et al., 2008) and it is possible that inter-kingdom signaling mediated by QseBC allows A. actinomycetemcomitans to detect and exploit the production of catecholamines by host cells to facilitate the acquisition of iron from lactoferrin or other host iron-binding proteins during infection.
In summary, we have shown that a combination of catecholamines and iron function as a signal to activate QseBC and that this response requires the EYRDD motif in the periplasmic domain of the QseC sensor and the QseB response regulator. Activation of QseC stimulates the expression of numerous gene products that are involved in anaerobic metabolism and respiration and may prime the organism for growth in an anaerobic niche in the host. Hence, QseBC may represent a therapeutic target, as suggested for QseBC of E. coli (Curtis et al., 2014) to control A. actinomycetemcomitans infections.
Acknowledgments
This study was supported by Public Health Services grant RO1DE14605 from the National Institute for Dental and Craniofacial Research (to DRD). The authors are grateful to Xiaohong Li, Nigel Cooper and the staff of the Microarray Core Facility at the University of Louisville for their assistance with the array studies. Drs Cooper and Li are supported by grant P20GM103436 from the Kentucky Biomedical Research Infrastructure Network. The authors report no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Anderson MT, Armstrong SK. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols and neuroendocrine catecholamines. J Bacteriol. 2006;188:5731–5740. doi: 10.1128/JB.00495-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. Norepinephrine mediates acquisition of transferrin-iron in Bordetella bronchiseptica. J Bacteriol. 2008;190:3940–3947. doi: 10.1128/JB.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson BL, Bearson SM, Uthe JJ, et al. Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect. 2008;10:807–816. doi: 10.1016/j.micinf.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Brown SW, Meyers RT, Brennan KM, et al. Catecholamines in a macrophage cell line. J Neuroimmunol. 2003;135:47–55. doi: 10.1016/s0165-5728(02)00435-6. [DOI] [PubMed] [Google Scholar]
- Burton CL, Chhabra SR, Swift S, et al. The growth response of Escherichia coli to neurotransmitters and related catecholamine drugs requires a functional enterobactin biosynthesis and uptake system. Infect Immun. 2002;70:5913–5923. doi: 10.1128/IAI.70.11.5913-5923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Russell R, Moreira CG, et al. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. MBio. 2014;5:e02165. doi: 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl MA, Rittirsch D, Nadeau BA, et al. Phagocyte-derived catecholamines enhance acute inflammatory injury. Nature. 2007;449:721–725. doi: 10.1038/nature06185. [DOI] [PubMed] [Google Scholar]
- Flierl MA, Rittirsch D, Huber-Lang M, Sarma JV, Ward PA. Catecholamines: crafty weapons in the inflammatory arsenal of immune/inflammatory cells or opening pandora's box? Mol Med. 2008;14:195–204. doi: 10.2119/2007-00105.Flierl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl MA, Rittirsch D, Nadeau BA, et al. Upregulation of phagocyte-derived catecholamines augments the acute inflammatory response. PLoS ONE. 2009;4:e4414. doi: 10.1371/journal.pone.0004414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KP, Chung WO, Lamont RJ, Demuth DR. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect Immun. 2001;69:7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KP, Gao L, Demuth DR. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect Immun. 2003;71:298–308. doi: 10.1128/IAI.71.1.298-308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone PP, Lyte M, Neal CP, Maggs AF, Haigh RD, Williams PH. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J Bacteriol. 2000;182:6091–6098. doi: 10.1128/jb.182.21.6091-6098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone PP, Haigh RD, Williams PH, Lyte M. Involvement of enterobactin in norepinephrine-mediated iron supply from transferrin to enterohaemorrhagic Escherichia coli. FEMS Microbiol Lett. 2003;222:39–43. doi: 10.1016/S0378-1097(03)00243-X. [DOI] [PubMed] [Google Scholar]
- Freestone PP, Sandrini SM, Haigh RD, Lyte M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 2008;16:55–64. doi: 10.1016/j.tim.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Guckes KR, Kostakioti M, Breland EJ, et al. Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc Natl Acad Sci USA. 2013;110:16592–16597. doi: 10.1073/pnas.1315320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic, E. coli (EHEC) PLoS Pathog. 2009;S5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch HF, Marz D, Kruger M. The effects of stress hormones on growth of selected periodontitis related bacteria. Anaerobe. 2013;24:49–54. doi: 10.1016/j.anaerobe.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Ji S, Hyun J, Park E, Lee BL, Kim KK, Choi Y. Susceptibility of various oral bacteria to antimicrobial peptides and to phagocytosis by neutrophils. J Periodontal Res. 2007;42:410–419. doi: 10.1111/j.1600-0765.2006.00962.x. [DOI] [PubMed] [Google Scholar]
- Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology. 2011;157:1851–1875. doi: 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth P, Trivedi U, Rumbaugh K, Whiteley M. Probing bacterial metabolism during infection using high-resolution transcriptomics. J Bacteriol. 2013;195:4991–4998. doi: 10.1128/JB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Rodríguez MD, Torres-Escobar A, Demuth DR. Construction of new cloning, lacZ reporter and scarless-markerless suicide vectors for genetic studies in Aggregatibacter actinomycetemcomitans. Plasmid. 2013a;69:211–222. doi: 10.1016/j.plasmid.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Rodríguez MD, Torres-Escobar A, Demuth DR. ygiW and qseBC are co-expressed in Aggregatibacter actinomycetemcomitans and regulate biofilm growth. Microbiology. 2013b;159:989–1001. doi: 10.1099/mic.0.066183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Rodríguez MD, Torres-Escobar A, Demuth DR. Transcriptional regulation of the Aggregatibacter actinomycetemcomitans ygiW-qseBC operon by QseB and integration host factor proteins. Microbiology. 2014;160:2583–3594. doi: 10.1099/mic.0.083501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany SC, Schwartz AB, Balashova NV, et al. Anti-leukemia activity of a bacterial toxin with natural specificity for LFA-1 on white blood cells. Leuk Res. 2010;34:777–785. doi: 10.1016/j.leukres.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xu Z, Zhou Y, et al. Global effects of catecholamines on Actinobacillus pleuropneumoniae gene expression. PLoS ONE. 2012;S7:e31121. doi: 10.1371/journal.pone.0031121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M, Septer AN, Carroll-Portillo A, et al. Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol. 2009;9:42. doi: 10.1186/1471-2180-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Novak EA, Shao H, Daep CA, Demuth DR. Autoinducer-2 and QseC control biofilm formation and in vivo virulence of Aggregatibacter actinomycetemcomitans. Infect Immun. 2010;78:2919–2926. doi: 10.1128/IAI.01376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permpanich P, Kowolik MJ, Galli DM. Resistance of fluorescent-labelled Actinobacillus actinomycetemcomitans strains to phagocytosis and killing by human neutrophils. Cell Microbiol. 2006;8:72–84. doi: 10.1111/j.1462-5822.2005.00601.x. [DOI] [PubMed] [Google Scholar]
- Rahamat-Langendoen JC, van Vonderen MG, Engstrom LJ, Manson WL, van Winkelhoff AJ, Mooi-Kokenberg EA. Brain abscess associated with Aggregatibacter actinomycetemcomitans: case report and review of literature. J Clin Periodontol. 2011;38:702–706. doi: 10.1111/j.1600-051X.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- Robertson PB, Lantz M, Marucha PT, Kornman KS, Trummel CL, Holt SC. Collagenolytic activity associated with Bacteroides species and Actinobacillus actinomycetemcomitans. J Periodontal Res. 1982;17:275–283. doi: 10.1111/j.1600-0765.1982.tb01154.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Sandrini SM, Shergill R, Woodward J, et al. Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J Bacteriol. 2010;192:587–594. doi: 10.1128/JB.01028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, James D, Lamont RJ, Demuth DR. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J Bacteriol. 2007;189:5559–5565. doi: 10.1128/JB.00387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Dzink JL, Smith CM. Chemically defined medium for oral microorganisms. J. Clin. Microbiol. 1985;22:303–305. doi: 10.1128/jcm.22.2.303-305.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele KH, O'Connor LH, Burpo N, Kohler K, Johnstone JW. Characterization of a ferrous iron-responsive two-component system in nontypeable Haemophilus influenzae. J Bacteriol. 2012;194:6162–6173. doi: 10.1128/JB.01465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend TR, Gillenwater JY. Urinary tract infection due to Actinobacillus actinomycetemcomitans. JAMA. 1969;210:558. [PubMed] [Google Scholar]
- Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell. 2000;103:113–125. doi: 10.1016/s0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Yew HS, Chambers ST, Roberts SA, et al. Association between HACEK bacteraemia and endocarditis. J Med Microbiol. 2014;63:892–895. doi: 10.1099/jmm.0.070060-0. [DOI] [PubMed] [Google Scholar]
- Zhang T, Kurita-Ochiai T, Hashizume T, Du Y, Oguchi S, Yamamoto M. Aggregatibacter actinomycetemcomitans accelerates atherosclerosis with an increase in atherogenic factors in spontaneously hyperlipidemic mice. FEMS Immunol Med Microbiol. 2010;59:143–151. doi: 10.1111/j.1574-695X.2010.00674.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


