Abstract
Objective
To assess candidate genes for association with osteoarthritis (OA) and identify promising genetic factors and, secondarily, to assess the candidate gene approach in OA.
Methods
A total of 199 candidate genes for association with OA were identified using Human Genome Epidemiology (HuGE) Navigator. All of their single-nucleotide polymorphisms (SNPs) with an allele frequency of >5% were assessed by fixed-effects meta-analysis of 9 genome-wide association studies (GWAS) that included 5,636 patients with knee OA and 16,972 control subjects and 4,349 patients with hip OA and 17,836 control subjects of European ancestry. An additional 5,921 individuals were genotyped for significantly associated SNPs in the meta-analysis. After correction for the number of independent tests, P values less than 1.58 × 10−5 were considered significant.
Results
SNPs at only 2 of the 199 candidate genes (COL11A1 and VEGF) were associated with OA in the meta-analysis. Two SNPs in COL11A1 showed association with hip OA in the combined analysis: rs4907986 (P = 1.29 × 10−5, odds ratio [OR] 1.12, 95% confidence interval [95% CI] 1.06−1.17) and rs1241164 (P = 1.47 × 10−5, OR 0.82, 95% CI 0.74−0.89). The sex-stratified analysis also showed association of COL11A1 SNP rs4908291 in women (P = 1.29 × 10−5, OR 0.87, 95% CI 0.82−0.92); this SNP showed linkage disequilibrium with rs4907986. A single SNP of VEGF, rs833058, showed association with hip OA in men (P = 1.35 × 10−5, OR 0.85, 95% CI 0.79−0.91). After additional samples were genotyped, association at one of the COL11A1 signals was reinforced, whereas association at VEGF was slightly weakened.
Conclusion
Two candidate genes, COL11A1 and VEGF, were significantly associated with OA in this focused meta-analysis. The remaining candidate genes were not associated.
The etiology of primary osteoarthritis (OA) is multifactorial and includes aging and mechanical, hormonal, and genetic factors (1). Investigations of many of these factors have produced contradictory results, making conclusions difficult. Some of the contradictory results are attributable to the design of the studies, but others can be attributable to undetermined differences between patients (1,2). It is also notable that susceptibility factors, including genetic factors, are not equally shared by the different joint regions affected by OA (3,4).
Previous attempts have been made to identify genetic factors involved in OA, using association studies of candidate genes, linkage studies in multicase families, or genome-wide association studies (GWAS) (2,5). Candidate genes were studied because of their important roles in the pathogenesis of OA or their altered expression in OA tissue. The results of most of these association studies have been inconclusive. The association of only one candidate gene, GDF5, was well supported by data from multiple studies and reached genome-wide significance (P < 5 × 10−8) in subsequent replication analysis (6,7). Linkage studies have provided some consistent results, but progression from these results to identification of the causal genes has proven to be difficult (2,5).
Recently, GWAS have identified 11 additional OA susceptibility loci with genome-wide significance levels. Two of them, in DVWA/COL6A4 and a region containing HLA class II/III genes, showed association in Asians but not in Europeans (8–10). The other 9 loci reached genome-wide significance in Europeans. They include a locus on chromosome 7q22 (which is located in a large linkage disequilibrium [LD] block that contains 6 genes) associated with knee OA (11,12); MCF2L associated with knee and hip OA (13); 5 loci that were identified in the arcOGEN GWAS (GNL3/GLT8D1 associated with knee and hip OA, ASTN2 associated with severe hip OA in women, FILIP1–SENP6 and PTHLH associated with hip OA, and CHTS11 associated with severe hip OA) (14); and DOT1L associated with joint space width of the hip (15) and with hip OA in men (16). Approximately 8 more loci are near this level of association (2,5,14).
All of the studies reaching genome-wide significance have used meta-analysis of data from multiple sample collections. This is a very efficient approach to increase power. In addition, it is also very useful for discovery of new associations when applied to GWAS, because each study provides information for most single-nucleotide polymorphisms (SNPs) in the genome, either directly or through imputation, and therefore add to the overall result (17). Another way to favor discovery of new loci is by focusing analysis on particular subsets of genes for which there is prior supporting evidence, thereby increasing the prior probability of association and reducing the burden of multiplicity and thus the stringency required for claiming association (18,19).
The aim of this study was to identify new OA genetic factors, using meta-analysis of GWAS and focused analysis of OA candidate genes. A secondary aim was to assess the validity of the candidate gene approach in OA. To this end, we performed a meta-analysis of 5,636 patients with knee OA and 4,349 patients with hip OA from 9 GWAS and explored the association with knee OA or hip OA with >24,000 SNPs corresponding to 199 previously reported OA candidate genes. Two candidate genes, COL11A1 and VEGF, were significantly associated with OA. The remaining 197 candidate genes were not significantly associated with OA.
PATIENTS AND METHODS
Sample collections
We performed a meta-analysis of 9 GWAS that included patients with knee or hip OA and control subjects of European descent: the deCODE study from Iceland (20), 3 collections from the Rotterdam Study (Rotterdam Study I, II, and III) (21), the Genetics Osteoarthritis and Progression (GARP) collection from The Netherlands (22), arcOGEN phase I (23) and TwinsUK (24) from the UK, the Framingham Osteoarthritis Study from the US (25), and EGCUT (Estonian Genome Center of the University of Tartu) from Estonia (26). Additional sample collections not involved in the GWAS were used for an extension study of significant results (Table 1). They included collections from the north of Spain (27–29), the center of Greece (30), and the Nottingham (31) and Genetics in Osteoarthritis and Lifestyle (GOAL) (32) studies from the UK.
Table 1.
Characteristics of the sample collections included in the GWAS meta-analysis and the extension study*
| Knee OA |
Hip OA |
||||
|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | OA definition | |
| Meta-analysis | |||||
| arcOGEN study | 1,643 | 4,896 | 1,728 | 4,896 | TJR and radiographic |
| deCODE study† | 1,312 | 2,318 | 1,423 | 2,318 | TJR and clinical |
| Framingham study | 417 | 1,667 | NA | Radiographic | |
| GARP study | 154 | 1,671 | 106 | 1,671 | Radiographic and clinical |
| Rotterdam Study I | 1,476 | 3,233 | 760 | 3,233 | Radiographic |
| Rotterdam Study II | 369 | 1,472 | 159 | 1,472 | Radiographic |
| Rotterdam Study III | 152 | 1,487 | 41 | 1,487 | Radiographic |
| TwinsUK | 113 | 228 | 68 | 228 | Radiographic |
| EGCUT study | 64 | 2,531 | Radiographic | ||
| Total | 5,636 | 16,972 | 4,349 | 17,836 | – |
| Extension study | |||||
| Spain‡ | – | – | 695 | 784 | TJR |
| Greece | – | – | 92 | 358 | TJR |
| Nottingham§ | – | – | 1,246 | 713 | TJR |
| GOAL§ | – | – | 1,270 | 763 | TJR and radiographic |
| Total | 3,303 | 2,618 | – | ||
*GWAS = genome-wide association study; OA = osteoarthritis; arcOGEN = Arthritis Research UK Osteoarthritis Genetics; TJR = total joint replacement; NA = not available; GARP = Genetics Osteoarthritis and Progression; EGCUT = Estonian Genome Center of the University of Tartu; GOAL = Genetics in Osteoarthritis and Lifestyle.
Effective sample size.
Samples were obtained from 3 collections (Santiago, Santander, and A Corunna).
Samples were genotyped only for rs833058 (VEGF).
All of these sample collections have been described in detail previously. Briefly, the deCODE study included patients with total knee replacement or total hip replacement, patients with OA, and population controls, which excluded all individuals on OA susceptibility lists (hand, hip, knee) obtained from hospitals and health care centers in Iceland (20). The Rotterdam Study, Framingham Osteoarthritis Study, and TwinsUK included patients with radiographic OA and controls from the same cohort without radiographic signs of OA, as defined according to standardized phenotypes, with patients having a Kellgren/Lawrence (K/L) score (33) of ≥2 and controls having a K/L score of <2 (34). The GARP cohort included patients with confirmed clinical and radiographic (K/L score of ≥2) OA at ≥2 joint sites (22). They were compared with population controls. The arcOGEN phase I cohort included cases of knee or hip OA, as determined by radiographic evidence of disease or clinical evidence of disease “to a level requiring joint replacement” (23); controls were derived from an early release of the Wellcome Trust Case Control Consortium 2 data. EGCUT included cases of radiographically confirmed OA (K/L score of >2) and controls that were free of any OA symptoms. All sample collections used for the extension study were derived from case−control studies. Patients were ascertained on the basis of having undergone hip joint replacement due to symptomatic and radiographically confirmed hip OA, except in GOAL, in which severe symptomatic hip OA was the selection criterion. All of the sample collections and the genetic studies received approval by the relevant ethics committees, and the samples were obtained with the written informed consent of the participants.
Gene and SNP selection
We used the Phenopedia tool of the Human Genome Epidemiology (HuGE) Navigator (35) to identify candidate genes that have been studied for their possible association with OA, without additional filtering. The query terms were as follows: osteoarthritis, spinal osteophytosis, and intervertebral disk displacement. The candidate genes derived from the spinal osteophytosis and intervertebral disk displacement lists showed a large degree of overlap with those from the osteoarthritis list (90% and 72%, respectively). The 17 nonoverlapping genes were considered to be of interest for the discovery of new OA loci. This database covers genetic studies published since 2000. Genes in chromosome X were excluded, because it is impossible to impute the genotypes needed for meta-analysis across different GWAS designs. FRAH1H was excluded because the bibliographic reference was incorrect. All of the genes with genome-wide significance in Europeans (P < 5 × 10−8) were also excluded. Duplicates were removed.
Map positions of loci encompassing the candidate genes and 50 kb downstream of their stop codon and upstream of their start codon were obtained from the Ensembl database. Overlapping loci were fused as a single locus. All SNPs with a minor allele frequency (MAF) of >5% in the CEU data set corresponding to the candidate gene loci were retrieved from HapMap (phases 1, 2, and 3; release 27) with in-house Perl programs interacting with the HapMart server. All SNPs were aligned according to the positive strand, to avoid ambiguities.
Genotyping and imputation of untyped SNPs
Genotyping technologies for the GWAS included in the meta-analysis were different and have been previously described in detail (11,20,23,25,36). Imputation of untyped SNPs was performed based on CEU data from HapMap (phases 1 and 2; release 22). Summary information on genotyping and imputation is shown in Supplementary Table 1 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38300/abstract). Genotyping of the extension study collections was performed at Hospital Clinico Universitario de Santiago, using single-base extension with a SNaPshot Multiplex Kit (Applied Biosystems) (Spanish and Greek collections) or was carried out by KBioscience, using a KASPar system (UK collections).
Statistical analysis
Each team contributing a GWAS performed association testing for knee OA and hip OA under a per-allele model. The lambda inflation factor was calculated per sex-specific effect size using the genomic control method (37), and the SEs were corrected by the square root of the lambda inflation factor (SEcorrected = SEobserved×√λ). Robust SEs were estimated to adjust for family relationships (deCODE and GARP studies). For meta-analysis, the effect size for each SNP (odds ratio [OR] per copy of minor allele as per HapMap) was calculated using fixed-effects inverse variance models, synthesizing all effect sizes and the corrected SEs.
Heterogeneity was assessed with the I2 statistic; when low or moderate heterogeneity was observed, no random-effects meta-analysis was performed (38). Meta-analysis of the GWAS was performed using METAL software (39), considering 6 strata with 2 joint levels (knee and hip) and 3 sex levels (all, women, and men). Two research centers (Ioannina, Greece and Erasmus MC Rotterdam, The Netherlands) performed both the quality control and meta-analyses for the whole GWAS. A quality control protocol was set up that included validation of the results file format, reports for range of values, and elimination of potential biases (i.e., extremely large beta values or SEs). Files were cross-validated between the 2 research centers after quality control and meta-analyses to check for inconsistencies. SNPs with a MAF of <1%, imputation quality of <0.30 (MACH program) or <0.40 (IMPUTE program) and beta values of >4 or <−4, and SNPs that were not available in >4 studies were excluded from further analysis.
The significance threshold for claiming association was determined considering the number of independent tests performed. This number was estimated using a modification of the simpleM algorithm (40) applied to the genotypes of the CEU collection in HapMap. The modification consisted of replacing the observed correlation matrix for the nearest positive semidefinite matrix, as implemented in the R software package corpcor (http://www.R-project.org), to correct for biases introduced by missing genotypes. It was estimated that the number of independent tests represented by these SNPs was 3,156 (see Supplementary Table 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38300/abstract). The number of independent tests was used to define a significance threshold of P = 1.58 × 10−5, according to Bonferroni multiplicity correction. No additional correction was performed for stratification according to joint and sex, because there is known heterogeneity across these strata in OA genetics (2–5) and, therefore, no correction of this type is used in OA genetic studies (6,8,9,11,13,14,16).
Results of the extension study were combined using a Mantel-Haenszel approach (40). Combination of the extension study data with the GWAS data was done using a fixed-effects model with R software (http://www.R-project.org). Power estimates were obtained with Power and Sample Size software (42). A full analysis of power is shown in Supplementary Figure 1 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38300/abstract). For example, the power to detect association with a SNP of MAF 20% and OR 1.15 for knee OA or 1.16 for hip OA was 80%, assuming no heterogeneity between the GWAS.
RESULTS
Systematic identification of candidate genes
A total of 199 genes (Figure 1) have been investigated for their association with OA in humans, according to the HuGe Navigator (35). The HuGe Navigator included 542 bibliographic references for the genetic studies of the candidate genes. Some of the genes are from loci that were associated with OA for the first time in GWAS (HLA class II/III, A2BP1, and LCRH1) or in genome-wide linkage studies (MATN3, DIO2, FRZB, and BMP5). These genes were included because the identification of many of them as putative susceptibility genes partially came from post hoc analyses of their potential biologic role, and because none has reached a genome-wide significant association in Europeans. The 199 genes were grouped in 158 nonoverlapping genome segments that contained 27,501 autosomal SNPs (MAF of >5%) with known genotypes in the CEU (Utah residents with ancestry from northern and western Europe) population of HapMap.
Figure 1.
Osteoarthritis candidate genes selected from Human Genome Epidemiology Navigator.
Meta-analysis of association of all SNPs in the candidate genes
The effect sizes for each of the 9 GWAS corresponding to SNPs in the candidate genes were obtained and combined in a meta-analysis. Genotypes were available for 25,839 of the 27,501 SNPs included in candidate genes after applying quality control filters (association results are available from the corresponding author). These genotypes had been directly typed or imputed. No significant associations were observed in the knee OA meta-analysis.
Meta-analysis of hip OA GWAS showed significant association at 2 candidate genes. Two SNPs, rs4907986 and rs1241164, in the 5′ and 3′ ends of COL11A1, respectively, were associated in the unstratified analysis (Table 2 and Figure 2A). They comprised 2 independent associations, as manifested by the low pairwise correlation coefficient (r2 = 0.09) between them. A SNP of COL11A1, rs2615977, was highlighted in the previously reported analysis of the arcOGEN phase I study (with P = 1.1 × 10−5) (23), which overlaps with the current meta-analysis. However, none of the 2 independent top associated SNPs showed strong correlation with rs2615977 (pairwise r2 <0.4), and rs2615977 was not among the most-associated SNPs in the meta-analysis. An analysis stratified by sex also showed association of COL11A1 SNP rs4908291 in women. This SNP showed LD with rs4907986 (r2 = 0.68) but not with rs1241164.
Table 2.
SNPs independently associated with hip osteoarthritis in all samples and in samples stratified by sex*
| Sample | Chr. | Position | Alleles | OR (95% CI) | P | I2 | P for heterogeneity |
|---|---|---|---|---|---|---|---|
| All subjects | |||||||
| COL11A1 | |||||||
| rs4907986 | 1 | 103322221 | T/C | 1.12 (1.06−1.17) | 1.29 × 10−5 | 0 | 0.63 |
| rs1241164 | 1 | 103129065 | T/C | 0.82 (0.74−0.89) | 1.47 × 10−5 | 0 | 0.92 |
| Women | |||||||
| COL11A1 | |||||||
| rs4908291 | 1 | 103345324 | A/T | 0.87 (0.82−0.92) | 1.29 × 10−5 | 33.2 | 0.16 |
| Men | |||||||
| VEGF | |||||||
| rs833058 | 6 | 43839832 | T/C | 0.85 (0.79−0.91) | 1.35 × 10−5 | 18.6 | 0.29 |
Single-nucleotide polymorphisms (SNPs) below the threshold of significance (P = 1.58 × 10−5) are shown. The odds ratios (ORs) and 95% confidence intervals (95% CIs) are relative to the first listed allele at each SNP. Chr. = chromosome.
Figure 2.
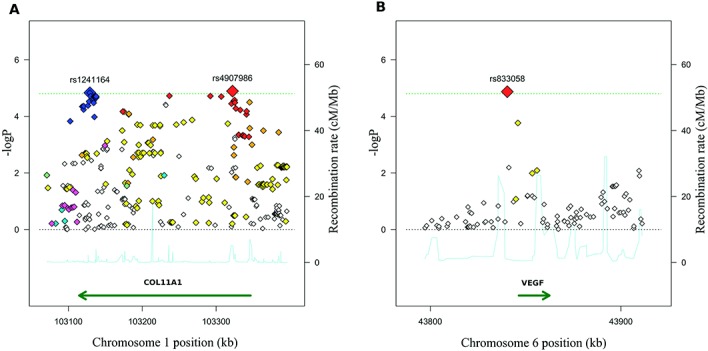
Regional association plot showing the association of COL11A1 (A) and VEGF (B) loci with hip osteoarthritis. Each single-nucleotide polymorphism (SNP) is shown as a diamond, located in the genome physical map (x-axis) and the –log10 scale of its P value of association (y-axis). A, Pairwise correlation of each SNP with rs1241164, with white diamonds representing minimal correlation, blue diamonds representing maximal correlation (r2 ≥0.8), and magenta diamonds representing intermediate correlation (r2 ≥0.5−<0.8), and pairwise correlation of each SNP with rs4907986, with white diamonds representing minimal correlation, red diamonds representing maximal correlation, and orange diamonds representing intermediate correlation. B, Pairwise correlation of each SNP with the top associated SNP (rs833058), with white diamonds representing minimal correlation and the red diamond representing maximal correlation. Dotted horizontal black lines show the 1.58 × 10−5 threshold for significance. Continuous blue lines represent the recombination rate (right y-axis).
The second candidate gene associated with hip OA was VEGF. Only a single SNP, rs833058, reached association over the required threshold in men (Table 2). No other SNP with strong or modest (r2 >0.5) LD with rs833058 was observed in the meta-analysis (Figure 2B). None of the other 197 candidate genes showed association with knee or with hip OA at the requested level of significance.
Extension and summary studies
We attempted to further establish the association of hip OA with 3 SNPs, 2 from COL11A1 representing the top SNPs of the 2 independent associations in the combined analysis, and the top SNP in VEGF. This part of the study was not intended as a replication of the results due to the relatively small number of independent samples that were available.
For the COL11A1 SNPs, 1,929 samples from Spanish and Greek individuals (787 patients with hip OA and 1,142 control subjects) were studied. No significant association was observed in this underpowered analysis; however, analysis of additional samples showed independent association (Kerkhof HJM, van Meurs JB: unpublished observations). In the combined analysis, the significance of rs1241164 association was clearer (P = 5.3 × 10−6) than before, given that the direction of change and effect size were similar in the meta-analysis and the extension analysis (Table 3). The second signal, corresponding to rs4907986, was slightly weakened in the combined analysis, because the risk allele identified in the meta-analysis showed the same direction only in women in the extension study, not in men.
Table 3.
Association of the 2 top independent SNPs in COL11A1 and the top SNP in VEGF with hip OA*
| GWAS meta-analysis |
Extension study |
GWAS meta-analysis plus extension study |
||||
|---|---|---|---|---|---|---|
| Gene, SNP, sex | ORM-H(95% CI) | P | ORM-H(95% CI) | P | OR (95% CI) | P |
| COL11A1 | ||||||
| rs1241164 | ||||||
| All | 0.82 (0.74–0.89) | 1.47 × 10−5 | 0.85 (0.68–1.06) | 0.15 | 0.82 (0.75–0.89) | 5.3 ×10−6 |
| Female | 0.79 (0.7–0.89) | 1.27 × 10−4 | 0.96 (0.70–1.32) | 0.81 | 0.81 (0.72–0.91) | 2.4 × 10−4 |
| Male | 0.84 (0.74–0.97) | 0.012 | 0.77 (0.56–1.06) | 0.11 | 0.83 (0.73–0.94) | 3.7 × 10−3 |
| rs4907986 | ||||||
| All | 1.12 (1.06–1.17) | 1.29 × 10−5 | 0.98 (0.85–1.12) | 0.77 | 1.09 (1.05–1.15) | 5.8 × 10−5 |
| Female | 1.13 (1.07–1.21) | 1.05 × 10−4 | 1.15 (0.95–1.39) | 0.13 | 1.13 (1.07–1.21) | 3.2 × 10−5 |
| Male | 1.1 (1.03–1.19) | 0.0069 | 0.95 (0.77–1.18) | 0.67 | 1.08 (1.01–1.16) | 0.015 |
| VEGF | ||||||
| rs833058 | ||||||
| All | 0.92 (0.88–0.97) | 0.0022 | 0.92 (0.85–0.99) | 0.03 | 0.92 (0.88–0.96) | 1.9 × 10−4 |
| Female | 0.99 (0.93–1.06) | 0.75 | 0.91 (0.81–1.01) | 0.07 | 0.97 (0.91–1.02) | 0.21 |
| Male | 0.85 (0.79–0.91) | 1.3 × 10−5 | 0.94 (0.84–1.06) | 0.31 | 0.87 (0.82–0.93) | 2.6 × 10−5 |
Single-nucleotide polymorphisms (SNPs) rs1241164 and rs4907986 were genotyped only in samples from Spain and Greece. The odds ratios (ORs) and 95% confidence intervals (95% CIs) are relative to the first listed allele for each SNP in Table 2. OA = osteoarthritis; GWAS = genome-wide association study; ORM-H = OR as determined using the Mantel-Haenszel approach. No significant heterogeneity (P < 0.05) was detected in any of the combined analyses.
For VEGF SNP rs833058, we genotyped 5,921 additional individuals (3,303 patients with hip OA and 2,618 control subjects). No significant association was observed in men (1,466 patients with hip OA and 1,263 control subjects), but the direction of change in the extension study was the same as that in the meta-analysis. Summary results in men showed association slightly below that in the meta-analysis (2.6 × 10−5). This SNP showed weak association in the combined analysis of men and women in the extension study (P = 0.03).
DISCUSSION
This focused analysis of candidate genes within a large meta-analysis of GWAS identified associations with hip OA in 2 genes: COL11A1, which showed 2 independent signals, and VEGF, which showed association in men. The main objective of the study was to highlight SNPs for further study and confirmation as new OA genetic factors. A secondary aim of the study was to assess the validity of the candidate gene approach in the study of OA. In this respect, we observed that none of the other 197 candidate genes showed association in our meta-analysis of GWAS.
Discovery of genetic associations has been greatly advanced by GWAS due to increased sample sizes, increased coverage of analyzed SNPs, and increases in quality control standards and in the requirements to claim association that have accompanied them. These positive characteristics of the GWAS are further potentiated by their combination through meta-analysis (17). The current meta-analysis had an unprecedented power to analyze most of the candidate genes (at least for an OR of >1.15 and allele frequencies of >0.2), and GWAS plus imputation provided very complete coverage of genetic variation in them.
In addition, focused analysis of candidate genes should increase the chances of uncovering associations worth pursuing, by the following 2 mechanisms: increased prior likelihood of association and more tolerant threshold to claim association (18,19). A series of studies of other diseases have capitalized on the first mechanism, either by considering candidate genes from previous genetic studies, as was done here, or by defining a set of genes of high relevance for the disease, using bibliographic analysis. The second mechanism, a more tolerant threshold for association, is related to a problem that affects all complex diseases: the effect sizes of most genetic factors are below an OR of 1.20, and therefore large sample sizes are required to identify association at the genome level. In this context, the use of focused analysis in a subset of genes allows the selection of SNPs not reaching genome-wide significance for further validation.
Another point that deserves comment is that small variations between sample collections result in large differences in statistical power. This is attributable to the low effect sizes of most genetic factors and the dramatic decrease in statistical power when the effect size approaches 1.0 (see Supplementary Figure 1 available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38300/abstract). It is very likely that this extreme sensitivity to variation of the effect size in the 1.0–1.15 range explains why confirmed genetic factors fail to show significant association even in some large studies, given that they are not large enough to offset these small fluctuations. In addition, there is a dramatic decrease in power for SNPs with a low MAF (<10%) (Supplementary Figure 1). Thus, we cannot exclude the possibility that some additional candidate genes may have associations with OA with an OR of ≤1.10 or if their minor allele frequencies are low.
COL11A1 codes for a minor component of the cartilage matrix, the importance of which has been shown by the chondrodysplasia mouse mutation, the skeletal abnormalities of Stickler syndrome, type II (OMIM ID 604841) and Marshall syndrome (OMIM ID 154780), and mutations in patients with fibrochondrogenesis and by association with susceptibility to lumbar disc herniation (http://omim.org/entry/120280). The 2 top SNPs associated with OA in our analysis were independent and different from those observed in a previous OA study (23). This raises the possibility of multiple variants with an effect in OA susceptibility.
VEGF was associated with hip OA only in men, according to our meta-analysis. This gene codes for a very important angiogenic factor that is involved in normal growth plate development, endochondral ossification, and articular cartilage formation (43). It is one of the overexpressed markers of hypertrophic chondrocytes. It contributes to OA changes in animal models by stimulating chondrocyte proliferation, apoptosis, and production of catabolic mediators (44). Although it has been considered an OA candidate gene, no previous study showed significant association.
For the remaining candidate genes, we did not observe association in spite of the large number of samples assembled (the largest ever studied for most candidate genes) and the more tolerant threshold for significance allowed by the focused analysis. This does not exclude SNPs of weak effect or those showing heterogeneity between the sample collections. Also, it is possible that some gene variants were not adequately covered, especially those with low frequency or multiallelic polymorphisms, as in ASPN (45) or BMP5 (46,47). Other reported associations have been described as specific for an OA subphenotype not included in our meta-analysis, such as the association of MATN3 with OA in the first carpometacarpal joint (48) or DIO2 in women with severe OA (49). In addition, GDF5, which has been also a candidate gene, was not included in this analysis because it is already confirmed as an OA susceptibility locus at the genome-wide significance level (6,7).
All of the previous caveats apply, but they do not negate the conclusion of a general lack of reproducibility of OA candidate genes. This is particularly true for genes highlighted in candidate gene studies of small size and showing large effect sizes. It has become clear that the effect sizes and ORs reported were widely overestimated, because we would have observed most associations with ORs >1.20, and most reported ORs in these studies were ∼2-fold. Our findings suggest that traditionally conducted candidate gene studies are unlikely to be fruitful in OA genetics. However, such candidate gene studies are likely to continue to be useful to validate or refine loci from hypothesis-free genome-wide studies.
In summary, our candidate gene meta-analysis of 9 OA GWAS highlighted the association of COL11A1 and VEGF with hip OA and showed a lack of association for the other 197 candidate genes.
Acknowledgments
We thank the Newcastle arm of arcOGEN and the Freeman Hospital arthroplasty surgical and nursing teams. We are grateful for the support of the National Institute of Health Research (NIHR) Newcastle Biomedical Research Centre (Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University) and the Northumberland, Tyne and Wear Comprehensive Local Research Network. Finally, we thank EGCUT technical personnel, especially Mr. V. Soo and S. Smit. Data analyses were carried out in part at the High Performance Computing Center, University of Tartu.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Gonzalez had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Rodriguez-Fontenla, Calaza, Arden, Chapman, Esko, Gomez-Reino Carnota, Hofman, Ntzani, Ollier, Ralston, Slagboom, Wallis, Wilkinson, Metspalu, Spector, Gonzalez.
Acquisition of data. Valdes, Arden, Blanco, Carr, Deloukas, Doherty, Esko, Garcés Aletá, Gomez-Reino Carnota, Helgadottir, Jonsdottir, Kerkhof, Kloppenburg, McCaskie, Ntzani, Ollier, Oreiro, Ralston, Ramos, Riancho, Rivadeneira, Styrkarsdottir, Thorsteinsdottir, Thorleifsson, Tsezou, Uitterlinden, Wallis, Wilkinson, Zhai, Felson, Ioannidis, Loughlin, Metspalu, Meulenbelt, Stefansson, van Meurs, Zeggini.
Analysis and interpretation of data. Rodriguez-Fontenla, Calaza, Evangelou, Arden, Esko, Ntzani, Panoutsopoulou, Slagboom, Wallis, Zhai, Zhu, Metspalu, van Meurs, Zeggini, Gonzalez.
ADDITIONAL DISCLOSURE
Mr. Helgadottir and Drs. Jonsdottir, Styrkarsdottir, Thorsteinsdottir, Thorliefsson, and Stefansson are employees of deCODE Genetics.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Data
REFERENCES
- 1.Blagojevic M, Jinks C, Jeffery A, Jordan KP. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2010;18:24–33. doi: 10.1016/j.joca.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Loughlin J. Genetics of osteoarthritis. Curr Opin Rheumatol. 2011;23:479–83. doi: 10.1097/BOR.0b013e3283493ff0. [DOI] [PubMed] [Google Scholar]
- 3.Hunter DJ, Demissie S, Cupples LA, Aliabadi P, Felson DT. A genome scan for joint-specific hand osteoarthritis susceptibility: The Framingham Study. Arthritis Rheum. 2004;50:2489–96. doi: 10.1002/art.20445. [DOI] [PubMed] [Google Scholar]
- 4.MacGregor AJ, Li Q, Spector TD, Williams FM. The genetic influence on radiographic osteoarthritis is site specific at the hand, hip and knee. Rheumatology (Oxford) 2009;48:277–80. doi: 10.1093/rheumatology/ken475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valdes AM, Spector TD. The contribution of genes to osteoarthritis. Med Clin North Am. 2009;93:45–66. doi: 10.1016/j.mcna.2008.08.007. x. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–33. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 7.Valdes AM, Evangelou E, Kerkhof HJ, Tamm A, Doherty SA, Kisand K, et al. The GDF5 rs143383 polymorphism is associated with osteoarthritis of the knee with genome-wide statistical significance. Ann Rheum Dis. 2011;70:873–5. doi: 10.1136/ard.2010.134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyamoto Y, Shi D, Nakajima M, Ozaki K, Sudo A, Kotani A, et al. Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nat Genet. 2008;40:994–8. doi: 10.1038/ng.176. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima M, Takahashi A, Kou I, Rodriguez-Fontenla C, Gomez-Reino JJ, Furuichi T, et al. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One. 2010;5:e9723. doi: 10.1371/journal.pone.0009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meulenbelt I, Chapman K, Dieguez-Gonzalez R, Shi D, Tsezou A, Dai J, et al. Large replication study and meta-analyses of DVWA as an osteoarthritis susceptibility locus in European and Asian populations. Hum Mol Genet. 2009;18:1518–23. doi: 10.1093/hmg/ddp053. [DOI] [PubMed] [Google Scholar]
- 11.Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, Arp P, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010;62:499–510. doi: 10.1002/art.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evangelou E, Valdes AM, Kerkhof HJ, Styrkarsdottir U, Zhu Y, Meulenbelt I, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis. 2011;70:349–55. doi: 10.1136/ard.2010.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day-Williams AG, Southam L, Panoutsopoulou K, Rayner NW, Esko T, Estrada K, et al. A variant in MCF2L is associated with osteoarthritis. Am J Hum Genet. 2011;89:446–50. doi: 10.1016/j.ajhg.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeggini E, Panoutsopoulou K, Southam L, Day-Williams A, Lopes M, Boraska V, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–23. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castano Betancourt MC, Cailotto F, Kerkhof HJ, Cornelis FM, Doherty SA, Hart DJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci U S A. 2012;109:8218–23. doi: 10.1073/pnas.1119899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evangelou E, Valdes AM, Castano-Betancourt MC, Doherty M, Doherty S, Esko T, et al. The DOT1L rs12982744 polymorphism is associated with osteoarthritis of the hip with genome-wide statistical significance in males. Ann Rheum Dis. 2013;72:1264–5. doi: 10.1136/annrheumdis-2012-203182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoury MJ, Bertram L, Boffetta P, Butterworth AS, Chanock SJ, Dolan SM, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170:269–79. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–7. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 21.Hofman A, Breteler MM, van Duijn CM, Krestin GP, Pols HA, Stricker BH, et al. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22:819–29. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riyazi N, Meulenbelt I, Kroon HM, Ronday KH, Hellio le Graverand MP, Rosendaal FR, et al. Evidence for familial aggregation of hand, hip, and spine but not knee osteoarthritis in siblings with multiple joint involvement: the GARP study. Ann Rheum Dis. 2005;64:438–43. doi: 10.1136/ard.2004.024661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panoutsopoulou K, Southam L, Elliott KS, Wrayner N, Zhai G, Beazley C, et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann Rheum Dis. 2011;70:864–7. doi: 10.1136/ard.2010.141473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spector TD, MacGregor AJ. The St. Thomas' UK Adult Twin Registry. Twin Res. 2002;5:440–3. doi: 10.1375/136905202320906246. [DOI] [PubMed] [Google Scholar]
- 25.Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metspalu A, Kohler F, Laschinski G, Ganten D, Roots I. The Estonian Genome Project in the context of European genome research. Dtsch Med Wochenschr. 2004;129(Suppl 1):S25–8. doi: 10.1055/s-2004-824840. In German. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Lopez J, Pombo-Suarez M, Liz M, Gomez-Reino JJ, Gonzalez A. Further evidence of the role of frizzled-related protein gene polymorphisms in osteoarthritis. Ann Rheum Dis. 2007;66:1052–5. doi: 10.1136/ard.2006.065938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rego-Perez I, Fernandez-Moreno M, Fernandez-Lopez C, Arenas J, Blanco FJ. Mitochondrial DNA haplogroups: role in the prevalence and severity of knee osteoarthritis. Arthritis Rheum. 2008;58:2387–96. doi: 10.1002/art.23659. [DOI] [PubMed] [Google Scholar]
- 29.Velasco J, Zarrabeitia MT, Prieto JR, Perez-Castrillon JL, Perez-Aguilar MD, Perez-Nunez MI, et al. Wnt pathway genes in osteoporosis and osteoarthritis: differential expression and genetic association study. Osteoporos Int. 2010;21:109–18. doi: 10.1007/s00198-009-0931-0. [DOI] [PubMed] [Google Scholar]
- 30.Kostopoulou F, Gkretsi V, Malizos KN, Iliopoulos D, Oikonomou P, Poultsides L, et al. Central role of SREBP-2 in the pathogenesis of osteoarthritis. PLoS One. 2012;7:e35753. doi: 10.1371/journal.pone.0035753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdes AM, Lories RJ, van Meurs JB, Kerkhof H, Doherty S, Hofman A, et al. Variation at the ANP32A gene is associated with risk of hip osteoarthritis in women. Arthritis Rheum. 2009;60:2046–54. doi: 10.1002/art.24627. [DOI] [PubMed] [Google Scholar]
- 32.Holliday KL, McWilliams DF, Maciewicz RA, Muir KR, Zhang W, Doherty M. Lifetime body mass index, other anthropometric measures of obesity and risk of knee or hip osteoarthritis in the GOAL case-control study. Osteoarthritis Cartilage. 2011;19:37–43. doi: 10.1016/j.joca.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Kellgren JH, Lawrence JS. Radiological assessment of osteoarthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerkhof HJ, Meulenbelt I, Akune T, Arden NK, Aromaa A, Bierma-Zeinstra SM, et al. Recommendations for standardization and phenotype definitions in genetic studies of osteoarthritis: the TREAT-OA consortium. Osteoarthritis Cartilage. 2011;19:254–64. doi: 10.1016/j.joca.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40:124–5. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 36.Valdes AM, Loughlin J, Timms KM, van Meurs JJ, Southam L, Wilson SG, et al. Genome-wide association scan identifies a prostaglandin-endoperoxide synthase 2 variant involved in risk of knee osteoarthritis. Am J Hum Genet. 2008;82:1231–40. doi: 10.1016/j.ajhg.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 38.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–9. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 41.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 42.Dupont WD, Plummer WD., Jr Power and sample size calculations: a review and computer program. Control Clin Trials. 1990;11:116–28. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 43.Stempel J, Fritsch H, Pfaller K, Blumer MJ. Development of articular cartilage and the metaphyseal growth plate: the localization of TRAP cells, VEGF, and endostatin. J Anat. 2011;218:608–18. doi: 10.1111/j.1469-7580.2011.01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludin A, Sela JJ, Schroeder A, Samuni Y, Nitzan DW, Amir G. Injection of vascular endothelial growth factor into knee joints induces osteoarthritis in mice. Osteoarthritis Cartilage. 2013;21:491–7. doi: 10.1016/j.joca.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura T, Shi D, Tzetis M, Rodriguez-Lopez J, Miyamoto Y, Tsezou A, et al. Meta-analysis of association between the ASPN D-repeat and osteoarthritis. Hum Mol Genet. 2007;16:1676–81. doi: 10.1093/hmg/ddm115. [DOI] [PubMed] [Google Scholar]
- 46.Wilkins JM, Southam L, Mustafa Z, Chapman K, Loughlin J. Association of a functional microsatellite within intron 1 of the BMP5 gene with susceptibility to osteoarthritis. BMC Med Genet. 2009;10:141. doi: 10.1186/1471-2350-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Fontenla C, Carr A, Gomez-Reino JJ, Tsezou A, Loughlin J, Gonzalez A. Association of a BMP5 microsatellite with knee osteoarthritis: case-control study. Arthritis Res Ther. 2012;14:R257. doi: 10.1186/ar4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefansson SE, Jonsson H, Ingvarsson T, Manolescu I, Jonsson HH, Olafsdottir G, et al. Genomewide scan for hand osteoarthritis: a novel mutation in matrilin-3. Am J Hum Genet. 2003;72:1448–59. doi: 10.1086/375556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meulenbelt I, Min JL, Bos S, Riyazi N, Houwing-Duistermaat JJ, van der Wijk HJ, et al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Hum Mol Genet. 2008;17:1867–75. doi: 10.1093/hmg/ddn082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


