Abstract
Background
Q fever is a widespread zoonotic disease caused by Coxiella burnetii. Ticks may act as vectors, and many epidemiological studies aim to assess C. burnetii prevalence in ticks. Because ticks may also be infected with Coxiella-like bacteria, screening tools that differentiate between C. burnetii and Coxiella-like bacteria are essential.
Methods
In this study, we screened tick specimens from 10 species (Ornithodoros rostratus, O. peruvianus, O. capensis, Ixodes ricinus, Rhipicephalus annulatus, R. decoloratus, R. geigy, O. sonrai, O. occidentalis, and Amblyomma cajennense) known to harbor specific Coxiella-like bacteria, by using quantitative PCR primers usually considered to be specific for C. burnetii and targeting, respectively, the IS1111, icd, scvA, p1, and GroEL/htpB genes.
Results
We found that some Coxiella-like bacteria, belonging to clades A and C, yield positive PCR results when screened with primers initially believed to be C. burnetii-specific.
Conclusions
These results suggest that PCR-based surveys that aim to detect C. burnetii in ticks by using currently available methods must be interpreted with caution if the amplified products cannot be sequenced. Future molecular methods that aim at detecting C. burnetii need to take into account the possibility that cross-reactions may exist with Coxiella-like bacteria.
Keywords: Q fever, tick-borne diseases, tick endosymbiont, false positive, surveillance, PCR primers
Q fever is a worldwide zoonosis caused by Coxiella burnetii, a ubiquitous intracellular bacterium that infects humans and a variety of animals. Livestock, especially small ruminants, are the main sources of human infections (1–3). In domestic ruminants, Q fever's major clinical manifestations are abortions and stillbirths, whose occurrence may translate into significant economic losses (1–3). In humans, C. burnetii infections range from asymptomatic to severe. Acute forms of the disease may result in high fevers and severe pneumonia or hepatitis, and chronic forms are strongly debilitating and may be fatal when endocarditis develops in patients with underlying heart disease (3, 4). Both animals and humans essentially become infected through the inhalation of airborne particles contaminated with C. burnetii (2, 5).
Ticks are historically known to be potential vectors for Q fever. Indeed, the first strain of C. burnetii was isolated from a Dermacentor andersoni tick in the 1930s (6–8). First assigned to the genus Rickettsia (in which infection by arthropods is the rule), it was later recognized as the Q fever etiologic agent and is now considered as C. burnetii reference strain (9). Because C. burnetii is frequently detected in field-sampled ticks (10–13) and because laboratory experiments have revealed that at least some tick species are competent vectors (6, 14, 15), it is currently considered that ticks may act as vectors and help transmit the bacterium among wildlife and, on occasion, domestic ruminants (2, 16).
Interestingly, ticks also frequently carry Coxiella-like bacteria that are likely involved in mutualistic symbioses with their arthropod hosts (16–19). To date, there has been no indication that these tick-carried Coxiella-like bacteria are transmitted to vertebrates, although Coxiella-like bacteria have sporadically been detected in pet birds (19, 20). Recent investigations based on multilocus phylogenetic analyses and whole genome sequencing data revealed that all known C. burnetii strains originate within the vast group of Coxiella-like endosymbionts and are the descendants of a Coxiella-like progenitor hosted by ticks (17).
Because epidemiologic studies that aim at assessing C. burnetii prevalence in ticks frequently rely on DNA detection by polymerase chain reaction (PCR), it is important to make sure that these screening methods are specific for C. burnetii. The objective of this study was to determine whether five molecular methods frequently used to detect or characterize C. burnetii cross-react with Coxiella-like bacteria present in ticks.
Materials and methods
Selection of a panel of 20 ticks infected with specific Coxiella-like bacteria
Ten tick species, previously shown to harbor specific Coxiella-like bacteria (17), were investigated.
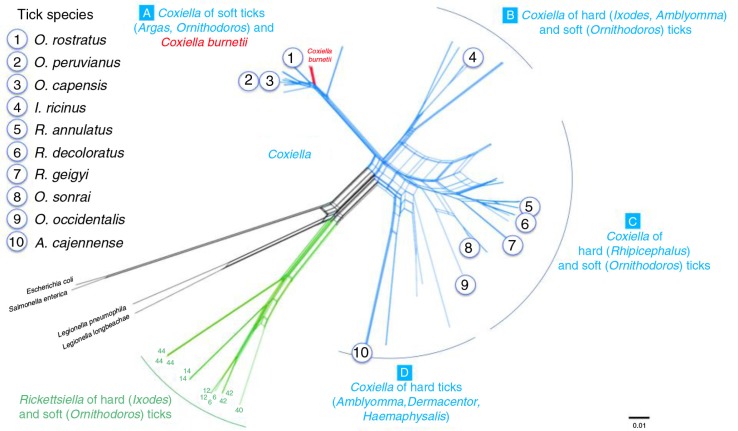
They were selected with the aim to represent the four clades (A–D) currently described for the Coxiella genus (17) (Fig. 1): clade A (Ornithodoros rostratus, O. peruvianus, O. capensis), clade B (Ixodes ricinus), clade C (Rhipicephalus annulatus, R. decoloratus, R. geigyi, O. sonrai, O. occidentalis), and clade D (Amblyomma cajennense). This panel included three tick species from which C. burnetii had previously been reported, namely O. sonrai (12), I. ricinus (21–23), and R. annulatus (24).
Fig. 1.
Genetic relatedness of the 10 tick species used in this study using as reference the phylogenetic network published by Duron et al. (17) with concatenated 16S rRNA, 23S rRNA, GroEL/htpB, rpoB, and dnaK sequences for 71 tick-borne Coxiella strains, 15 C. burnetii reference strains, and several bacterial outgroups.
Two tick specimens were examined for each species. They were either obtained from breeding colonies or sampled from their host species or habitats and they were processed as previously described (17). Briefly, the ticks were first washed with sterile water to avoid external bacterial contamination. Then, DNA was individually extracted using the DNeasy Blood & Tissue Kit (Qiagen) following manufacturer's instructions. DNA template quality was verified via PCR amplification of 18S ribosomal RNA or cytochrome oxidase 1 arthropod primers. Nested PCR assays were conducted using primers designed to amplify bacteria from the Coxiellacae family (i.e. Coxiella and its sister genus Rickettsiella) and to target the rpoB (DNA-directed RNA polymerase beta) gene and the GroEL/htpB (60 kDa chaperone heat shock protein B) gene as described elsewhere (17, 25). Sequencing of the PCR products obtained showed that each tick species was infected by a specific Coxiella-like bacterium that was genetically related to, but distinct from, C. burnetii. None of these tick DNA templates was found infected with C. burnetii on the basis of multilocus DNA sequencing (17).
Selection of qPCR primers thought to be specific for C. burnetii
The 20 tick specimens were tested using quantitative PCR (qPCR) methods using primers that are usually considered to be specific for C. burnetii (Table 1). We used TaqMan Universal PCR Master Mix (UMM 2×) following the amplification protocol: 1 cycle at 50°C for 2 min and 1 cycle at 95°C for 10 min, followed by 40 PCR cycles of 95°C for 15 s and 60°C for 1 min. Two of the targeted markers – the multicopy IS1111 insertion sequence (26, 27) and the icd (isocitrate dehydrogenase) housekeeping gene (26) – are frequently used in surveys that aim to estimate the prevalence of C. burnetii infection in ticks (16). The following genes were also targeted: scvA (small-cell-variant protein A), which is likely involved in chromatin condensation when the bacterium is ‘sporulating’, and porine p1, which encodes an outer membrane protein (28). Finally, we focused on a specific region of the GroEL/htpB, distinct from the region targeted to amplify the genome of Coxiella-like bacteria (17) and considered to be specific to C. burnetii (29). Nine Mile phase II genomic DNA (RSA 493 isolate) was used as a reference. In silico comparisons of the primers and probes with currently published sequences of R. turanicus (GenBank accession number: CP011126) and A. americanum (GenBank accession number: CP007541) suggested that mismatches with these symbionts were unlikely (Table 1).
Table 1.
Details about the qPCR methods used in the study
| Gene | Function | Primer designation | Primers and probe sequences (5′–3′) | Fragment length (bp) | Reference | % covering with the endosymbiont of R. turanicus b | % covering with the endosymbiont of A. americanum c |
|---|---|---|---|---|---|---|---|
| IS1111 | Insertion sequence | Forward primer Reverse primer Probe |
Confidentiala
Confidentiala Confidentiala |
76 | (1) | 58 0 0 |
63d
0 63d |
| icd | Isocitrate dehydrogenase | Forward primer Reverse primer Probe |
GACCGACCCATTATTCCCT CGGCGTAGATCTCCATCCA CGCCCGTCATGAAAAACGTGGTC |
139 | (2) | 84 0 0 |
0 0 0 |
| p1 | Porine | Qp1-F Qp1-R Probe |
CGGCGATTGGCGTTTC GGTTGCGGTAATGCCGTTAA AACTGTTCAAAATCCGAAACGAGTCGCA |
68 | (3) | 0 12d 50d |
0 0 0 |
| scvA | Chromatin condensation | QscvA-F QscvA-R Probe |
TGGAAAGACAAAATGTCCAACAA GGTTAGAAGCACCCGGTCGT ACGTGGAAAAGACCAACG |
69 | (3) | 52d
0 67d |
0 0 0 |
| GroEL/htpB | Heat shock protein | HtpB-1 HtpB-2 Probe |
TGGCTCAAGCGATTTTGGTT TTATCAATACCCCGTTTCAAATCC AAAGCCGTTATTGCTGGAATGAACCCC |
82 | (4) | 65d
92d 70d |
0 0 0 |
The detailed protocol used for the amplification of IS1111 will be soon published by Sidi-Boumedine et al. (in preparation) and remains meanwhile confidential;
GenBank accession number: CP011126;
GenBank accession number: CP007541;
sequence positions are distant from each other on the endosymbiont complete genome.
Results
We found that some Coxiella-like bacteria, belonging to clades A and C, yield positive PCR results when screened with primers initially believed to be C. burnetii-specific (Table 2). Overall, DNA was amplified for at least one marker in 6 of the 10 tick species studied. The most frequently amplified marker was IS1111, which was detected in five different species, whereas GroEL/htpB and scvA were amplified from three species. Porine p1 was solely amplified from a R. geigyi specimen, which was also positive for htpB, scvA, and IS1111, and displayed a particularly low C t value (C t =33) for IS1111. Conversely, icd was not detected in any of our samples. Interestingly, we observed intraspecific variation: one of the Coxiella-like endosymbiont from R. decoloratus was positive for IS1111, whereas the other was positive for scvA. Unfortunately, because all PCR products were poorly concentrated, sequencing was unsuccessful.
Table 2.
C t values obtained using qPCR for both specimens of the 10 tick species tested
| Coxiella-like clade | Tick species | IS1111 | icd | GroEL/htpB | p1 | scvA |
|---|---|---|---|---|---|---|
| A | O. rostratus | –/– | –/– | –/– | –/– | –/– |
| O. peruvianus | 39/37 | –/– | 30/31 | –/– | –/– | |
| O. capensis | –/– | –/– | 35/35 | –/– | –/38 | |
| B | I. ricinus | –/– | –/– | –/– | –/– | –/– |
| C | R. annulatus | 37/38 | –/– | –/– | –/– | –/– |
| R. decoloratus | –/36 | –/– | –/– | –/– | 39/– | |
| R. geigyi | 37/33 | –/– | –/38 | –/35 | –/38 | |
| O. sonrai | 39/36 | –/– | –/– | –/– | –/– | |
| O. occidentalis | –/– | –/– | –/– | –/– | –/– | |
| D | A. cajennense | –/– | –/– | –/– | –/– | –/– |
The sign ‘/’ is used to separate the results obtained for the first and the second tick specimen; ‘–’ indicates that no amplification was observed.
Discussion
The marker we most frequently detected in ticks infected with Coxiella-like endosymbionts was the IS1111 transposable element, which is routinely targeted during epidemiological surveys examining C. burnetii prevalence in ticks (16). We thus showed that C. burnetii detection assays based only on IS1111 may lead to misidentification with Coxiella-like endosymbionts. The recent work of Duron (30) corroborates this finding: several genetically divergent IS1111 copies were found widespread in many Coxiella-like endosymbionts, therefore showing that IS1111 can no longer be considered specific to C. burnetii. These findings may explain why surveys based on IS1111 screening occasionally report prevalence levels >10% (23, 31–33).
Our results also showed that the use of a combination of primers targeting different markers, as performed in some studies (11, 21, 34–36), is not sufficient to guarantee the specificity of C. burnetii detection. Indeed, up to four of our markers were detected in a same Coxiella-like endosymbiont. Interestingly, icd, which is frequently used as a PCR target in epidemiological studies (16, 34), was not amplified from our panel of Coxiella-like infected ticks. However, Reeves et al. (37) were able to amplify a 612-bp icd fragment, displaying 93% homology with C. burnetii, from a Coxiella-like bacterium that infects ticks from the O. capensis complex in South Carolina, USA. This result contrasts with our observation that icd was not amplified from the endosymbiont of O. capensis ticks sampled from Cape Verde and highlights the fact that the amplification of a specific genetic marker strongly depends on the PCR method (PCR, nested PCR, or qPCR) and the primer sequences used.
More generally, negative results may be due to low detection sensitivity, which is supported by high detection thresholds for most of the genes. In particular, intraspecific variation may be due to the individual ticks having a low bacterial burden, sex- or stage-specific differences, and the presence of PCR inhibitors. In our study, it is possible that the IS1111-based PCR method was the most sensitive of the tests used, because several copies of this gene are likely present in the genome of Coxiella-like bacteria, as is the case for C. burnetii. This hypothesis is supported by the observation that GroEL/htpB, porine p1, and scvA were detected in the endosymbiont of a R. geigyi specimen that displayed a low Ct value for IS1111.
Standardizing methodology across laboratories is essential to allow comparisons among studies. Although remarkable progress has recently been made in designing new PCR-based techniques to detect C. burnetii, these advances have overlooked that an important genetic diversity actually exists within the Coxiella genus (16). In this context, it may not be surprising that the PCR primers routinely used to target C. burnetii actually cross-react with Coxiella-like bacteria. Therefore, in the future, molecular methods aiming at detecting C. burnetii should make sure that no cross-reaction exists not only with other abortive agents but also with Coxiella-like organisms. Recent full-genome sequencing data indeed not only highlighted obvious genetic similarities of C. burnetii with Coxiella-like bacteria but also revealed some mutations specific to Coxiella-like bacteria (18, 38). This pattern likely explains why PCR cross-reactions with Coxiella-like bacteria are partial and variable between markers. Interestingly, identical IS1111 copies were found in C. burnetii and some Coxiella-like bacteria (30), suggesting that the risk of detecting Coxiella-like bacteria with IS1111 primers designed to detect C. burnetii is very high and must not be underestimated.
PCR-based surveys that aim to detect C. burnetii in ticks must be interpreted with caution if the amplified DNA products are not sequenced. Unfortunately, the ratio of bacterial DNA to tick DNA is frequently low, which makes it challenging to obtain PCR products concentrated enough for direct sequencing via conventional PCR. Additionally, currently available qPCR methods, such as those used in this study, often yield very short DNA fragments that are difficult to concentrate for sequencing purposes and that correspond to rather uninformative sequences. Therefore, there is an urgent need to develop a multiplex qPCR or microchip method that would make it possible to directly differentiate Coxiella-like bacteria from C. burnetii in tick samples and to detect co-infections. Pending development of such a test, useful alternative methods include the sequencing of the 16S rRNA, rpoB, and GroEL genes of Coxiella bacteria, after amplification by nested PCR, as previously described (17, 25).
Acknowledgements
We thank Elodie Rousset for reading and providing comments on the manuscript; Aurélien Joulie, Sébastien Masseglia, and Elise Yang for their involvement in the molecular analyses; and Karen McCoy, Christine Chevillon, Abel Biguezoton, Marcelo Labruna, Patrick Durand, and François Renaud for collecting the ticks.
Conflict of interest and funding
This work was supported by the LABEX ECOFECT (LEGOXiNET project, ANR-11-LABX-0048) of Université de Lyon, which is part of the French Stimulus Initiative program (Investissements d'Avenir; ANR-11-IDEX-0007), operated by the French National Research Agency (ANR) and the FONDECYT project (project number 1130948).
References
- 1.Rodolakis A. Q fever in dairy animals. Rickettsiology and rickettsial diseases – Fifth International Conference. Ann N Y Acad Sci. 2009;1166:90–3. doi: 10.1111/j.1749-6632.2009.04532.x. [DOI] [PubMed] [Google Scholar]
- 2.EFSA. Scientific Opinion on Q fever. EFSA J. 2010;8 1595. [Google Scholar]
- 3.European Centre for Disease Prevention and Control. Risk assessment on Q fever. ECDC Technical report. 2010. p. 40. Panel with representatives from the Netherlands, France, Germany, UK, United States. Asher, A., Bernard, H., Coutino, R., Durat, G., De Valk, H., Desenclos, J-C., Holmberg, J., Kirkbridge, H., More, S, Scheenberger, P., van der Hoek, W., van der Poel, C., van Steenbergen, J., Villanueva, S., Coulombier, D., Forland, F., Giesecke, J., Jansen, A., Nilsson, M., Guichard, C., Mailles, A., Pouchol, E., Rousset, E. doi: http://dx.doi.org/10.2900/28860. Available from: http://www.ecdc.europa.eu [cited 13 November 2011].
- 4.Anderson A, Bijlmer H, Fournier PE, Graves S, Hartzell J, Kersh GJ, et al. Diagnosis and management of Q Fever – United States, 2013: recommendations from CDC and the Q Fever Working Group. MMWR Recomm Rep. 2013;62:1–28. [PubMed] [Google Scholar]
- 5.Tissot-Dupont H, Amadei MA, Nezri M, Raoult D. Wind in November, Q fever in December. Emerg Infect Dis. 2004;10:1264–9. doi: 10.3201/eid1007.030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis GE, Cox HR. A filter-passing infectious agent isolated from ticks. I. Isolation from Dermacentor andersoni, reactions in animals, and filtration experiments. Public Health Rep. 1938;53:2259–67. [Google Scholar]
- 7.Dyer RE. A filter-passing infectious agent isolated from ticks. III. Human infection. Public Health Rep. 1938;53:2277–82. [Google Scholar]
- 8.McDade JE. Historical aspects of Q fever. In: Marrie TJ, editor. Q fever, Volume 1: the disease. Boca Raton, Florida: CRC Press; 1990. pp. 5–22. [Google Scholar]
- 9.Hechemy KE. History and prospects of Coxiella burnetii research. In: Toman R, Heinzen RA, Samuel JE, Mege J-L, editors. Coxiella burnetii: recent advances and new perspectives in research of the Q Fever bacterium. Dordrecht: Springer; 2012. pp. 1–11. [Google Scholar]
- 10.Toma L, Mancini F, Di Luca M, Cecere JG, Bianchi R, Khoury C, et al. Detection of microbial agents in ticks collected from migratory birds in central Italy. Vector-Borne Zoonotic Dis. 2014;14:199–205. doi: 10.1089/vbz.2013.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper A, Stephens J, Ketheesan N, Govan B. Detection of Coxiella burnetii DNA in wildlife and ticks in Northern Queensland, Australia. Vector-Borne Zoonotic Dis. 2013;13:12–16. doi: 10.1089/vbz.2011.0853. [DOI] [PubMed] [Google Scholar]
- 12.Mediannikov O, Fenollar F, Socolovschi C, Diatta G, Bassene H, Molez JF, et al. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis. 2010;4:8. doi: 10.1371/journal.pntd.0000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannou I, Sandalakis V, Kassinis N, Chochlakis D, Papadopoulos B, Loukaides F, et al. Tick-borne bacteria in mouflons and their ectoparasites in cyprus. J Wildl Dis. 2011;47:300–6. doi: 10.7589/0090-3558-47.2.300. [DOI] [PubMed] [Google Scholar]
- 14.Siroky P, Kubelova M, Modry D, Erhart J, Literak I, Spitalska E, et al. Tortoise tick Hyalomma aegyptium as long term carrier of Q fever agent Coxiella burnetii – evidence from experimental infection. Parasitol Res. 2010;107:1515–20. doi: 10.1007/s00436-010-2037-1. [DOI] [PubMed] [Google Scholar]
- 15.Daiter AB. Transovarial and transspermal transmission of Coxiella burnetii by the tick Hyalomma asiaticum and its role in Q-rickettsiosis ecology. Parazitologiya. 1977;11:403–11. [PubMed] [Google Scholar]
- 16.Duron O, Sidi-Boumedine K, Rousset E, Moutailler S, Jourdain E. The importance of ticks in Q fever transmission: what has (and has not) been demonstrated? Trends Parasitol. 2015;31:536–52. doi: 10.1016/j.pt.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Duron O, Noël V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii . PLoS Pathog. 2015;11:e1004892. doi: 10.1371/journal.ppat.1004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith TA, Driscoll T, Gillespie JJ, Raghavan R. A Coxiella-like endosymbiont is a potential vitamin source for the lone star Tick. Genome Biol Evol. 2015;7:831–8. doi: 10.1093/gbe/evv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong J. Coxiella-like endosymbionts. In: Toman R, Heinzen RA, Samuel JE, Mege J-L, editors. Coxiella burnetii: recent advances and new perspectives in research of the Q fever bacterium. Dordrecht: Springer; 2012. pp. 365–79. [Google Scholar]
- 20.Shivaprasad HL, Cadenas MB, Diab SS, Nordhausen R, Bradway D, Crespo R, et al. Coxiella-like infection in psittacines and a toucan. Avian Dis. 2008;52:426–32. doi: 10.1637/8192-120707-Reg. [DOI] [PubMed] [Google Scholar]
- 21.Sprong H, Tijsse-Klasen E, Langelaar M, De Bruin A, Fonville M, Gassner F, et al. Prevalence of Coxiella burnetii in ticks after a large outbreak of Q fever. Zoonoses Public Health. 2012;59:69–75. doi: 10.1111/j.1863-2378.2011.01421.x. [DOI] [PubMed] [Google Scholar]
- 22.Reye AL, Stegniy V, Mishaeva NP, Velhin S, Hubschen JM, Ignatyev G, et al. Prevalence of tick-borne pathogens in Ixodes ricinus and Dermacentor reticulatus ticks from different geographical locations in Belarus. PLoS One. 2013;8:e54476. doi: 10.1371/journal.pone.0054476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnet S, de la Fuente J, Nicollet P, Liu X, Madani N, Blanchard B, et al. Prevalence of tick-borne pathogens in adult Dermacentor spp. ticks from nine collection sites in France. Vector-Borne Zoonotic Dis. 2013;13:226–36. doi: 10.1089/vbz.2011.0933. [DOI] [PubMed] [Google Scholar]
- 24.Reye AL, Arinola OG, Hubschen JM, Muller CP. Pathogen prevalence in ticks collected from the vegetation and livestock in Nigeria. Appl Environ Microbiol. 2012;78:2562–8. doi: 10.1128/AEM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duron O, Jourdain E, McCoy KD. Diversity and global distribution of the Coxiella intracellular bacterium in seabird ticks. Ticks Tick Borne Dis. 2014;5:557–63. doi: 10.1016/j.ttbdis.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 26.de Bruin A, de Groot A, de Heer L, Bok J, Wielinga PR, Hamans M, et al. Detection of Coxiella burnetii in complex matrices by using multiplex quantitative PCR during a major Q fever outbreak in the Netherlands. Appl Environ Microbiol. 2011;77:6516–23. doi: 10.1128/AEM.05097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joulie A, Laroucau K, Bailly X, Prigent M, Gasqui P, Lepetitcolin E, et al. Circulation of Coxiella burnetii in a naturally infected flock of dairy sheep: shedding dynamics, environmental contamination, and genotype diversity. Appl Environ Microbiol. 2015;81:7253–60. doi: 10.1128/AEM.02180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol. 2004;186:7344–52. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes I, Rousset E, Dufour P, Sidi-Boumedine K, Cupo A, Thiery R, et al. Evaluation of the recombinant heat shock protein B (HspB) of Coxiella burnetii as a potential antigen for immunodiagnostic of Q fever in goats. Vet Microbiol. 2009;134:300–4. doi: 10.1016/j.vetmic.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Duron O. The IS1111 insertion sequence used for detection of Coxiella burnetii is widespread in Coxiella-like endosymbionts of ticks. FEMS Microbiol Lett. 2015;362 doi: 10.1093/femsle/fnv132. fnv132. Available from: http://femsle.oxfordjournals.org/content/362/17/fnv132. [DOI] [PubMed] [Google Scholar]
- 31.Szymanska-Czerwinska M, Galinska EM, Niemczuk K, Zasepa M. Prevalence of Coxiella burnetii infection in foresters and ticks in south-eastern Poland and comparison of diagnostic methods. Ann Agric Environ Med. 2013;20:699–704. [PubMed] [Google Scholar]
- 32.Pastiu AI, Matei IA, Mihalca AD, D'Amico G, Dumitrache MO, Kalmar Z, et al. Zoonotic pathogens associated with Hyalomma aegyptium in endangered tortoises: evidence for host-switching behaviour in ticks? Parasit Vectors. 2012;5:301. doi: 10.1186/1756-3305-5-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loftis AD, Reeves WK, Szumlas DE, Abbassy MM, Helmy IM, Moriarity JR, et al. Population survey of Egyptian arthropods for rickettsial agents. Ann N Y Acad Sci. 2006;1078:364–7. doi: 10.1196/annals.1374.072. [DOI] [PubMed] [Google Scholar]
- 34.Michelet L, Delannoy S, Devillers E, Umhang G, Aspan A, Juremalm M, et al. High-throughput screening of tick-borne pathogens in Europe. Front Cell Infect Microbiol. 2014;4:103. doi: 10.3389/fcimb.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Park HS, Jang WJ, Koh SE, Park TK, Kang SS, et al. Identification of the Coxiella sp. detected from Haemaphysalis longicornis ticks in Korea. Microbiol Immunol. 2004;48:125–30. doi: 10.1111/j.1348-0421.2004.tb03498.x. [DOI] [PubMed] [Google Scholar]
- 36.Tozer SJ, Lambert SB, Strong CL, Field HE, Sloots TP, Nissen MD. Potential animal and environmental sources of Q fever infection for humans in Queensland. Zoonoses Public Health. 2014;61:105–12. doi: 10.1111/zph.12051. [DOI] [PubMed] [Google Scholar]
- 37.Reeves WK, Loftis AD, Sanders F, Spinks MD, Wills W, Denison AM, et al. Borrelia, Coxiella, and Rickettsia in Carios capensis (Acari: Argasidae) from a brown pelican (Pelecanus occidentalis) rookery in South Carolina, USA. Exp Appl Acarol. 2006;39:321–9. doi: 10.1007/s10493-006-9012-7. [DOI] [PubMed] [Google Scholar]
- 38.Gottlieb Y, Lalzar I, Klasson L. Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-Like endosymbionts in ticks. Genome Biol Evol. 2015;7:1779–96. doi: 10.1093/gbe/evv108. [DOI] [PMC free article] [PubMed] [Google Scholar]