Abstract
Objective
Polymorphisms in the transcription factor IRF5 are associated with an increased risk of developing RA. This study was done to determine the role of IRF5 in arthritis development.
Methods
K/BxN serum transfer arthritis was induced in mice deficient in IRF5, or lacking IRF5 only in myeloid cells, and arthritis severity was evaluated. K/BxN arthritis was also induced in mice deficient in TRIF, TLR2, TLR3, TLR4 and TLR7 to determine pathways through which IRF5 might promote arthritis. In-vitro studies were performed to determine the role of IRF5 in IL-1 receptor and TLR signaling.
Results
Arthritis severity was reduced in IRF5-deficient, TRIF-deficient, TLR3-deficient and TLR7-deficient mice. The expression of multiple genes regulating neutrophil recruitment or function and bioactive IL-1β formation was reduced in the joints during active arthritis in IRF5-deficient mice. In vitro studies showed that TLR7 and the TRIF-dependent TLR3 pathway induce pro-inflammatory cytokine production in disease relevant cell types in an IRF5-dependent manner.
Conclusion
IRF5 contributes to disease pathogenesis in inflammatory arthritis. This is likely due at least in part to the role of IRF5 in mediating pro-inflammatory cytokine production downstream of TLR7 and TLR3. As TLR7 and TLR3 are both RNA-sensing TLRs, this suggests that endogenous RNA ligands present in the inflamed joint promote arthritis development. These findings may be relevant to human RA as RNA capable of activating TLR7 and TLR3 is present in synovial fluid and TLR7 and TLR3 are upregulated in the joints of RA patients.
Rheumatoid arthritis (RA) is an autoimmune disease that affects approximately 0.5-1% of adults worldwide. The etiology of RA is unknown but involves both environmental and genetic factors. Autoantibodies against citrullinated self-proteins and rheumatoid factors may precede clinical disease by many years [1]. The precise events that trigger the onset of joint disease are not known but the established synovitis comprises a broad range of cells from the innate and adaptive immune systems that interact with each other and with joint-resident cells including fibroblast-like synoviocytes and chondrocytes. Synovial inflammation also induces the differentiation of osteoclasts that resorb articular bone, resulting in bone erosions [1]. The final common pathway of cartilage destruction involves proinflammatory cytokine induction of matrix metalloproteinases and other proteinases. Over the past decade the advent of biologic therapies that inhibit these cytokines, in particular TNF-α and IL-6, has revolutionized the management of RA . However, the mechanisms responsible for inducing the proinflammatory cytokines in RA are not fully characterized.
Polymorphisms in the transcription factor interferon regulatory factor 5 (IRF5) have been identified in human genetic studies as risk factors for rheumatoid arthritis [2, 3], as well as for other autoimmune diseases including systemic lupus erythematosus, scleroderma, Sjögren’s disease and ulcerative colitis [4]. The IRF5 polymorphisms that are associated with an increased risk of developing systemic autoimmune disease cause the expression of novel IRF5 isoforms and/or an increased level of IRF5 expression by promoting the stability of the IRF5 mRNA or protein [4]. Although IRF5 is only one of many genes that have been linked to RA and by itself confers only a modest increase in risk, understanding the role of IRF5 in RA may help identify biological pathways of importance in RA pathogenesis. IRF5 has diverse functions that include the induction of type I interferons and proinflammatory cytokines downstream of Toll-like receptors (TLRs), nucleotide-binding oligomerization domain 2 (NOD2), and retinoic acid inducible gene I (RIG-I) or following viral infection [4].
The K/BxN mouse model has proven to be a particularly valuable model for dissecting the immunopathologic mechanisms involved in the effector phase of inflammatory arthritis [5]. This model has many features in common with human RA including synovial pannus formation and bone and cartilage erosion [6]. K/BxN mice produce antibodies against glucose-6-phosphate isomerase (GPI) which form immune complexes with GPI in the joints and thereby initiate arthritis, similar mechanistically to the immune complex-induced synovitis that is thought to contribute to human RA [7]. The inflammatory arthritis can be reproduced in a wide range of mouse strains with the passive transfer of serum from K/BxN mice [5]. The innate immune system plays a central role in the serum transfer model but T and B lymphocytes are not required. The disease is dependent on Fcγ receptors, macrophages, neutrophils, the alternative complement pathway, the C5a receptor and CXCR2 [6]. As regards cytokines in this model, IL-1 is absolutely required for disease, TNF-α plays a variable role and IL-6 is not required [6].
In this report, we used the K/BxN serum transfer model to determine the role of IRF5 in the effector phase of inflammatory arthritis. In addition to demonstrating an important role for IRF5 in promoting disease, these studies also led to the identification of TLR7 and TLR3 as receptors involved in disease pathogenesis. This suggests that the induction of the proinflammatory cytokines that play a central role in the pathogenesis of RA might be driven in part through the activation of RNA-sensing TLRs by endogenous RNA ligands.
MATERIALS AND METHODS
Mice
C57BL/6 wild-type and TLR3-deficient (TLR3−/−) mice were from the Jackson Laboratory. Mice with combined deficiency of TLR2 and TLR4 (TLR2−/−4−/− mice) were provided by Lee Wetzler. IRF5−/− mice were provided by Tadatsugu Taniguchi and Tak Mak. The IRF5−/− mice used in Figures 1-4 may have contained a Dock2 mutation [8, 9]. The IRF5−/−mice used in Figure 5 did not contain the Dock2 mutation (confirmed by PCR [9]). IFNAR1−/− mice were obtained from Jonathan Sprent. TLR7−/− mice were provided by Shizuo Akira. K/BxN T cell receptor transgenic mice were provided by Diane Mathis and Christophe Benoist. C57BL/6 mice homozygous for the loxP flanked IRF5 allele (IRF5flox/flox; The Jackson Laboratory #17311) were crossed with mice expressing Cre under control of the murine lysozymeM gene promoter (LyzMcre; The Jackson Laboratory #4781), thereby generating LyzMcre IRF5flox/flox mice. All mice used were either made in the C57BL/6 background or backcrossed at least 9 generations to C57BL/6, except for the LyzMcre which were backcrossed 6 generations. All mice were maintained at the Boston University Laboratory Animal Sciences Center or the Center for Comparative Medicine at Northwestern University in accordance with the American Association for the Accreditation of Laboratory Animal Care regulations. All studies were approved by the IACUC at Boston University or Northwestern University.
Figure 1.
Arthritis is ameliorated in IRF5-deficient mice following transfer of K/BxN serum. C57BL/6 wildtype (WT; n = 10) and IRF5-deficient (IRF5−/−; n = 10) mice were injected intraperitoneally (i.p) with 150 μl of K/BxN serum on day 0 and day 2. (A) Change in ankle thickness (left) and clinical score (right) were measured on days 0, 2, 4 and 7. (B) Ankles (2 per mouse) isolated on day 7 after serum transfer were prepared as described in Materials and Methods, after which ankle sections were scored for various histologic features (left panel). Representative images are shown in the panels on the right; B, bone C, cartilage S, synovium ; long arrow indicates a bone erosion and arrowhead indicates inflammatory cells. Data are expressed as mean ± SEM. * = P < 0.05; ** = P < 0. 005; *** = P < 0. 0001.
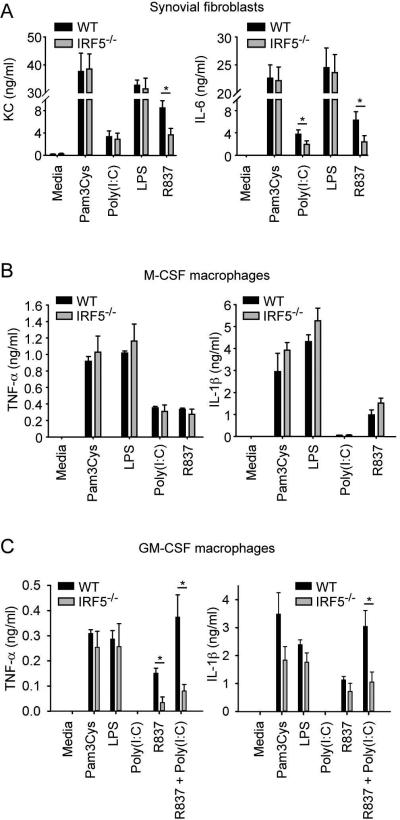
Figure 4.
TLR7 and TLR3 promote inflammatory cytokine and chemokine production through IRF5 and TLR3 synergizes with TLR7 to enhance these effects. (A) Synovial fibroblasts from C57BL/6 WT and IRF5−/− mice were stimulated, or not stimulated (media), with the TLR2 ligand Pam3Cys (100 ng/ml), the TLR3 ligand Poly(I:C) (10 ug/ml), the TLR4 ligand LPS (100 ng/ml) and the TLR7 ligand R837 (1 ug/ml). Supernatants were collected after 24 h and KC and IL-6 measured by ELISA. Data represent mean ± SEM of 5 independent experiments. Bone marrow-derived M-CSF macrophages (B) and GM-CSF macrophages (C) from C57BL/6 WT and IRF5−/− mice were stimulated, or not stimulated (media), with Pam3Cys (100 ng/ml), Poly(I:C) (10 ug/ml), LPS (100 ng/ml), R837 (0.3 ug/ml) or R837 (0.3 ug/ml) plus Poly(I:C) (10 ug /ml). On the TNF-α plate, supernatants were collected 24 h after the addition of stimuli. On the IL-1β plate, nigericin (40 μM) was added 16 h after addition of the stimuli and supernatants collected 2 h thereafter. TNF-α and IL-1β were measured by ELISA. Data represent mean ± SEM of 4 independent experiments (B) and 5 independent experiments (C). * = P < 0.05.
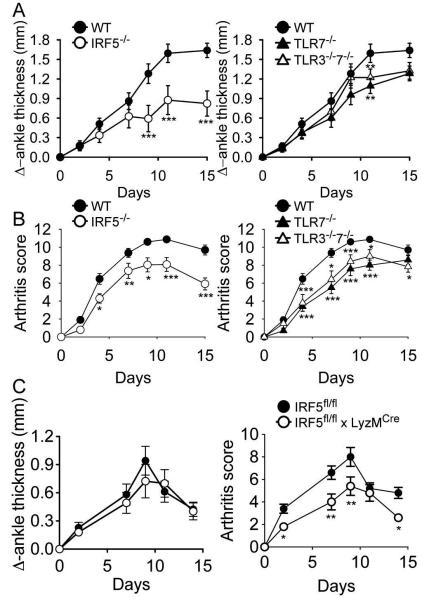
Figure 5.
Mice deficient in IRF5 or TLR7 develop less severe arthritis but no additional protection is seen in mice deficient in both TLR7 and TLR3. C57BL/6 wildtype (WT; n = 20), IRF5-deficient (IRF5−/−; n = 12), TLR7-deficient (TLR7−/−; n = 13), and TLR3 and 7 double-deficient (TLR3−/−7−/−; n = 12) mice were injected i.p with 150 μl of K/BxN serum on day 0 and day 2. Change in ankle thickness (A) and clinical score (B) were measured on days 0, 2, 4, 7, 9, 11 and 15. The IRF5−/− data are shown in the left hand panels and the TLR7−/− and TLR3−/−7−/− data are shown in the right hand panels for clarity. There were no statistically significant differences between the IRF5−/−, TLR7−/− and the TLR3−/−7−/− cohorts except for day 15 where the change in ankle thickness and clinical score were lower in the IRF5−/− cohort than in the other 2 cohorts (P < 0.05). (C) IRF5flox/flox mice (wild-type; n = 5) and LyzMcre IRF5flox/flox mice (n = 5) were injected i.p with 75 μl of K/BxN serum on day 0. * = P < 0.05; ** = P < 0.01; *** = P < 0. 001 versus WT.
Reagents
Polyinosinic-polycytidylic acid (poly(I:C), Lipopolysaccharide (LPS), R837 (Imiquimod) and Pam3Cys-Ser-Lys4 (Pam3Cys) were from InvivoGen. Mouse recombinant TNF-α and interferon-γ were from eBioscience. Mouse recombinant IL1-β was from R&D Systems. Nigericin was from Invitrogen.
Serum transfer protocol and arthritis measurement
Serum was collected from K/BxN mice at 8 weeks of age, pooled, aliquoted and stored at −20° C. To induce arthritis, 150 μl of serum were injected intraperitoneally on day 0 and again on day 2, as previously described [5]. For experiments performed at Northwestern University (TLR3−/− and IRF5flox/flox), 75 μl of serum were injected intraperitoneally on day 0, as previously described [10]. The thickness of both ankles was measured using calipers (Pro-Max 152mm caliper from Ted Pella, Inc. or from Fowler Tools of Canada). In addition, a clinical index for arthritis severity was determined using a 0–3 point scale for each paw (thus 0–12 total score) as described, [5, 10].
Histology
Ankle sections were decalcified, embedded in paraffin, and stained with H&E, Safranin O and methyl green. Histopathologic scoring was performed as previously described [10, 11]. A pathologist blinded to the study findings and sample identity (GKH) evaluated ankle sections by examining at least 3 sections per ankle and 3 fields per section at 400X magnification.
Preparation of cells for in vitro assays
M-CSF macrophages and GM-CSF macrophages were generated as described [12], and plated overnight at 1 x 106 cells/ml in flat bottom 96 well plates before stimulation, with duplicate wells for all stimulation conditions. To generate synovial fibroblasts, synovial tissues were obtained from microdissected ankle joints. The synovial fibroblasts were grown as previously described [13]. Synovial fibroblasts from passages 3 through 6 were used for experiments. For in vitro assays, synovial fibroblasts were seeded at 105 cells/ml in flat bottom 96 well plates, starved overnight of FCS, and then the assays were performed in media containing 2% FCS. Mouse embryonic fibroblasts (MEFs) from C57BL/6 WT mice were purchased from Jackson Laboratory. MEFs from IRF5−/− mice were prepared by the Transgenic Core Facility at Boston University using standard techniques.
Cytokine, chemokine and nitric oxide measurement
For the in vitro studies, IL-1β, TNF-α and KC levels were measured using commercial ELISAs (R&D Systems). IL-6 levels were measured using an in-house ELISA as previously described [14]. Nitric oxide production was determined by the Griess method. The levels of cytokines and chemokines in serum were determined using a Luminex based assay according to the manufacturer’s specifications (Affymetrix).
RNA isolation
The mouse joints, either freshly harvested (for Nanostring analysis) or previously snap-frozen and stored at −80 degrees (for RT-PCR analysis), were placed in RLT lysis buffer (Qiagen) with β-mercaptoethanol and disrupted using the TissueLyser (Qiagen) at 30 Hz for 2 minutes. Samples were then spun at 12,000 rpm for 1 minute and the supernatant was removed from the bone fragment pellet and collected for RNA isolation. RNA was isolated using the RNeasy® Fibrous Tissue Mini Kit (Qiagen) according to the manufacturer’s protocol and resuspended in RNAse-free water (Qiagen). RNA concentration was measured using a Nanodrop 2000 Spectrophotometer (Thermo Scientific).
Real-time PCR
RNA from the right wrist joints was reverse transcribed into cDNA using Thermoscript reverse transcriptase (Invitrogen). cDNA was diluted 1:10 and PCR was performed using Taqman primers specific for murine Irf5 and Gapdh (Applied Biosystems). Data was analyzed using the 2−ΔΔ Ct method and is expressed as ‘Fold Change’ (expression relative to housekeeping gene and normalized to reference sample).
Nanostring analysis
RNA (100 ng) from the left ankle joints of C57BL/6 (n=4) and IRF5−/− (n=5) arthritic mice was probed using NanoString technology and the Mouse Immunology code set (NanoString Technologies). Gene expression was normalized to the expression of fourteen housekeeping genes and then the C57BL/6 and IRF5−/− mice were compared and cluster analysis was performed using nSolver Analysis Software 2.5.34 as recommended by the manufacturer (NanoString Technologies).
Statistical analysis
Data are depicted as mean ± SEM. Statistical significance of differences was determined by the paired two-tailed Student’s t test for the in vitro experiments. For the in vivo experiments, groups were compared using repeated measures ANOVA with post-hoc analysis using Bonferroni correction. P values < 0.05 were considered significant.
RESULTS
Arthritis is ameliorated in IRF5-deficient mice
To determine whether IRF5 is involved in disease pathogenesis, we administered K/BxN serum to C57BL/6 wildtype (WT) and IRF5-deficient (IRF5−/−) mice and evaluated the arthritis at days 2, 4 and 7 after serum administration. Consistent with previous studies in this model [5], ankle thickness and the overall arthritis score increased progressively over this time period in the WT mice, both indicative of worsening disease severity. In contrast, the increase in ankle thickness and the overall arthritis score were substantially lower in IRF5−/− mice, although mild disease was still evident (Figure 1 A). The clinical data were confirmed by histologic analysis which showed that all the measured histologic parameters of arthritis were less severe in the IRF5−/− mice (Figure 1 B). Overall, these data demonstrate that the severity of arthritis is reduced in IRF5−/− mice.
The protective effect of IRF5 deficiency is mediated through reduced proinflammatory cytokine production and not through an effect on IL-1 receptor signaling
IL-1 is required for disease development in the K/BxN model [15]. As the intracellular adaptor molecule MyD88 is absolutely required for IL-1 receptor (IL-1R) signaling [16] and IRF5 specifically associates with MyD88 [17, 18], we hypothesized that IRF5 might play a role in pathogenesis by participating in IL-1R signaling. To test this hypothesis we measured responses to IL-1 in several different cell types derived from WT and IRF5−/− mice and used TNF-α-induced responses as a control. There was no difference between WT and IRF5−/− mice in IL-1- or TNF-α−induced responses in mouse embryonic fibroblasts (Figure 2 A), bone marrow-derived macrophages (Figure 2 B), or synovial fibroblasts (Figure 2 C).
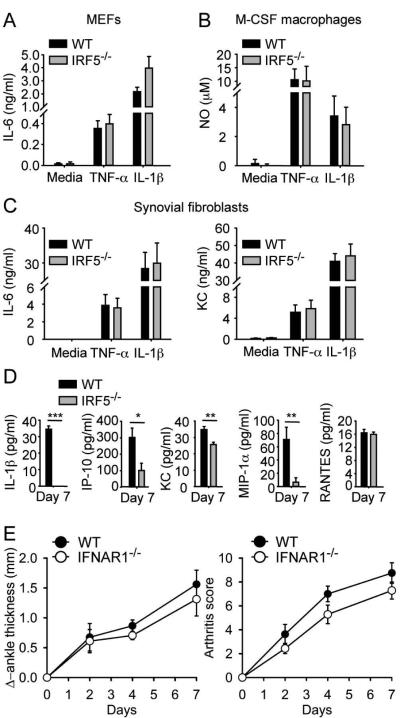
Figure 2.
IRF5 deficiency does not impair IL-1 receptor signaling, but IRF5-deficient mice have reduced serum IL-1β and chemokine levels after K/BxN serum transfer. (A) Mouse embryonic fibroblasts (MEFs), (B) M-CSF bone marrow-derived macrophages and (C) synovial fibroblasts from C57BL/6 wildtype (WT) and IRF5-deficient (IRF5−/−) mice were stimulated, or not stimulated (media), with TNF-α (10 ng/ml) and IL-1β (10 ng/ml). Supernatants were collected after 24 h and IL-6 and KC measured by ELISA and nitric oxide (NO) measured by the Griess method. Data represent mean ± SEM of 5 independent experiments (A and C) and 4 independent experiments (B). (D) Serum cytokines were measured on day 7 after K/BxN serum injection in the C57BL/6 WT (n = 10) and IRF5−/− (n = 10) mice shown in Figure 1. Data represent mean ± SEM. * = P < 0.05; ** = P < 0. 01; *** = P < 0. 001. (E) C57BL/6 WT ( n = 8) and IFNAR1-deficient (IFNAR1−/−(n = 7)) mice were injected i.p. with 150 μl of K/BxN serum on day 0 and day 2. Change in ankle thickness (left) and clinical score (right) were measured on days 0, 2, 4 and 7.
An important aspect of IRF5 signaling involves the production of cytokines and chemokines [4]. We therefore measured serum cytokine and chemokine levels on day 7 after K/BxN serum transfer in the WT and IRF5−/− mice. Levels of keratinocyte chemoattractant (KC; CXCL1), interferon gamma-induced protein 10 kDa (IP-10; CXCL10) and macrophage inflammatory protein 1 alpha (MIP-1α; CCL3) were lower in the sera of IRF5−/− mice (Figure 2 D). The reduction in KC is noteworthy in regard to this model as absence of CXCR2, the receptor for KC, attenuates arthritis severity [19]. More importantly, IL-1β was present in the sera of WT mice but was not detected in the sera of IRF5−/− mice. As IL-1β is absolutely required for disease in the K/BxN model [15], this suggested that an important mechanism through which IRF5 might be contributing to disease is through the production of IL-1β. It is likely that the elevated serum levels of IL-1β are due largely to IL-1β production in the joint as the pathologic manifestations in the K/BxN serum-transfer model are joint-specific [15]. However it is possible that systemic IL-1β production contributes to the total IL-1β level. Although IRF5 also plays a role in the production of type I interferons, this was not a factor in pathogenesis as the severity of arthritis was not reduced in mice that lacked the IFNAR1 chain of the type I IFN receptor (IFNAR1−/− mice) and were therefore unable to respond to type I IFN (Figure 2 E) [20].
From these studies we conclude that the protective effects of IRF5 deficiency in this model are likely mediated through a reduction in proinflammatory cytokine production and not through regulation of type I IFN production or IL-1R signaling.
The severity of arthritis is significantly reduced in mice deficient in TLR3 or TLR7 signaling, but is not reduced in mice deficient in both TLR2 and TLR4
TLRs are a family of innate pattern recognition receptors that recognize pathogen-associated molecular patterns (PAMPS) and thereby initiate anti-pathogen immune responses [21]. However, it is now evident that TLRs can also recognize self-ligands under some circumstances and thereby induce autoimmune or auto-inflammatory disease [22]. As IRF5 mediates proinflammatory cytokine production induced by TLR signaling [17], we next tested the possibility that TLRs might contribute to the development of arthritis. Previous studies in the K/BxN model had shown that TLR4 and TLR9 do not play a role within the first 7 days of disease (the time-frame evaluated in Figure 1) although TLR4-deficient mice have less severe disease at later time points [23, 24]. However, microbial and endogenous TLR2 and TLR4 ligands can induce arthritis in mouse models [25]. We therefore compared disease development in WT mice and mice deficient in both TLR2 and TLR4 (TLR2−/−4−/− mice). The arthritis was at least as severe in the TLR2−/−4−/− mice as in the WT mice (Figure 3 A), demonstrating that endogenous TLR2 and TLR4 ligands do not contribute to disease within the first 7 days after serum transfer. However, disease severity was decreased in TRIF-mutant and TLR7−/− mice (Figure 3 A). TRIF is required for TLR3 signaling and for signaling through the MyD88-independent TLR4 pathway but as TLR4 does not play a role within the time-frame of this study, the effect of TRIF deficiency very likely represents the effect of functional TLR3 deficiency. To confirm this, we evaluated disease development in TLR3−/− mice and found a similar reduction in disease severity as was observed in the TRIF-mutant mice (Figure 3 B). TLR3 recognizes double-stranded RNA and TLR7 recognizes single-stranded RNA [21]. We conclude from these data that both TLR3 and TLR7 are involved in disease pathogenesis, presumably as a consequence of activation by endogenous RNA-containing ligands.
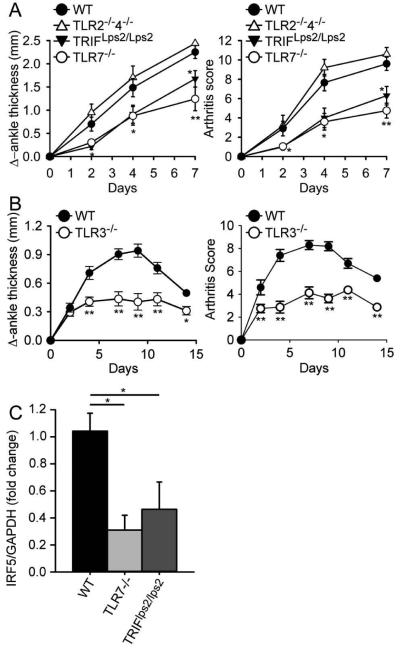
Figure 3.
TLR7-deficient ,TRIF-mutant and TLR3-deficient mice develop less severe arthritis following transfer of K/BxN serum. (A) C57BL/6 wildtype (WT; n = 10), TLR7-deficient (TLR7−/−; n = 10), TRIF-mutant (TRIFLps2/Lps2; n = 10), and TLR2 and 4 double-deficient (TLR2−/−4−/−; n = 5) mice were injected i.p with 150 μl of K/BxN serum on day 0 and day 2. Change in ankle thickness (left) and clinical score (right) were measured on days 0, 2, 4 and 7. (B) C57BL/6 wildtype (WT; n = 10) and TLR3-deficient (TLR3−/−; n =8) mice were injected i.p with 75 μl of K/BxN serum on day 0. Change in ankle thickness (left) and clinical score (right) were measured on days 0, 2, 4, 7, 9, 11 and 14. (C) IRF5 expression levels were measured by RT-PCR in wrist joints isolated 7 days after K/BxN serum administration from C57BL/6 wildtype (WT; n = 4), TLR7-deficient (TLR7−/−; n = 5) and TRIF-mutant (TRIFLps2/Lps2; n = 5) mice described in (A). * = P < 0.05; ** = P < 0.01; *** = P < 0. 001 versus WT.
To explore whether there is a functional link between IRF5 and TLR3/7 in joint inflammation, we measured expression levels of IRF5 in joint tissue from the TRIF-mutant and TLR7−/− mice. We found that IRF5 expression levels were substantially reduced in both the TRIF-mutant and TLR7−/− mice as compared with WT mice (Figure 3 C), suggesting either a direct effect of TRIF and TLR7 on IRF5 expression or a reduced recruitment of IRF5-expressing cells to the joints in TRIF-mutant and TLR7−/− mice.
TLR3 and TLR7 promote inflammatory cytokine and chemokine production through IRF5 and TLR3 synergizes with TLR7 to enhance these effects
We next examined how TLR3, TLR7 and IRF5 might interact to induce the production of cytokines relevant not only to this model (IL-1β, KC, and, to a variable degree, TNF-α) [15, 19], but also to human RA (TNF-α, IL-1β, and IL-6) [1]. Synovial fibroblasts are believed to play an important role in RA pathogenesis [26]. We found that KC and IL-6 production induced by the TLR7 ligand R837 was substantially IRF5-dependent in synovial fibroblasts whereas KC and IL-6 production induced by TLR2 and TLR4 ligands was IRF5-independent (Figure 4 A). TLR3-induced IL-6 production was also IRF5-dependent although TLR3-induced KC production was not (Figure 4 A).
Synovial macrophages have been identified as a major source of TNF-α, IL-1 and IL-6 in the human rheumatoid joint [27]. Macrophages are also required for disease in the K/BxN model [28]. In particular, macrophage colony-stimulating factor (M-CSF; also known as CSF1)-independent macrophages are required whereas M-CSF-dependent macrophages are not required [29]. To evaluate the roles of TLR3, TLR7 and IRF5 in these different macrophage subtypes we utilized adherent cell populations resulting from culture of bone marrow cells with granulocyte/macrophage colony-stimulating factor (GM-CSF) or M-CSF that have been likened to M1- or M2-polarized macrophages, respectively [30]. In M-CSF macrophages, TLR ligands induced both TNF-α and IL-1β and this did not require IRF5 (Figure 4 B). Similarly in GM-CSF macrophages, induction of TNF-α and IL-1β by TLR2 and TLR4 ligands did not require IRF5 (Figure 4 C). However, in contrast to M-CSF macrophages, induction of TNF-α by a TLR7 ligand in GM-CSF macrophages was mostly IRF5-dependent. Notably, there was a marked synergy between TLR3 and TLR7 ligands for both TNF-α and IL-1β production that was also mostly IRF5-dependent. The TLR3 ligand poly (I:C) alone did not induce either TNF-α or IL-1β production but it increased TLR7-induced responses approximately 3-fold in WT GM-CSF macrophages, but not in IRF5−/− GM-CSF macrophages (Figure 4 C).
Overall, these in vitro data demonstrate that TLR7 is able to induce inflammatory cytokine production in synovial fibroblasts and M1-type macrophages, both disease relevant cell types, in an IRF5-dependent manner. In addition, the data demonstrate IRF5-dependent synergy between TLR7 and TLR3 for inflammatory cytokine production suggesting that similar synergy might occur in vivo in the presence of endogenous RNA ligands. As IL-1β, TNF-α and KC all contribute to disease in the K/BxN model it is likely that IRF5 promotes arthritis, at least in part, through the regulation of multiple cytokines and chemokines produced by various cell types.
Mice deficient in IRF5 or TLR7 develop less severe arthritis but no additional protection is seen in mice deficient in both TLR7 and TLR3
After completion of the studies described above it was found that the IRF5 knockout mouse line, used by many investigators as well as ourselves, had a potentially confounding mutation in the Dock2 gene [8, 9]. We therefore thought it was important to repeat the in vivo experiments using IRF5 knockout mice in which we had bred out the Dock2 mutation. We also wanted to determine whether the protective effect of IRF5 deficiency would be maintained beyond the 7 day time point and whether combined TLR3 and TLR7 deficiency would confer additional protection in vivo.
To do this, we compared arthritis severity using the K/BxN model in WT mice, IRF5−/− mice (without the Dock2 mutation), TLR7−/− mice, and TLR3/TLR7 double-deficient (TLR3−/−7−/−) mice. Confirming our initial finding, ankle thickness and clinical arthritis score were reduced in IRF5−/− mice and TLR7−/− mice compared with WT, and this reduction was maintained for the 15 day duration of the study (Figure 5 A and B). The TLR3−/−7−/− mice exhibited a similar degree of protection as the TLR7−/− mice. At the 15 day time point, IRF5−/− mice had significantly less disease than both the TLR7−/− and the TLR3−/−7−/− mice suggesting the possibility that IRF5 may also promote disease through a non-TLR3/7 pathway later in the disease process. To examine which IRF5-expressing cell type(s) might be involved in disease pathogenesis we also evaluated mice lacking IRF5 expression only in myeloid cells, predominantly in monocyte/macrophages and neutrophils (Figure 5 C). We found a reduction in disease severity as measured by clinical arthritis score but only a trend toward a reduction in ankle thickness, suggesting that IRF5 expression in both myeloid cells and non-myeloid cells is required for the full development of arthritis in the K/BxN model.
The expression of genes involved in proIL-1β processing and of genes regulating neutrophil recruitment or function are reduced in the joints of IRF5-deficient mice with arthritis
To further investigate mechanisms by which IRF5-deficiency reduces arthritis severity, we compared mRNA expression in the joints of WT and IRF5−/− mice on day 7 after the induction of arthritis in the K/BxN model using a Nanostring Mouse Immunology gene expression panel comprising 561 genes. We found a total of 18 genes with reduced expression, and 6 genes with increased expression, in the IRF5−/− mice using a threshold statistical significance of p ≤ 0.02 (Figure 6). In the K/BxN model, bioactive IL-1β, which is required for disease pathogenesis, is produced by processing of proIL-1β by either caspase 1 or neutrophil serine proteases [31, 32]. We found that mRNA expression of both caspase 1 (Casp1) and cathepsin G (Ctsg), one of the 3 major neutrophil serine proteases, was reduced in IRF5−/− mice (Figure 6). Five other genes whose expression was reduced in the IRF5−/− mice were also either genes expressed predominantly by neutrophils or genes with important roles in neutrophil function (Figure 6). Ppbp encodes pro-platelet basic protein (also known as neutrophil-activating peptide 2 or CXCL7), a chemokine produced mainly by platelets and which is important for neutrophil chemotaxis [33]. Camp (also known as LL-37) encodes the cathelicidin antimicrobial peptide, which is produced in large amounts by neutrophils and which can bind specifically to dsRNA, ssRNA and DNA facilitating their uptake by diverse cells and promoting the activation of TLR3, TLR7 and TLR9 respectively [34]. S100A9 is an abundant cytoplasmic protein in neutrophils and other myeloid cells and is required for generation of the NADPH oxidative burst in neutrophils [35]. Extracellular S100A9 also promotes neutrophil chemotaxis [36]. Lactoferrin is an iron-binding protein that is released from activated neutrophils and may be a survival factor for neutrophils in rheumatoid synovial fluid [37]. Thus, the expression of multiple genes regulating neutrophil recruitment or function is reduced during active arthritis in IRF5-deficient mice. This likely reduces arthritis severity as neutrophils are essential for the initiation and progression of disease in the K/BxN model [38]. Further studies will be required to determine the extent to which the IRF5-dependent effects on neutrophil recruitment or function are due to neutrophil-intrinsic IRF5 or to a neutrophil-extrinsic effect of IRF5, for instance the regulation of neutrophil chemoattractants such as KC or pro-platelet basic protein.
Figure 6.
Gene expression in the joints of wildtype and IRF5-deficient mice during active arthritis. Gene expression was analyzed using Nanostring technology on RNA isolated from the joints of C57BL/6 wildtype (WT; n = 4) and IRF5-deficient (IRF5−/−; n = 5) mice 7 days after the i.p. injection of 150 μl of K/BxN serum on day 0 and day 2. Genes that differed between WT and IRF5−/− mice using a threshold statistical significance of p ≤ 0.02 are shown. Samples are arranged by hierarchical clustering and are displayed as a heat map.
DISCUSSION
The major conclusion from this report is that IRF5 contributes to the pathogenesis of the effector phase of inflammatory arthritis in the K/BxN model and that this is likely mediated mostly through the role of IRF5 in TLR7 and TLR3 signaling. The implications of this finding are that endogenous RNA ligands are likely involved in disease pathogenesis and that TLR activation by these ligands may be responsible in part for the production of the proinflammatory cytokines that are central to the progression of inflammatory arthritis.
TLR7 has also been implicated in disease pathogenesis in the collagen-induced arthritis model [39, 40]. The extent to which the findings in the K/BxN or collagen-induced arthritis models apply to human RA or other human inflammatory arthropathies remains to be determined. However, both TLR3 and TLR7 are highly expressed in synovial tissue from patients with RA and stimulation of TLR3 increased TNF-α release in human rheumatoid synovial membrane cultures [25, 41]. In addition, potential TLR3 and TLR7 ligands are found in human synovial fluid and sera from RA patients. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts through TLR3 [42]. Abundant free nucleosomes are present in the joints of RA patients indicative of the release of apoptotic cell material [43], and dsRNA is present in the synovial fluid of rheumatoid arthritis patients with an erosive disease course [44]. Ligation of TLR7 by single stranded RNA present in rheumatoid arthritis synovial fluid induces transcription of TNFα in monocytes [45].
The putative endogenous RNA ligands could originate from a number of potential sources. Endogenous ligands released from apoptotic or necrotic cells are thought to contribute to the pathogenesis of certain autoimmune or inflammatory conditions through engagement of TLRs [22]. For example, endogenous RNA released from necrotic cells is a ligand for TLR3 and TLR3 acts as a sensor of tissue necrosis during acute inflammatory events [46]. Self-RNA released from dying cells can activate TLR7 in dendritic cells following internalization of the RNA complexed either with anti-RNA autoantibodies or the antimicrobial peptide LL37 [14, 47]. It is thus possible that in arthritis, immunostimulatory RNA is released from cells dying as a consequence of tissue injury induced by immune complexes or other components of the inflammatory response in the joint.
IRF5 is involved in many TLR7-driven responses [9, 14, 18] and our report provides further evidence of this in cell types relevant to inflammatory arthritis. Regarding the role of IRF5 in TLR3 signaling, IRF5 has been shown to play an important role in promoting synergy between MyD88-dependent and TRIF-dependent TLR pathways in peritoneal macrophages [48]. In particular, IRF5-dependent synergy between TLR3 and TLR9 ligands was observed for the induction of IL-12p40, IL-23p19 and IL-6 but not for TNF-α. We extended these studies to include an analysis of the role of IRF5 in TLR3 and TLR7 synergy and found a marked IRF5-dependent synergy between TLR3 and TLR7 in pro-inflammatory cytokine production by M1-type macrophages. The mechanism whereby IRF5 promotes this synergy is not completely clear. IRF5 can bind directly to MyD88 and this interaction is required for IRF5 activation by TLR7 [17]. However, exactly how IRF5 participates in TLR3 signaling is less well understood. Although we and others have previously reported that poly(I:C) responses are IRF5-dependent [14, 17, 48] (and Figure 4 A), no direct interaction between IRF5 and TRIF has yet been demonstrated and poly(I:C) did not induce IRF5 activation in HEK cell transfection experiments [18]. Further work will be required to define the precise role of IRF5 in TLR3 signaling.
In summary, this report demonstrates that TLR7 and TLR3 contribute to disease development in the K/BxN model of inflammatory arthritis, suggesting a role for endogenous RNA ligands in pathogenesis. The requirement for IRF5 in mediating these TLR signaling pathways provides a possible explanation for how the gain of function polymorphisms in IRF5 associated with rheumatoid arthritis might contribute to disease. However, the limitations of this model need to be emphasized as it is mostly dependent on innate immune responses [49]. IRF5 also plays an important role in adaptive immune responses including B cell function and the generation of effective TH1 and TH17 T cell responses [17, 50]. As dysregulated T and B cell responses are strongly implicated in rheumatoid arthritis pathogenesis [1], IRF5 could potentially also play a role in human rheumatoid arthritis by driving pathogenic adaptive immune responses.
Acknowledgments
This work was supported by National Institutes of Health grants PO1 AR050256 (I.R.R), R01 AR055952 (E.M.G), KO1 AR055965 (T.A.), KO1 AR060169 (C.M.C.), PO1 HL108795 (H.P.), R01 AR050250 (H.P.), R01 AR054796 (H.P.) and R01 AR051089 (R.L.). P. Duffau was supported by grants from Société Française de Médecine Interne and CHU de Bordeaux. C. Richez was supported by a grant from Société Française de Rhumatologie. A Watkins was supported by a National Institutes of Health/NIAID ImmunologyTraining Grant 5T32AI007309-22. K. Yasuda was supported by a National Institutes of Health/NIDDK NephrologyTraining Grant 2T32DK007053. P. Monach was supported by an Arthritis Investigator Award from the Arthritis Foundation. Harris Perlman was also supported by funds provided by the United States-Israel Binational Science Foundation (2013247), Rheumatology Research Foundation (Agmt 05/06/14) and the Solovy-Arthritis Research Society Chair in Medicine to Harris Perlman.
Footnotes
The authors have no conflicting financial interests.
REFERENCES
- 1.McInnes IB, O'Dell JR. State-of-the-art: rheumatoid arthritis. Ann Rheum Dis. 2010;69:1898–1906. doi: 10.1136/ard.2010.134684. [DOI] [PubMed] [Google Scholar]
- 2.Dieguez-Gonzalez R, Calaza M, Perez-Pampin E, de la Serna AR, Fernandez-Gutierrez B, Castaneda S, et al. Association of interferon regulatory factor 5 haplotypes, similar to that found in systemic lupus erythematosus, in a large subgroup of patients with rheumatoid arthritis. Arthritis Rheum. 2008;58:1264–1274. doi: 10.1002/art.23426. [DOI] [PubMed] [Google Scholar]
- 3.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cham CM, Ko K, Niewold TB. Interferon regulatory factor 5 in the pathogenesis of systemic lupus erythematosus. Clin Dev Immunol. 2012;2012:780436. doi: 10.1155/2012/780436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monach P, Hattori K, Huang H, Hyatt E, Morse J, Nguyen L, et al. The K/BxN mouse model of inflammatory arthritis: theory and practice. Methods Mol Med. 2007;136:269–282. doi: 10.1007/978-1-59745-402-5_20. [DOI] [PubMed] [Google Scholar]
- 6.Ditzel HJ. The K/BxN mouse: a model of human inflammatory arthritis. Trends Mol Med. 2004;10:40–45. doi: 10.1016/j.molmed.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Shih F, Atkinson JP. Systemic humoral autoimmunity but joint-specific inflammation: the syndrome of rheumatoid arthritis. Arthritis Rheum. 2007;56:2823–2828. doi: 10.1002/art.22858. [DOI] [PubMed] [Google Scholar]
- 8.Purtha WE, Swiecki M, Colonna M, Diamond MS, Bhattacharya D. Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc Natl Acad Sci U S A. 2012;109:E898–904. doi: 10.1073/pnas.1118155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda K, Nundel K, Watkins AA, Dhawan T, Bonegio RG, Ubellacker JM, et al. Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int Immunol. 2013;25:295–306. doi: 10.1093/intimm/dxs114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misharin AV, Cuda CM, Saber R, Turner JD, Gierut AK, Haines GK, 3rd, et al. Nonclassical Ly6C(−) monocytes drive the development of inflammatory arthritis in mice. Cell Rep. 2014;9:591–604. doi: 10.1016/j.celrep.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scatizzi JC, Bickel E, Hutcheson J, Haines GK, 3rd, Perlman H. Bim deficiency leads to exacerbation and prolongation of joint inflammation in experimental arthritis. Arthritis Rheum. 2006;54:3182–3193. doi: 10.1002/art.22133. [DOI] [PubMed] [Google Scholar]
- 12.Watkins AA, Yasuda K, Wilson GE, Aprahamian T, Xie Y, Maganto-Garcia E, et al. IRF5 Deficiency Ameliorates Lupus but Promotes Atherosclerosis and Metabolic Dysfunction in a Mouse Model of Lupus-Associated Atherosclerosis. J Immunol. 2015;194:1467–1479. doi: 10.4049/jimmunol.1402807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Case JP, Sano H, Lafyatis R, Remmers EF, Kumkumian GK, Wilder RL. Transin/stromelysin expression in the synovium of rats with experimental erosive arthritis. In situ localization and kinetics of expression of the transformation-associated metalloproteinase in euthymic and athymic Lewis rats. J Clin Invest. 1989;84:1731–1740. doi: 10.1172/JCI114356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasuda K, Richez C, Maciaszek JW, Agrawal N, Akira S, Marshak-Rothstein A, et al. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol. 2007;178:6876–6885. doi: 10.4049/jimmunol.178.11.6876. [DOI] [PubMed] [Google Scholar]
- 15.Ji H, Pettit A, Ohmura K, Ortiz-Lopez A, Duchatelle V, Degott C, et al. Critical roles for interleukin 1 and tumor necrosis factor alpha in antibody-induced arthritis. J Exp Med. 2002;196:77–85. doi: 10.1084/jem.20020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 17.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 18.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs JP, Ortiz-Lopez A, Campbell JJ, Gerard CJ, Mathis D, Benoist C. Deficiency of CXCR2, but not other chemokine receptors, attenuates autoantibody-mediated arthritis in a murine model. Arthritis Rheum. 2010;62:1921–1932. doi: 10.1002/art.27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 21.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 22.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu Rev Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 23.Choe JY, Crain B, Wu SR, Corr M. Interleukin 1 receptor dependence of serum transferred arthritis can be circumvented by toll-like receptor 4 signaling. J Exp Med. 2003;197:537–542. doi: 10.1084/jem.20021850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu HJ, Sawaya H, Binstadt B, Brickelmaier M, Blasius A, Gorelik L, et al. Inflammatory arthritis can be reined in by CpG-induced DC-NK cell cross talk. J Exp Med. 2007;204:1911–1922. doi: 10.1084/jem.20070285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh FG, Midwood KS. Intrinsic danger: activation of Toll-like receptors in rheumatoid arthritis. Rheumatology (Oxford) 2012;51:7–23. doi: 10.1093/rheumatology/ker257. [DOI] [PubMed] [Google Scholar]
- 26.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Q, Pope RM. Toll-like receptor signaling: a potential link among rheumatoid arthritis, systemic lupus, and atherosclerosis. J Leukoc Biol. 2010;88:253–262. doi: 10.1189/jlb.0310126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon S, Rajasekaran N, Jeisy-Walder E, Snapper SB, Illges H. A crucial role for macrophages in the pathology of K/B x N serum-induced arthritis. Eur J Immunol. 2005;35:3064–3073. doi: 10.1002/eji.200526167. [DOI] [PubMed] [Google Scholar]
- 29.Bruhns P, Samuelsson A, Pollard JW, Ravetch JV. Colony-stimulating factor-1-dependent macrophages are responsible for IVIG protection in antibody-induced autoimmune disease. Immunity. 2003;18:573–581. doi: 10.1016/s1074-7613(03)00080-3. [DOI] [PubMed] [Google Scholar]
- 30.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 31.Joosten LA, Netea MG, Fantuzzi G, Koenders MI, Helsen MM, Sparrer H, et al. Inflammatory arthritis in caspase 1 gene-deficient mice: contribution of proteinase 3 to caspase 1-independent production of bioactive interleukin-1beta. Arthritis Rheum. 2009;60:3651–3662. doi: 10.1002/art.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer-Hoffert U, Wiedow O. Neutrophil serine proteases: mediators of innate immune responses. Curr Opin Hematol. 2011;18:19–24. doi: 10.1097/MOH.0b013e32834115d1. [DOI] [PubMed] [Google Scholar]
- 33.Ghasemzadeh M, Kaplan ZS, Alwis I, Schoenwaelder SM, Ashworth KJ, Westein E, et al. The CXCR1/2 ligand NAP-2 promotes directed intravascular leukocyte migration through platelet thrombi. Blood. 2013;121:4555–4566. doi: 10.1182/blood-2012-09-459636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahlenberg JM, Kaplan MJ. Little peptide, big effects: the role of LL-37 in inflammation and autoimmune disease. J Immunol. 2013;191:4895–4901. doi: 10.4049/jimmunol.1302005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markowitz J, Carson WE., 3rd Review of S100A9 biology and its role in cancer. Biochim Biophys Acta. 2013;1835:100–109. doi: 10.1016/j.bbcan.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 37.Wong SH, Francis N, Chahal H, Raza K, Salmon M, Scheel-Toellner D, et al. Lactoferrin is a survival factor for neutrophils in rheumatoid synovial fluid. Rheumatology (Oxford) 2009;48:39–44. doi: 10.1093/rheumatology/ken412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wipke BT, Allen PM. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol. 2001;167:1601–1608. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- 39.Alzabin S, Kong P, Medghalchi M, Palfreeman A, Williams R, Sacre S. Investigation of the role of endosomal Toll-like receptors in murine collagen-induced arthritis reveals a potential role for TLR7 in disease maintenance. Arthritis Res Ther. 2012;14:R142. doi: 10.1186/ar3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen SY, Shiau AL, Li YT, Lin YS, Lee CH, Wu CL, et al. Suppression of collagen-induced arthritis by intra-articular lentiviral vector-mediated delivery of Toll-like receptor 7 short hairpin RNA gene. Gene Ther. 2012;19:752–760. doi: 10.1038/gt.2011.173. [DOI] [PubMed] [Google Scholar]
- 41.Huang QQ, Pope RM. The role of toll-like receptors in rheumatoid arthritis. Curr Rheumatol Rep. 2009;11:357–364. doi: 10.1007/s11926-009-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brentano F, Schorr O, Gay RE, Gay S, Kyburz D. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via Toll-like receptor 3. Arthritis Rheum. 2005;52:2656–2665. doi: 10.1002/art.21273. [DOI] [PubMed] [Google Scholar]
- 43.Monach PA, Hueber W, Kessler B, Tomooka BH, BenBarak M, Simmons BP, et al. A broad screen for targets of immune complexes decorating arthritic joints highlights deposition of nucleosomes in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2009;106:15867–15872. doi: 10.1073/pnas.0908032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bokarewa M, Tarkowski A, Lind M, Dahlberg L, Magnusson M. Arthritogenic dsRNA is present in synovial fluid from rheumatoid arthritis patients with an erosive disease course. Eur J Immunol. 2008;38:3237–3244. doi: 10.1002/eji.200838362. [DOI] [PubMed] [Google Scholar]
- 45.Chamberlain ND, Kim SJ, Vila OM, Volin MV, Volkov S, Pope RM, et al. Ligation of TLR7 by rheumatoid arthritis synovial fluid single strand RNA induces transcription of TNFalpha in monocytes. Ann Rheum Dis. 2013;72:418–426. doi: 10.1136/annrheumdis-2011-201203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavassani KA, Ishii M, Wen H, Schaller MA, Lincoln PM, Lukacs NW, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J Exp Med. 2008;205:2609–2621. doi: 10.1084/jem.20081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206:1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouyang X, Negishi H, Takeda R, Fujita Y, Taniguchi T, Honda K. Cooperation between MyD88 and TRIF pathways in TLR synergy via IRF5 activation. Biochem Biophys Res Commun. 2007;354:1045–1051. doi: 10.1016/j.bbrc.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 49.Ji H, Ohmura K, Mahmood U, Lee DM, Hofhuis FM, Boackle SA, et al. Arthritis critically dependent on innate immune system players. Immunity. 2002;16:157–168. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 50.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]