Abstract
Background
In this work, we aimed to identify molecular epidermal growth factor receptor (EGFR) tissue biomarkers in patients with ovarian cancer who were treated within the phase III randomized European Organisation for Research and Treatment of Cancer-Gynaecological Cancer Group (EORTC-GCG) 55041 study comparing erlotinib with observation in patients with no evidence of disease progression after first-line platinum-based chemotherapy.
Methods
Somatic mutations in KRAS, BRAF, NRAS, PIK3CA, EGFR, and PTEN were determined in 318 (38 %) and expression of EGFR, pAkt, pMAPK, E-cadherin and Vimentin, and EGFR and HER2 gene copy numbers in 218 (26 %) of a total of 835 randomized patients. Biomarker data were correlated with progression-free survival (PFS) and overall survival (OS).
Results
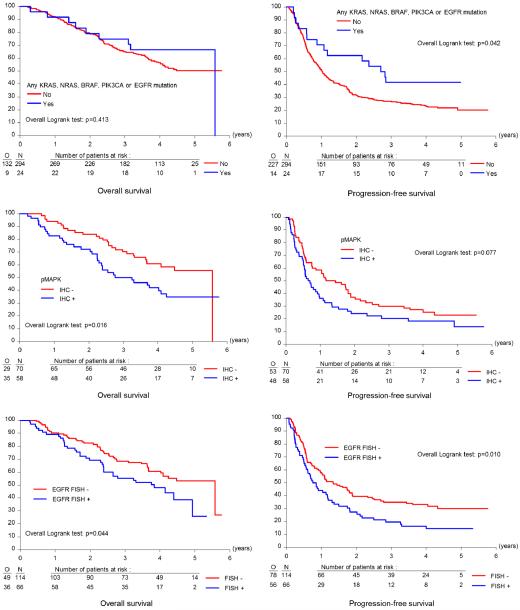
Only 28 mutations were observed among KRAS, BRAF, NRAS, PIK3CA, EGFR, and PTEN (in 7.5 % of patients), of which the most frequent were in KRAS and PIK3CA. EGFR mutations occurred in only three patients. When all mutations were pooled, patients with at least one mutation in KRAS, NRAS, BRAF, PIK3CA, or EGFR had longer PFS (33.1 versus 12.3 months; HR 0.57; 95 % CI 0.33 to 0.99; P=0.042) compared to those with wild-type tumors. EGFR overexpression was detected in 93 of 218 patients (42.7 %), and 66 of 180 patients (36.7 %) had EGFR gene amplification or high levels of copy number gain. Fifty-eight of 128 patients had positive pMAPK expression (45.3 %), which was associated with inferior OS (38.9 versus 67.0 months; HR 1.81; 95 % CI 1.11 to 2.97; P=0.016). Patients with positive EGFR fluorescence in situ hybridization (FISH) status had worse OS (46.1 months) than those with negative status (67.0 months; HR 1.56; 95 % CI 1.01 to 2.40; P=0.044) and shorter PFS (9.6 versus 16.1 months; HR 1.57; 95 % CI 1.11 to 2.22; P=0.010). None of the investigated biomarkers correlated with responsiveness to erlotinib.
Conclusions
In this phase III study, increased EGFR gene copy number was associated with worse OS and PFS in patients with ovarian cancer. It remains to be determined whether this association is purely prognostic or is also predictive.
1 Introduction
A major focus of cancer therapy research over the past decade has been in the targeting of cellular processes affecting cell proliferation, differentiation, growth, and survival. One of the best studied among these is the epidermal growth factor receptor (EGFR), given its dysregulation in the vast majority of human tumors of epithelial origin [1]. EGFR is a member of the ErbB family consisting of four tyrosine kinase (TK) receptors: EGFR/ErbB-1, HER-2/neu (ErbB-2), HER-3 (ErbB-3), and HER-4 (ErbB-4) [2]. Binding of specific ligands such as epidermal growth factor (EGF) and transforming growth factor α (TGF-α) to the EGFR results in the dimerisation of the receptor, tyrosine auto-phosphorylation, with subsequent initiation of the intracellular signaling pathways cascade. Downstream signaling pathways include the ras-raf-mitogen-activated protein kinase (Ras/Raf/MAPK) and the phosphatidylinositol 3-kinase (PI3K/AKT) pathways, which are all involved in cell proliferation and survival [2].
EGFR overexpression is observed in up to 98 % of advanced epithelial ovarian cancers (EOCs) [3, 4] and has been associated with a worse prognosis [3, 5], although data are conflicting [6, 7]. The targeting of EGFR or its downstream pathways, therefore, appears to be a promising strategy in EOC. Monoclonal antibodies (mAbs), such as cetuximab and panitumumab, bind competitively to the extracellular domain of EGFR, leading to internalization and degradation of the receptor, while tyrosine kinase inhibitors (TKIs) like erlotinib and gefitinib compete with ATP for binding to the receptor’s intracellular TK domain, thereby inhibiting TK activity. Both molecular strategies have been investigated in EOC (Table 1).
Table 1.
Clinical trials reporting administration of anti-EGFR agents to patients with epithelial ovarian cancer
| Study | Treatment regimen | Phase | Patient population | No. of pts | Response |
|---|---|---|---|---|---|
| Erlotinib | |||||
| Gordon et al. [8] | 150 mg/d | II | Recurrent EGFR-positive | 34 | PR: 2 (6 %) SD: 15 (44 %) PD: 17 (50 %) |
| Blank et al. [9] | Paclitaxel (175 mg/m2) and carboplatin (AUC6) every 3 weeks and erlotinib 150 mg/d |
II | Primary a: after optimal CRS, b: after suboptimal CRS, c: before CRS |
56 a: 28, b: 23, c: 5 |
CR (a): 8 (29 %) CR (b): 3 (13 %) |
| Hirte et al. [10] | Carboplatin (AUC5) every 3 weeks and erlotinib 150 mg/d |
II | Recurrent a: platinum-sensitive b: platinum-resistant |
50 a: 33 b: 17 |
CR (a): 3 (9 %) PR (a): 14 (42 %) PR (b): 1 (6 %) |
| Gefitinib | |||||
| Schilder et al. [11] | 500 mg/d | II | Recurrent | 27 | PR: 1 (3.7 %) PFS>6 m: 14.8 % |
| Posadas et al. [12] | 500 mg/d | II | Recurrent | 24 | CR: 0 PR: 0 SD: 9 (38 %) |
| Wagner et al. [13] | Tamoxifen 40 mg/d and gefitinib 500 mg/d |
II | Recurrent | 56 | SD: 16 (28.6 %) |
| Pautier et al. [14] | Paclitaxel (175 mg/m2) and carboplatin (AUC5) every 3 weeks and gefitinib 500 mg/d |
II | Recurrent a: platinum-resistant b: platinum-sensitive |
68 a: 26 b: 42 |
CR (a): 1 (3.8 %) CR (b): 10 (23.8 %) PR(a): 4 (15.4 %) PR (b): 16 (38.1 %) |
| Cetuximab | |||||
| Schilder et al. [15] | Initial dose 400 mg/m2 and 250 mg/m2
weekly for two 3-week cycles |
II | Recurrent | 25 | PR: 1 (4 %) SD: 9 (36 %) |
| Secord et al. [16] | Initial dose 400 mg/m2, followed by weekly infusion of 250 mg/m2 and carboplatin (AUC6) every 3 weeks |
II | Recurrent platinum-sensitive EGFR positive |
26 | CR: 3 (11.5 %) PR: 6 (23 %) SD: 8 (30.8 %) |
AUC area under the curve, CR complete response, CRS cytoreductive surgery, No. of pts number of evaluable patients, PR partial response, SD stable disease
Although several clinical prognostic factors have been identified in EOC (e.g., age at diagnosis, extent of disease, amount of residual disease after initial surgery, tumor grade, and tumor histological subtype) [17, 18], molecular prognostic markers, or even predictive biomarkers for targeted agents, are still lacking. In other disease entities such as non-small cell lung cancer (NSCLC) and metastatic colorectal cancer (mCRC)—where anti-EGFR therapies have been widely studied—several predictive biomarkers for anti-EGFR agents have been identified. EGFR protein expression as determined by immunohistochemistry (IHC) was the first biomarker claimed to predict a response in NSCLC. However, data have been conflicting, and its association with sensitivity to anti-EGFR therapies remains unclear [19–21]. Activating mutations in EGFR exons 18 to 21 [22–24] and an increased EGFR gene copy number may increase sensitivity to anti-EGFR agents [19, 20, 25–28], whereas deregulation of downstream targets of the EGFR pathway (i.e., mutations in the KRAS, NRAS, BRAF, or PIK3CA genes or loss of PTEN protein expression) have emerged as an important negative predictive factor for the efficacy of anti-EGFR mAbs [29–34]. However, following the initial response, NSCLC patients harboring activating EGFR mutations may become resistant to TKIs due to an acquired secondary EGFR kinase domain mutation, T790M [35]. While an increased HER2 gene copy number in NSCLC may affect sensitivity to EGFR TKIs [36–39], preclinical and clinical studies in mCRC have shown cetuximab resistance in cases with HER2 gene amplification [40, 41]. Additionally, in NSCLC, TKI responsiveness may be predicted by EGFR downstream proteins such as activated (phosphorylated) AKT [19]. Furthermore, epithelial-mesenchymal transition (EMT), a key player in cancer progression and metastasis, which is characterized by a loss in expression of E-cadherin and a gain in vimentin expression, is associated with resistance to gefitinib and erlotinib in NSCLC [42, 43]. Thus far, the extensive data investigating responsiveness to anti-EGFR agents have focused mainly on NSCLC and mCRC; data for ovarian cancer, on the other hand, are scarce.
Studies with EGFR-targeted agents in EOC suggest that only a subgroup of ovarian cancer patients might benefit from these therapies (Table 1). However, Gordon et al. reported 44 % stable disease in a phase II study in patients with refractory recurrent EGFR-positive epithelial ovarian cancer treated with erlotinib [8]. This finding led to the European Organisation for Research and Treatment of Cancer-Gynaecological Cancer Group (EORTC-GCG) 55041 trial, in which ovarian cancer patients with no evidence of disease progression following first-line platinum-based chemotherapy were treated with erlotinib as maintenance therapy. Unfortunately, in this unselected patient population, erlotinib did not improve progression-free or overall survival. The aim of the present explorative translational biomarker study based on the prospective EORTC-GCG 55041 trial [44] was to determine the frequency of alterations of components of the EGFR pathways and to correlate biomarker data with the efficacy of erlotinib.
2 Materials and Methods
2.1 Patient Population
In the EORTC-GCG 55041 phase III trial, eligible patients were those with histologically confirmed high-risk FIGO stage I (grade 3, or aneuploid grade 1 or 2, or clear cell) or stages II–IV epithelial ovarian, primary peritoneal, or fallopian tube cancer [44]. All patients underwent first-line platinum-based chemotherapy and showed no signs of progression at the end of chemotherapy according to the Response Evaluation Criteria In Solid Tumors (RECIST) criteria [45] and/or the Gynecological Cancer Intergroup (GCIG) criteria in the case of CA125-based evaluation at the end of first-line treatment [46]. Overall, 835 patients were randomized 1:1 to maintenance erlotinib 150 mg orally daily for 2 years or observation. From the 835 patients registered in the trial, 527 patients(63 %) consented to optional translational research. Prospective bio-banking was performed, and formalin-fixed paraffin-embedded (FFPE) tissue sampled before and/or during first-line chemotherapy was requested from the participating centers for this translational study. The FFPE histological tissue was accepted independent of the site of biopsy—for example, surgical specimens from primary ovarian tumors, lymph nodes, or distant metastases. Patients gave written informed consent by signing the separate translational research informed consent form before any study-specific procedure. The translational research study and the informed consent forms were approved by the ethics committees of the institutions involved. The study was conducted in accordance with IHC good clinical practice guidelines and the Declaration of Helsinki, and was registered at ClinicalTrials.gov, number NCT00263822.
2.2 Molecular Tissue Biomarker Analyses
The translational analyses were performed at the Department of Oncology, Division of Gynecological Oncology, at the Catholic University Leuven (Leuven, Belgium), the Vesalius Research Center at the Flanders Institute for Biotechnology (VRC/VIB3) (Leuven, Belgium), and the University of Colorado Comprehensive Cancer Center (Aurora, CO, USA). Tumor tissues were sent as FFPE blocks, eppendorf tubes or glass slides. All tumor tissue samples were checked for quality, tissue integrity, and tumor content on a 5-μm hematoxylin and eosin (H&E) stained section. Tissue microarrays (TMAs) for the IHC analyses were constructed using a manual tissue puncher/arrayer (Beecher Instruments, Silver Spring, MD, USA). Tumor DNA was extracted using a phenol-chloroform method after macrodissection of the marked tumor region on the H&E slide. Biopsies with a low estimated tumor percentage on H&E and those failing to yield sufficient DNA after extraction were excluded (n=27).
2.2.1 Immunohistochemistry
Immunohistochemical analyses were performed on the constructed TMAs or individual slides using methods and assessment criteria described elsewhere [25]. The following primary antibodies were used for IHC: EGFR (Cell Signaling Technology, Inc., Beverly, MA, USA) diluted at 1:50, phosphorylated Akt (Ser473) at 1:50, phosphorylated p44/42 MAPK (Thr202/Tyr204; E10) monoclonal antibody (Cell Signaling Technology, Inc.) at 1:50, vimentin antibody V9 (Dako/Agilent Technologies, Carpinteria, CA, USA), and E-cadherin antibody H-108 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The percentage of total tumor cells within each staining intensity category [0 (no staining), 1+ (weak), 2+ (moderate), 3+ (strong)] was reported. A hybrid (H)-score was then generated based on the fraction of staining cells in each intensity category. The H-score was calculated by completing the formula (% cells of 0 intensity × 0)+(% of 1+ intensity × 1)+(% of 2+ intensity × 2)+ (% of 3+ intensity × 3), producing a final H-score with a range of 0–300 [25]. For statistical analyses, the immunohistochemical results of EGFR, E-cadherin, pMAPK, and pAkt were analyzed in a binary fashion due to the skewed distribution of the results.
2.2.2 Fluorescence In Situ Hybridization (FISH) Analyses
We analyzed the EGFR gene copy number per cell using the LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen Probe (Abbott Molecular, Des Plaines, IL, USA), as previously described [19, 25]. Tumor specimens were classified into six FISH strata according to the frequency of cells with each EGFR gene copy number and referred to the chromosome 7 centromere, as follows: (1) disomy (three or four copies in <10 % of cells), (2) low trisomy (three copies in 10 % to <40 % of cells and four copies in <10 % of cells), (3) high trisomy (three copies in ≥40 % of cells and four copies in <10 % of cells), (4) low polysomy (four copies in 10 % to <40 % of cells), (5) high polysomy (four or more copies in ≥40 % of cells), and (6) gene amplification (presence of loose or tight EGFR gene clusters with ≥4 copies, EGFR gene-to-CEP 7 ratio≥2, or 15 copies of EGFR per cell in ≥10 % of cells). The high polysomy and gene amplification categories were considered to indicate a high EGFR copy number (EGFR-FISH positive), and the other categories were considered to indicate no significant increase in the EGFR copy number (EGFR-FISH negative), as previously described [34, 47].
Dual-target, dual-color HER2 FISH assays were performed using the PathVysion HER-2 DNA probe kit (Abbott Molecular), which includes the LSI HER-2 SpectrumOrange and the CEP 17 SpectrumGreen probes. The reference slide (stained with H&E) was the adjacent section on which the dominant tumor foci were identified, and copy numbers of the HER2 gene and chromosome 17 centromere probes were assessed and recorded independently in at least 50 non-overlapping nuclei with intact morphology. Analysis was performed blinded to the patients’ clinical characteristics. Based on the mean number of copies of the HER2 gene and chromosome 17 centromere per cell, patients were classified into three strata: negative for HER2 amplification when the mean HER2/mean CEP17 ratio was <1.8, positive for HER2 amplification when then mean HER2/mean CEP17 ratio was >2.2, and equivocal when the mean HER2/mean CEP17 ratio was between 1.8 and 2.2, in which case a second observer independently scored 50 tumor cells, and the final classification was assessed based on the results in all 100 cells.
2.2.3 Somatic Mutation Profiling
The COSMIC (Catalogue Of Somatic Mutations In Cancer) database [48] was queried for the most frequent mutations occurring in commonly mutated oncogenes related to the EGFR pathway, which resulted in coverage of >97 %, >94 %, >97 %, >79 %, >65 %, and >7 % for KRAS, BRAF, NRAS, PIK3CA, EGFR, and PTEN, respectively. DNA was aliquoted into 384-well plates and genotyped centrally at the Vesalius Research Center (Leuven, Belgium). iPLEX technology was used on a MassARRAY Compact Analyser (Sequenom Inc., San Diego, CA, USA), as described by De Roock et al. [31]. DNA of a sample was considered of sufficient quality when more than 75 % of mutations were reliably genotyped. A sample was considered wild-type for a given gene when the most frequently mutated sites in this gene did not show a mutation.
2.2.4 EGFR Sequencing
Targeted re-sequencing of EGFR was performed to identify potential somatic mutations that were not assessed in the Sequenom hotspot mutation panel. In particular, EGFR was sequenced using the TruSeq Custom Amplicon Kit on an Illumina MiSeq sequencer (Illumina, Inc., San Diego, CA, USA). Custom oligo probes targeting EGFR exons were designed using DesignStudio (Illumina, Inc.). Amplicons were generated in an extension-ligation reaction across the region of interest and amplified in a subsequent PCR, which also incorporates two unique sample-specific indexes, following the manufacturer’s instructions. After pooling, the samples were sequenced on an Illumina MiSeq in a 2x150-bp paired-end sequencing run using a v2 flow cell. Data were mapped with the Burrows-Wheeler Aligner (bwa-0.5) [49] to the human reference genome hg19. The Genome Analysis Toolkit (GATK version 1.6; Broad Institute) was further used to process the data, and variants and indels were called by the Unified Genotyper and Dindel, respectively [50, 51]. The mutations were further filtered [52] and manually curated in IGV. EGFR re-sequencing was successful if the average coverage per sample was >100x.
2.3 Statistical Analyses
All statistical analyses for the translational study of the EORTC-GCG 55041 trial were performed centrally at the headquarters of the EORTC in Brussels (Belgium). The sample size of the trial (n=835) was based on the primary endpoint, progression-free survival (PFS), to detect an increase of 25 % in the median PFS from 15 to 18.75 months [44]. Secondary endpoints were overall survival (OS), toxicity, occurrence of rash, and quality of life. OS and PFS were defined as the time from randomization to the date of the event (death and death or progression, respectively). Time-to-event analyses were investigated via the Kaplan-Meier method with hazard ratios (HR) obtained through Cox regression and compared via a nonparametric log-rank test stratified for treatment (where possible). Predictiveness for treatment effect was assessed via Cox regression using an interaction test with the allocated protocol treatment (erlotinib versus observation). All tests were nonparametric two-sided tests, evaluated at the 5 % significance level. No correction for multiplicity was made, and the 5 % level was used as a screening measure rather than a formal hypothesis test.
3 Results
3.1 Patient Characteristics
Tumor tissues were available for 358 of the 527 patients (68 %) who consented to optional translational research. Mutation data were available for 318 patients and FISH and IHC data for 218 patients. The main reason for the different numbers was due to the fact that DNA extraction was possible in most samples, where IHC or FISH analysis was not achievable due to technical reasons. Detailed patient characteristics are summarized in Table 2. With regard to important baseline parameters (e.g., age, stage, histological type and grade) no significant imbalances between the overall study population and the translational study population were apparent, except for histological grade (with more high-grade tumors and fewer patients with unknown histological grade in the translational study population).
Table 2.
Baseline patient characteristics: overall study population (n=835) and translational study population (mutation analysis n=318, FISH and immunohistochemical analysis n=218)
| Parameter | Translational |
Overall |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| Mutation group (n=318) |
FISH/IHC group (n=218) |
(n=835) |
Mutation vs overall | FISH/IHC vs overall | ||||
| No. | % | No. | % | No. | % | |||
| Age (years) | 0.300 | 0.532 | ||||||
| Median | 59.5 | 59.0 | 59.0 | |||||
| Range | 31–85 | 31–85 | 19–85 | |||||
| WHO performance status | 0.987 | 0.134 | ||||||
| 0 | 213 | 67 | 137 | 62.8 | 559 | 66.9 | ||
| 1 | 105 | 33 | 81 | 37.2 | 276 | 33.1 | ||
| FIGO stage | 0.561 | 0.128 | ||||||
| I | 25 | 7.9 | 21 | 9.6 | 57 | 6.8 | ||
| II | 27 | 8.5 | 19 | 8.7 | 62 | 7.4 | ||
| III | 212 | 66.7 | 144 | 66.1 | 563 | 67.4 | ||
| IV | 54 | 17 | 33 | 15.1 | 152 | 18.2 | ||
| Unknown | 0 | 0 | 1 | 0.5 | 1 | 0.1 | ||
| Histological grade | 0.001 | 0.002 | ||||||
| Grade 1 | 12 | 3.8 | 9 | 4.1 | 58 | 6.9 | ||
| Grade 2 | 58 | 18.2 | 38 | 17.4 | 152 | 18.2 | ||
| Grade 3 | 172 | 54.1 | 127 | 58.3 | 388 | 46.5 | ||
| Unknown | 76 | 23.9 | 44 | 20.2 | 237 | 28.4 | ||
| Histological type | 0.055 | 0.285 | ||||||
| Serous | 205 | 64.5 | 134 | 61.5 | 521 | 62.4 | ||
| Mucinous | 7 | 2.2 | 6 | 2.8 | 14 | 1.7 | ||
| Clear cell | 23 | 7.2 | 18 | 8.3 | 51 | 6.1 | ||
| Endometrioid | 28 | 8.8 | 16 | 7.3 | 61 | 7.3 | ||
| Undifferentiated | 8 | 2.5 | 7 | 3.2 | 21 | 2.5 | ||
| Other/Unknown | 47 | 14.8 | 37 | 17 | 167 | 20 | ||
| Treatment arm | 0.995 | 0.795 | ||||||
| Erlotinib | 160 | 50.3 | 108 | 49.5 | 420 | 50.3 | ||
| Observation | 158 | 49.7 | 110 | 50.5 | 415 | 49.7 | ||
| First-line chemotherapy | 0.612 | 0.693 | ||||||
| Platinum alone | 12 | 3.8 | 11 | 5 | 36 | 4.3 | ||
| Platinum doublet or triplet | 306 | 96.2 | 207 | 95 | 799 | 95.7 | ||
| Response at end of first-line chemotherapy* | 0.221 | 0.550 | ||||||
| Complete remission | 246 | 77.4 | 163 | 74.8 | 602 | 72.1 | ||
| Partial remission | 65 | 20.4 | 49 | 22.5 | 204 | 24.4 | ||
| Stable disease | 7 | 2.2 | 6 | 2.8 | 29 | 3.5 | ||
| Overall Survival (months) | 0.272 | 0.588 | ||||||
| Median | 66.99 | 50.99 | 51a–59b | |||||
| Progression-free survival (months) | 0.923 | 0.776 | ||||||
| Median | 12.8 | 13.04 | 12.4b-12.7a | |||||
3.2 Frequency of Alterations in Molecular EGFR Pathway Biomarkers and Clinical Outcome
Table 3 summarizes the correlation between biomarker results and both PFS and OS. The distribution of selected baseline patient characteristics between the molecular marker results are summarized in Online Resource 1.
Table 3.
Correlation of biomarker results with overall survival (OS) and progression-free survival (PFS)
| Biomarker | n | OS |
PFS |
||||
|---|---|---|---|---|---|---|---|
| Mos. | HR (95 % CI) | P | Mos. | HR (95 % CI) | P | ||
| KRAS | |||||||
| Wild-type | 309 | NR | 12.5 | ||||
| Mutation | 9 | 67.0 | 1.08 (0.43–2.69) | 0.876 | 14.1 | 0.90 (0.42–1.91) | 0.784 |
| PIK3CA | |||||||
| Wild-type | 306 | 67.0 | 12.4 | ||||
| Mutation | 12 | NR | 0.53 (0.17–1.65) | 0.262 | NR | 0.36 (0.15–0.86) | 0.017 |
| EGFR | |||||||
| Wild-type | 315 | NR | 12.9 | ||||
| Mutation | 3 | 37.9 | 1.80 (0.53–6.15) | 0.345 | 4.01 | 2.09 (0.67–6.54) | 0.195 |
| KRAS, NRAS, BRAF, PIK3CA or EGFR | |||||||
| Wild-type | 294 | NR | 12.3 | ||||
| Mutation | 24 | 67.0 | 0.75 (0.38–1.49) | 0.413 | 33.1 | 0.57 (0.33–0.99) | 0.042 |
| EGFR IHC | |||||||
| Negative | 125 | 51.0 | 11.7 | ||||
| Positive | 93 | 46.1 | 1.23 (0.85–1.78) | 0.273 | 13.0 | 1.03 (0.76–1.39) | 0.835 |
| EGFR FISH | |||||||
| Negative | 114 | 67.0 | 16.1 | ||||
| Positive | 66 | 46.1 | 1.56 (1.01–2.40) | 0.044 | 9.6 | 1.57 (1.11–2.22) | 0.010 |
| pAkt-IHC | |||||||
| Negative | 108 | 49.1 | 11.6 | ||||
| Positive | 27 | 53.7 | 0.89 (0.48–1.63) | 0.696 | 13.8 | 0.65 (0.39–1.09) | 0.100 |
| pMAPK-IHC | |||||||
| Negative | 70 | 67.0 | 15.7 | ||||
| Positive | 58 | 38.9 | 1.81 (1.11–2.97) | 0.016 | 8.1 | 1.42 (0.96–2.10) | 0.077 |
| E-cadherin-IHC | |||||||
| Negative | 64 | 67.0 | 10.5 | ||||
| Positive | 71 | 51.0 | 0.99 (0.61–1.62) | 0.970 | 13.1 | 0.87 (0.59–1.28) | 0.491 |
| Vimentin-IHC | |||||||
| Negative | 30 | NR | 13.8 | ||||
| Intermediate | 58 | 49.0 | 1.61 (0.78–3.30) | 13.1 | 0.88 (0.53–1.45) | ||
| Positive | 40 | 39.6 | 2.07 (0.99–4.34) | 0.139 | 9.1 | 1.18 (0.70–1.98) | 0.430 |
| HER2-FISH | |||||||
| Negative | 30 | 49.1 | 22.1 | ||||
| Equivocal | 23 | 51.0 | 1.07 (0.48–2.35) | 8.8 | 2.13 (1.13–4.01) | ||
| Positive | 53 | 67.0 | 0.97 (0.49–1.89) | 0.965 | 13.7 | 1.51 (0.87–2.63) | 0.061 |
CI confidence interval, FISH fluorescence in situ hybridization, HR hazard ratio, IHC immunohistochemistry, Mos. months, NR not reached, OS overall survival, PFS progression-free survival. Bold entries indicate statistically significant p values (P <0.05)
3.2.1 EGFR Protein Expression
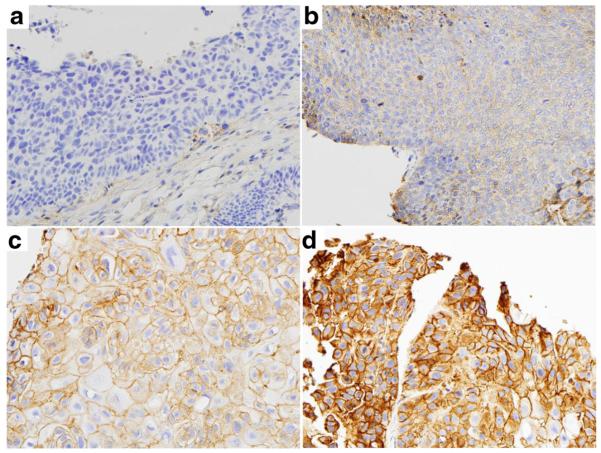
EGFR protein expression was evaluated by immunohistochemistry in 218 patients. Of these, 42.7 % had EGFR membranous staining (EGFR IHC-positive) (Fig. 1). EGFR protein expression had no impact on OS (HR: 1.23, P=0.273) or PFS (HR: 1.03, P=0.835).
Fig. 1.
EGFR immunohistochemical staining. A hybrid (H)-score was generated based on the fraction of staining cells in each intensity category [0 (no staining), 1+ (weak), 2+ (moderate), 3+ (strong)]. The H-score was calculated by completing the formula (% cells of 0 intensity × 0)+(% of 1+ intensity × 1)+(% of 2+ intensity × 2)+(% of 3+ intensity × 3), with the overall score ranging from 0 to 300. Panels illustrate specimens graded with score 0 (a), score 80 (low expression, b), score 170 (moderate expression, c) and score 280 (high expression, d) (image magnification 40x)
3.2.2 EGFR Gene Copy Number
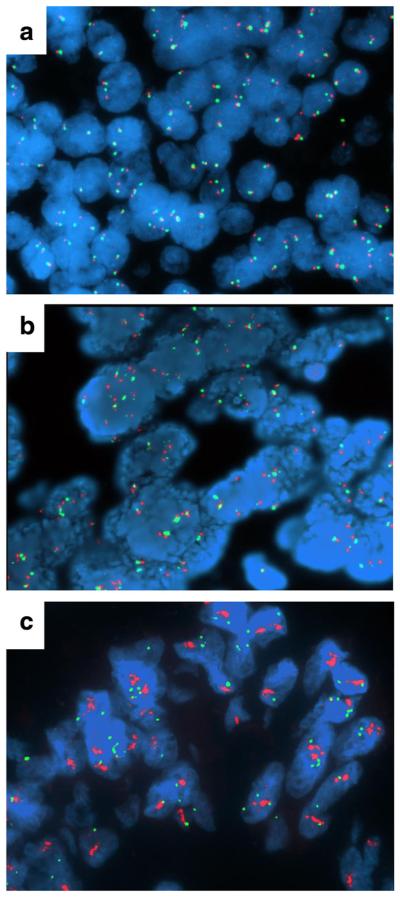
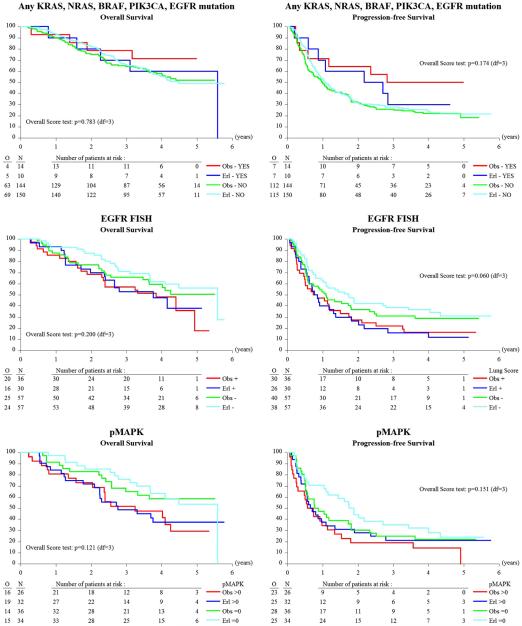
We assessed EGFR gene copy number by FISH in 180 patients. Patients with high polysomy and gene amplification categories were combined and designated EGFR FISH-positive, and all other categories were categorized as EGFR FISH-negative, as previously described [19]. Representative images of these two classes are shown in Fig. 2. FISH-positive patients represented 36.7 % of the total group, with 63.3 % FISH-negative. Next, we correlated EGFR FISH categories with clinical outcome (Fig. 3). Compared to FISH-negative patients, EGFR FISH-positive patients had statistically significantly shorter OS (median 46.1 versus 67.0 months, HR=1.56 [95 % CI 1.01-2.40], P=0.044) and shorter PFS (median 9.6 versus 16.1 months, HR = 1.57 [95 % CI1.11–2.22], P=0.010). However, EGFR gene copy number could not predict responsiveness to erlotinib (Fig. 4).
Fig. 2.
EGFR determined by fluorescence in situ hybridization. FISH was performed with the EGFR (red)/CEP7 (green) probe (Abbott Molecular, Des Plaines, IL, USA). Panels illustrate specimens representing low gain in gene copy number per cell (EGFR FISH-negative) (a), high (high polysomy=b; gene amplification=c) gain in gene copy number per cell (EGFR FISH-positive)
Fig. 3.
Kaplan-Meier curves for overall survival and progression-free survival. Data were analyzed according to presence of mutations (top), pMAPK immunohistochemistry (middle), and EGFR gene copy number (bottom)
Fig. 4.
Kaplan-Meier curves for overall survival and progression-free survival according to treatment allocation. Data were analyzed according to presence of mutations (top), pMAPK immunohistochemistry (middle), and EGFR gene copy number (bottom)
3.2.3 pAkt and pMAPK Expression by IHC
Immunohistochemical evaluation of pAkt and pMAPK was successful in 135 and 128 patients, respectively, with 27 cases (20 %) for pAkt and 58 cases (45.3 %) for pMAPK classified as positive. Positive pMAPK expression was seen mainly in serous tumors. While no effect of pAkt IHC positivity was seen on OS (HR: 0.89, P=0.696) or PFS (HR: 0.65, P=0.100), pMAPK IHC positivity was significantly associated with worse OS (HR: 1.81, P=0.016) (Fig. 3). pMAPK did not seem to have predictive value (Fig. 4).
3.2.4 Epithelial-Mesenchymal Transition (EMT) Status by IHC
Changes in EMT status including the expression of E-cadherin as epithelial marker and vimentin as mesenchymal marker were successfully evaluated by IHC in 135 and 128 patients, respectively. The protein expression of E-cadherin was considered high in 52.6 % of patients, while vimentin was categorized as low (≤30), intermediate (30–130), or high (>130). High protein expression of vimentin was observed in 31.3 %. Neither E-cadherin nor vimentin IHC correlated with outcome or were predictive (Table 3).
3.2.5 HER2 Gene Copy Number Status by FISH
HER2 FISH was assessable in 106 patients with available tumor tissue and classified as positive (>2.2) in 50 % of cases. No significant correlation between HER2 copy number status and outcome was observed (Table 3).
3.2.6 Mutation Analysis
Hotspot mutation profiling of 20, 2, 20, 33, 15, and 8 somatic mutations in KRAS, BRAF, NRAS, PIK3CA, EGFR, and PTEN, respectively, using Sequenom MassARRAY was technically successful for 318 cases (Online Resource 2). Overall, only 28 mutations were observed among 24 samples (7.5 %). Of the genes analyzed, PIK3CA and KRAS were the most frequently mutated. PIK3CA mutations were present in 3.8 % (12/318) of the samples, most of which were located in exon 9 (4/12; 33 %) or exon 20 (4/12; 33 %). KRAS mutations were detected in nine samples (2.8 %), nearly all within codon 12 (8/9, 88.9 %). EGFR mutations occurred in only three patients. EGFR re-sequencing was successful in 59 of 64 samples. The average coverage of EGFR exons for these samples was 590x. No additional somatic mutations were detected in any of the EGFR exons. Mutations in PTEN were not detected. Mutations occurred less frequently in serous cancers (7/206 [3 %] in serous versus 17/112 [15 %] in non-serous cancers; see Online Resource 1). The limited frequency of mutated samples in this analysis prevents a meaningful evaluation of clinical outcomes in relation to individual mutations. As such, patients with at least one mutation in the EGFR signaling cascades were pooled. Patients with at least one mutation in KRAS, NRAS, BRAF, PIK3CA, or EGFR had longer PFS (33.1 versus 12.3 months, mean difference=20.8 months, P=0.042) compared to those with wild-type tumors (Fig. 3), but among the former, there was no significant benefit of erlotinib over observation (Fig. 4). No significant difference in OS was seen when all mutations were pooled.
4 Discussion
Most data that have added to our understanding of molecular responsiveness to anti-EGFR agents have been derived from well-conducted trials in mCRC and NSCLC. To date, only limited data on EGFR pathway biomarkers are available for epithelial ovarian cancer and, even more specifically, in patients being treated with anti-EGFR therapies. The EORTC-GCG 55041 trial is the first randomized phase III trial investigating the use of maintenance erlotinib in patients with ovarian, peritoneal or fallopian tube cancer. In this project, we investigated the status of EGFR and related pathways using immunohistochemistry, FISH, hotspot mutation analysis, and DNA sequencing to determine the frequency of these alterations in patients with ovarian cancer and to correlate these biomarker data with outcome and with the efficacy of erlotinib.
In this study, EGFR overexpression was found with IHC in 39.4 % of patients, but we could not validate EGFR expression as a poor prognostic marker. The prognostic role of EGFR expression in EOC remains controversial. While some studies associate EGFR overexpression with poor clinical outcome [3, 5], others have shown no effect [6, 7]. However, we found a statistically significant correlation with PFS and OS for EGFR copy number status. Patients who were EGFR FISH-positive had worse OS (46.1 months) than those who were EGFR FISH-negative (67.0 months) (HR: 1.56; 95 % CI 1.01–2.40; P=0.044). The median PFS was 9.6 months he EGFR FISH-positive patients and 16.1 months for EGFR FISH-negative patients (HR: 1.57; 95 % CI 1.11–2.22; P=0.010). However, EGFR amplification was not predictive of erlotinib responsiveness, suggesting that EGFR copy number status may be a prognostic rather than predictive factor for erlotinib. Results reported by Lassus et al. also support the assumption that EGFR copy number serves as a prognostic biomarker in EOC, showing that an increased EGFR copy number was associated with shorter OS and PFS [53].
In contrast to mCRC and NSCLC, EGFR mutation did not predict responsiveness to erlotinib treatment, nor did gain-of-function mutations in the EGFR signaling cascades (KRAS, BRAF, NRAS, and PIK3CA). The limited frequency of mutated samples, however, prevents meaningful evaluation of clinical outcomes in relation to these mutations. However, when pooling all mutations together, patients with at least one mutation in either KRAS, NRAS, BRAF, PIK3CA or EGFR had a longer PFS (33.1 months) compared to those with wild-type tumors (12.3 months) (HR: 0.57; 95 % CI 0.33-0.99; P=0.042).
With the exception of pMAPK expression, which was associated with shorter overall survival, none of the investigated biomarkers demonstrated prognostic significance or were predictive of erlotinib efficacy in our translational study population. Some limitations to the study, however, must be taken into consideration. Although there was a compelling body of basic and preclinical evidence for launching this study, it failed to show a benefit of erlotinib over standard management (observation) [44], making it very difficult to evaluate predictive biomarkers. In addition, 26 % of the patients stopped erlotinib due to side effects. Moreover, obtaining a full analysis data set with complete results for all tissue samples submitted to molecular analysis remains a challenge. The different techniques often require fresh tissue or a sufficient amount of adequate tissue or tumor cells. Thus, tissue quality and technical performance may also significantly affect biostatistical results. Finally, EOC represents a genetically complex disease, rendering simple translation of the biomarker data difficult.
Although the EGFR pathway appears to play an important role in ovarian cancer, particularly in tumor development, tumor cell survival, and metastasis, it is not yet clear how this pathway can be exploited to yield a therapeutic benefit. In the future, rather than using an unselected patient population, it will be important to select patients based only on molecular characteristics for randomized studies investigating EGFR-targeted treatments. In the event that findings are similar to those in breast cancer for HER2 and trastuzumab or in NSCLC for EGFR and gefitinib, where increased copy number is a poor prognostic feature but a good predictor of response, EGFR copy number status may represent a criterion for selecting EOC patients for those clinical trials.
In conclusion, in this translational study on the EORTC-GCG 55041 trial, the presence of EGFR mutations or gain-of-function mutations in the EGFR signaling cascades (KRAS, BRAF, NRAS, and PIK3CA), increased EGFR copy number, and positive EGFR protein overexpression were not predictive of erlotinib efficacy. However, increased EGFR copy number seems to have a prognostic role. Therefore, EGFR FISH analysis may be interesting for further investigation as a criterion for selecting patients for prospective studies on erlotinib or other anti-EGFR therapies in EOC.
Supplementary Material
Acknowledgments
We thank all patients for their participation in the EORTC-GCG 55041 trial. In addition, the active commitment of the EORTC, the study coordinators, and investigators providing tumor tissue is gratefully acknowledged, as it enabled this translational study. We are grateful to Mrs. Katrien Drijkoningen and Mrs. Godelieve Verbist for their expert technical support. This publication was supported by the EORTC Charitable Trust and by an unrestricted educational grant from Roche. Evelyn Despierre was supported by a grant for the NOCI project “Pharmacogenomic and immunohistochemical evaluation of the EGFR pathway in ovarian cancer in patients treated with erlotinib: combined prognostic and predictive assessment on the EORTC 55041-study,” which was competitively selected during the EORTC Groups Annual Meeting in 2009. FISH analyses were supported in part by NCI grant CCSG P30 CA046934 to the University of Colorado Comprehensive Cancer Center.
We acknowledge all collaborators who actively contributed to this study, including C. Abraham (Hôpital André Mignot, Le Chesnay, France), F. Amant (University Hospital Leuven, Leuven, Belgium), R. Anderson (University of Colorado, Boulder, CO, USA), A. Azzedine (Centre Hospitalier, Avignon, France), C. Benedetto (University of Turin, Turin, Italy), G. Bertelli (Singelton Hospital, Swansea, UK), P. Berteloot (University Hospital Leuven, Leuven, Belgium), D. Berton-Rigaud (Centre René Gauducheau, Saint-Herblain, France), N. Biglia (University of Turin, Turin, Italy), N. Bonichon-Lamichhane (Clinique Tivoli, Bordeaux, France), P. Bougnoux (Centre Hospitalier Universitaire [CHU], Tours, France), E. Bourbouloux (Centre René Gauducheau, Saint-Herblain, France), C. Bourcier (Centre Hospitalier Départemental [CHD] Les Oudairies, La Roche-Sur-Yon, France), M. Buck (Sir Charles Gairdner Hospital, Nedlands, Australia), M. Campone (Centre René Gauducheau, Saint-Herblain, France), E.M. Canuto (University of Turin, Turin, Italy), A. Casado Herraez (Hospital Universitario San Carlos, Madrid, Spain), I. Cauvin (Centre Hospitalier, Chambéry, France), L. Chauvenet (Hôpital Hôtel-Dieu, Paris, France), A. Chevalier-Place (Centre Oscar Lambret, Lille, France), P.-H. Cottu (Institut Curie, Paris, France), J. Cretin (Clinique de Valdegour, Nîmes, France), I. Cumin (Centre Hospitalier de Bretagne Sud, Lorient, France), H. Curé (Institut Jean Godinot, Reims, France), F. Dalenc (Institut Claudius Régaud, Toulouse, France), S. Danese (University of Turin, Turin, Italy), A. Davis (The Canberra Hospital, Garran (woden), Australia), P. Debruyne (AZ St. Augustinus, Kortrijk, Belgium), G. Delplanque (Groupe Hospitalier Saint-Joseph, Paris, France), R. Delva (Centre Paul Papin, Angers, France), De Valk (Onze Lieve Vrouw Gasthuis, Amsterdam), V. D'Hondt (Hôpitaux Universitaires Jules Bordet), D. Dramais (Centre Hospitalier, Valence, France), X. Durando (Centre Jean Perrin, Clermont-Ferrand, France), C. El Kouri (Centre Catherine de Sienne, Nantes, France), C. Esteban (Hospital Virgen de la Salud, Toledo, Spain), M. Fabbro (Institut du Cancer de Montpellier Val d'Aurelle, Montpellier, France), C. Falandry (Centre Hospitalier Lyon Sud, Pierre-Bénite, France), B. Filleul (Hopital de Jolimont, Haine-Saint-Paul, Belgium), A. Floquet (Institut Bergonie, Bordeaux, France), P. Fumoleau (Centre Georges François Leclerc, Dijon, France), M. Garcia-Varella (University of Colorado, Boulder, CO, USA), C. Garnier (Groupe Hospitalier Mutualiste, Institut Daniel Hollard, Grenoble, France), E. Gilby (Royal United Hospital, Bath, UK), L. Gladieff (Centre Claudius Regaud, Toulouse, France), F. Goffin (Centre Hospitalier Regional de la Citadelle, Liége, Belgium), M.-C. Gouttebel (Centre Hospitalier, Valence, France), J.A. Green (Clatterbridge Cancer Centre NHS Foundation Trust, Bebington, Wirral, UK), J.-P. Guastalla (Centre Léon Bérard, Lyon, France), A.-C. Hardy-Bessard (Clinique Armoricaine de Radiologie, Saint-Brieuc, France), F. Hirsch (University of Colorado, Boulder, CO, USA), A. Hughes (Gateshead Hospitals, Queen Elizabeth Hospital, Gateshead, UK), D. Jaubert (Clinique Tivoli, Bordeaux, France), M.-C. Kaminsky (Centre Alexis Vautrin, Vandoeuvre-lés-Nancy, France), D. Katsaros (Clinica Universita, Torino, Italy), Frederic Kridelka (Centre Hospitalier Regional de la Citadelle, Liége, Belgium), R. Largillier (Centre Antoine Lacassagne, Nice, France), D. Lebrun-Jezekova (Institut Jean Godinot, Reims, France), B. Leduc (Centre Hospitalier, Brive-la-Gaillarde, France), M. Leheurteur (Centre Jean Perrin, Clermont-Ferrand, France), A. Lesoin (Centre Oscar Lambret, Lille, France), K. Leunen (University Hospital Leuven, Leuven, Belgium), N. Levasseur (Groupe Hospitalier Saint-Joseph, Paris, France), C. Leyronnas (Groupe Hospitalier Mutualiste, Institut Daniel Hollard, Grenoble, France), J.-F. Llory (Institut d'Oncologie Hartmann, Levallois Perret, France), A. Lortholary (Centre Catherine de Sienne, Nantes, France), F. Mayer (Centre Georges Francois Leclerc, Dijon, France), D. Mayeur (Hôpital André Mignot, Le Chesnay, France), C. Mendiola (Hospital Universitario 12 de Octubre, Madrid, Spain), L. Mignot (Institut Curie, Paris, France), J. Morgan (Ipswich Hospital NHS Trust, Ipswich, Suffolk, UK), M.-A. Mouret-Reynier (Centre Jean Perrin, Clermont-Ferrand, France), P. Neven (University Hospital Leuven, Leuven, Belgium), Paola Modaffari (Ospedale Mauriziano Umberto I, Torino, Italy),T. Petit (Centre Paul Strauss, Strasbourg, France),E. Picardo (University of Turin, Turin, Italy), J. Plaza (Centre Hospitalier Général, Montbelliard, France), M. Pluvio-Coronado (Universitario San Carlos Madrid, Madrid, Spain), F. Priou (CHD Les Oudairies, La Roche-Sur-Yon, France), E. Pujade-Lauraine (Hôpital Hôtel-Dieu, Paris, France), I. Ray Coquard (Centre Leon Berard, Lyon, France), N. Reed (NHS Greater Glasgow and Clyde Beatson West of Scotland Cancer Centre-Gartnavel General Hospital, Glasgow, UK), I. Rigault de la Longrais (University of Turin, Turin, Italy), S. Scholl (Institut Curie, Paris, France), I. Sillet-Bach (Centre Hospitalier, Brive-La-Gaillarde, France),C. Steer (Murray Valley Private Hospital, Wodonga, Australia), J. Summers (Mid Kent Oncology Centre, Maidstone, Kent, UK), V. Trillet-Lenoir (Centre Hospitalier Lyon Sud, Pierre-Bénite, France), P. Van Dam (AZ St. Augustinus, Wilrijk, Belgium), M.E.L. Van Der Burg (Erasmus Medical Center, Rotterdam, the Netherlands), E. Vanlerenberghe (Centre Oscar Lambret, Lille, France), J.-M. Vannetzel (Institut d'Oncologie Hartmann, Levallois Perret, France),I. Vergote (University Hospital Leuven, Leuven, Belgium), J.A. Vidart Aragon (Universitario San Carlos Madrid, Madrid, Spain), J. Waters (East Kent Hospitals University NHS Foundation Trust, Queen Elizabeth The Queen Mother Hospital, Margate, UK), B. Weber (Centre Alexis Vautrin, Vandoeuvre-Les-Nancy, France), G. Yazbek (Institut Jean Godinot, Reims, France), P. Zola (Ospedale Mauriziano Umberto I, Torino, Italy).
Footnotes
Conflict of Interest Marileila Varella-Garcia is co-inventor on a patent held by the University of Colorado to use EGFR copy number as biomarker for selection of lung cancer patients for targeted therapy. Evelyn Despierre, Ignace Vergote, Ryan Anderson, Corneel Coens, Dionyssios Katsaros, Fred R. Hirsch, Bram Boeckx, Annamaria Ferrero, Isabelle Ray-Coquard, Els MJJ Berns, Antonio Casado, Diether Lambrechts, and Antonio Jimeno declare no conflict of interest.
References
- 1.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358(11):1160–74. doi: 10.1056/NEJMra0707704. doi:10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 3.Skirnisdóttir I, Sorbe B, Seidal T. The growth factor receptors HER-2/neu and EGFR, their relationship, and their effects on the prognosis in early stage (FIGO I-II) epithelial ovarian carcinoma. Int J Gynecol Cancer. 2001;11(2):119–29. doi: 10.1046/j.1525-1438.2001.011002119.x. [DOI] [PubMed] [Google Scholar]
- 4.Fischer-Colbrie J, Witt A, Heinzl H, Speiser P, Czerwenka K, Sevelda P, Zeillinger R. EGFR and steroid receptors in ovarian carcinoma: comparison with prognostic parameters and outcome of patients. Anticancer Res. 1997;17(1B):613–9. [PubMed] [Google Scholar]
- 5.Berchuck A, Rodriguez GC, Kamel A, Dodge RK, Soper JT, Clarke-Pearson DL, Bast RC., Jr Epidermal growth factor receptor expression in normal ovarian epithelium and ovarian cancer. I. Correlation of receptor expression with prognostic factors in patients with ovarian cancer. Am J Obstet Gynecol. 1991;164(2):669–74. doi: 10.1016/s0002-9378(11)80044-x. [DOI] [PubMed] [Google Scholar]
- 6.Baekelandt M, Kristensen GB, Tropé CG, Nesland JM, Holm R. Epidermal growth factor receptor expression has no independent prognostic significance in advanced ovarian cancer. Anticancer Res. 1999;19(5C):4469–74. [PubMed] [Google Scholar]
- 7.Elie C, Geay JF, Morcos M, Le Tourneau A, Girre V, Broët P, Marmey B, Chauvenet L, Audouin J, Pujade-Lauraine E, Camilleri-Broët S, GINECO Group Lack of relationship between EGFR-1 immunohistochemical expression and prognosis in a multicentre clinical trial of 93 patients with advanced primary ovarian epithelial cancer (GINECO group) Br J Cancer. 2004;91(3):470–5. doi: 10.1038/sj.bjc.6601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon AN, Finkler N, Edwards RP, Garcia AA, Crozier M, Irwin DH, Barrett E. Efficacy and safety of erlotinib HCl, an epidermal growth factor receptor (HER1/EGFR) tyrosine kinase inhibitor, in patients with advanced ovarian carcinoma: results from a phase II multicenter study. Int J Gynecol Cancer. 2005;15(5):785–92. doi: 10.1111/j.1525-1438.2005.00137.x. [DOI] [PubMed] [Google Scholar]
- 9.Blank SV, Christos P, Curtin JP, Goldman N, Runowicz CD, Sparano JA, Liebes L, Chen HX, Muggia FM. Erlotinib added to carboplatin and paclitaxel as first-line treatment of ovarian cancer: a phase II study based on surgical reassessment. Gynecol Oncol. 2010;119(3):451–6. doi: 10.1016/j.ygyno.2010.08.008. doi:10.1016/j.ygyno.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirte H, Oza A, Swenerton K, Ellard SL, Grimshaw R, Fisher B, Tsao M, Seymour L. A phase II study of erlotinib (OSI-774) given in combination with carboplatin in patients with recurrent epithelial ovarian cancer (NCIC CTG IND.149) Gynecol Oncol. 2010;118(3):308–12. doi: 10.1016/j.ygyno.2010.05.005. doi:10.1016/j.ygyno.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Schilder RJ, Sill MW, Chen X, Darcy KM, Decesare SL, Lewandowski G, Lee RB, Arciero CA, Wu H, Godwin AK. Phase II study of gefitinib in patients with relapsed or persistent ovarian or primary peritoneal carcinoma and evaluation of epidermal growth factor receptor mutations and immunohistochemical expression: a Gynecologic Oncology Group Study. Clin Cancer Res. 2005;11(15):5539–48. doi: 10.1158/1078-0432.CCR-05-0462. [DOI] [PubMed] [Google Scholar]
- 12.Posadas EM, Liel MS, Kwitkowski V, Minasian L, Godwin AK, Hussain MM, Espina V, Wood BJ, Steinberg SM, Kohn EC. A phase II and pharmacodynamic study of gefitinib in patients with refractory or recurrent epithelial ovarian cancer. Cancer. 2007;109(7):1323–30. doi: 10.1002/cncr.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner U, du Bois A, Pfisterer J, Huober J, Loibl S, Lück HJ, Sehouli J, Gropp M, Stähle A, Schmalfeldt B, Meier W, Jackisch C, AGO Ovarian Cancer Study Group Gefitinib in combination with tamoxifen in patients with ovarian cancer refractory or resistant to platinum-taxane based therapy–a phase II trial of the AGO Ovarian Cancer Study Group (AGO-OVAR 2.6) Gynecol Oncol. 2007;105(1):132–7. doi: 10.1016/j.ygyno.2006.10.053. [DOI] [PubMed] [Google Scholar]
- 14.Pautier P, Joly F, Kerbrat P, Bougnoux P, Fumoleau P, Petit T, Rixe O, Ringeisen F, Carrasco AT, Lhommé C. Phase II study of gefitinib in combination with paclitaxel (P) and carboplatin (C) as second-line therapy for ovarian, tubal or peritoneal adenocarcinoma (1839IL/0074) Gynecol Oncol. 2010;116(2):157–62. doi: 10.1016/j.ygyno.2009.10.076. doi:10.1016/j.ygyno.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 15.Schilder RJ, Pathak HB, Lokshin AE, Holloway RW, Alvarez RD, Aghajanian C, Min H, Devarajan K, Ross E, Drescher CW, Godwin AK. Phase II trial of single agent cetuximab in patients with persistent or recurrent epithelial ovarian or primary peritoneal carcinoma with the potential for dose escalation to rash. Gynecol Oncol. 2009;113(1):21–7. doi: 10.1016/j.ygyno.2008.12.003. doi:10.1016/j.ygyno.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Secord AA, Blessing JA, Armstrong DK, Rodgers WH, Miner Z, Barnes MN, Lewandowski G, Mannel RS, Gynecologic Oncology Group Phase II trial of cetuximab and carboplatin in relapsed platinum-sensitive ovarian cancer and evaluation of epidermal growth factor receptor expression: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;108(3):493–9. doi: 10.1016/j.ygyno.2007.11.029. doi:10.1016/j.ygyno.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24):2519–29. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 18.Bristow RE, Tomacruz RS, Armstrong DK, Trimble EL, Montz FJ. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J Clin Oncol. 2002;20(5):1248–59. doi: 10.1200/JCO.2002.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 19.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn PA, Jr, Varella-Garcia M. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97(9):643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, Bunn PA, Jr, Franklin WA, Crowley J, Gandara DR, Southwest Oncology Group Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23(28):6838–45. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 21.Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L, Klimstra DS, Fridman D, Kelsen DP, Saltz LB. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23(9):1803–10. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 22.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 23.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 24.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, Barón AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798–807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 26.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6(5):279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 27.Cappuzzo F, Finocchiaro G, Rossi E, Jänne PA, Carnaghi C, Calandri C, Bencardino K, Ligorio C, Ciardiello F, Pressiani T, Destro A, Roncalli M, Crino L, Franklin WA, Santoro A, Varella-Garcia M. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19(4):717–23. doi: 10.1093/annonc/mdm492. [DOI] [PubMed] [Google Scholar]
- 28.Sartore-Bianchi A, Moroni M, Veronese S, Carnaghi C, Bajetta E, Luppi G, Sobrero A, Barone C, Cascinu S, Colucci G, Cortesi E, Nichelatti M, Gambacorta M, Siena S. Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol. 2007;25(22):3238–45. doi: 10.1200/JCO.2007.11.5956. [DOI] [PubMed] [Google Scholar]
- 29.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–12. doi: 10.1200/JCO.2008.18.0786. doi:10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 30.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97(8):1139–45. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 32.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9(10):962–72. doi: 10.1016/S1470-2045(08)70206-7. doi:10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 33.Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–9. doi: 10.1200/JCO.2010.33.5091. doi:10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 34.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13(10):2890–6. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 35.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, Settleman J, Haber DA. The T790M "gate-keeper" mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7(4):874–9. doi: 10.1158/1535-7163.MCT-07-2387. doi:10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 36.Cappuzzo F, Ligorio C, Toschi L, Rossi E, Trisolini R, Paioli D, Magrini E, Finocchiaro G, Bartolini S, Cancellieri A, Hirsch FR, Crino L, Varella-Garcia M. EGFR and HER2 gene copy number and response to first-line chemotherapy in patients with advanced non-small cell lung cancer (NSCLC) J Thorac Oncol. 2007;2(5):423–9. doi: 10.1097/01.JTO.0000268676.79872.9b. [DOI] [PubMed] [Google Scholar]
- 37.Daniele L, Macrí L, Schena M, Dongiovanni D, Bonello L, Armando E, Ciuffreda L, Bertetto O, Bussolati G, Sapino A. Predicting gefitinib responsiveness in lung cancer by fluorescence in situ hybridization/chromogenic in situ hybridization analysis of EGFR and HER2 in biopsy and cytology specimens. Mol Cancer Ther. 2007;6(4):1223–9. doi: 10.1158/1535-7163.MCT-06-0719. [DOI] [PubMed] [Google Scholar]
- 38.Cappuzzo F, Varella-Garcia M, Shigematsu H, Domenichini I, Bartolini S, Ceresoli GL, Rossi E, Ludovini V, Gregorc V, Toschi L, Franklin WA, Crino L, Gazdar AF, Bunn PA, Jr, Hirsch FR. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. J Clin Oncol. 2005;23(22):5007–18. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 39.Hirsch FR, Varella-Garcia M, Cappuzzo F. Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene. 2009;28(Suppl 1):S32–7. doi: 10.1038/onc.2009.199. doi:10.1038/onc.2009.199. [DOI] [PubMed] [Google Scholar]
- 40.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, Ercan D, Rogers A, Roncalli M, Takeda M, Fujisaka Y, Philips J, Shimizu T, Maenishi O, Cho Y, Sun J, Destro A, Taira K, Takeda K, Okabe T, Swanson J, Itoh H, Takada M, Lifshits E, Okuno K, Engelman JA, Shivdasani RA, Nishio K, Fukuoka M, Varella-Garcia M, Nakagawa K, Jänne PA. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3(99):99ra86. doi: 10.1126/scitranslmed.3002442. doi:10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A, Saletti P, De Dosso S, Spitale A, Tejpar S, Kalogeras KT, Mazzucchelli L, Frattini M, Cappuzzo F. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108(3):668–75. doi: 10.1038/bjc.2013.4. doi:10.1038/bjc.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr, Raben D. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol Cancer Ther. 2007;6(6):1683–91. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- 43.Uramoto H, Iwata T, Onitsuka T, Shimokawa H, Hanagiri T, Oyama T. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res. 2010;30(7):2513–7. [PubMed] [Google Scholar]
- 44.Vergote IB, Jimeno A, Joly F, Katsaros D, Coens C, Despierre E, Marth C, Hall M, Steer CB, Colombo N, Lesoin A, Casado A, Reinthaller A, Green J, Buck M, Ray-Coquard I, Ferrero A, Favier L, Reed NS, Curé H, Pujade-Lauraine E. Randomized phase III study of erlotinib versus observation in patients with no evidence of disease progression after first-line platin-based chemotherapy for ovarian carcinoma: a European Organisation for Research and Treatment of Cancer-Gynaecological Cancer Group, and Gynecologic Cancer Intergroup study. J Clin Oncol. 2014;32(4):320–6. doi: 10.1200/JCO.2013.50.5669. doi:10.1200/JCO.2013.50.5669. [DOI] [PubMed] [Google Scholar]
- 45.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 46.Vergote I, Rustin GJ, Eisenhauer EA, Kristensen GB, Pujade-Lauraine E, Parmar MK, Friedlander M, Jakobsen A, Vermorken JB. Re: new guidelines to evaluate the response to treatment in solid tumors [ovarian cancer]. Gynecologic Cancer Intergroup. J Natl Cancer Inst. 2000;92(18):1534–5. doi: 10.1093/jnci/92.18.1534. [DOI] [PubMed] [Google Scholar]
- 47.Varella-Garcia M. Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006;1:19. doi: 10.1186/1746-1596-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. Database issue. doi:10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. doi:10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21(6):961–73. doi: 10.1101/gr.112326.110. doi:10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, Van Loo P, Van Den Bossche M, Catthoor K, Sabbe B, Despierre E, Vergote I, Hilbush B, Lambrechts D, Del-Favero J. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol. 2011;30(1):61–8. doi: 10.1038/nbt.2053. doi:10.1038/nbt.2053. [DOI] [PubMed] [Google Scholar]
- 53.Lassus H, Sihto H, Leminen A, Joensuu H, Isola J, Nupponen NN, Butzow R. Gene amplification, mutation, and protein expression of EGFR and mutations of ERBB2 in serous ovarian carcinoma. J Mol Med (Berl) 2006;84(8):671–81. doi: 10.1007/s00109-006-0054-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.