Abstract
Mechanism-based inactivation (MBI) of CYP450 enzymes is a unique form of inhibition in which the enzymatic machinery of the victim is responsible for generation of the reactive metabolite. This precondition sets up a time-dependency for the inactivation process, a hallmark feature that characterizes all MBI. Yet, MBI itself is a complex biochemical phenomenon that operates in different modes, namely covalent binding to apoprotein, covalent binding of the porphyrin group, and also complexation of the catalytic iron. Using lapatinib as a recent example of toxicological interest, we present an example of a mixed-function MBI that can confound clinical drug-drug interactions manifestation. Lapatinib exhibits both covalent binding to the apoprotein, and formation of a metabolite-intermediate (MI) complex in an enzyme-selective manner (CYP3A4 versus CYP3A5), each with different reactive metabolites. The clinical implication of this effect is also contingent upon genetic polymorphisms of the enzyme involved as well as the co-administration of other substrates, inhibitors or inducers, culminating in drug-drug interactions. This understanding recapitulates the importance of applying isoform-specific mechanistic investigations to develop customized strategies to manage such outcomes.
Section 1: Introduction to mechanism-based inactivation of CYP450
Mechanism-based inactivation (MBI) is a unique phenomenon in drug metabolism with widespread implications in pharmacology, toxicology and therapeutics; yet, it is frequently misinterpreted. A common reductionistic view of MBI is that of “suicide inhibition” of enzymes. This term describes the action of a substrate binding irreversibly to the target enzyme, leading to permanent inhibition of its enzymatic function. In this process, the substrate is consumed, thereby representing a “kamikaze” act of suicide. However, the essence of MBI is characterized by an additional metabolic conversion of the substrate that utilizes the intrinsic enzymatic function of the host enzyme. The substrate gains chemical reactivity through this bioactivation, which subsequently primes itself for irreversible binding to the enzyme. For this reason, the time-dependent nature of MBI possesses a unique kinetic dimension (Riley et al., 2007); the longer the exposure of the mechanism-based inactivator to the enzyme, the greater the extent of inhibition. Separately, the prerequisite of bioactivation of the substrate to form a reactive metabolite before MBI can take place is dependent on the presence of drug metabolizing enzymes. This effect, therefore, carries an unequivocal significance in the context of drug therapy and drug safety among different types of inhibitors.
Various subtypes of MBI are caused by the different moieties in drug metabolizing enzymes that are amenable to irreversible binding and inhibition. This subject has been thoroughly reviewed in recent years and will be only briefly mentioned here (Ortiz de Montellano, 2005; Masubuchi and Horie, 2007). Intuitively, all elements contributing to the active site biochemistry can be targeted to achieve an irreversible disruption of the enzyme. This includes (1) covalent modification of key amino acids in the apoprotein by the generated reactive metabolites, especially those residues carrying nucleophilic side chains like cysteine, lysine and glutamine (in some literature, this is simply referred to as MBI); (2) alkylation or degradation of the porphyrin ring of the heme group; (3) a quasi-irreversible binding (i.e. tight, but reversible in vitro) to the prosthetic heme iron via a coordinate bond or a metabolite-intermediate (MI) complex. Each of these mechanisms sets up a unique biochemical signature that can be experimentally differentiated; while all three subtypes demonstrate time-dependent inhibition, alkylation of the porphyrin ring tends to generate a reduction in the carbon monoxide (CO)-CYP450 differential spectrum (Rock, 2010), while formation of an MI complex produces a characteristic spectrum in the Soret region (400–500 nm), which can be reversed by the addition of ferricyanide (Riley et al., 2007). Regardless of these subtle differences, these inactivation processes arise from the initial metabolism of the substrate, alluding to a profound specificity and an increased risk that the resulting reactive metabolite will bind within the active site (Ortiz de Montellano, 2005).
Nonetheless, MBI of CYP450 enzymes can cause extensive toxicological concerns (Masubuchi and Horie, 2007). Firstly, drug-drug interactions are a major clinical consequence of inactivation. Dysregulation of CYP450 enzymes such as CYP3A4, the main isoform responsible for the metabolism of multiple drugs, can have tremendous impact on the accumulation and disposition of the inactivator and of any co-administered drugs that are also substrates or inhibitors of the same enzyme. Such interactions are not limited to drugs alone, and food-drug interactions are also well documented. Bergamottin, found in grapefruit juice, is a stereotypical mechanism-based inactivator of CYP3A4 and was recently shown to adduct a glutamine residue in the active site (Lin et al., 2012). By virtue of CYP3A4’s wide range of substrates and the predominance of this enzyme in the liver, the effect of bergamottin on co-administered drugs is is clinically relevant. Secondly, the covalently modified CYP450 enzymes can occasionally trigger immune-mediated hypersensitivity as a secondary toxicological effect. Tienilic acid is a classic example, being a mechanism-based inactivator for CYP2C9 (Lopez-Garcia et al., 1994). The onset of hepatitis with tienilic acid intake was found to be associated with an increase in anti-liver-kidney-microsome antibodies (Homberg et al., 1984; Beaune et al., 1987) presumably arising from an immunogenic response towards the modified CYP2C9. Finally, MBI has been shown to trigger heme degradation, causing the release of heme precursors and breakdown products into the system (Marks et al., 1988). Acute porphyria has been reported to occur as a consequence of exposure to mechanism-based inactivators such as carbamazepine (Thunell et al., 2007). Intuitively, these associations of MBI with toxicity underpin the bioactivation of substrate as a molecular trigger for the adverse outcomes. For this reason, MBI can be considered as a biomarker for reactive metabolite formation. In some companies, the observation of MBI motivates further efforts to detect reactive metabolite in drug candidates (disclosed by personal communication).
Evidently, the toxicological effects of MBI arise from molecular mechanisms that are independent from the intended pharmacological action of the drugs. Due consideration about the possibility of MBI should, therefore, be applied for all small molecule drugs, particularly if they have been shown to be extensively metabolized. In this review, we will showcase an example of a relatively new tyrosine kinase inhibitor, lapatinib, on which our group, and other laboratories, has recently gathered evidence and mechanistic insights into how it acts as an MBI. This example unveils a growing complexity of the subject as lapatinib displays a dichotomous mode of inactivation, depending on which isoform of CYP450 is involved, and is further exacerbated by a potential effect of genetic variations on the inhibition. The knowledge gained from this exercise will chart the way for investigating other drugs with similar propensities.
Section 2: Pharmacology and toxicology of lapatinib
Lapatinib is the very first orally available EGFR/HER2 dual kinase inhibitor approved by the FDA. It is now used as a first line treatment for HER2-positive breast cancers or metastatic breast cancer patients in combination with capecitabine or letrozole (Gomez et al., 2008; Jelovac and Wolff, 2012). While lapatinib has demonstrated success in the pharmacotherapy of breast cancer, several issues remain in its clinical use. As an effective drug for managing the disease, patients can be put on lapatinib for long term treatment, which elevates the concern of toxicity arising from chronic exposure. Reports of hepatotoxicity surfaced, including incidences of fatalities, which prompted a black box warning label (European Medicines Agency Website http://www.emea.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/000795/WC500044960.pdf) since 2008. There is a delayed onset of the toxicities which occurs at about 6–8 weeks after the initiation of therapy (Teo et al., 2012). While high grade toxicity is rare, a larger subset of the population (10%) may experience notable transient transaminitis according to a recent report (Azim et al., 2013).
From these clinical observations, lapatinib-induced hepatotoxicity carries some classical features of idiosyncratic toxicity (Uetrecht, 2007). It occurs rarely, is seen at high doses (1.25 to 1.5 g per day), and may not be linked to the mechanism of action for the drug (trastuzumab, the anti-HER2 monoclonal antibody, does not exhibit a similar toxicity profile). Moreover, studies by Spraggs et al. revealed a strong association to an HLA allele (DQA1*02:01) and invoked the role of immune-mediated response for sub-population susceptibility (Spraggs et al., 2011). Putting all these observations together suggests that the generation of a reactive intermediate during the disposition of lapatinib can be a susceptibility factor. This potential toxicological implication set off a series of experiments to expound on this possibility.
Section 3: MBI of CYP3A5
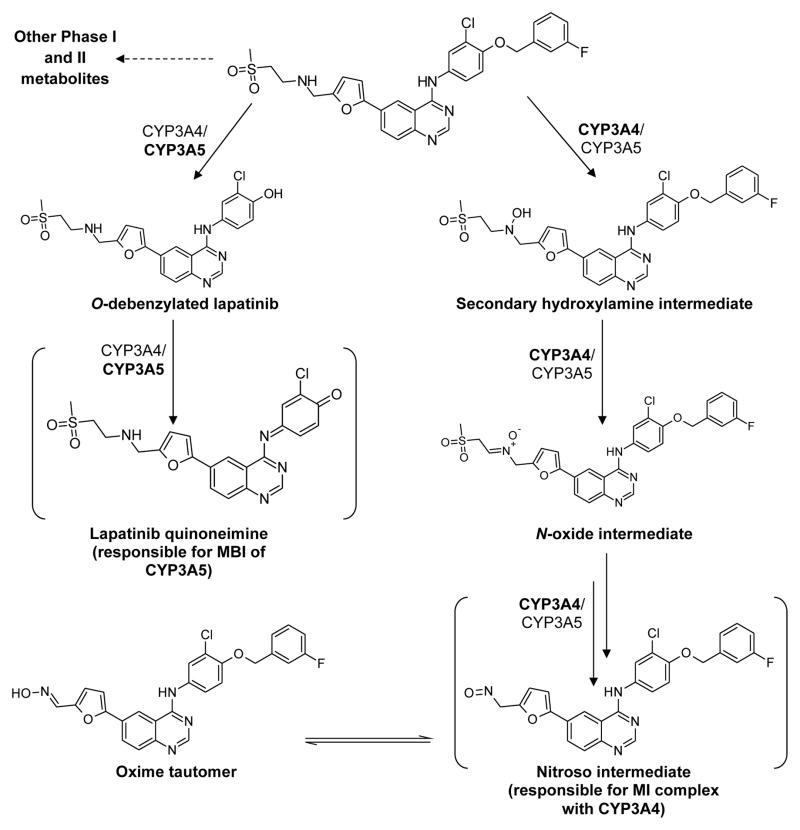
According to the literature, lapatinib is extensively metabolized by multiple CYP450 enzymes including CYP3A4/5, CYP2C8 and CYP2C18 (Frampton, 2009). Yet, the contribution of each enzyme to the generation of specific metabolites remains elusive. In characterizing the disposition of lapatinib, it was demonstrated that lapatinib undergoes metabolism via two key pathways: O-debenzylation and N-hydroxylation (Figure 1). O-Debenzylation is catalyzed predominantly by CYP3A5; while N-hydroxylation is catalyzed primarily by CYP3A4 (Teng et al., 2010; Castellino et al., 2012; Barbara et al., 2013).
Figure 1.
Summary of the multiple metabolism pathways and MBI of lapatinib. All metabolites except for the putative quinoneimine and nitroso intermediates have been directly detected through LC/MS analysis. Only the intermediates directly responsible for inactivation of the respective enzymes are displayed. The predominant biotransforming enzyme is indicated in bold.
A consequence of O-debenzylation of lapatinib is the unmasking of a p-hydroxyaniline chemical feature, similar to that found in acetaminophen, that is susceptible to oxidation to form a reactive quinoneimine species (Figure 1). This metabolite is analogous to the N-acetyl-p-benzoquinoneimine (NAPQI) generated by CYP2E1 from acetaminophen. Indeed, recombinant CYP3A4 and CYP3A5 are both capable of oxidizing O-debenzylated lapatinib to a quinoneimine, which has been indirectly detected as a glutathione (GSH) adduct (Hardy, 2014). Furthermore, CYP3A5 undergoes time-, concentration- and NADPH-dependent inactivation by lapatinib. Coupling these observations with the detection of a lapatinib-GSH adduct suggests that the putative reactive metabolite responsible for the inactivation is the quinoneimine via covalent binding to the active site (Teng et al., 2010). Interestingly, substrate-specific inactivation was observed when testosterone, but not midazolam, was used as a probe substrate, indicating that lapatinib covalently binds to a specific region of the large active site of CYP3A5, uniquely blocking the binding of testosterone but not midazolam (Barbara et al., 2013). Further studies to characterize lapatinib provide mechanistic evidence that the inactivation is mechanism-based: enzyme activity does not recover post-dialysis, a Soret peak is lacking and the inactivation is irreversible when potassium ferricyanide is added (Barbara et al., 2013). However, and unusually for MBI, the addition of an exogeneous nucleophile (GSH) is able to reverse the inactivation, suggesting that at least some of the reactive quinoneimine metabolite can exit the active site and interact with other nucleophiles (Teng et al., 2010). Although contrary to the current dogma of MBI, this is not unprecedented. For instance, CYP2E1 is covalently inactivated by the acetaminophen regioisomer 3′-hydroxyacetanilide in a time-dependent manner, and yet the addition of exogeneous GSH protects CYP2E1 from inactivation (Harrelson et al., 2012).
Despite being able to generate the quinoneimine, CYP3A4 as opposed to CYP3A5, does not covalently bind with the quinoneimine, as evidenced by the absence of lapatinib adducts to the apoprotein or heme of CYP3A4 in intact protein studies (Takakusa et al., 2011). This lends further credence to the idea that the quinoneimine can leave the active site without inactivating the enzyme. We believe that the topology of the enzyme active site plays a critical role in determining whether covalent binding by a reactive intermediate occurs. Since CYP3A4 and CYP3A5 share approximately 83% homology (Takakusa et al., 2011), it is conceivable that the remaining 17% difference could account for the differential presentation of the quinoneimine intermediate to CYP3A4 or CYP3A5, translating to different manifestations of inactivation. The site of adduction of lapatinib to CYP3A5 is currently under investigation in our laboratory, and these studies should yield insights into enzyme-substrate interactions for the CYP3A family. Two additional O-debenzylated metabolites, hydroxylated at the alkylamine chain or quinazoline ring were reported by Barbara et al. and Castellino et al. respectively (Castellino et al., 2012; Barbara et al., 2013), and these metabolites are being examined for their potential to form quinoneimines and their contributions to the reported inactivation.
Section 4: MI Complex formation with CYP3A4
Since lapatinib does not form a covalent adduct with CYP3A4, the drug’s time-, concentration- and NAPDH-dependent inactivation of CYP3A4 occurs via a different mechanism. N-Hydroxylation and/or N-dealkylation of lapatinib by CYP3A4 were proposed mechanisms that would generate a nitroso reactive metabolite with the ability to form an MI complex via a strong coordinate bond that is virtually irreversible under physiological conditions (Takakusa et al., 2011). Barbara et al. further clarified that the pathway to nitroso and MI complex formation is via formation of a secondary hydroxylamine, rather than via the traditional pathway involving N-dealkylation and the formation of a primary hydroxylamine (Barbara et al., 2013) (Figure 1). In contrast to the inactivation observed with CYP3A5, a clear Soret peak and reversibility of the inactivation with potassium ferricyanide substantiated the role of an MI complex in the inactivation process (Takakusa et al., 2011; Barbara et al., 2013). The evidence supporting MBI of CYP3A5 and formation of the CYP3A4 MI complex is summarized in Table 1. Collectively, the strength of the inactivation by lapatinib determined in vitro using human liver microsomes gives a kinact/KI value of around 0.012 μM−1min−1 (Teng et al., 2010), which is considered to be a moderately strong inactivator compared to other potent inactivators such as paroxetine, which inactivates CYP2D6 with a kinact/KI value of around 0.21 μM−1min−1, or ritonavir, which inactivates CYP3A4 with a kinact/KI value of 1.18 μM−1min−1(Obach et al., 2007).
Table 1.
Summary of mechanistic studies performed with recombinant CYP3A4 and CYP3A5 (Takakusa et al., Barbara et al., Chan et al.)
| Mechanistic studies | CYP3A4 | CYP3A5 | Interpretation |
|---|---|---|---|
| GSH trapping | Yes | Yes | Indicates formation of electrophilic metabolite capable of covalent binding |
| GSH protection from inactivation | Data not available | Yes | Indicates electrophilic metabolite release from active site |
| Reversal of inactivation via dialysis | Data not available | No | Indicates non-covalent inactivation, likely via MI complex of the heme iron |
| Formation of Soret peak | Yes | No | Indicates coordination of heme iron through a metabolite-intermediate complex |
| Reduced CO difference spectroscopy | Yes | Yes | Decrease in absorbance at 450 nm indicates alkylation of heme or adduction of the apoprotein vicinal to the heme |
| Reversal of inactivation by ferricyanide | Yes | No | Reversal indicates a non-covalent inactivation; Absence of reversal indicates covalent adduction |
| Mass shift of intact protein by mass spectrometry | No | Data not available | Indicates covalent binding to apoprotein or heme |
Overall, the differential effects of lapatinib on CYP3A4 and CYP3A5 is an interesting case study that suggests that despite the extent of homology and shared substrate specificity between these enzymes, they may interact distinctly, with preference for a particular pathway, presumably because of the different conformations and orientations a particular substrate can adopt within the active site of the enzymes. Such behavior may not be anomalous, as other tyrosine kinase inhibitors exhibit differential inhibition of CYP3A4 and CYP3A5. For example, pre-incubation of 10 μM imatinib with NADPH and recombinant CYP3A4 enhances the inhibition of midazolam hydroxylation but does not show any effect on CYP3A5 (Filppula et al., 2012). Moving forward, unambiguous determination of the differential inhibition of CYP450 enzymes could be aided by the development of more-selective substrates and inhibitors to be used together with liver microsomes. While recombinant enzymes permit mechanistic investigations on a single CYP450 isoform, they do not represent a holistic and integrated system such as human liver microsomes. Recent work in this area has yielded a CYP3A4 selective inactivator, CYP3cide (Walsky et al., 2012), a CYP3A4 selective probe substrate, bufalin (Ge et al., 2013) and a CYP3A5 selective probe substrate, T-5 (Li et al., 2014), all of which provide additional tools to investigators to aid in the delineation of the differential interactions of these two enzymes with pharmacological compounds in complex systems such as liver microsomes. We are hopeful that, eventually, sufficient information will be accumulated to allow construction of computational models for in silico docking studies to accurately predict the interaction of candidate compounds with these enzymes at the discovery stage, permitting the design of structural features to limit undesirable interactions that may precipitate toxicity or drug-drug interactions.
Section 5a: Potential clinical implications of CYP3A inactivation by lapatinib: effect of concomitant administration of CYP3A inhibitors, inducers or substrates
The complexity of CYP3A inactivation by lapatinib can be further aggravated in clinical circumstances of drug-drug interaction, where the effect of lapatinib inhibition of the enzyme is perturbed by other substrates or inhibitors. Two opposing scenarios can emerge: lapatinib acting as the “perpetrator” of enzyme inactivation affecting the mass balance of the second drug or metabolite, or lapatinib as the “victim”. Teo et al. reported that concomitant use of lapatinib with the CYP3A4 inducer dexamethasone correlated with an increased incidence of hepatotoxicity in metastatic breast cancer patients compared to patients treated with lapatinib alone. Dexamethasone was also shown to increase the cytotoxicity of lapatinib in TAMH cells (transforming growth factor α mouse hepatocytes) (Teo et al., 2012). Moreover, studies were undertaken using the human hepatoma cell line HepaRG to further examine the link between CYP3A4 induction and generation of reactive, potentially toxic metabolites of lapatinib. Induction of CYP3A4 by prototypical inducers, such as dexamethasone and rifampin, potentiated the cytotoxicity of lapatinib, which was correlated with increased formation of O-debenzylated lapatinib and cysteine adducts of the putative quinoneimine metabolite. Furthermore, the O-debenzylated metabolite of lapatinib was more cytotoxic to HepaRG cells than was lapatinib itself (Hardy et al., 2014). For the opposing scenario, CYP3A inducers such as carbamazepine reduce lapatinib exposure by increasing its clearance, necessitating dose adjustments, and conversely inhibitors such as ketoconazole increases lapatinib exposure (Smith et al., 2009). Collectively, these findings suggest that clinical use of lapatinib with CYP3A4 inducers or inhibitors may put patients at increased risk for drug toxicity and/or diminished therapeutic effects.
There is an emerging interest in investigating combinatorial therapy between lapatinib and other breast cancer agents to augment clinical efficacy or overcome resistance to breast cancer treatment, given the heterogeneous nature of breast cancer. This has pertinent implications, particularly when the co-administered drug is a CYP3A substrate. Clinical examples of such interactions include the CYP3A substrates vinorelbine and pazopanib for which decreased clearance was documented, and increased SN-38 exposure after irinotecan administration when these drugs were co-administered with lapatinib in Phase I trials (Midgley et al., 2007; Rezai et al., 2011; de Jonge et al., 2013). While pharmacokinetic studies of such intentional co-administration permit prospective dose adjustments to arrest such interactions, in other cases of polypharmacy the presence of potential interactions may not be immediately obvious and can lead to unintended toxicities or reduced efficacy.
Section 5b: The effect of genetic variability on CYP3A inactivation by lapatinib
In view of the potential clinical implication of CYP3A inactivation by laptinib, genetic variability of CYP3A becomes a relevant consideration. In addition to different binding modes of lapatinib for CYP3A4 versus CYP3A5 active sites, another factor that complicates the drug’s metabolism and MBI behavior is the extent of genetic polymorphism found in the general population. This remains an area of continued investigation. While CYP3A4 is consistently the major CYP450 expressed in adult human liver and small intestine (Guengerich, 1999), CYP3A5’s disposition is highly variable because of polymorphic expression (Kuehl et al., 2001; Lamba et al., 2002). Individuals with the CYP3A5*1 allele express high levels of the functional protein, whereas the CYP3A5*3 variant allele results in low to undetectable levels of CYP3A5. The frequency of having at least one CYP3A5*1 allele is 10–30% in Caucasians, 30–40% in Asians of different ethnic groups, and 50–70% in African-Americans (Kuehl et al., 2001; Lamba et al., 2002). In CYP3A5*1 carriers, CYP3A5 may represent up to 50% of the total CYP3A content and contribute significantly to the overall CYP3A catalytic activity (Kuehl et al., 2001).
This inherent variability draws attention to the impact of CYP3A5 polymorphism in the bioactivation of lapatinib, and consequently, the reciprocal inactivation of the enzyme through MBI. This may have important implications for the identification of potential patient-specific factors that may increase the risk of idiosyncratic lapatinib-induced hepatotoxicity as well as drug-drug interactions. This potential was evaluated recently in a two-stage study that aimed to ascertain the effect of polymorphism on metabolism and reactive metabolite formation, and, therefore, its effect on inactivation of the drug metabolizing enzymes. First, the kinetic parameters of lapatinib O-debenzylation were examined using cDNA-expressed CYP450 enzymes (Supersomes™) (BD Biosciences, Woburn, MA) and individual genotyped human liver microsomal preparations. Both CYP3A4 and CYP3A5 Supersomes™ turned over lapatinib rapidly and with high affinity, albeit CYP3A4 exhibited an overall 2.5-fold higher catalytic efficiency (Vmax/Km) than CYP3A5. However, GSH trapping experiments unexpectedly revealed that CYP3A5 Supersomes™ were seven times more efficient than CYP3A4 for generation of GSH adducts from lapatinib. These observations accentuated the different binding modes of lapatinib between CYP3A4 and CYP3A5 and, hence, revealed a different extent of O-debenzylated metabolite and quinoneimine reactive metabolite formation. In an orthogonal experiment using individually genotyped human liver microsomal preparations, the rates of O-debenzylation of lapatinib were comparable between carriers of the CYP3A5*1 wild-type allele and CYP3A5 non-expressers (CYP3A5*3/*3). On the other hand, the generation of GSH adducts was significantly higher (P= 0.03) in human liver microsomes from CYP3A5*1 expressers than in those from CYP3A5*3/*3 livers (Hardy, 2014). These data show that CYP3A5 is a quantitatively important contributor to hepatic lapatinib bioactivation in vitro. Further studies are required to elucidate the quantitative significance of the CYP3A5 polymorphism in lapatinib bioactivation in vivo.
While CYP3A5 appears to play a significant role in the generation of the quinoneimine reactive intermediate, elucidating the effect of this intermediate further downstream remains crucial, in particular on the magnitude of CYP3A inactivation by lapatinib. To ascertain the influence of CYP3A5 polymorphism on the inactivation as measured by kinact/KI values, inactivation studies on a panel of 12 human liver microsomal preparations genotyped for their CYP3A5 expression status were carried out. By using testosterone 6β-hydroxylation as a marker of remaining CYP3A activity, a clear trend, albeit not one of statistical significance, of higher mean kinact/KI values in CYP3A5*1/*1 genotypes was observed at nearly two-fold higher than CYP3A5*3/*3 carriers (manuscript in preparation). In other words, CYP3A5*1/*1 carriers experienced more potent CYP3A inactivation than did CYP3A5*3/*3 carriers. This result was corroborated by a similar trend of higher levels of GSH adduct formation derived from lapatinib in CYP3A5*1/*1 genotypes in the same panel of 12 donors (Hardy, 2014). The author hypothesized that because CYP3A5*1/*1 carriers possess a higher baseline CYP3A activity than do CYP3A5*3/*3 carriers, inactivation by lapatinib effects a quantitatively greater decrease in CYP3A activity among the CYP3A5*1/*1 carriers. This gives rise to steeper inactivation curves, whereby at saturation, CYP3A5*1/*1 carriers have higher kinact values and correspondingly larger kinact/KI ratios. This postulation is supported by the significant correlation between the kinact/KI ratios and GSH adduct formation with baseline testosterone 6β-hydroxylation values of each donor. Despite the limitations of a small sample size (n=12), this pertinent finding illustrates the need for further investigations into inactivators of other polymorphically-expressed CYP450 enzymes, for example paroxetine with CYP2D6, to define a potential gene-response relationship determining inter-individual susceptibility to inactivation.
Inasmuch as CYP450 polymorphisms have been demonstrated to affect the pharmacological activity of a xenobiotic, they may also modulate the susceptibility of an individual to inactivation and its clinical consequences. Using lapatinib as an example, the work by Chan et al. marks the first description of such potential impact of CYP450 polymorphic expression on this phenomenon (manuscript in preparation). This finding extends our understanding of the clinical impact of lapatinib-induced CYP3A inactivation, in which drug-drug interaction potential is not only bystander-drug-specific, but possibly patient-specific. While it may be tempting to suggest a blanket ban on co-administering CYP3A substrates with lapatinib, such an approach may unnecessarily deprive patients of vital therapeutic options. Clearly, while clinical caution is warranted in light of such findings, these observations reiterate the importance of careful investigation of the complex interplay between inactivator, substrate and patient characteristics and suggest that patient stratification on the basis of CYP450 genetic background may further refine pharmacotherapeutic decisions and enhance the quality of patient care.
Section 6: Future perspective
Overall, this case study with lapatinib illustrates the reality of the multiple modes of inactivation for even structurally similar CYP3A4 and CYP3A5 when exposed to the same substrate. Such phenomenon could be exacerbated by the large binding pocket exhibited for this subfamily, allowing the substrate to adopt alternative binding poses to access the catalytic center for oxidation to take place (Dabrowski et al., 2002). It is plausible that other CYP3A substrates may exhibit similar behavior, therefore requiring an isoform-specific investigation to determine the true clinical impact of enzyme inhibition.
Secondly, MBI of CYP3A through lapatinib metabolism highlights the significance of different reactive metabolites. O-debenzylation of lapatinib promotes the formation of an electrophilic quinoneimine that binds nucleophilic residues like lysines and cysteines, whereas N-hydroxylation and subsequent nitroso intermediate formation generates nucleophiles with potential to chelate iron, leading to a similar inactivation outcome. These independent metabolic processes need to be unequivocally considered for other substrates that carry multiple reactive functionalities (Riley et al., 2007), thereby expanding the range of influence for other concomitantly administered CYP450 substrates.
Ultimately, demonstrable toxicological consequences of mechanism-based inactivator formation, as observed experimentally, must be subjected to appropriate in vitro-in vivo extrapolations. A number of key considerations can affect the in vivo presentation of a drug: (1) Plurality of metabolic pathways for the disposition of the drug of concern. Multiple routes of elimination (Phase I vs. II; hepatic clearance vs. renal clearance) reduce the flux through any particular pathway affected by MBI formation. Alternative routes for drug clearance can help to cushion some of the changes to overall metabolism of either the inhibitor or the co-administered drug, thereby softening the effect on drug-drug interaction. On that note, this propensity also depends on the extent that hepatic clearance is a limiting factor for overall clearance. Generally speaking, a hepatically cleared drug of low hepatic extraction ratio would be more susceptible to the effect of MBI as its clearance is limited by both the fraction unbound passaging through the liver as well as the intrinsic metabolic capacity (Verbeeck, 2008). For this reason, not all predicted drug-drug interactions will be clinically significant. (2) Secondly, the partition ratio alters the balance of MBI and of the amount of reactive metabolite released for binding to other hepatic proteins (Rock, 2010). This ratio is a quantitative guide that defines the likelihood of inhibition and should be discussed in conjunction with the actual presentation of the drug to CYP450 enzymes for metabolism. A drug presented in higher concentration as a free drug to the liver can exert a greater mass effect to elicit stronger MBI risk, especially when the CYP450 enzyme of concern is of a lower natural abundance. For example, MBI of CYP3A5 would exert a greater effect in an individual with the CYP3A5*1/*1 genotype and impose bigger challenges for the clearance of drugs primarily cleared via this pathway. (3) Accordingly, the total daily dose burden is another key determinant that will affect exposure to the toxicant since saturation of competing metabolic pathways or generation of reactive metabolites exceeding the detoxification capacity may trigger the toxicological manifestation. (4) Finally, it is important to appreciate the antigenicity of the resultant drug-CYP450 adduct and how such an immune response may differ between individuals. While tienilic acid remains the only well-characterized case of an MBI-mediated immune-hepatitis event, its future occurrence cannot be undermined, especially with more potent inactivators being discovered in recent years.
In summary, lapatinib embodies the complexity of MBI of CYP450 and the associated diversity of toxicological implications that comes with its understanding. While MBI has not fully explained the onset of idiosyncratic hepatotoxicity from usage of lapatinib, existing groundwork has been laid to further explore the effect of this MBI on drug-drug interactions. Studies are underway to investigate the interaction between lapatinib and the high prevalence of CYP3A5 genetic polymorphism to identify patient-specific toxicity risk factors that may affect patient selection. These studies should advance the field by providing insight into the role of CYP3A5 polymorphism in the hepatotoxic potential of lapatinib to improve patient safety and prevent unnecessary adverse reactions.
Acknowledgments
This project is funded by the Singapore Ministry of Education’s (MOE) Academic Research Grants R -148-000-187-112 (HKH) and R-148-000-135-112 (ECYC) and National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) Grant GM32165 (KDH) and the UNCF-Merck Science Initiative (KDH). CYC is supported by the NUS President Graduate Fellowship. This review is written in memory of Professor Sidney D. Nelson, who has inspired each of the co-authors both intellectually and personally with his transformative thinking and compelling reasoning.
References
- Agency EM. Assessment Report for Tyverb. 2007. [Google Scholar]
- Azim HA, Jr, Agbor-Tarh D, Bradbury I, Dinh P, Baselga J, Di Cosimo S, Greger JG, Jr, Smith I, Jackisch C, Kim SB, Aktas B, Huang CS, Vuylsteke P, Hsieh RK, Dreosti L, Eidtmann H, Piccart M, de Azambuja E. Pattern of Rash, Diarrhea, and Hepatic Toxicities Secondary to Lapatinib and Their Association With Age and Response to Neoadjuvant Therapy: Analysis From the NeoALTTO Trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013 doi: 10.1200/JCO.2013.50.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara JE, Kazmi F, Parkinson A, Buckley DB. Metabolism-dependent inhibition of CYP3A4 by lapatinib: evidence for formation of a metabolic intermediate complex with a nitroso/oxime metabolite formed via a nitrone intermediate. Drug metabolism and disposition: the biological fate of chemicals. 2013;41:1012–1022. doi: 10.1124/dmd.113.051151. [DOI] [PubMed] [Google Scholar]
- Beaune P, Dansette PM, Mansuy D, Kiffel L, Finck M, Amar C, Leroux JP, Homberg JC. Human anti-endoplasmic reticulum autoantibodies appearing in a drug-induced hepatitis are directed against a human liver cytochrome P-450 that hydroxylates the drug. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:551–555. doi: 10.1073/pnas.84.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino S, O’Mara M, Koch K, Borts DJ, Bowers GD, MacLauchlin C. Human metabolism of lapatinib, a dual kinase inhibitor: implications for hepatotoxicity. Drug metabolism and disposition: the biological fate of chemicals. 2012;40:139–150. doi: 10.1124/dmd.111.040949. [DOI] [PubMed] [Google Scholar]
- Dabrowski MJ, Schrag ML, Wienkers LC, Atkins WM. Pyrene.pyrene complexes at the active site of cytochrome P450 3A4: evidence for a multiple substrate binding site. Journal of the American Chemical Society. 2002;124:11866–11867. doi: 10.1021/ja027552x. [DOI] [PubMed] [Google Scholar]
- de Jonge MJ, Hamberg P, Verweij J, Savage S, Suttle AB, Hodge J, Arumugham T, Pandite LN, Hurwitz HI. Phase I and pharmacokinetic study of pazopanib and lapatinib combination therapy in patients with advanced solid tumors. Investigational new drugs. 2013;31:751–759. doi: 10.1007/s10637-012-9885-8. [DOI] [PubMed] [Google Scholar]
- Filppula AM, Laitila J, Neuvonen PJ, Backman JT. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. British journal of pharmacology. 2012;165:2787–2798. doi: 10.1111/j.1476-5381.2011.01732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton JE. Lapatinib: a review of its use in the treatment of HER2-overexpressing, trastuzumab-refractory, advanced or metastatic breast cancer. Drugs. 2009;69:2125–2148. doi: 10.2165/11203240-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Ge GB, Ning J, Hu LH, Dai ZR, Hou J, Cao YF, Yu ZW, Ai CZ, Gu JK, Ma XC, Yang L. A highly selective probe for human cytochrome P450 3A4: isoform selectivity, kinetic characterization and its applications. Chem Commun (Camb) 2013;49:9779–9781. doi: 10.1039/c3cc45250f. [DOI] [PubMed] [Google Scholar]
- Gomez HL, Doval DC, Chavez MA, Ang PC, Aziz Z, Nag S, Ng C, Franco SX, Chow LW, Arbushites MC, Casey MA, Berger MS, Stein SH, Sledge GW. Efficacy and safety of lapatinib as first-line therapy for ErbB2-amplified locally advanced or metastatic breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:2999–3005. doi: 10.1200/JCO.2007.14.0590. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annual review of pharmacology and toxicology. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- Hardy KD, Wahlin MD, Papageorgiou I, Unadkat JD, Rettie AE, Nelson SD. Studies on the role of metabolic activation in tyrosine kinase inhibitor-dependent hepatotoxicity: induction of CYP3A4 enhances the cytotoxicity of lapatinib in HepaRG cells. Drug metabolism and disposition: the biological fate of chemicals. 2014;42:162–171. doi: 10.1124/dmd.113.054817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy KDWM, Rettie AE. Role of CYP3A5 in the Metabolic Activation of Lapatinib. 53rd Annual Meeting of the Society of Toxicology; Pheonix, Arizona, USA: Oxford University Press; 2014. p. S143. [Google Scholar]
- Harrelson JP, Stamper BD, Chapman JD, Goodlett DR, Nelson SD. Covalent modification and time-dependent inhibition of human CYP2E1 by the meta-isomer of acetaminophen. Drug metabolism and disposition: the biological fate of chemicals. 2012;40:1460–1465. doi: 10.1124/dmd.112.045492. [DOI] [PubMed] [Google Scholar]
- Homberg JC, Andre C, Abuaf N. A new anti-liver-kidney microsome antibody (anti-LKM2) in tienilic acid-induced hepatitis. Clinical and experimental immunology. 1984;55:561–570. [PMC free article] [PubMed] [Google Scholar]
- Jelovac D, Wolff AC. The adjuvant treatment of HER2-positive breast cancer. Current treatment options in oncology. 2012;13:230–239. doi: 10.1007/s11864-012-0186-4. [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature genetics. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Advanced drug delivery reviews. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Li X, Jeso V, Heyward S, Walker GS, Sharma R, Micalizio GC, Cameron MD. Characterization of T-5 N-oxide formation as the first highly selective measure of CYP3A5 activity. Drug metabolism and disposition: the biological fate of chemicals. 2014;42:334–342. doi: 10.1124/dmd.113.054726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HL, Kenaan C, Hollenberg PF. Identification of the residue in human CYP3A4 that is covalently modified by bergamottin and the reactive intermediate that contributes to the grapefruit juice effect. Drug metabolism and disposition: the biological fate of chemicals. 2012;40:998–1006. doi: 10.1124/dmd.112.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia MP, Dansette PM, Mansuy D. Thiophene derivatives as new mechanism-based inhibitors of cytochromes P-450: inactivation of yeast-expressed human liver cytochrome P-450 2C9 by tienilic acid. Biochemistry. 1994;33:166–175. doi: 10.1021/bi00167a022. [DOI] [PubMed] [Google Scholar]
- Marks GS, McCluskey SA, Mackie JE, Riddick DS, James CA. Disruption of hepatic heme biosynthesis after interaction of xenobiotics with cytochrome P-450. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1988;2:2774–2783. doi: 10.1096/fasebj.2.12.3044903. [DOI] [PubMed] [Google Scholar]
- Masubuchi Y, Horie T. Toxicological significance of mechanism-based inactivation of cytochrome p450 enzymes by drugs. Critical reviews in toxicology. 2007;37:389–412. doi: 10.1080/10408440701215233. [DOI] [PubMed] [Google Scholar]
- Midgley RS, Kerr DJ, Flaherty KT, Stevenson JP, Pratap SE, Koch KM, Smith DA, Versola M, Fleming RA, Ward C, O’Dwyer PJ, Middleton MR. A phase I and pharmacokinetic study of lapatinib in combination with infusional 5-fluorouracil, leucovorin and irinotecan. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2007;18:2025–2029. doi: 10.1093/annonc/mdm366. [DOI] [PubMed] [Google Scholar]
- Obach RS, Walsky RL, Venkatakrishnan K. Mechanism-based inactivation of human cytochrome p450 enzymes and the prediction of drug-drug interactions. Drug metabolism and disposition: the biological fate of chemicals. 2007;35:246–255. doi: 10.1124/dmd.106.012633. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. Cytochrome P450 structure, mechanism, and biochemistry. Kluwer Academic/Plenum Publishers; New York: 2005. pp 1 online resource (xx, 689 p) ill. [Google Scholar]
- Rezai K, Urien S, Isambert N, Roche H, Dieras V, Berille J, Bonneterre J, Brain E, Lokiec F. Pharmacokinetic evaluation of the vinorelbine-lapatinib combination in the treatment of breast cancer patients. Cancer chemotherapy and pharmacology. 2011;68:1529–1536. doi: 10.1007/s00280-011-1650-8. [DOI] [PubMed] [Google Scholar]
- Riley RJ, Grime K, Weaver R. Time-dependent CYP inhibition. Expert opinion on drug metabolism & toxicology. 2007;3:51–66. doi: 10.1517/17425255.3.1.51. [DOI] [PubMed] [Google Scholar]
- Rock DW, LC . In: Characterization of Cytochrome P450 Mechanism-Based Inhibition. Gad SC, editor. John Wiley & Sons; New York: 2010. p. 56. [Google Scholar]
- Smith DA, Koch KM, Arya N, Bowen CJ, Herendeen JM, Beelen A. Effects of ketoconazole and carbamazepine on lapatinib pharmacokinetics in healthy subjects. British journal of clinical pharmacology. 2009;67:421–426. doi: 10.1111/j.1365-2125.2009.03370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraggs CF, Budde LR, Briley LP, Bing N, Cox CJ, King KS, Whittaker JC, Mooser VE, Preston AJ, Stein SH, Cardon LR. HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:667–673. doi: 10.1200/JCO.2010.31.3197. [DOI] [PubMed] [Google Scholar]
- Takakusa H, Wahlin MD, Zhao C, Hanson KL, New LS, Chan EC, Nelson SD. Metabolic intermediate complex formation of human cytochrome P450 3A4 by lapatinib. Drug metabolism and disposition: the biological fate of chemicals. 2011;39:1022–1030. doi: 10.1124/dmd.110.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng WC, Oh JW, New LS, Wahlin MD, Nelson SD, Ho HK, Chan EC. Mechanism-based inactivation of cytochrome P450 3A4 by lapatinib. Molecular pharmacology. 2010;78:693–703. doi: 10.1124/mol.110.065839. [DOI] [PubMed] [Google Scholar]
- Teo YL, Saetaew M, Chanthawong S, Yap YS, Chan EC, Ho HK, Chan A. Effect of CYP3A4 inducer dexamethasone on hepatotoxicity of lapatinib: clinical and in vitro evidence. Breast cancer research and treatment. 2012;133:703–711. doi: 10.1007/s10549-012-1995-7. [DOI] [PubMed] [Google Scholar]
- Thunell S, Pomp E, Brun A. Guide to drug porphyrogenicity prediction and drug prescription in the acute porphyrias. British journal of clinical pharmacology. 2007;64:668–679. doi: 10.1111/j.0306-5251.2007.02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht J. Idiosyncratic drug reactions: current understanding. Annual review of pharmacology and toxicology. 2007;47:513–539. doi: 10.1146/annurev.pharmtox.47.120505.105150. [DOI] [PubMed] [Google Scholar]
- Verbeeck RK. Pharmacokinetics and dosage adjustment in patients with hepatic dysfunction. European journal of clinical pharmacology. 2008;64:1147–1161. doi: 10.1007/s00228-008-0553-z. [DOI] [PubMed] [Google Scholar]
- Walsky RL, Obach RS, Hyland R, Kang P, Zhou S, West M, Geoghegan KF, Helal CJ, Walker GS, Goosen TC, Zientek MA. Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug metabolism and disposition: the biological fate of chemicals. 2012;40:1686–1697. doi: 10.1124/dmd.112.045302. [DOI] [PubMed] [Google Scholar]