Summary
Background
Membrane-associated guanylate kinases (MAGUKs), such as Discs-Large (DLG), play critical roles in synapse maturation by regulating the assembly of synaptic multiprotein complexes. Previous studies have revealed a genetic interaction between DLG and another PDZ scaffolding protein, SCRIBBLE (SCRIB), during the establishment of cell polarity in developing epithelia. A possible interaction between DLG and SCRIB at synaptic junctions has not yet been addressed. Likewise, the biochemical nature of this interaction remains elusive, raising questions regarding the mechanisms by which the actions of both proteins are coordinated.
Results
Here we report the isolation of a new DLG-interacting protein, GUK-holder, that interacts with the GUK domain of DLG and which is dynamically expressed during synaptic bouton budding. We also show that at Drosophila synapses DLG colocalizes with SCRIB and that this colocalization is likely to be mediated by direct interactions between GUKH and the PDZ2 domain of SCRIB. We show that DLG, GUKH, and SCRIB form a tripartite complex at synapses, in which DLG and GUKH are required for the proper synaptic localization of SCRIB.
Conclusions
Our results provide a mechanism by which developmentally important PDZ-mediated complexes are associated at the synapse.
Introduction
A precise spatial arrangement of proteins at both the pre- and postsynaptic membranes underlies the highly efficient signal transmission at synaptic junctions. Recent studies have identified synaptic scaffolding molecules, which by virtue of their ability to simultaneously bind several proteins, play crucial roles in the orchestration of structural and functional building blocks [1, 2]. In particular, membrane-associated guanylate kinases (MAGUKs), such as PSD-95, have emerged as central elements in the formation of heteromultimeric scaffolds underneath the membranes of glutamatergic synapses in both vertebrates and invertebrates. At the mammalian postsynaptic density, MAGUKs can bind ionotropic glutamate receptors [3–5], components of second messenger cascades [6, 7], and cell adhesion molecules [8] via their PDZ and SH3 domains. Similarly, at insect neuromuscular junctions (NMJs), the PSD-95 ortholog DLG can concurrently bind a cell adhesion molecule (Fasciclin II [FasII]) [9] and a Shaker K+ channel [10]. Such arrangements may contribute to the functional coupling of the respective MAGUK binding partners. For example, simultaneous binding of NMDA receptors and a synaptic Ras-GTPase activating protein (SynGAP) to PSD-95 could allow for the cooperative coupling of synaptic activity and Ras-mediated signaling pathways [6].
While the PDZ and SH3 domains of MAGUKs are known to bind components required for synapse function, the significance of the guanylate kinase-like (GUK) domain has remained puzzling. Several studies suggest that it might act as a protein interaction domain. For example, in mammals, this domain binds to GKAP/SAPAPs [11, 12], which are in turn linked to Shank/ProSAP [13, 14]. It has also been reported to bind MAP1A [15] to a kinesin-like protein [16], to SPAR, an actin cytoskeleton regulator [17], and to interact intra-molecularly with the SH3 domain [18–20].
In Drosophila, dlg mutants in which the GUK domain is absent exhibit abnormalities in synapse structure [21]. Moreover, transgenic DLG lacking the GUK domain fails to localize at synapses when expressed in a dlg mutant background [22]. These findings imply that the GUK domain is required for a synaptic function and targeting of DLG. To gain further insight on how the GUK domain of DLG exerts its various functions, we searched for proteins interacting with this domain. We report the isolation of GUK-holder, a novel synaptic protein containing a WH1/EVH1-like domain in its N-terminal half and a PDZ binding motif at its C terminus. We demonstrate that GUKH is expressed in a dynamic fashion during synaptic bouton formation. In addition, we show that it also binds to a PDZ domain of Scribble (SCRIB), a tumor suppressor protein that has previously been shown to genetically interact with DLG in developing epithelia [23, 24], thus physically linking DLG to SCRIB. Indeed, our coimmunoprecipitation analyses together with immunocytochemical studies on wild-type and mutant larvae provide strong evidence that DLG, GUKH, and SCRIB exist in a tripartite complex at the NMJ. Most notably, we found that normal GUKH function was required for the synaptic localization of SCRIB.
Results
Identification of GUKH, a Novel DLG-Interacting Partner
To understand the functional significance of the GUK domain of DLG, we searched for binding partners of this domain using a yeast two-hybrid screen [25]. We used the GUK domain of DLG (amino acids 765–960; [26]) as bait to screen a late embryonic stage Drosophila cDNA library. Thirty-eight interacting clones were recovered from this screen, and from these, nine were overlapping cDNAs representing a single novel gene, which we named GUK-holder (GUKH) (Figure 1).
Figure 1. GUKH Is a Novel Synaptic Protein that Interacts with DLG.
(A) Schematic representation of chromosomal region 91E according to the BDGP database [27] (for a more detailed representation, see http://flybase.bio.indiana.edu/.bin/fbgrmap?spp=fly&chr=3R&self=1&range=4670844). The actual gukh gene covers three adjacent conceptual genes (represented by arrows). Arrowheads mark P insertions gukhJ3 and gukh2 that affect gukh expression. The P element in gukhJ3 was mapped to a large 3′ intron of conceptual gene CG17836.
(B) Exon-intron organization of gukh and deduced protein structure. Exons E1–E6 are indicated by boxes, with coding regions in black. At least two isoforms (S and L) can be generated due to alternative splicing of exon 5. The L isoform of GUKH is shown in alignment to E1–E6. All clones that were isolated in the yeast two-hybrid screen map to E6 and are indicated by black lines. “P” indicates a PEST sequence within the GUK-holding domain. Gray lines below the protein diagram indicate regions with homology to Kelch.
(C) Interaction between the C terminus of GUKH (GUKH-C) and the GUK domain of DLG, as demonstrated with the yeast two-hybrid assay. “+” and “−” signs denote the strength of the interaction (see Table 1, legend).
(D) GUKH was immunoprecipitated with GUKH antibodies from larval body wall muscle extracts, and the immunoblots were probed sequentially with anti-GUKH, anti-DLG, and anti-eGFP antibodies. The lanes represent extracts from wild-type (lane1), wild-type expressing eGFP-GUKH-C (lane 2), and dlgXI-2 (lane 3). Note that in the wild-type expressing GUKH-C, both endogenous (110 kDa) and eGFP-tagged transgenic GUKH-C (90 kDa) are immunoprecipitated with the GUKH antibody. In dlgXI-2 mutants, levels of endogenous GUKH are normal, but DLG coimmunoprecipitation is not observed.
To characterize the gukh transcription unit, we performed a database analysis and identified several overlapping expressed sequence tag (EST) clones. Further sequencing of these EST clones and alignment with the genomic region indicated that the gukh transcription unit covers three conceptual genes predicted by the BDGP database (CG5456, CG14288, and CG6003), thereby comprising at least six exons spread over a 38 Kb region (Figures 1A and 1B). The deduced protein sequence comprises 1044 residues, with a calculated molecular weight of 111.4 kDa (Figure 1B; long isoform, “L”). In addition, we found that some EST clones represent an alternatively spliced transcript missing the fifth exon, suggesting the existence of a C-terminally truncated isoform (Figure 1B, short isoform, “S”; 534 amino acids, 57.6 kDa).
The predicted GUKH protein exhibited no signal sequences or transmembrane domains, consistent with it being intracellular. A homology search revealed a region with similarity to the WH1/EVH1 domain of the Drosophila homolog of Suppressor of cAR (SCAR; 32% identity; 54% similarity) [27] and its murine ortholog WAVE-1 [28, 29]. Moreover, a region of moderate homology to the Drosophila actin binding protein Kelch is found within the C-terminal half of GUKH [30]. This region of GUKH also includes a predicted PEST sequence. The DLG binding region of GUKH maps to the C-terminal third of the protein, as deduced from the overlapping cDNAs obtained from the yeast two-hybrid screen. Notably, GUKH terminates in the potential PDZ binding motif tETAL ([31]; Figure 1B).
To determine the precise regions of interaction between the two proteins, deletion constructs of the DLG GUK domain and of the GUKH C terminus were generated and assayed for binding using the yeast two-hybrid assay (Figure 1C). Nearly the entire GUK domain is necessary for an interaction with GUKH, as deletion of more than 15 residues from either end resulted in a loss of binding. A construct encompassing the last 156 amino acids of GUKH (amino acids 888–1044) was sufficient to mediate binding to the GUK domain of DLG, defining this region as the GUK-holding domain (Figure 1B).
GUKH Interacts with DLG In Vivo
We generated an affinity-purified polyclonal antiserum directed against the last 238 C-terminal amino acids of GUKH (GUKH-C). In Western blots from body wall muscle extracts, the antibody detected a single band of ~110 kDa, consistent with the predicted size of the L isoform of GUKH. Moreover, anti-GUKH immunoreactivity was reduced at the NMJs or CNS of the hypomorphic gukh mutants gukhJ3E1 and gukh2, eliminated in gukh2EM9 embryos, and enhanced upon overexpression of a gukh transgene, confirming the specificity of the antibody.
To establish that DLG and GUKH interact in vivo, we performed a coimmunoprecipitation of body wall muscle extracts using GUKH antibody. In wild-type, DLG-specific bands at 97 and 116 kDa coprecipitated with the 110 kDa GUKH band, suggesting that GUKH exists in the same complex as DLG (Figure 1D, lane 1). In contrast, DLG was not coimmunoprecipitated from dlgXI-2 mutants that lack the GUK domain (Figure 1D, lane 3). Together, these results strongly suggest that GUKH binds to the GUK domain of DLG in vivo.
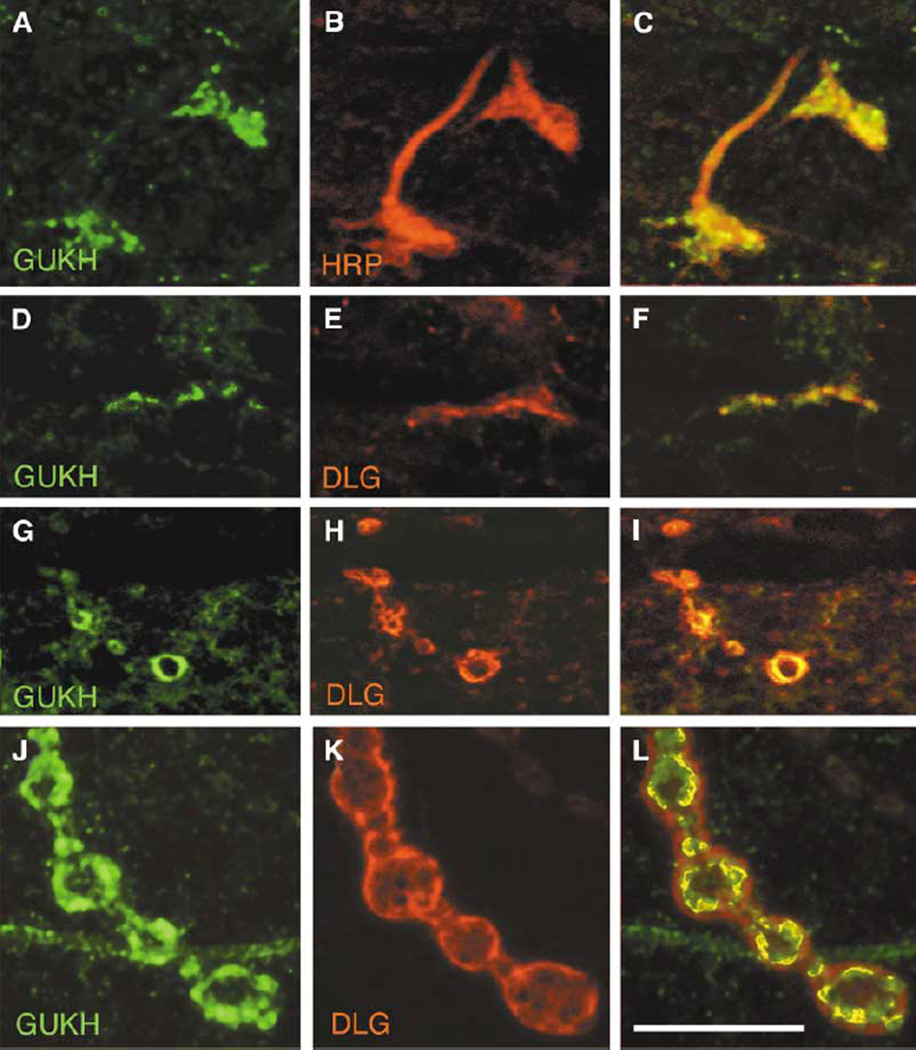
We next addressed the question of a possible interaction between DLG and GUKH at synaptic sites by examining the colocalization of the proteins through development. GUKH makes its first appearance presynaptically at the NMJ during embryonic stage 17 where it overlaps with the neuronal marker, anti-HRP (Figures 2A–2C), and with DLG (Figures 2D–2F; [32]). At this time, GUKH is distributed throughout the developing boutons. During the first larval instar, the protein becomes enriched at the rim of the boutons in colocalization with DLG (Figures 2G–2I). This pattern is maintained through late larval development (Figures 2J–2L). Interestingly, a similar developmental pattern of expression is observed for DLG [32]. However, while DLG immunoreactivity is found throughout a large extent of the postsynaptic junctional region (SSR), typically, GUKH immunoreactivity is distributed in interrupted patches along the synapse border that usually extend a short way into the bouton interior. These observations together with the protein interaction studies provide strong evidence for a direct interaction between GUKH and DLG at the NMJ.
Figure 2. GUKH Is Expressed at Synaptic Boutons Where It Colocalizes with DLG at Bouton Borders.
(A and D) GUKH immunoreactivity at the NMJ in a stage 17 embryo shown in preparations double labeled with (B) anti-HRP and (E) anti-DLG. (C) and (F) are merged images from (A) and (B) and from (D) and (E), respectively. Note that at this stage GUKH immunoreactivity is presynaptic. (G and J) GUKH immunoreactivity in (G) a mid first instar and (J) a third instar larval NMJ double stained with (H and K) anti-DLG. (I) and (L) are merged images from (G) and (H) and from (J) and (K), respectively. Scale bar represents 12 µm in (A)–(F) and 9 µm in (G)–(I).
GUKH immunoreactivity is also found in the embryonic and larval CNS and asymmetrically distributed in neuroblasts (see the Supplementary Material available with this article online for a short description of GUKH expression at these sites).
Differential Localization of GUKH and DLG during Bouton Budding
Comparison of GUKH and DLG distribution at synaptic boutons revealed that, although both proteins colocalize at bouton borders, they also show distinctly complementary patterns during bouton budding. Larval NMJs expand during development to compensate for an increase in muscle size [33, 34]. This expansion involves an enhancement in bouton and active zone number, which serves to maintain synaptic strength despite the changes in muscle size. The process of NMJ expansion occurs by the formation of new boutons that bud off from existing boutons, as has been described both in vivo and in fixed NMJs [35].
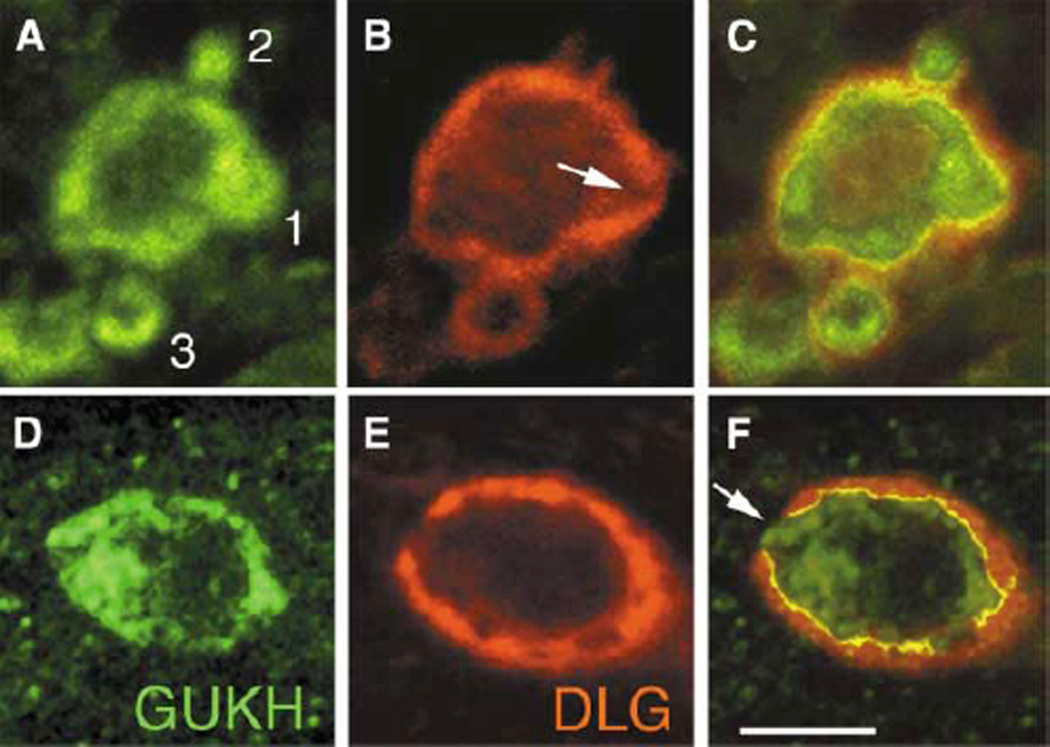
We found that GUKH was enriched at budding boutons where it filled the entire bud, in contrast to its more peripheral distribution in the mature boutons (Figure 3). To compare the distribution of GUKH and DLG during bouton budding, we acquired complete confocal Z series of synaptic boutons and analyzed their expression in single slices (n = 39 buds). Figures 3A–3C show a single slice from the midline of a bouton in a first instar larva. Analysis of GUKH and DLG expression during this process revealed that the distribution of both proteins changed at different stages of bouton budding, consistent with a strikingly dynamic expression.
Figure 3. GUKH and DLG Are Dynamically Expressed during Bouton Budding.
(A–C) First instar NMJs showing three budding boutons (“1,” “2,” and “3”), double labeled with (A and D) anti-GUKH and (B and E) anti-DLG. (C) and (F) are merged images from (A) and (B) and from (D) and (E), respectively. Numbers 1–3 in (A) indicate stages of bouton budding (1, protrusion; 2, bud separation; 3, new bouton formation). Arrow in (B) and (F) points to low DLG levels at the site of bouton protrusion. Scale bar represents 1.5 µm in (A)–(C) and 3 µm in (D)–(E).
During the stage of protrusion, GUKH was highly enriched in the core of the protruding bud (Figure 3A, #1). At the same stage, DLG immunoreactivity decreased at the site of protrusion and became strong at the borders immediately adjacent to the site of low DLG (Figure 3B). Throughout this stage, GUKH and DLG colocalized at the bouton border, except for the leading edge of the protrusion, where DLG was low (Figure 3C).
Once the bud separated from the parent bouton, GUKH remained enriched in the bud but disappeared from the neck of the bud (Figure 3A, #2). In contrast, DLG completely disappeared from the distal border of the bud and became highly enriched at the neck of the bud (Figure 3B). As the bud takes on a distinctly bouton-like morphology (Figure 3A, # 3), the distribution of GUKH and DLG is similar to a mature bouton, i.e., both proteins localize at the periphery of the bouton. However, GUKH is still substantially enriched at the distal border of the nascent bouton (Figure 3A, #3). Similar observations were made in NMJ from older larva, and Figures 3D–3F show a very early stage of bud formation as a nub of GUKH immunoreactivity protrudes from the surrounding DLG. Thus, GUKH and DLG appear to be dynamically localized during bouton budding, overlapping at the edges but being complementary at the buds.
GUKH Interacts with Scribble, Another PDZ Protein
The interaction of DLG and GUKH is reminiscent of the interaction between the GUK domain of mammalian MAGUKs and GKAP [11, 12]. Moreover, while GUKH and GKAP do not share significant sequence homology, both proteins terminate in a similar tS/TXV/L/I PDZ binding motif (i.e., tETAL versus tQTRL; [31]). In fact, GKAP proteins link the GUK domain of PSD-95/SAP90 to the PDZ domain of Shank/ProSAP [14, 36]. By analogy, we inferred that GUKH might link DLG to other PDZ domain-containing proteins. Recent studies have revealed that at epithelia DLG exists in a complex with Scribble (SCRIB), a protein comprising 16 leucine-rich repeats followed by four PDZ domains [23, 24]. However, the molecular nature of this interaction remained elusive. In this study, we performed a coimmunoprecipitation assay on body wall muscle extracts using a SCRIB-specific antibody [23]. We found that anti-SCRIB efficiently coimmunoprecipitated DLG from wild-type but not from a severe hypomorphic scrib allele (Figure 4A, lanes 1 and 4). This indicates that, similar to the case in epithelia, DLG and SCRIB may exist in a complex at the NMJ. In line with this finding, SCRIB exhibits striking colocalization with DLG at type I boutons (Figure 4B).
Figure 4. DLG and SCRIB Colocalize at the NMJ; and GUKH, DLG, and SCRIB Exist in a Tripartite Complex.
(A) Body wall muscle extracts were immunoprecipitated with anti-SCRIB antibodies, and the immunoblots were sequentially probed with anti-SCRIB (top), anti-GUKH (middle), and anti-DLG (bottom). Lanes correspond to body wall muscle extracts from wild-type (lane 1), gukhJ3E1/Df (lane 2), dlgXI–2 (lane 3), and scrib2/Df (lane 4). Molecular weights at the right of each blot are expressed in kDa.
(B) Type I synaptic boutons of wild-type third instar body wall muscles stained with (top) anti-SCRIB and (middle) anti-DLG. (Bottom) Merged image of the top and middle panels showing DLG and SCRIB colocalization.
We next assessed whether GUKH might provide a physical link between DLG and SCRIB. Indeed, we detected GUKH in anti-SCRIB immunoprecipitates from wild-type but not from scrib mutant extracts. (Figure 4A, lanes 1 and 4). Moreover, immunoprecipitation of DLG by anti-SCRIB antibodies from a hypomorphic gukh allele (see below) was dramatically reduced (Figure 4A, lane 2).
To investigate the possibility that the interaction between GUKH and SCRIB might be direct, we used the yeast two-hybrid assay, which showed that GUKH specifically interacted with the PDZ2 domain of SCRIB but not with either its PDZ3–4 or its LRR motifs (Table 1). The interaction between GUKH and the PDZ2 domain of SCRIB was mediated by the C terminus of GUKH, as just the ten last amino acids of GUKH were sufficient for this interaction. Deletion of the last 23 amino acids of GUKH (GUKH-ΔC) prevented the interaction with the PDZ2 domain of SCRIB. Moreover, when the ten amino acid peptide contained a mutation (L→A) at the C-terminal residue, it failed to interact with PDZ2 (Table 1). In addition, the last ten amino acids of Shaker K+ channel, which strongly binds to PDZ1–2 of DLG [10], failed to bind PDZ2 of SCRIB, demonstrating a degree of ligand specificity (Table 1). In contrast, constructs encompassing PDZ1–2 or PDZ3 of DLG failed to bind GUKH (Table 1). Together, the localization, immunoprecipitation, and yeast two-hybrid studies strongly suggest that DLG, GUKH, and SCRIB may form a tripartite complex in which GUKH serves as a physical link between DLG and SCRIB.
Table 1.
Yeast Two-Hybrid of DLG, GUKH, and SCRIB
| GUKH | GUKHΔC | PLPPSFETAL | PLPPSFETAA | ALAVSIETDV | No Insert | |
|---|---|---|---|---|---|---|
| GUK (DLG) | +++ | − | − | |||
| PDZ1–2 (DLG) | − | +++ | − | |||
| PDZ3 (DLG) | − | − | ||||
| LRR (SCRIB) | − | |||||
| PDZ1–2 (SCRIB) | +++ | +++ | ||||
| PDZ3–4 (SCRIB) | − | − | − | − | ||
| PDZ1 (SCRIB) | +++ | +++ | ||||
| PDZ2 (SCRIB) | +++ | − | ++ | − | − | − |
| No insert | − | − | − | − | − |
+++, positive β-gal reaction within 40 min. ++, positive β-gal reaction between 40 and 60 min. +, positive β-gal reaction between 60 and 80 min. +, positive β-gal reaction after 80 min.
Isolation of gukh Mutants
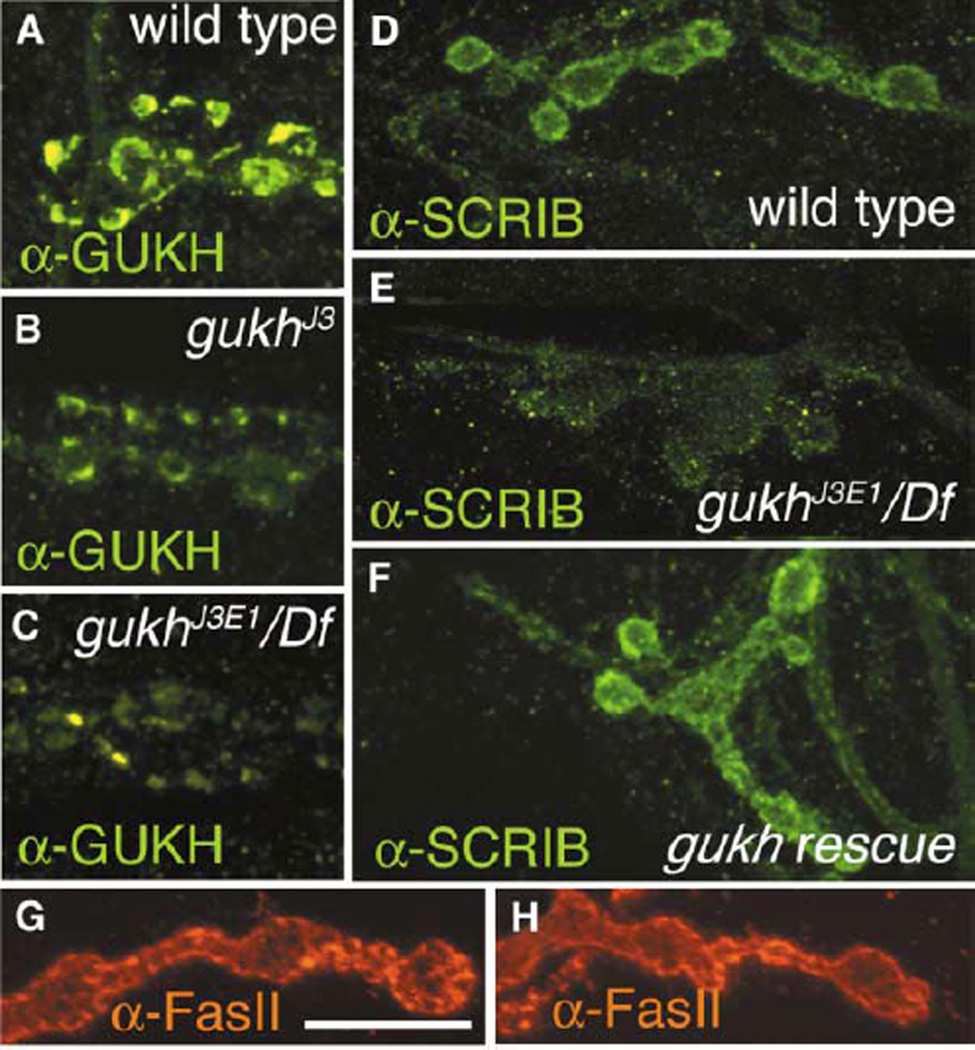
The gukh gene was mapped to position 91E on the right arm of the third chromosome by in situ hybridization in agreement with the database. Within this region, we identified two homozygous viable P element insertions (see Experimental Procedures) subsequently referred to as gukhJ3 and gukh2 that exhibited a moderate but significant decrease in GUKH immunoreactivity at larval NMJs or CNS. Using inverse PCR, we determined that in gukhJ3 and gukh2 the P elements were inserted ~60 kb and 380 bp, respectively, upstream from the transcriptional start site of gukh (Figure 1A). Several genes are predicted to lie in-between the P insertion in gukhJ3 and the first exon of gukh. We suggest that both P insertions affect regulatory elements required for proper gukh expression. To test whether the P insertions were responsible for the reduction of GUKH immunoreactivity, we generated additional alleles by P element excision. One new allele, gukhJ3E1, resulted from an imprecise excision which caused a deletion of ~5 kb. Both homozygous and hemizygous gukhJ3E1 flies exhibited decreased viability and, most notably, a further reduction in GUKH immunoreactivity as compared to gukhJ3 (Figures 5A–5C; Table 2). In contrast, synaptic GUKH immunoreactivity was reverted to wild-type levels in another allele, gukhrev, in which the P element was excised precisely. In the case of gukh2, imprecise excision of the P element resulted in complete elimination of GUKH immunoreactivity in the embryo (data not shown), but this mutation was lethal prior to hatching.
Figure 5. gukh Is Required for the Synaptic Localization of SCRIB.
(A–C) GUKH immunoreactivity at type I boutons of (A) wild-type, (B) gukhJ3, and (C) gukhJ3E1/Df, showing a decrease in immunoreactivity levels in the gukh mutants.
(D–F) Type I synaptic boutons immunostained with anti-SCRIB antibodies, showing its synaptic localization in (D) wild-type, (E) its abnormal clustering at synapses of gukhJ3E1 mutants, and (F) the rescue of this abnormal clustering in gukh mutants by presynaptic expression of GUKH-C.
(G and H) Anti-FasII staining at (G) wild-type and (H) gukhJ3E1/Df mutant boutons. Note that FasII staining is not altered in gukh mutants. Scale bar, 17 µm.
Table 2.
Intensity of SCRIB and GUKH Staining in gukh Mutants
| Genotype | SCRIB | n | Genotype | GUKH | n |
|---|---|---|---|---|---|
| Wild-type | 3.7 ± 0.12 | 52/4 | wild-type | 4.4 ± 0.38 | 111/5 |
| gukhJ3E1 | 2.0 ± 0.06 | 46/3 | gukhJ3E1 | 3.4 ± 0.35 | 47/4 |
| gukhJ3E1 + presynaptic GUKH-C | 3.0 ± 0.2 | 38/4 | gukhJ3E1/Df | 2.8 ± 0.26 | 97/6 |
| gukhJ3E1 + postsynaptic GUKH-C | 2.4 ± 0.15 | 55/4 | gukhrev | 4.1 ± 0.27 | 143/9 |
Numbers under SCRIB and GUKH column represent signal to noise ratio (see Experimental Procedures). Numbers under n represent number of boutons/number of larvae.
GUKH Is Required to Localize the PDZ-Containing Protein SCRIBBLE
To understand the role of GUKH at synapses, we examined the morphology of gukhJ3, gukhJ3E1, gukhJ3E1/Df(3R)Cha7, and gukh2 NMJs. No noticeable defects in synaptic bouton number and morphology were found upon examining preparations stained with the presynaptic marker anti-HRP. Similarly, immunocytochemical analysis of the distribution of several synaptic proteins, including FasII (Figures 5G and 5H), DLG, synapsin, cysteine string protein (CSP), and synaptotagmin and CaMKII revealed no significant changes in their distribution in gukh mutants.
In contrast, dramatic changes in the synaptic distribution of SCRIB were observed in gukh mutants. In wild-type larvae, SCRIB tightly colocalizes with DLG at type I boutons (Figure 4B). Interestingly, SCRIB immunoreactivity was much less intense, appearing dramatically mislocalized or not as tightly concentrated at the rim of type I boutons in gukhJ3E1 homozygotes, in gukhJ3E1/Df, and in gukh2 (Figure 5E; Table 2). The decrease in synaptic SCRIB localization in both the P element insertion allele (gukhJ3) and in the more severe excision allele (gukhJ3E1) was specific, as targeted expression of a UAS-gukh-c transgene rescued the mislocalization of SCRIB (Figure 5F), and synaptic SCRIB localization was restored in gukhrev and gukh2revEM30 (Table 2).
To determine if GUKH is required pre- or postsynaptically to maintain normal SCRIB localization at synaptic boutons, we used Gal4 drivers BG487 to target muscle-specific GUKH-C expression and C380 to drive transgenic expression in the motorneurons [37]. The UAS-gukh-c transgene encodes an amino-terminally truncated variant of GUKH (aa 652–1044), which lacks the WH1 domain but still contains the DLG and SCRIB binding motifs. As indicated by increased immunoreactivity, GUKH-C became localized to type I boutons upon both pre- and postsynaptic expression (data not shown). Surprisingly, we found that driving GUKH-C in motorneurons was sufficient to rescue the abnormal SCRIB localization in gukhJ3E1 mutants (Figure 5F; Table 2). However, driving GUKH-C in the muscles alone was much less effective in rescuing SCRIB localization at type I boutons. The reduced rescue observed with postsynaptic expression might be due to the lack of the amino-terminal region of the transgene.
We next determined whether mutations in dlg affect the synaptic localization of SCRIB or GUKH. In dlgX1–2 mutants, SCRIB was mislocalized to an extent similar to that observed in gukh mutants (Figures 6B and 6C, top), and this effect was enhanced in dlg;gukh double mutants (data not shown). Thus, both DLG and GUKH are required for normal SCRIB localization at NMJs. This relationship is unidirectional, since both DLG and GUKH immunoreactivities remained unaltered at NMJs in scrib2 mutant larvae (Figure 6D, middle and bottom). A simple explanation for the mislocalization of SCRIB in dlgX1–2 mutants would be that DLG recruits GUKH to the NMJ. However, we found that GUKH immunoreactivity was normal in dlgX1–2 mutants (Figure 6C, middle).
Figure 6. Relative SCRIB, GUKH, and DLG Signal at Type I Synaptic Boutons.
Representative images of synaptic boutons in wild-type (A), gukhJ3E1/Df (B), dlgXI–2 (C), and scrib2/Df (D) stained with anti-SCRIB (top), anti-GUKH (middle), and anti-DLG (bottom). Scale bar, 7.5 µm.
Discussion
Our previous studies have demonstrated that DLG is essential to cluster Shaker K+ channels and the cell adhesion molecule FasII, and mutations in dlg result in abnormal development of synapse structure [9, 10, 21]. In our quest to understand the nature of the enigmatic GUK domain of DLG, in this study we identified GUKH, a novel synaptic protein that binds to this domain of DLG. We further demonstrated that both DLG and GUKH are required for synaptic localization of another scaffolding protein, SCRIB.
Together, our yeast two-hybrid, coimmunoprecipitation, and colocalization studies provide compelling evidence that GUKH interacts with DLG in vivo. This interaction is mediated by a region near the C terminus of GUKH. However, as revealed by genetic analysis, the synaptic localization of GUKH does not depend on DLG. This suggests that domains other than the DLG interacting motif may mediate its synaptic localization. For instance, the single WH1-like domain of GUKH might interact directly or indirectly with the synaptic cytoskeleton. WH1 domains in other proteins bind F-actin, actin-associated proteins such as zyxin, vinculin, and profilin [38, 39], or the spectrin-bound scaffolding protein Shank/ProSAP [36, 40]. Association of GUKH with cytoskeletal elements might also be mediated by those sequences that exhibit moderate similarity to the actin binding protein Kelch [30].
The GUK domain of DLG and related MAGUKs is enzymatically inactive and may have evolved as a protein-protein interaction domain. A number of vertebrate GUK domain binding partners, including GKAP, MAP1A, the kinesin GAKIN, and the Rap-specific GTPase activating protein SPAR, have been identified [11, 15, 16]. Although these proteins are structurally quite diverse, a common theme appears to be their association with the cytoskeleton.
While an association of GUKH with the actin-based synaptic cytoskeleton currently remains hypothetical, we could demonstrate that the C-terminal tETAL motif specifically binds to the second PDZ domain of SCRIB. Anatomical and biochemical experiments suggest that in vivo, DLG, SCRIB, and GUKH may exist in the same complex at the NMJ. Alternatively, the three proteins could interact pairwise, forming separate heterodimers. Since GUKH was found to still localize normally at dlg mutant NMJs, we propose that DLG and GUKH act in concert rather than in a hierarchical manner to recruit SCRIB. As a possible mechanism, binding to the GUK domain of DLG could cause sterical changes in GUKH, such that the tETAL motif becomes available for interaction with SCRIB. A caveat to this study is that we used hypomorphic gukh mutants, and therefore, a requirement of GUKH in DLG localization cannot be ruled out.
The presence of multiple protein-protein interaction domains in both DLG and SCRIB suggests that GUKH may link two different multiprotein complexes in a defined spatial arrangement. This is reminiscent of the coupling of NMDA receptors and metabotropic glutamate receptors at mammalian PSDs through a quaternary complex formed by PSD-95, GKAP, Shank/ProSAP, and Homer [36].
While DLG and SCRIB are colocalized along the rims of synaptic boutons, which, as previously demonstrated for DLG, comprises both the presynaptic membrane and the SSR, GUKH intersected that region only in a narrow strip. Yet, in budding boutons, GUKH displayed a complementary pattern to DLG. These observations suggest that GUKH may not be continuously bound to DLG but rather may be involved in transient interactions. The process of bouton budding is a dynamic process that is characterized by equally dynamic changes in both GUKH and DLG distribution. The accumulation of GUKH at the core of budding boutons and the disappearance of DLG at the border of buds suggest that both proteins serve different roles during this process. Interestingly, FasII, a molecule that mediates synapse stabilization but that also imposes an adhesive constraint on synaptic growth [34], faithfully resembles the changes in distribution of DLG during budding [35], consistent with a role of DLG in synaptic localization [9]. The presence of GUKH at budding regions may represent a role for this protein in destabilizing regions of the synaptic bouton, thereby allowing for bud formation.
In contrast to GUKH, SCRIB was expressed throughout the SSR in exact colocalization with DLG. Nonetheless, SCRIB localization at distal regions of the SSR was also affected in gukh mutants. In fact, considering the hypomorphic character of the gukh alleles that were used in this study, the effect on SCRIB localization appears remarkably strong. This observation might indicate that GUKH activity is required only temporarily and/or in a locally restricted fashion to prime a secondary mechanism by which SCRIB becomes associated with the SSR, e.g., through a more direct interaction with DLG. Interestingly, presynaptic expression of GUKH-C was largely sufficient to restore postsynaptic SCRIB localization at gukh mutant NMJs. Together, these observations suggest a second, more indirect mechanism by which GUKH contributes to the recruitment of SCRIB to the postsynaptic SSR and which may involve trans-synaptic signaling.
Conclusion
Our studies provide evidence for one mechanism by which scaffolding proteins with different interaction domains may be linked to form a network of multiprotein complexes. GUKH, in physically linking DLG and SCRIB, can therefore bring together these complexes and their associated proteins. Since a single protein forms this link, it would be a straightforward point at which to also separate the complexes, along with their actions, to regulate different aspects of synapse formation. Examples would be during synapse stabilization and during synapse growth through bouton budding. Thus, our work provides a means by which macromolecular complexes can mediate and finely tune various structural changes at the highly dynamic structure of the synapse.
Experimental Procedures
Flies
We used the following fly strains: (1) mutations in gukh (P[hs neo]142; gukhJ3, a P[Gal4] derivative of P[hs neo]142 obtained by exchanging P[hs neo]142 by P[Gal4] according to [41]; gukhJ3E1 and gukhrev, generated by excision of gukhJ3; gukh2 (w1118; P{w+ mGT=GT1}CG5456BG02660); gukh2EM9 generated by excision of gukh2; gukh2revEM30, Df(3R)Cha7 and Df(3R)Dl-KX23 (deficiencies of the 91E region); (2) a mutation in dlg (dlgXI-2) [9, 21]; (3) mutations in scrib (scrib1, scrib2, and Df(3R)T1-x, a deficiency of the scrib region); [23, 24]; (4) Gal4 driver strains (BG487 and C380) [37]; (5) UAS constructs (UAS-GUKH-C and UAS-eGFP-GUKH-C), generated by germline transformation, and UAS-SCRIB [23]; (6) the wild-type strain Canton S (CS).
Yeast Two-Hybrid Analysis
The Clontech Matchmaker LexA Two-Hybrid System was used according to the manufacturer’s instructions. A late embryonic cDNA library cloned into the pB42AD plasmid was transformed into yeast strain EGY48 carrying the reporter plasmid p8op-lacZ (EGY48p[8op-lacZ]). A bait consisting of the DLG GUK domain (amino acids 765–961) inserted into the pLexA vector was used to screen 1.625 × 107 cDNAs. Positive clones were selected on the basis of activation of the LEU2 and LacZ reporter genes [10]. Strength of interaction was determined by time elapsed from application of the colorimetric substrate (X-Gal) to the onset of the color reaction.
Immunocytochemistry and Generation of GUKH Antibodies
The following antibodies were used [37]: rabbit or rat anti-DLGPDZ [37] (1:40,000 and 1:1,000); anti-FasII [37] (1:4000); anti-SCRIB [24] (1:1000); anti-GUKH (1:400); anti-synaptotagmin (1:200) [42]; anti-CSP (DCSP-2; 1:200) [43]; anti-synapsin (synorf1; 1:10) [44]; anti-HRP (Sigma; 1:400); FITC or Texas red-conjugated secondary antibodies (Jackson Labs; 1:200). The GUKH antibody was generated from amino acids 806–1044 of GUKH by immunization of rabbits and rats with His-tagged protein. The rabbit antiserum was affinity purified (Sigma-Genosys, Inc). Immunocytochemistry for each antibody and genotype was performed in a minimum of ten samples. For quantification of staining intensities (Table 2), samples were processed simultaneously and imaged using the confocal microscope, using identical acquisition parameters. Background intensity (noise) was measured in NIH Image by tracing a line of 100 µm across the muscle in a region devoid of boutons and measuring mean intensity (in 0–256 grayscale). Signal intensity at boutons was determined by tracing four lines at 90 to each other from the center of the bouton to the end of the bouton rim and measuring the maximum signal intensity (in a 0–256 grayscale). The mean of the four measurements was divided by the noise to obtain number in Table 2. Statistical analysis was performed using Student’s t test.
Immunoprecipitations
Preparations (30 to 50) (consisting of body wall muscles, NMJs, and CNS) were processed for immunoprecipitation as in [9]. Immunoprecipitations were performed with anti-SCRIB (5 µl undiluted) or anti-GUKH (10 µl undiluted).
Inverse PCR
Genomic DNA from gukhJ3 was digested with Sau3AI and self-ligated. A 600 bp P element flanking fragment was then amplified from the self-ligated genomic DNA by using the P[GawB] forward and reverse primers CGCTGTCTCACTCAGACTCAATAC and AACC CTTAGCATGTCCGTGG and sequenced.
Supplementary Material
Acknowledgments
We thank the Central Microscopy Facility (supported by National Science Foundation grant BBS8714235) at the University of Massachusetts, Amherst, for confocal support. We thank Dr. Konrad Zinsmaier for the CSP antibody; Dr. Erich Buchner for the synapsin antibody; and Dr. Hugo Bellen for the synaptotagmin antibody. This work was supported by grants from the National Institutes of Health (RO1 NS 37061 and NS30072 to V.B. and NIH F31 NS10861-01 to L.S.G.) and a Human Frontiers Research grant.
Footnotes
Supplementary Material including additional methodological details and figures showing GUKH immunoreactivity during embryonic development, GUKH immunoreactivity in chordotonal organs and epithelial cells, and GUKH expression in the larval CNS and eye disc can be found online at http://images.cellpress.com/supmat/supmatin.htm.
References
- 1.Garner CC, Kindler S, Gundelfinger ED. Molecular determinants of presynaptic active zones. Curr. Opin. Neurobiol. 2000;10:321–327. doi: 10.1016/s0959-4388(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 2.Koh YH, Gramates LS, Budnik V. Drosophila larval neuromuscular junction: molecular components and mechanisms underlying synaptic plasticity. Microsc. Res. Tech. 2000;49:14–25. doi: 10.1002/(SICI)1097-0029(20000401)49:1<14::AID-JEMT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 3.Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron. 1998;21:727–739. doi: 10.1016/s0896-6273(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 4.Kim E, Cho KO, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 5.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 6.Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 7.Furuyashiki T, Fujisawa K, Fujita A, Madaule P, Uchino S, Mishina M, Bito H, Narumiya S. Citron, a Rho-target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J. Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 9.Thomas U, Kim E, Kuhlendahl S, Koh YH, Gundelfinger ED, Sheng M, Garner CC, Budnik V. Synaptic clustering of the cell adhesion molecule fasciclin II by discs-large and its role in the regulation of presynaptic structure. Neuron. 1997;19:787–799. doi: 10.1016/s0896-6273(00)80961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J. Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J. Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J. Biol. Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 13.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 14.Boeckers TM, Winter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED. Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem. Biophys. Res. Commun. 1999;264:247–252. doi: 10.1006/bbrc.1999.1489. [DOI] [PubMed] [Google Scholar]
- 15.Brenman JE, Topinka JR, Cooper EC, McGee AW, Rosen J, Milroy T, Ralston HJ, Bredt DS. Localization of postsynaptic density-93 to dendritic microtubules and interaction with microtubule-associated protein 1A. J. Neurosci. 1998;18:8805–8813. doi: 10.1523/JNEUROSCI.18-21-08805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanada T, Lin L, Tibaldi EV, Reinherz EL, Chishti AH. GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. J. Biol. Chem. 2000;275:28774–28784. doi: 10.1074/jbc.M000715200. [DOI] [PubMed] [Google Scholar]
- 17.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 18.McGee AW, Bredt DS. Identification of an intra-molecular interaction between the SH3 and guanylate kinase domains of PSD-95. J. Biol. Chem. 1999;274:17431–17436. doi: 10.1074/jbc.274.25.17431. [DOI] [PubMed] [Google Scholar]
- 19.Shin H, Hsueh YP, Yang FC, Kim E, Sheng M. An intramolecular interaction between Src homology 3 domain and guanylate kinase-like domain required for channel clustering by postsynaptic density-95/SAP90. J. Neurosci. 2000;20:3580–3587. doi: 10.1523/JNEUROSCI.20-10-03580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Reissner C, Kuhlendahl S, Coblentz B, Reuver S, Kindler S, Gundelfinger ED, Garner CC. Intramolecular interactions regulate SAP97 binding to GKAP. EMBO J. 2000;19:5740–5751. doi: 10.1093/emboj/19.21.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, Gorczyca M. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 1996;17:627–640. doi: 10.1016/s0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas U, Ebitsch S, Gorczyca M, Koh YH, Hough CD, Woods D, Gundelfinger ED, Budnik V. Synaptic targeting and localization of discs-large is a stepwise process controlled by different domains of the protein. Curr. Biol. 2000;10:1108–1117. doi: 10.1016/s0960-9822(00)00696-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 24.Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403:676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 25.Bartel PL, Chien C-T, Sternglanz R, Fields S. Using the 2-hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular Interactions in Development: a Practical Approach. Oxford: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 26.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila ncodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 27.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 29.Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson DN, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 32.Guan B, Hartmann B, Kho YH, Gorczyca M, Budnik V. The Drosophila tumor suppressor gene dlg, is involved in structural plasticity at a glutamatergic synapse. Curr. Biol. 1996;6:695–706. doi: 10.1016/s0960-9822(09)00451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorczyca M, Augart C, Budnik V. Insulin-like receptor and insulin-like peptide are localized at neuromuscular junctions in Drosophila. J. Neurosci. 1993;13:3692–3704. doi: 10.1523/JNEUROSCI.13-09-03692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 35.Zito K, Parnas D, Fetter RD, Isacoff EY, Goodman CS. Watching a synapse grow: noninvasive confocal imaging of synaptic growth in Drosophila. Neuron. 1999;22:719–729. doi: 10.1016/s0896-6273(00)80731-x. [DOI] [PubMed] [Google Scholar]
- 36.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 37.Koh YH, Popova E, Thomas U, Griffith LC, Budnik V. Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell. 1999;98:353–363. doi: 10.1016/s0092-8674(00)81964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 39.Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 40.Bockers TM, Mameza MG, Kreutz MR, Bockmann J, Weise C, Buck F, Richter D, Gundelfinger ED, Kreienkamp HJ. Synaptic Scaffolding Proteins in Rat Brain. Ankyrin repeats of the multidomaIN Shank protein family interact with the cytoskeletal protein alpha-fodrin. J. Biol. Chem. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- 41.Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Littleton JT, Stern M, Schulze K, Perin M, Bellen HJ. Mutational analysis of Drosophila synaptotagmin demonstrates its essential role in Ca(2+)-activated neurotransmitter release. Cell. 1993;74:1125–1134. doi: 10.1016/0092-8674(93)90733-7. [DOI] [PubMed] [Google Scholar]
- 43.Zinsmaier KE, Eberle KK, Buchner E, Walter N, Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]
- 44.Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO, Reifegerste R, Reisch D, Schaupp M, Buchner S, Buchner E. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J. Neurosci. 1996;16:3154–3165. doi: 10.1523/JNEUROSCI.16-10-03154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.