Abstract
Withania somnifera, a multipurpose medicinal plant is a rich reservoir of pharmaceutically active triterpenoids that are steroidal lactones known as withanolides. Though the plant has been well-characterized in terms of phytochemical profiles as well as pharmaceutical activities, limited attempts have been made to decipher the biosynthetic route and identification of key regulatory genes involved in withanolide biosynthesis. This scenario limits biotechnological interventions for enhanced production of bioactive compounds. Nevertheless, recent emergent trends vis-à-vis, the exploration of genomic, transcriptomic, proteomic, metabolomics, and in vitro studies have opened new vistas regarding pathway engineering of withanolide production. During recent years, various strategic pathway genes have been characterized with significant amount of regulatory studies which allude toward development of molecular circuitries for production of key intermediates or end products in heterologous hosts. Another pivotal aspect covering redirection of metabolic flux for channelizing the precursor pool toward enhanced withanolide production has also been attained by deciphering decisive branch point(s) as robust targets for pathway modulation. With these perspectives, the current review provides a detailed overview of various studies undertaken by the authors and collated literature related to molecular and in vitro approaches employed in W. somnifera for understanding various molecular network interactions in entirety.
Keywords: Withania somnifera, withanolides, tissue culture, elicitor, medicinal plant, molecular cloning, secondary metabolites, pathway engineering
Introduction
Plants have long been known to allocate substantial resources toward developing chemical solutions to enhance survival strategies in the form of varied natural products. These natural products have been scrupulously used as pharmaceuticals, additives, pesticides, agrochemicals, fragrance and flavor ingredients, food additives, and pesticides. A great majority of these plant derived natural products have long been the basis of many traditional medicines and still they continue to provide mankind with new remedies. Since time immemorial medicinal plants and their extracts have been used by humans for the treatment of different ailments and diseases. For instance, oils of Cedrus species (Cedar), Cupressus sempervirens (Cypress), Glycyrrhiza glabra (Licorice), Commiphora species (Myrrh), and Papaver somniferum (Poppy juice), all of which are still in use today for the treatment of ailments ranging from coughs and colds to parasitic infections and inflammation. Many of the modern drugs against various ailments are also based on the chemical structures of such plant derived chemical products. During the period of 2005–2007 the Food and Drug Administration introduced 13 new drugs of natural origin into the market and more than 100 natural product-based drugs are in clinical studies (Li and Vederas, 2011). Today, all drugs used in western medicine, around 40–45% are natural products or compounds derived from them, and of these, 25% are obtained from plants. Moreover, the dominant role of natural products like vinca alkaloid derivatives (etoposide, teniposide, etoposide phosphate) acting against cancer (60%) and infectious diseases (75%) is all the more identified (Mander and Liu, 2010). In recent years, from a health perspective, protective dietary constituents in the form of plant derived natural compounds have become progressively significant part of human nutrition research (Pandey and Rizvi, 2009; Choi et al., 2012).
Such vast range of chemical entities in the form of natural products that do not contribute directly in growth and development of a plant are termed as secondary metabolites. Plant secondary metabolites like phenylpropanoids, terpenoids, and alkaloids play significant role in plant survival under specialized ecological conditions, e.g., biotic and abiotic stresses. In contrast with primary metabolites, secondary metabolites are often restricted in distribution and in many instances a specific secondary metabolite is associated with a specific taxonomic groups or a plant species. Medicinal plants have long been the basis of herbal drugs for prevention and treatment of various ailments and secondary metabolites are attributed with the medicinal properties (Croteau et al., 2000; Rao and Ravishankar, 2002). These herbal drugs have thus been in use for thousands of years in different cultures due to their potency, efficacy, low cost, and fewer side-effects. With an ever-increasing global demand for herbal medicine, there is not only requirement for large quantity of raw material of medicinal plants, but also of appropriate quality where active principles are available in desired concentrations (Shahid et al., 2013). Additionally, due to the complex chemical structures, it is often difficult to synthesize complex natural compounds through synthetic chemistry as the whole process is economically prohibitive. Thus, plants remain as a sole sustainable natural resource of many medicinally important secondary metabolites.
The biosynthesis of plant secondary metabolites is tightly regulated by spatial and temporal cues that limit the levels of targeted secondary metabolites in plant tissues (Dhar et al., 2013). Furthermore, many secondary metabolites are often species specific in distribution and the plant species in question may be distributed to specific geographical location. Cumulatively, these issues may limit proper exploitation of plants for large scale production of economically important secondary metabolites. For obvious reasons, a desirable aspect is to improve the level of secondary metabolites in native plant species as well as to develop alternative technologies to produce high value bioactive compounds in microbial or yeast heterologous hosts by using synthetic biology approaches. In this regard, molecular biotechnological interventions and in vitro approaches offer attractive possibilities for metabolic engineering of plant secondary metabolites. However, biogenesis of several important plant secondary metabolites at the level of pathway steps and their regulation is poorly understood. These issues can be attributed to the lack of functional genomics platforms comprising of genome resources, mutants, and transformation systems for medicinal plants. Hence, refinement in tools and techniques to carry out comprehensive studies of medicinal plant secondary metabolism is imperative.
Herein, we provide a comprehensive review of the detailed studies carried out by the authors and other significant contributions collated from the available literature to elucidate different aspects of biosynthesis of steroidal lactone compounds known as withanolides from W. somnifera, a medicinal plant of immense repute. It also entails many important aspects related to enhancement of withanolide production in corroboration with molecular deciphering and in vitro approaches to understand the regulation of withanolide production.
Withania somnifera
Withania somnifera (Solanaceae) commonly known as ashwagandha or Indian ginseng, is a valued medicinal plant known since antiquity (~3000 years; Winters, 2006). It is a commended genus described in the Indian Ayurvedic system of medicine and also enlisted as an important herb in Unani and Chinese traditional medicinal systems. It displays an efficient reproductive behavior of mixed mating which enables it to maximize benefits of both selfing as well as outcrossing. Its breeding behavior also guarantees wide chemotypic variability (Lattoo et al., 2007). W. somnifera is widely distributed around the world and is mainly adapted to xeric and drier regions of tropical and subtropical domains, ranging from the Canary Islands, the Mediterranean region and Northern Africa to Southwest Asia (Mirjalili et al., 2009).
Traditionally, W. somnifera is recommended to enhance physiological endurance, overall vitality, strength and general health. It is frequently equated with Korean ginseng (Panax ginseng) for its restorative bioactivities. It also aids to offset impotency, chronic fatigue, weakness, bone weakness, dehydration, premature aging, muscle tension, and emaciation. All parts of the plant, like leaves, stem, flower, root, seeds, and bark are used medicinally. Root of Withania is an important ingredient of more than 200 formulations in traditional systems of medicine like Ayurveda, Siddha, and Unani. These systems are being used for ages in the management of numerous physiological ailments (Sukanya et al., 2010). The bitter leaves of the plant have characteristic odor, used as an antihelmantic and infusion is given in fever. In Ayurveda, berries and tender leaves are prescribed to be applied externally to tumors, tubercular glands, carbuncles, and ulcers (Gauttam and Kalia, 2013).
Withanolides
W. somnifera is known to structure a wide-range of low molecular weight secondary metabolites for example terpenoids, flavonoids, tannins, alkaloids, and resins. It has been extensively studied for its chemical constituents that include compounds of diverse chemical structures viz. withanolides, alkaloids, flavonoids, tannin (Elsakka et al., 1990; Attaurrahman et al., 1991; Arshad Jamal et al., 1995; Choudhary et al., 2010). Of these, withanolides are credited with widely acclaimed remedying properties. Withanolides, with as many as 40 reported structures represent a collection of naturally occurring C-28 steroidal lactone triterpenoids assembled on an integral or reorganized ergostane structure, in which C-22 and C-26 are oxidized to form a six-membered lactone ring (Glotter, 1991; Ray and Gupta, 1994). The elementary structure is labeled as the withanolide skeleton chemically nomenclatured as 22-hydroxy ergostane-26-oic acid 26, 22-lactones (Misra et al., 2008). The withanolides are generally polyoxygenated and believed to be produced via enzyme system capable of catalyzing oxidation of all carbon atoms in a steroid nucleus (Kirson et al., 1971; Figure 1).
Figure 1.
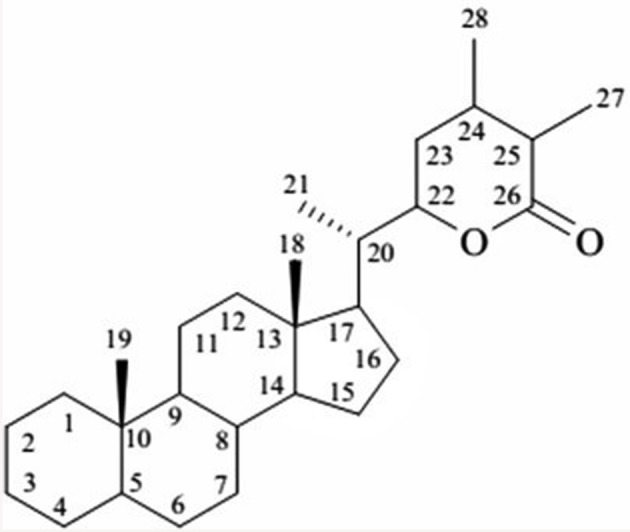
The basic structure of withanolide.
The characteristic feature of withanolides and ergostane-type steroids is one C-8 or C-9 side chain with a either six or five membered lactone or lactol ring. This lactone ring could be attached through a carbon-carbon bond or through an oxygen bridge with the carbocyclic part of the molecule (Kirson et al., 1971; Glotter, 1991). Various kinds of structural rearrangements involving oxygen substituents like bond scission, new bond formation, ring aromatization among others can result in formation of novel structural variants often described as modified withanolides or ergostane type steroids (Misico et al., 2011). They are distributed in distinct amounts and ratios in fruits and vegetative parts of the plant (Sangwan et al., 2008). However, withanolides are mainly localized to leaves, and their concentration is generally low i.e., ranges from 0.001 to 0.5% of dry weight basis (DWB). Many factors such as growth rate, geographical, and environmental conditions are known to modulate the content of withanolides (Dhar et al., 2013). Apart from Withania, withanolides are also distributed to other Solanaceous genera like Iochroma, Acnistus, Deprea, Datura, Lycium, Dunalis, Nicandra, Jaborosa, Physalis, Salpichroa, Tubocapsicum, Discopodium, Trechonaetes, and Witheringia. Moreover, their distribution is not restricted completely to Solanaceous plants, withanolides have been reported to be isolated from Taccaceae, Leguminosae (Glotter, 1991), Dioscoreaceae (Kim et al., 2011), Myrtaceae, and Lamiaceae families including reports of isolation from marine organisms also (Chao et al., 2011).
Pharmacology
During last two decades, there has been a notable surge in the pharmacological based research of this plant as evidence for anti-tumor, anti-arthritic, anti-aging, and neuroprotective properties (Budhiraja and Sudhir, 1987; Ray and Gupta, 1994; Jayaprakasam et al., 2003; Choudhary et al., 2005; Kuboyama et al., 2005; Kaileh et al., 2007; Singh et al., 2011) and also positive influences on the endocrine, cardiopulmonary, and central nervous systems have been reported (Mishra et al., 2000). Anabalagan and Sadique (1981) reported efficient anti-inflammatory activity of W. somnifera as compared to hydrocortisone. W. somnifera extracts have revealed anti-inflammatory effects in a range of rheumatological conditions (al-Hindawi et al., 1992). W. somnifera preparations are reported to influence the cholinergic and GABA-ergic neurotransmission (GABA: γ-amino-butyric acid) accounting for various central nervous system related disorders (Kulkarni and George, 1996; Tohda et al., 2005). The active principles, sitoindosides VII–X and withaferin A (WS-3) significantly reduces the lipid peroxidation and increased levels of the superoxide dismutase, catalase, glutathione peroxidase, and ascorbic acid activity, thus possessing a free radical scavenging property (Bhattacharya et al., 1997; Panda and Kar, 1997). Further, normal and cyclophosphamide-treated mice showed enhanced levels of interferon-γ, interleukin-2, and granulocyte macrophage colony stimulating factor by exhibiting immuno-potentiating and myeloprotective effects (Davis and Kuttan, 1999). Further, toxicity studies have shown withanolides to be safer compounds with minute or no related toxicity (Mishra et al., 2000). Withanolides have been noticed to enhance the ability of macrophage to “eat” pathogens (Davis and Kuttan, 2000). WS-3 acts defending in definite types of cancers as an inhibitor of angiogenesis (Mohan et al., 2004). Other studies reveal that withanolides possess potent antimicrobial potential against pathogenic bacteria, including Salmonella, an organism associated with food poisoning (Owais et al., 2005). Withanoside VI and withanolide A (WS-1) facilitate the regeneration of axons and dendrites by reconstructing pre- and post-synapses in neurodegenerative diseases and preventing pathogenesis and neuronal death. It is found to ameliorate the memory deficit in mice (Kuboyama et al., 2005, 2006). Mechanistically, withanolides act as anti-inflammatory agents by inhibiting lymphocyte proliferation, complement system, land delayed-type hypersensitivity (Rasool and Varalakshmi, 2006). WS-3 is a promising agent for the treatment of the inflammatory cascade of cardiovascular diseases as a potent inhibitor of the pro-inflammatory transcription factors (Kaileh et al., 2007). It exhibits in vivo anti-cancer activity against pancreatic cancer by inhibiting Hsp90 chaperone activity and another potential withanolide isolated from roots act as an effective agent to protect against skin carcinoma induced by UV-B (Mathur et al., 2004; Yu et al., 2010). Withanolidesulfoxide have been isolated and identified to inhibit COX-2 enzyme in various tumor cell lines and to suppress their proliferation (Mulabagal et al., 2009). Withanolide D can be used with traditional chemotherapeutic agents as it augments the ceramide accretion by triggering neutral-sphingomyelinase 2, modulate phosphorylation of the JNK and p38MAPK and induced apoptosis in both myeloid and lymphoid cells along with primary cells derived from leukemia patients (Mondal et al., 2010). WS-3 and withanolide D have been demonstrated to hamper angiogenesis, Notch-1, NFκB in cancer cells and trigger apoptosis in breast cancer cells (Kaileh et al., 2007; Koduru et al., 2010; Hahm et al., 2011). The oral administration of withanolides and withanosides reversed behavioral deficits, plaque pathology, accumulation of β-amyloid peptides (Aβ) in the animal models of Alzheimer disease through higher expression of low-density lipoprotein receptor-related protein in brain micro-vessels and the Aβ-degrading protease neprilysin (Sehgal et al., 2012). Recently, WS-3 was found to activate Cdc2 protein in prostate cancer cell lines which result in arrest of the cell cycle and leads to cell death (Roy et al., 2013) and the root extract have been observed to possess neuroprotective effect against β-amyloid and HIV-1Ba-L (clade B) induced neuro-pathogenesis (Kurapati et al., 2013). The other withanolides and alkaloids like withasomine, cuseohygrine, and anahygrine remain to be the promising lead-compounds for the development of the new anti-inflammatory drugs (Mirjalili et al., 2009).
Biosynthesis of withanolides
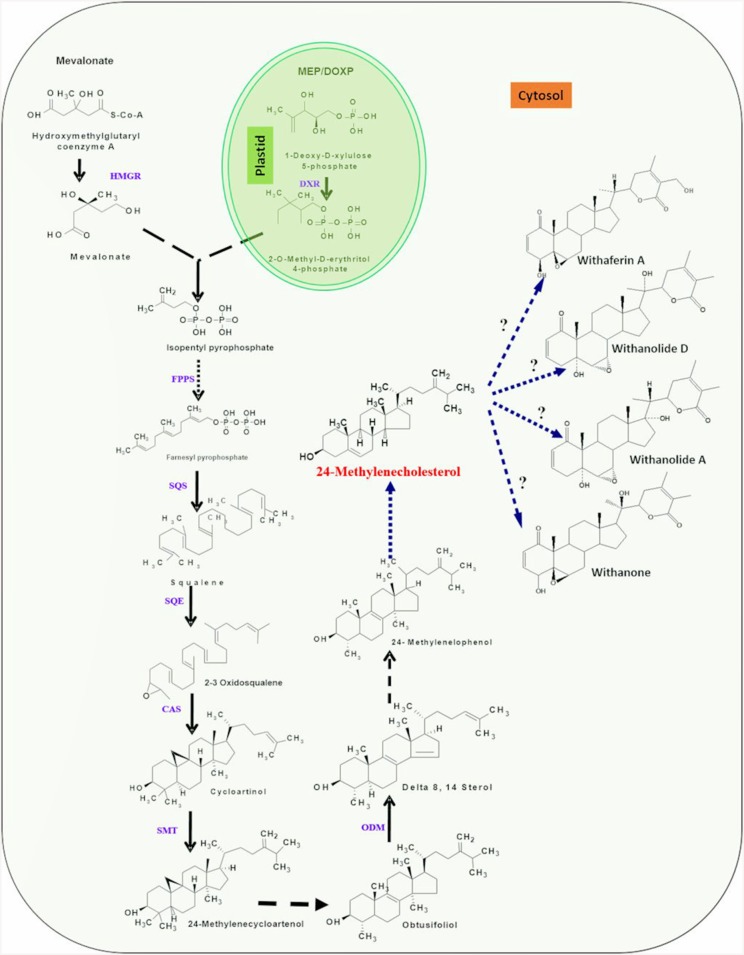
Chemically, withanolides are 30-carbon compounds called triterpenoids. Triterpenoid backbone, like other terpenoid compounds is biosynthesized by metabolic pathway requiring isoprene units (isopentenylnpyrophosphate; IPP and dimethyl allyl pyrophosphate; DMAPP) as precursors. Therefore, isoprenogenesis could be one of the key upstream metabolic processes governing flux of isoprene units for synthesis of metabolic intermediate(s) of triterpenoid pathway committed to withanolide biosynthesis (Bhat et al., 2012). Dual autonomous pathways for the isoprenoid precursor biosynthesis co-exist in plant cell including the classical cytosolic mevalonic acid (MVA) pathway and the alternative route, plastidial methylerythritol phosphate (MEP) pathway (Newman and Chappell, 1999). Plastidial MEP pathway synthesizes IPP and DMAPP required for production of photosynthesis associated isoprenoids (carotenoids and side chains of chlorophylls, plastoquinones, and phylloquinones) and hormones (gibberellins and abscisic acid). In plants, MVA-derived isoprenoid end products comprise of sterols (modulators of membrane architecture and plant growth and developmental processes), brassinosteroids (steroid hormones), dolichol (involved in protein glycosylation), and the prenyl groups necessary for protein prenylation and cytokinin biosynthesis (Lichtenthaler, 1999; Rodriguez-Concepción et al., 2004). However, now there is growing evidence that a considerable cross-talk between the two pathways of isoprenogenesis exists and exchange of isoprene units may occur at different sub-cellular locations (Chaurasiya et al., 2012). A brief overview of the two metabolic routes for isoprene biosynthesis is elucidated under following sub-sections (Figure 2).
Figure 2.
An overview of putative withanolide biosynthesic pathway. DXP, 1-deoxy-D-xylulose 5-phosphate; HMBDP, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; IPP, Isopentylpyrophosphate; DMPP, Dimethylalyl diphosphate; IPP isomerase, Isopentylpyrophosphate isomerase; FPPS, farnesyldiphosphate synthase; SQS, Squalene synthase; SQE/CPR, Squalene epoxidase/cytochrome P450 reductase; CAS, Cycloartenol synthase; SMT-1, Sterol methyl transferase/cytochrome P450 reductase; ODM/CPR, Obtusifoliol-14-demethylase/cytochrome P450 reductase. First three highlighted (yellow) steps indicating involvement of P450 monooxygenases and CPR. Single dark arrows represent one step, two or more dark arrows represent multiple steps and dashed arrow represents unknown steps.
Mevalonate pathway
MVA pathway involves seven enzymes for the synthesis of precursor molecules i.e., IPP and DMAPP for terpenoid biosynthesis. First step involves the condensation of two molecules of acetyl-CoA into acetoacetyl (AcAc)-CoA by the enzyme AcAc-CoA thiolase (Vranová et al., 2013) to form 3-hydroxy-3-methylglutaryl-coenzyme (HMG-CoA). In the second step, HMG-CoA synthase facilitate condensation of AcAc-CoA with one molecule of acetyl-CoA to form HMG-CoA (Nagegowda et al., 2004). Subsequently, HMG-CoA reductase (HMGR), a nicotinamide adenine dinucleotide (phosphate)-dependent (NAD(P)H) enzyme that catalyzes a double reduction reaction involving four electron transfers, results in biosynthesis of mevalonate from HMG-CoA (Benveniste, 2002). Conversion of mevalonate to IPP encompass two phosphorylations and decarboxylation events involving mevalonate kinase, phosphomevalonate kinase, and mevalonate diphosphate decarboxylase enzymes, respectively. Further, IPP derived from cytosolic MVA pathway, is acted upon by isopentenyl diphosphate isomerase, a divalent, metal ion-requiring enzyme, to form DMAPP (Hunter, 2007).
Methylerythritol phosphate pathway
First step of the MEP pathway is catalyzed by 1-deoxy-D-xylulose 5-phosphate synthase (DXS), converting the precursors pyruvate and glyceraldehyde 3-phosphate into 1-deoxy-Dxylulose 5-phosphate (DXP; Sprenger et al., 1997; Cordoba et al., 2009). DXP reductoisomerase transforms DXP into MEP which is further converted to 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate by the consecutive enzymatic action of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, and (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HMBPP). The last step is the branching of HMBPP to IPP and DMAPP catalyzed by the simultaneous enzymatic action of a single enzyme, (E)-4-hydroxy-3-methyl but-2-enyl diphosphate reductase (HDR). Although, HDR in MEP pathway produces both IPP and DMAPP, albeit at 85:15 ratio, the plastid localized isopentenyl diphosphate isomerase is involved in substrate optimization by catalyzing IPP isomerization. The head-to-tail condensation of IPP leads to formation of farnesyl pyrophosphate (FPP). FPP is the main precursor for triterpenoids (Kuzuyama, 2002; Chaurasiya et al., 2012) and is synthesized by catalytic action of the enzyme farnesyl diphosphate synthase (FPPS). It serves as a substrate for first committed reaction of several branched pathways leading to synthesis of compounds that are essential for plant growth and development as well as of pharmaceutical interest (Newman and Chappell, 1999). FPPS catalyzed reaction occurs in two consecutive steps; condensation of IPP with DMAPP to form 10-C intermediate geranyl diphosphate (GPP) and condensation of GPP with another molecule of IPP which results into 15-C FPP (Ohnuma et al., 1996). Squalene is believed to be a metabolic intermediate for biosynthesis of diverse triterpenoids. Its biosynthesis takes place by a reaction requiring squalene synthase enzyme that catalyzes head-to-head condensation of two molecules of FPP in NADPH dependent manner to produce squalene. The squalene undergoes epoxidation at one of its terminal double bonds by squalene epoxidase yielding squalene 2,3-epoxide. A ring closure reaction acting upon squalene 2,3-epoxide, catalyzed by cycloartenol synthase enzyme leads to the biosynthesis of cycloartenol that may further get converted into a variety of different steroidal triterpenoidal skeletons (Zhao et al., 2013; Dhar et al., 2014). The 24-methylenecholestrol, believed to be biosynthesized from cycloartenol has been proposed to be a central intermediate in the metabolic route toward withanolide biosynthesis. The hydroxylation at C-22 and δ-lactonization between C-22 and C-26 of 24-methylenecholestrol are believed to be important reactions leading to withanolides biosynthesis (Figure 2). In addition, it has also been suggested that α, β-unsaturated ketone in ring A of common withanolides may be produced through the sequential reactions. There is a scanty understanding with respect to enzymes and genes involved in these downstream reactions and withanolide biosynthesis.
De novo tissue-specific withanolide biosynthesis
Phytochemical data generated over the years support considerable qualitative overlap of leaf and root withanolides in W. somnifera. Gradient of withanolide concentration with higher in leaves and lower in roots and radiotracer studies using 24-methylene cholesterol as a precursor also hints toward a possible import of withanolides from leaves to roots. Nevertheless, investigations reveal incorporation of 14C from [2–14C]-acetate and [U-14C]-glucose into WS-1 in in vitro cultured roots and native/orphan roots of W. somnifera. This study showed these primary metabolites being integrated into WS-1, thus indicative of de novo synthesis of root-specific WS-1 from primary isoprenogenic precursors rather than hinting toward an import from leaves (Sangwan et al., 2007).
Another interesting study reveals the analogy in the qualitative and quantitative profile of withanolide accumulation in leaf and root tissues of two morpho-chemovariants, suggesting de novo tissue-specific withanolide biosynthesis. Two genetic stocks designated as WS-Y-08 (25-30 cm tall with yellow berries) and WS-R-06 (100–125 cm tall with red berries) showed appreciable variation in their competence to synthesize and accumulate different withanolides. Three major withanolides viz. WS-1, withanone (WS-2), and WS-3 were assayed from leaf and root tissues harvested at five developmental stages. Additionally, transcript profiles of five withanolide biosynthetic pathway genes namely squalene synthase (WsSQS; GenBank Accession Number GU474427), squalene epoxidase (WsSQE; GenBank Accession Number GU574803), cycloartenol synthase (WsOSC/CS; GenBank Accession Number HM037907), cytochrome P450 reductase 1 (WsCPR1; GenBank Accession Number HM036710), cytochrome P450 reductase 2 (WsCPR2; GenBank Accession Number GU808569; Table 1) were also examined in the harvested tissues. The aim was to compare gene expression with that of metabolite flux at different phenophases. Relative transcript abundance demonstrated significant deviation in leaf and root tissues that was mostly parallel with the divergence in withanolide pool. Leaves in comparison to roots showed elevated gene expression in corroboration with improved concentration of all the three withanolides. This relative dynamics of all the three withanolides at quantitative and qualitative levels in the two withanolide richest tissues possibly indicate toward de novo tissue-specific biosynthesis (Dhar et al., 2013).
Table 1.
Putative pathway genes from Withania somnifera.
| Genes involved | Expression system used | Abiotic/Biotic factors used | Effect on gene/Expression and withanolide levels | Reference |
|---|---|---|---|---|
| 3-Hydroxy-3-methylglutaryl coenzyme A reductase | E. coli | Salicylic acid, methyl-jasmonate, and mechanical injury | http://link.springer.com/article/10.1007%2Fs00709-012-0450-2 | |
| WsSGTL1/Sterolglycosyltrnasferases | A. thaliana | Salt, heat, and cold | Increase in WsSGTL1 HSP70, HSP90, RD29, SOS3, and LEA4-5 levels | http://www.ncbi.nlm.nih.gov/pubmed/23646175 |
| 1-Deoxy-D-xylulose-5-phosphate synthase | Salicylic acid, methyl jasmonate, mechanical injury | Maximum expression in flower and young leaves | http://www.ncbi.nlm.nih.gov/pubmed/22526204 | |
| 1-Deoxy-D-xylulose-5-phosphate reductase | Salicylic acid, methyl jasmonate, mechanical injury | Differentially expressed in different tissues | http://www.ncbi.nlm.nih.gov/pubmed/22526204 | |
| WsCYP98A | E. coli | Methyl jasmonate salicylic acid, gibberellic acid | Methyl jasmonate salicylic acid increased and gibberellic acid decreased WsCYP98 expression | http://www.biomedcentral.com/1472-6750/14/89 |
| WS-3 accumulation increased upon methyl jasmonate elicitation; gibberellic acid treatment decreased WS-3 and WS-1 accumulation | ||||
| WsCYP76A | E. coli | Methyl jasmonate, salicylic acid, gibberellic acid | Methyl jasmonate, salicylic acid as positive regulators and gibberellic acid acted as a negative regulator for WsCYP98A | http://www.biomedcentral.com/1472-6750/14/89 |
| Methyl jasmonate elicitation increased WS-3 accumulation; gibberellic acid decreased WS-3 and WS-1 accumulation | ||||
| WsCPR1 | E. coli | Methyl jasmonate, salicylic acid | The expression remained unchanged in WsCPR1; increase in WS-1 levels, less in WS-2 levels | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3578826/ |
| WsCPR2 | E. coli | Methyl jasmonate, salicylic acid | The treatments with methyl jasmonate, salicylic acid resulted in induction of WsCPR2 the expression levels | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3578826/ |
| WsSQS | E. coli | Methyl jasmonate, salicylic acid | Withanolides were regulated by methyl-jasmonate, salicylic acid and 2,4-D, treatments | http://www.ncbi.nlm.nih.gov/pubmed/22425978 |
| WsSQE | E. coli | Methyl jasmonate, salicylic acid | Biosynthesis of withanolide was up-regulated by methyl-jasmonate, salicylic acid and 2,4-D | http://www.ncbi.nlm.nih.gov/pubmed/23065254 |
| Oxidosqualene cyclases (OSCs) WsOSC/LS, WsOSC/BS, WsOSC/CS | S. pombe | Methyl jasmonate, gibberellic acid and yeast extract | Upon methyl jasmonate treatment transcript levels of WsOSC/LS were down regulated, unchanged in case of WsOSC/CS, and upregulated in case of Bs. YE down regulated WsOSC/LS and WsOSC/BS transcripts. gibberellic acid induced WsOSC/BS and down regulated WsOSC/LS transcripts | http://www.jbc.org/content/early/2014/04/25/jbc.M114.571919 |
| WsDWF1 | E. coli | Methyl jasmonate, gibberellic acid and 2,4-D | The treatments with methyl jasmonate, 2,4-D and salicylic acid resulted increased expression of WsDWF1 | |
| Transcripts | ||||
| WsFPPS | Methyl jasmonate, salicylic acid, mechanical injury | Maximum expression was found in flower and young leaf and NMITLI-101 chemotype. Significant elevation in response to salicylic acid, methyl jasmonate and mechanical injury | http://link.springer.com/article/10.1007%2Fs10725-011-9578-x#page-1 |
Gene elucidation of withanolide biosynthetic pathway
Synthesis of withanolides chemically, is complex due to the stereo-chemical ring closure, occurrence of chiral centers, rigid trans-lactone groups, and high energy epoxy ring (Neumann et al., 2008). Thus, making synthetic production economically unworkable due to minimal yields at high costs. Therefore, it demands viable alternatives for the production of withanolides in large quantities for commercial exploitation. Genetic manipulation with genes encoding enzymes involved in withanolide biosynthesis seems to be a viable approach that may be useful in developing genotypes of W. somnifera with enhanced levels of withanolides as well as for withanolide producing alternative microbial/yeast hosts. This whole approach entails expression of metabolic circuitries in heterologous host(s). It requires comprehensive knowledge about complete genetic architecture of withanolide biosynthesis including enzymatic and regulatory genes of the pathway. Nevertheless, withanolide biosynthetic pathway still remains in its putative stage at molecular level. Hence, it is of fundamental research value to elucidate the withanolide biosynthetic pathway encompassing the delineation of regulatory aspects of their biosynthesis also. Inception of pathway exploration has gained much interest by researchers over the past few years vis-à-vis withanolides. There have been several endeavors by many workers including comprehensive investigations carried out by our group related to isolation, cloning, characterization and regulation of several pathway genes, promoters, elicitations in corroboration with metabolite production and substrate pool diversion for enhanced withanolide yields. These different aspects are covered in the ensuing text.
3-hydroxy-3-methylglutaryl coenzyme A reductase
3-Hydroxy-3-methylglutaryl coenzyme A reductase (3-HMGR) is a NADH dependent rate limiting enzyme involved in the conversion of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) into mevalonic acid; chief precursor of IPP and DMAPP in the isoprenoid biosynthetic pathway. Plant HMGR have been located in the subcellular organelles like endoplasmic reticulum (ER), mitochondria, and plastids with catalytic domains present in cytosolic portions of cell (Kim et al., 2015). Owing to its rate limiting nature this enzyme is a target of various cholesterol lowering drugs such as statins (Istvan and Deisenhofer, 2001). Functionally it has been characterized in both mammalian and plant systems. In plants HMGR is encoded by multigene family which exhibits variegated temporal and spatial expression pattern. In Arabidopsis HMGR isozymes are encoded by HMGR-1 and HMGR-2. Loss of function of HMGR-1 leads to the generation of dwarf phenotype, sterility and senescence, corresponding to the diminished sterol biosynthesis (Kim et al., 2015). Considering its important role in isoprenoid biosynthesis HMGR has been explicitly studied and characterized in many plant and other plant species which include Catharanthus roseus, Ginkgo biloba, Taxus media, Salvia miltiorrhiza, etc. (Akhtar et al., 2013).
Studies carried out have shown the existence of a relationship between withanolide biosynthesis and HMGR-1 expression. In this study a positive correlation between high transcript levels of HMGR and optimum accumulation of withanolides was found in the root tissue of W. somnifera (Table 1; Akhtar et al., 2013). It may be attributed to the enhanced biosynthesis of substrate pool or precursors like IPP and DMAPP for various biosynthetic pathways including withanolide biosynthetic pathway. Also reports of HMGR-1 mutants generating diminished sterol content in A. thaliana and mevinolin directed inhibition of HMGR leading to significant decrease in total ginsenoside in P. ginseng adventitious roots (Kim et al., 2015) suggest a link between HMGR-1 expression and sterol biosynthesis. Positive elicitation i.e. increased expression of WsHMGR in response to salicylic acid (SA) and methyl jasmonate (MJ) indicates presence of cis regulatory elements in promoter region which may regulate the expression of WsHMGR in various biosynthetic pathways including withanolide biosynthetic pathway. Young leaves expressed high levels of WsHMGR transcripts than in mature leaves (Chaurasiya et al., 2007). These results correlated positively with the enhanced levels of withanolide production in young leaves relative to that of mature leaves of W. somnifera. HMGR has been demonstrated as an accelerator of isoprenoid biosynthesis. There was two-fold increase in the biosynthesis of β-carotene in E. coli (Akhtar et al., 2013) by tandem expression of WsHMGR and PAC Beta gene. It suggested that HMGR provides enhanced progenitor substrate pool for various biosynthetic pathways including withanolide biosynthesis.
1-deoxy-D-xylulose-5-phosphate reductoisomerase and 1-deoxy-D-xylulose-5-phosphate synthase
Plausibly, withanolides, the signature secondary metabolites of W. somnifera, are biosynthesized through metabolic deviation from sterol pathway at the level of 24-methylene cholesterol (Sangwan et al., 2004). Isoprenoid precursor for the same is synthesized by MVA pathway and MEP pathway wherein MEP pathway is the plastid-derived alternative route for isoprenoid biosynthesis. MEP pathway contributes about 30% in biosynthesis of withanolide precursor isoprenoids (Tuli et al., 2009) in which the first step is a condensation of pyruvate with Dglyceraldehyde-3-phosphate to form 1-deoxy-D-xylulose-5-phosphate (DXP), catalyzed by DXP synthase (DXS). DXP acts as a precursor for IPP and DMAPP biosynthesis (Julliard and Douce, 1991; Julliard, 1992; Himmeldirk et al., 1996). Subsequently, conversion of DXP to MEP, is catalyzed by DXP reductoisomerase (DXR). Though, DXR is the opening committed step for terpenoid biosynthesis through the MEP, DXS, the first enzyme of this pathway, also is significant for isoprenoid biosynthesis in several organisms, including bacteria and plants (Estévez et al., 2001; Guevara-García et al., 2005). To understand the significance of MEP pathway in isoprenoid biosynthesis in Withania, full-length cDNAs of WsDXS and WsDXR were cloned and characterized (Table 1). Saptial expression analysis revealed elevated level of WsDXS and WsDXR transcripts in young leaf that correlates with the reported enhanced rates of withanolide biosynthesis in young than in mature leaf of W. somnifera (Chaurasiya et al., 2007). Lower root expression of WsDXS and WsDXR than leaf also hinted toward their plastid-localization. This hints toward root being less active site for isoprenoid biosynthesis utilizing substrate from MEP pathway. Though, leaves and roots both are involved in withanogenesis independently, leaves are possibly the prime site for the same. Further, gene expression level of WsDXR and WsDXS were corroborated with qualitative and quantitative withanolide variations in chemotypes NMITLI-101, NMITLI-118, and NMITLI-135 possessing WS-3, WS-2, and withanolide D as the main withanolides in leaf tissue and withanolide A as the main withanolide in the root tissue. WsDXS expressed maximally in leaf of NMITLI-118 and 135 which was concurrent with high accretion of withanolides in these chemotypes. Conversely, expression of WsDXR transcript was observed approximately equivalent in leaves of all three chemotypes. Consequently, indicating that enzymes contribute in similar manner during isoprenoid biosynthesis in different chemotypes (Gupta et al., 2013a).
Farnesyl diphosphate
MVA and MEP pathway are attributed with most of the bioactive molecules synthesized in Withania. In these biosynthetic routes, farnesyl diphosphate (FPP), acts as a substrate for foremost committed reaction of numerous branched pathways and is synthesized by the enzyme farnesyl diphosphate synthase (FPPS) in two successive steps. Firstly, condensation of IPP with DMAPP structures 10-C intermediate geranyl diphosphate (GPP). Further condensation of GPP with another molecule of IPP forms FPP. FPPS is an important enzyme for biosynthesis of isoprenoid that synthesize sesquiterpene precursors for vital metabolites including sterols, dolichols, ubiquinones, and carotenoids in addition to substrates for farnesylation and geranylgeranylation of proteins. Overexpression of ginseng farnesyl diphosphate synthase in Centella asiatica hairy roots also enhanced phytosterol and triterpene biosynthesis (Kim et al., 2010). FPPS as well caters an important role in incipient steps of triterpenoid precursor production related to withanolide biosynthesis. Consequently, highlighting the significance of FPPS in any pathway engineering attempt for enhancing a desired isoprenoid of primary or secondary importance. FPPS has been characterized from a range of different plant species like Arabidopsis (Closa et al., 2010), Artemisia (Matsushita et al., 1996), Hevea (Takaya et al., 2003), maize (Cervantes-Cervantes et al., 2006), etc. As a step toward elucidating the significance of FPPS as the key entry point enzyme of the withanolide biosynthesis in W. somnifera, full-length FPPS cDNA was isolated and characterized as it constitutes a key step en route to biosynthesis of the progenitor(s) of withanolide biosynthesis (Table 1). Significance of WsFPPS gene in synthesis of sesqui- and higher isoprenoids counting the metabolites obtained from them was displayed by the constitutive expression of WsFPPS in all parts of the plant. Higher expression level of WsFPPS in young leaf as compared to the mature leaves corroborated with the reported enhanced withanolide biosynthesis in young leaf of W. somnifera (Chaurasiya et al., 2007; Gupta et al., 2011).
Squalene synthase
Squalene synthase (SQS) (farnesyl diphosphate: farnesyl diphosphate farnesyl transferase, EC 2.5.1.21) catalyzes one of the initial enzymatic steps of phytosterol biosynthetic pathway, facilitating condensation of two farnesyl pyrophosphate molecules to squalene. SQS routes carbon flux from isoprenoid pathway toward the phytosterol biosynthesis resulting in formation of endproducts like brassinosteroids, withanolides, and triterpenoids (Abe et al., 1993). SQS has been reported to be active in ER, it anchors to it via carboxyterminal portion. The cytosolic portion is anchored via amino terminal of protein (Robinson et al., 1993). SQS plays a key regulatory function in phytosterol biosynthesis. Overexpression of SQS genes in P. ginseng (Lee et al., 2004) and Eleutherococcus senticosus (Seo et al., 2005) led to the improved accretion of phytosterols and triterpenes thus highlighting the significant regulatory function of SQS in plants. Although ample evidence is available regarding the role of SQS in phytosterol biosynthesis, scanty is identified about the biosynthetic pathway of withanolides, genes involved in the withanolide biosynthesis and regulatory elements of promoter region governing the gene expression in W. somnifera. Thus, for pathway intensification leading to enhancement of withanolide accumulation in W. somnifera Bhat et al. (2012) investigated the significance of squalene synthase in withanolide biosynthesis. Characterization of WsSQS including tissue specific expression analysis and regulatory studies at promoter level substantiated WsSQS as an imperative gene target involved in withanolide biosynthesis (Table 1). WsSQS demonstrated increased expression pattern in leaves that was in consonance with the elevated production of withanolides in leaves of W. somnifera. Additionally, biosynthesis of withanolides and mRNA abundance of WsSQS were enhanced through diverse signaling molecules including methyl-jasmonate, salicylic acid, and 2,4-D that was regular with the expected results of WsSQS promoter. Thus, hinting toward the unraveling of a key committed step of the withanolide biosynthetic pathway (Bhat et al., 2012).
Squalene epoxidase
Squalene epoxidase (SE) (EC 1.14.99.7) is a rate limiting enzyme in the sterol biosynthetic pathway, catalyzing the conversion of squalene into 2,3-oxidosqualene by carrying out stereospecific epoxidation reaction (Ryder, 1992; He et al., 2008). This enzyme requires cytosolic (S105) fraction, molecular oxygen, NADPH-cytochrome c reductase, NADPH and flavine adenine dinucleotide (FAD) for its activity (Abe et al., 2007). SQE, a lightly bound FAD flavin, attains electrons from NADPH-cytochrome reductase, instead of binding the nicotinamide cofactor directly which differentiates it from other flavin mono-oxygenases. It is mainly located in the endoplasmic reticulum and lipid droplets but protein located in the endoplasmic reticulum is active (Leber et al., 1998). Additionally, SQE activity can result in the formation of 6,7-oxidosqualene, 10,11-oxidosqualene, and dioxidosqualene (Bai and Prestwich, 1992). SQE has been found to be main precursor for all identified angiosperm cyclic triterpenoids, that comprise membrane sterols, non-steroidal triterpenoids, brassinosteroid, and phytohormones. Being a rate limiting enzyme SQE has a cascading influence on the upregulation of downstream genes (Han et al., 2010). Thus, genetic manipulation of SQE in host plant offers an exciting prospect for production of desired therapeutic triterpenoid molecules (Takemura et al., 2010). This has been demonstrated in P. ginseng for heightened synthesis of triterpene saponins and phytosterols using squalene synthase (Lee et al., 2004). Against this backdrop, Razdan et al. (2013) has reported the substantial notice to comprehend the regulatory function of SE in withanolides biosynthesis. Toward this goal, WsSQE gene along with its promoter was isolated from W. somnifera and several cis-regulatory elements of promoter region were revealed (Table 1). This paves a way to recognize the regulatory function of SQE in withanolides biosynthesis as WsSQE also displayed maximum expression in withanolide richest leaf tissue. Keeping in view the prospect of pathway intensification, significance of WsSQE as a robust target lies in its rate limiting nature (Razdan et al., 2013). This significance can be utilized with an efficient Agrobacterium mediated transformation system in W. somnifera for homologous modulation of withanolide biosynthesis.
Cycloartenol synthase
Withanolides are synthesized via both MVA and MEP pathways which direct the flux of the isoprene (C5) units for the synthesis of triterpenoid pathway intermediates which are further committed to withanolide biosynthesis. Sterols, withanolides, and various triterpenoids are synthesized through a common 30-carbon intermediate 2,3-oxidosqualene in a highly regio and stereo-specific step catalyzed by a family of genes called oxidosqualene cyclases (OSCs) (Phillips et al., 2006). Plants produce a variety of triterpenoid skeletons structured by numerous OSC enzymes broadly belonging to two groups i.e., protosteryl and dammarenyl cations based on the nature of their supposed catalytic intermediates (Phillips et al., 2006). Both these cations as backbones impart discrete stereochemistry and ring configurations to various triterpenes. The protosteryl cation with chair-boat-chair (C-B-C) configuration forms cycloartenol, lanosterol, cucurbitadienol, and parkeol tetracyclic triterpene structures. The majority of the pentacyclic triterpenes are however, derived from the dammarenyl cation by D-ring expansion to form lupeol or further E-ring expansion to form β-amyrin (Xu et al., 2004). Similarly, partitioning of the common substrate, 2,3-oxidosqualene in W. somnifera takes place between OCS-cycloartenol synthase [(S)-2,3-epoxysqualene mutase (cyclizing, cycloartenol forming), EC 5.4.99.8] (CAS) and other OSCs. CAS forms cycloartenol, a pentacyclic triterpene with nine chiral centers and functions as the precursor to phytosterols and apparently to withanolides and other diverse OSCs structure diverse triterpenoids like lupeol, beta amyrin, etc., (Rees et al., 1969). This partitioning constitutes a metabolic branching point leading to the division of 2,3-oxidosqualene between sterol/withanolides and range of triterpenoids (Figure 1). Thus, making genes covering the branches of these sub-dividing point prospective candidates for perturbation. Such manipulations hold significant possibility of impacting respective branch flux by redirecting the precursor reservior in the direction of preferred secondary compound and concurrently reduce the flux via competitive biosynthetic routes. Against this backdrop, in W. somnifera three members of OSC superfamily viz. β-amyrin synthase (WsOSC/BS; GenBank Accession Number JQ728553), lupeol synthase (WsOSC/LS; GenBank Accession Number JQ728552), and WsOSC/CS covering three branches of a sub-dividing junction leading to withanolides, sterols and a suite of triterpenoids have been characterized (Table 1). Regulatory studies of WsOSCs involving plant-derived methyl jasmonate and giberrelic acid and microbe-derived yeast extract elicitations displayed differential transcriptional and translational profiles that were clearly reflected in visible variations in withanolide quantity. MJ elicitation considerably augmented WS-3 accretion over a period of 48 h that was in consonance with studies involving MJ-induced up-regulation of WsSQS, WsSQE, and WsCPR2 mRNA also led to enhanced withanolide accumulation. It may be attributed to increased synthesis of 2,3-oxidosqualene produced by induced upstream genes. As a consequence, WsOSC/CS is able to utilize an increased precursor pool for withanolide biosynthesis. Although the OSC mRNA expression model in case of gibberellic acid (GA3) coincided with MJ treatment, the total withanolide accumulation demonstrated a regular drop with increasing time course. This may be attributed mainly to the decrease in WsOSC/CS protein concentration as evident from the Western blot study. Nevertheless, transcript abundance of WsOSC/BS showed a rise that hinted toward the decrease in the total substrate availability for WsOSC/CS, but at the protein level, WsOSC/BS expression declined with increasing time intervals, thus possibly substantiating the drop in WS-3 concentration caused by decreased WsOSC/CS protein availability. Interestingly, microbe-derived exogenous yeast extract (YE) elicitor played a role of negative regulator for the two competitive OSCs of WsOSC/CS (WsOSC/BS and WsOSC/LS) at both the protein and mRNA levels, whereas WsOSC/CS showed no change in its transcript or protein expression in response to YE. However, there was significant increase in withanolide concentration with YE in comparison with MJ treatment. The down-regulation of WsOSC/BS and WsOSC/LS is possibly indicative of differential channeling of common substrate among the three branch OSCs. Plausibly, this leads to rearrangement of metabolic fluxes wherein bulk of 2,3-oxidosqualene substrate pool shifts toward WsOSC/CS, leading to much improved withanolide yields. The characterization and validation of WsOSCs seem important for strategizing the enhanced production of withanolides (Dhar et al., 2014).
Cytochrome P450 reductase and monooxygenases
Cytochrome P450 enzymes, member of one of the key functionally diverse protein super-families. It is essential in a variety of metabolic molecular circuitries. P450s are heme thiolate-proteins, catalyse enormously varied reactions like hydroxylations, dealkylations, sulfoxidations, epoxidations, reductive dehalogenations, peroxidations, and different types of isomerization for the synthesis of a number of primary and secondary metabolites indispensable for plant growth and development (Guengerich, 2001a; Hrycay and Bandiera, 2012). P450 monooxygenases comprise a substrate explicit class of enzymes which are highly regio and stereo-specific. Gene annotation has shown that about 1% of the entire genes in the plant's genome are cytochrome P450s. Arabidopsis genome contains 244 genes and 28 pseudo-gene representing cytochrome P450s (Nelson et al., 2004). P450s are ER localized with their catalytic functioning depending on source of electrons through NADPH cytochrome P450 reductase (CPR: diflavoenzyme).
CPRs (EC 1.6.2.4) possess a N-terminal positioned flavin mononucleotide (FMN) binding domain linked to NADPH binding domain via flavin adenine dinucleotide (FAD) domain are membrane bound proteins localized to ER. These are responsible for shuttling electrons obtained from NADPH through FAD and FMN domains into the heme iron-center of the various P450 enzymes. CPR genes have been isolated from numerous species of yeast, animals and insects. Between all, only one form is identified to network with several P450s (Simmons et al., 1985). Conversely, many CPR paralogs varies based on the vascular plant species. Ro et al. (2002) have categorized CPRs into Class I and class II groups on the basis of N-terminal anchoring sequences. CPR1 belonging to class I expresses constitutively whereas CPR2 in class II, is expressed in stress or on wound elicitation. Plants encode several CPRs that reflects the range of P450s (Werck-Reichhart et al., 2000; Feldmann, 2001) and their role in primarily confronting the elevated requirement of electrons during various stresses or varied expression at different plant developmental stages (Mizutani and Ohta, 2010).
Additionally, P450 monooxygenases also correspond to a highly regio and stereo-specific class of fixed substrate-specific enzymes that play a decisive function in secondary metabolism and mostly aid in functionalizing core structures of molecules like withanolides. Due to their regio and stereo-specific catalyzing flexibility, these are possible targets for industrial biocatalysis. P450s have been useful in industry for the examination of new medicine, drugs or xenobiotics (Guengerich, 2002, 2011; Miners, 2002). P450s are considered as the most versatile biological catalysts in nature due to the notable diversity of chemical reactions catalyzed and vast substrates attacked (Sono et al., 1996; Guengerich, 2001b; Coon, 2005). Consequently, accentuating their identification and characterization for biosynthetic pathway elucidation. Due to the polyphyletic nature of plant P450s these are commonly categorized in two main clades, A-type and non-A-type. Plant specific metabolism and biosynthesis of diverse natural products involves A-type P450s (Bak et al., 2011). The molecular and biochemical characteristics of cytochrome P450 reductases and monooxygenases in relation to pathway engineering, emphasizes the significance of this family of genes as robust gene targets of both primary and secondary metabolite biosynthesis.
Molecular and biochemical studies started to reveal biosynthetic routes for several withanolides in W. somnifera encompasses this important family of genes. Two A-types P450 WsCYP98A (GenBank Accession Number HM585369) and WsCYP76A (GenBank Accession Number KC008573) and two paralogs of cytochrome P450 reductases from W. somnifera have been isolated, sequenced and heterologously expressed in E. coli (Rana et al., 2013, 2014) (Table 1). All the four CYPs at transcript level are spatially regulated displaying variance in tissue specificity. Expression of WsCPR2 is coincident with the elevated withanolides content in W. somnifera leaves. This probably indicates involvement of WsCPR2 to confront increased reductive demand of diverse P450 monooxygenases for carrying the withanolides biosynthesis. Elicitation studies showed exogenous elicitors acting as both positive and negative regulators of mRNA transcripts of Ws monooxygenases and reductases. MJ and SA resulted in abundant WsCYP98A and WsCYP76A expression. Increased mRNA levels also agreed with the high accumulation of elicitation driven withanolides biosynthesis. There was appreciable enhancement in WS-1 and WS-3 in response to elicitations. In MJ treated samples, there was a 2.5-fold increase in WS-1 and a significant 4.2-fold enhancement of WS-3. SA treated samples showed marked increase of 1.7- and 3.2-fold in WS-1 and WS-3, respectively. Conversely, GA3 decreased the expression of WsCYP98A as WsCYP76A in addition to the gradual decline in WS-3 whereas WS-1 showed an increase up to 24 h followed by a decrease in WS-1 after 48 h (Rana et al., 2014). Whereas, MJ and SA elicitation induced only WsCPR2 reductase while as WsCPR1 expression showed no change along with significant increase in WS-1 and WS-3 (Rana et al., 2013).
Additionally, four more CYP genes from W. somnifera christened as WSCYP93Id, WSCYP93Sm, WSCYP734B, and WSCYP734 belonging to CYP83B1 and CYP734A1 family and CYP71 and CYP72 clans, also displayed variance in expression in different tissues and in response to different treatments. Interestingly, all the four CYPs showed maximal expression in leaf tissue that was co-incident with the tissue specific secondary metabolite profile of W. somnifera. Furthermore, similar expression profile of WSCYP93Id, WSCYP93Sm, WSCYP734B and WSCYP734 in different W. somnifera chemotypes hinted toward their specialized role in biosynthesis of chemotype-specific metabolites. Light and auxin led to enhancement in expression of all the four CYPs. However, WSCYP734B displayed predominant responsiveness to light and auxin proposing its association with withasteroid/brassinosteroid regulation in planta. MJ and SA elicitations showed an increasing m-RNA abundance trend of the CYPs with increasing concentration of the elicitor. Functional validation of WSCYP93Id in E. coli using withanolides as substrates revealed change of withaferinA to a hydroxylated product. Relationship chart drawn for WSCYP93Id, WSCYP93Sm, WSCYP734B, and WSCYP734 sequences proposed that various withanolides like withanolide A, withanolide D, withaferin A, and withanone are possibly the ensuing yields of metabolic changes involving downstream biosynthetic genes by means of WSCYP enzymes (Srivastava et al., 2015).
Likewise, these results cumulatively, give a better understanding of the regulatory role of CPRs for increased production of withanolides by means of Agrobacterium mediated transformation system. This can lead to homologous intensification of overall metabolite flux with higher transcript levels of key regulatory genes correspondingly up-regulating downstream genes.
Glucosyltransferases
Plant metabolism involves glycosylation as a common modification reaction that is perpetually related with secondary metabolism. Enzymes leading to glucoside formation are called as uridine diphosphate glycosyltransferases (UGTs), members of family 1 of the glycosyltransferase superfamily, which contains over 80 families of enzymes (Campbell et al., 1997; Coutinho et al., 2003) and their functioning involves transferring a uridine diphosphate (UDP)-activated glucose to an equivalent acceptor molecule. UGTs use UDP-activated sugars as donors and allocate their sugar moiety to many acceptors. Plant family 1 UGTs catalyse the glycosylation of surplus bioactive natural compounds. This is frequently the concluding step for biosynthesis of various natural products (Jones and Vogt, 2001), to improve their stability and solubility, and to facilitate storage and build-up in plant cells. Over the years, many UGT gene sub-families have evolved for molecular glycosylation (Vogt and Jones, 2000; Jones and Vogt, 2001). UGTs functioning in secondary metabolism carry a conserved 44 amino acid residue motif (60–80% identity) called as the plant secondary product glucosyltransferase box (PSPG), validated to include the UDP–sugar binding moiety (Hughes and Hughes, 1994; Offen et al., 2006). Nevertheless, UGTs show comparatively meager levels of sequence identity, particularly in the regions associated with acceptor binding. This might be important for the recognition of many acceptors and synthesis of huge number of products.
Many of the pharmacological properties of W. somnifera are attributed to its distinctive steroidal compounds, called glycowithanolides (Matsuda et al., 2001; Singh et al., 2001; Misra et al., 2005; Lal et al., 2006). However, there exists scarcity in the information related to the metabolic step(s) leading to their glyco-transformations due to non-availability of the relevant enzymes and genes.
The first study on sterol glucosyltransferases (SGTs) from W. somnifera reported three different (SGTs) SGTL1, SGTL2, and SGTL3 comprising conserved SGT family domains (Table 1). Among these, SGTL1 was cloned in full length (DQ356887) and was found to be ubiquitously expressing in different parts of the plant. Deduced amino acid sequence of SGTL1 showed the presence of transmembrane domains and preference for membrane sterol glucosylation. Moreover, partially purified recombinant SGT displayed specificity to sterols with hydroxyl group at C-3 position. Functional recruitment of SGTL1 under environmental challenge(s) has also been reported in response to stress (Sharma et al., 2007). Further characterization of new sterol glucosyltransferases in W. somnifera can contribute to the disclosure of functions of various glycosterol in plants.
DIMINUTO/DWARF1 (DIM/DWF1)
DWF1 is an important gene of sterol biosynthesis and regulates the metabolic flux by carrying out isomerization, reduction, and epoxidation, respectively, of their immediate metabolic precursors. DWF1 coding for key enzyme is involved in isomerization and reduction of 24-methylene cholesterol to campesterol and isofucosterol to sitosterol (Klahre et al., 1998). GFP-DIM-DWF1 transient expression studies have shown that DWF1 is an integral membrane protein and is located to ER and not present in nucleus. Sequence homology with flavine adenine dinucleotide-binding domain (conserved in oxidoreductases) indicates that DWF1 has possibly a catalytic rather than a regulatory function. Mutant studies have established the functional role of DIM protein in plant phytosterol biosynthesis. Other homologs of DWF1 have been characterized from Arabidopsis thaliana, Pisum sativum, Zea mays, etc. and their function has been elucidated by mutant studies
In one of the reports Arabidopsis DIMINUTO/DWARF1 gene was found to encode for protein involved in phytosterol biosynthesis. Analysis of A. thaliana DIM mutant has revealed the dwarf phenotype with decreased fertility can be restored by addition of exogenous brassinolide. 24-Methylene cholestrol was found to accumulate in dim mutants. All of these mutants were deficient in campesterol, indicating toward the mutation in DIM/DWF1 severely affects the phytosterol biosynthesis in plants (Klahre et al., 1998).
Reports of some site directed mutants have revealed complete loss of function of DWF1 in plants due to loss of calmodulin binding, similarly complementation studies revealed that fractional loss of calmodulin binding led to partial dwarf phenotype (Du et al., 2009).
DWF1 gene homologs have been isolated from pea and maize, and their function have been elucidated by mutant studies The mutants have been shown to accumulate 24-methylene cholesterol and are severely dwarfed with reduced fertility. Biochemical and mutant studies have revealed the function of DIM homolog in pea known as LKB. The homolog for DIM in pea is LKB, it was observed that mutation in DIM gene directly correlated with the altered phenotype such as decreased enzyme activity, truncated internodal length, epinastic leaves, and thickened stem, and accumulation of 24-methylene cholesterol. Upon exogenous application of brassinolide the phenotype reverted to the normal type. Northern analysis showed the ubiquitous presence of LKB gene in the plant. Role of DWF1 is well-established in the brassinolide biosynthetic pathway. Analysis of dim mutants has revealed that DIM mutants lead to impaired biosynthesis of campesterol and consequent expression of a dwarf phenotype. 24-Methylene cholestrol is a shared precursor of both withanolide and brassinolide biosynthetic pathway (Choe et al., 1999).
DWF1 is a multifunctional enzyme which may carry out isomerization and reduction of post 24-methylene cholestrol intermediates upto withanolide biosynthesis, similar to its role in brassinolide biosynthesis. Increase in the transcript levels of DWFI in response to MJ, 2,4-dichlorophenoxyacetic acid (2,4-D) and SA and corresponding increase in the withanolide content indicates toward the involvement of DWF1 in withanolide biosynthesis along with other biosynthetic pathway genes. Increase in the withanolide content may be due to the increase in the 24-methylenecholestrol substrate pool which may be a consequence of increased expression of upstream genes of MVA pathway such as WsSQS, WsSQE, WsOSC/CS, WsOSC/LS, WsOSC/BS, WsCPR1, and WsCPR2 upon elicitation with abiotic factors such as MJ, 2,4-D, and SA. The presence of cis regulatory inducible elements present in the promoter region of genes involved in the withanolide biosynthetic pathway may be the reason behind the effect of elicitors.
Highest amount of DWF1 transcript level were found in leaves when compared to root and stalk tissues that corroborates well with the high levels of withanolides accumulation in leaves as explained in earlier studies. Also it supports the de novo synthesis of withanolides i.e., synthesis of withanolides in various parts of plant via complete metabolic pathway rather than transfer from any other plant tissue. The molecular cloning and characterization of DIM (GenBank Accession Number KP318739) from W. somnifera and transcript profiling data entails one of the recent studies carried out by the authors.
Tissue-specific transcriptome analysis
More recently, for identification of putative biosynthetic pathway genes, there has been a huge swing toward the “omics” approach, that takes advantage of sequencing technologies to obtain genomic and transcriptomic sequence resource. For species where genomic data are unavailable, transcriptome sequencing through the use of differential expression studies is considered as a main way discovering novel genes in non-model organisms. Against this backdrop, to aid the basic understanding of withanolide biogenesis, transcriptome sequencing for Withania leaf (101L) and root (101R) synthesizing WS-3 and WS-1, respectively, has also been reported (Gupta et al., 2013b). Pyrosequencing results have yielded 834,068 and 721,755 reads assembled into 89,548 and 114,814 unique sequences from 101 L and 101 R. These are presumed to be involved in synthesis of tissue-specific withanolides. Annotations revealed all the genes involved in triterpenoid backbone biosynthesis that incorporated MVA and MEP pathways up to 24-methylene cholesterol, the apparent precursor for withanogenesis. Biosynthesis of 24-methylene cholesterol is followed by various secondary conversions including transfer of diverse moieties or oxidation/reduction reactions for structuring of tissue specific withanolides (Chaurasiya et al., 2012). Using gene Ontology and KEGG analyses, members of cytochrome P450, glycosyltransferase, and methyltransferase gene families with restricted presence or differential leaf and root expression have been reported. Quantification of reads for specific contig showed 305 contigs encoding CYP450s comprising of 12 and 36 unique CYPs for leaf and root tissues that may be responsible for the tissue specific difference in the activities counting withanolide biosynthesis. Unigene resource generated in this study also may be of immense value for interpretation of withanolide biosynthetic pathway and for search of tissue specific molecular mechanism fundamental for structuring definite withanolides (Gupta et al., 2013b).
Comparative proteome analysis
Proteomic approach encompassing research centered on two dimensional electrophoresis (2-DE) and mass spectroscopy (MS) presents a new system for identifying known and unknown genes due to its ability to investigate hundreds of proteins simultaneously (Singh et al., 2015). This feasibility of proteomic analysis would make a considerable contribution in understanding the complex metabolic networks of withanolide biosynthesis in W. somnifera that could be a significant addition to the genomic knowledge resource. As a step forward, comprehensive 2-DE and MS analysis of in vitro grown adventitious roots and in vivo root samples of W. somnifera was conducted. The study showed a high similarity in protein spots of in vitro and in vivo root samples. Thus, suggesting that in vitro roots may have a analogous developmental route as that of in vivo roots though these are developed independent of shoot organs (Senthil et al., 2013). Cumulative proteome examination of leaf and seed tissues of W. somnifera differentiated the proteome on the basis of differential expression, count, and function of identified and characterized tissue-specific proteins. Relative examination of the two tissues further hinted that several proteins of common housekeeping pathways, while a few were tissue specific associated with definite metabolic complement (Dhar et al., 2012). Further, studies on low, abundant and poor soluble proteins would help in characterization of unknown pathway genes that are responsible for the production of withanolides.
Characterization of genes, their high throughput metabolic profiling, sequence resource and proteome information not only provides an insight into the withanolide biosynthetic pathway but also offers molecular wherewithals for biotechnological improvement of W. somnifera. However, in-depth knowledge about withanogenesis still remains elusive and all these results in totality could be useful to reveal various underlying signal transduction pathways to identify specific transcription factors in addition to uncharacterized downstream pathway genes. Further, such biosynthetic genes along with the transcription factors can become prospective targets for pathway engineering.
Tissue culture approaches for withanolide production
Immense therapeutic value of W. somnifera attracts exploration of all possible approaches covering both recombinant DNA techniques and in vitro methods for obtaining chemotypes with desired enhanced chemoprofiles. Recombinant DNA or cell fusion techniques are viable alternatives, but are hampered by the lack of genetic and biochemical knowledge regarding the biosynthesis of secondary metabolites (Evans and Sharp, 1986). Thus, demanding immediate biotechnological advances to enhance the yield at a reduced time gap. On the other hand, in search of alternatives, in vitro techniques present a feasible option for the production of these therapeutically valuable compounds. Tissue culture techniques deliver unceasing, consistent, and renewable source of valued plant pharmaceuticals utilized for the large-scale culture of the plant cells necessary for extraction of secondary metabolites. Substantial work has been reported using different types of in vitro strategies for W. somnifera with main emphasis on manipulation of plant growth regulator adjuvants and cultural conditions for withanolide accumulation. The prospect of application of in vitro methods to produce cell/organ/root cultures for enhanced withanolide production is reviewed below.
Cell suspension culture
Various withanolides correspond to a very minor percentage of total withanolide content in the native plant. Investigation of condition adjusted cultures for resourceful in vitro biogeneration of such pharmacologically promising withanolides is important. Withanolide D, WS-1, WS-3, and WS-2 production have been described in organogenic cultures (Roja et al., 1991; Banerjee et al., 1994; Ray et al., 1996; Vitali et al., 1996; Ray and Jha, 1999; Furmanowa et al., 2001; Sangwan et al., 2007; Murthy et al., 2008). Several reports on accumulation of withanolide D and WS-3 in transformed roots/shooty teratomas cultures are also present, however WS-1 was reported to be absent in these cultures (Banerjee et al., 1994; Ray and Jha, 1999). Successful establishment of cell suspension cultures of W. somnifera for biogenesis of WS-1 has been reported and optimized for its enhanced accumulation.
Highest WS-1 content (1.27 mg g−1 DWB) was observed in suspension cultures supplemented with 2.0 mg L−1 2,4-D, and 2.0 mg L−1 2,4-D + 0.5 mg L−1 kinetin (KN). Thus, revealing that combination of 2,4-D with kinetin is the most suitable for enhanced WS-1 production. It has also been reported a combination of 1.0 ppm benzylaminopurine plus 0.5 ppm kinetin responsible for highest accumulation of WS-1 (14.3 mg per 100 g fresh weight and 238 mg per 100 g DWB, i.e., 0.24%) with the shoot cultures of W. somnifera (Table 2; Sangwan et al., 2007). Growth kinetics study of W. somnifera cell suspension cultures revealed maximum accumulation of biomass (11.02 g L−1 of DWB) and withanolide A (2.03 mg g−1 DWB) at the end of the fourth week. Therefore, making it clear that the biomass growth is closely concurrent with WS-1 amassing. Inoculum density of 10 g L1 was found to be the most suitable for maximum biomass (10.88 g L−1 DWB) and highest production of withanolide A (2.42 mg g−1 DWB). Among the different medias like Murashige and Skoog (MS), B5, NN, and N6, highest accumulation of WS-1 (2.39 mg g−1 DWB) was observed with full strength MS medium suspension culture supplemented with 3% sucrose with an initial medium pH of 6.0 (Table 2; Nagella and Murthy, 2010).
Table 2.
In vitro studies in Withania somnifera in relation to withanolides production.
| In vitro strategy used | Effect on withanolide levels | References |
|---|---|---|
| CELL SUSPENSION CULTURE | ||
| (a) Supplementation with 2.0 mg L−1 2,4-D and 2.0 mg L−1 2,4-D + 0.5 mg L−1 kinetin (b) Supplementation with 1.0 ppm benzylaminopurine plus 0.5 ppm kinetin (c) Murashige and Skoog medium suspension culture supplemented with 3% sucrose |
(a) 1.27 mg g−1 DWB of WS-1 (b) 14.3 mg/100 gm fresh weight and 238 mg/100 gm DWB of WS-1 (c) 2.39 mg g−1 DWB of WS-1 |
https://www.jstage.jst.go.jp/article/cpb/55/9/55_9_1371/_article http://www.sciencedirect.com/science/article/pii/S0960852410005559 |
| IN VITRO SHOOT CULTURE | ||
| (a) Supplementation with 1.00 ppm benzylaminopurine and 0.50 ppm kinetin (b) Elicitation with salicylic acid at 100 μM in combination with 0.6 mg L−1 6-benzyladenine and 20 mg L−1 spermidine |
(a) 14.3 mg per 100 g FWB and 238 mg per 100 g DWB of WS-1 (b) 1.14− to 1.18-fold higher withanolide production |
http://jlc.jst.go.jp/JST.JSTAGE/cpb/55.1371?from=Google http://link.springer.com/article/10.1007/s11738-012-1112-x#page-1 |
| ROOT CULTURE | ||
| (a) MS-based liquid medium supplemented with 40 g/L sucrose for hairy roots (b) Adventitious root cultures elicited with 150 μM salicylic acid |
(a) 2.7-fold higher WS-1 (b) 64.65 mg g−1 DWB of WS-1 (48-fold), 33.74 mg g−1 DWB of withanolide B (29-fold), 17.47 mg g−1 DWB WS-3 (20-fold), 42.88 mg g−1 DWB WS-2 (37-fold), 5.34 mg g−1 DWB 12-deoxy withastramonolide (nine-fold), 7.23 mg g−1 DWB withanoside V (seven-fold), and 9.45 mg g−1 DWB withanoside IV |
http://onlinelibrary.wiley.com/doi/10.1111/j.1744-7909.2008.00680.x/full http://link.springer.com/article/10.1007/s12010-012-9809-2#page-1 |
| SOMACLONAL VARIANTS | ||
| Somaclonal variants | Somaclonal variant regenerated with 0.516% (DWB) of 12-deoxywithastramonolide | http://link.springer.com/article/10.1007/s11627-012-9458-8 |
Investigations have also been carried on the biotransformation capacity of cell suspension cultures generated from W. somnifera leaf using WS-1, WS-3, and WS-2 as precursor substrates. Interestingly, there was a noticeable inter-conversion of WS-1 to WS-2, and vice versa involving substitution of 20-OH group to 17-OH in WS-1 (Sabir et al., 2011). It displays the potential of suspension cultures of W. somnifera for the production of withanolides with multifactorial modulations.
In vitro shoot culture
Root specific production of WS-1 makes root cultures, particularly hairy roots, and its bioreactor based upscaling the foremost way for its in vitro production. However, presence of WS-1 has not been detected in Agrobacterium rhizogene-transformed hairy roots of W. somnifera (Banerjee et al., 1994; Ray and Jha, 1999). Withanolides detected in these hairy root cultures have been reported to be predominantly produced by the aerial parts. Therefore, WS-1 biogenesis was explored in W. somnifera shoot cultures that are the tissue culture complements of the aerial parts of the native plant. Shoot cultures were initiated using explants from the two experimental lines of W. somnifera (Ashwagandha)-RS Selection-1 (RS-Sel-1) and RS Selection-2 (RS-Sel-2) on MS medium with different plant growth regulators. RS-Sel-1 raised shoot culture supplemented with benzylaminopurine (BAP) 1.00 ppm and KN 0.50 ppm showed the highest concentration of WS-1 (14.3 mg per 100 g fresh weight and 238 mg per 100 g dry weight, i.e., 0.24%) in the green shoots (Table 2). Investigative quantities of green shoot cultures (0.24% DWB) was more as compared to the isolation yields of dried roots of field-grown plants. Solid mass/shooty teratoma of RS-Sel-1 raised shoot culture also displayed the highest concentration of WS-1 production (3.7 mg per 100 g fresh weight; 46.2 mg per 100 g dry weight) with BAP 1.00 ppm and kinetin 0.50 ppm. Comparatively, RS-Sel-1 proved to have superior biogenesis/accumulation of WS-1 than RS-Sel-2. Radioactivity fed shoot cultures as well led to isolation of almost pure radiolabeled WS-1 and pointed toward de novo biosynthesis of WS-1 in the in vitro shoot cultures (Sangwan et al., 2007).
The effect of hormones, culture conditions and elicitations on the production of withanolides in multiple shoot cultures of W. somnifera has also been reported. Elicitation with salicylic acid at 100 μM in combination with 0.6 mg L−1 6-benzyladenine and 20 mg L−1 spermidine for 4 h at the fourth week in 20 ml liquid medium reported 1.14− to 1.18-fold higher withanolide production in comparison to the elicitation treatment with MJ at 100 μM after 5 weeks of culture (Table 2; Sivanandhan et al., 2013). Hence, confirming in vitro shoot culture biosystems as an alternative amenable to fine-tuning for harvesting therapeutically valuable WS-1 in comparison to field grown plants. Shoot cultures also represent a suitable system for functional genomic studies of withanolides.
Root culture
Hairy (transformed) roots mediated with A. rhizogenes holds immense potential for studies on secondary metabolite biosynthesis as rapid growth and extensive branching with genetic stability is their characteristic feature. They also exhibit capability of synthesizing root specific secondary metabolites (Giri and Narasu, 2000; Hu and Du, 2006). Consequently, hairy roots in numerous aromatic and medicinal plants for the production of significant secondary compounds have been induced (Le Flem-Bonhomme et al., 2004; Zhao et al., 2004; Santos et al., 2005). Murthy et al. (2008) reported transformation of W. sominifera with A. rhizogenes strain R1601 and obtained transformed hairy roots from cotyledons and leaf explants. Four clones of hairy roots differing in morphology were established. MS-based liquid medium supplemented with 40 g/L sucrose proved to be optimum for biomass building. WS-1 content was found to be 2.7-fold higher in transformed roots (line 3) in comparison to non-transformed roots (Table 2; Murthy et al., 2008).
Leaf derived callus of W. somnifera has also been used for development of adventitious root cultures in MS half-strength medium supplemented with 0.5 mg L−1 indole-3-butyric acid (IBA) and 0.1 mg L−1 indole-3-acetic acid (IAA) with 2% sucrose. These adventitious root cultures were further elicited with MJ and SA autonomously to investigate the improvement in the productivity of withanolides. Root biomass (11.70 g FWB) on 30-day-old adventitious root cultures treated with 150 μM SA for 4 h resulted in the production of 64.65 mg g−1 DWB WS-1 (48-fold), 33.74 mg g−1 DWB withanolide B (29-fold), 17.47 mg g−1 DWB WS-3 (20-fold), 42.88 mg g−1 DWB WS-2 (37-fold), 5.34 mg g−1 DWB 12-deoxy withastramonolide (nine-fold), 7.23 mg g−1 DWB withanoside V (seven-fold), and 9.45 mg g−1 DWB withanoside IV (nine-fold) following elicitation of 10 days (40th day of culture) in comparison to untreated cultures (Table 2). Withanolide production was found to be dependent on biomass, culture age of the adventitious roots, elicitation concentration and time period involved (Sivanandhan et al., 2012). Thus, highlighting the considerable potential of transformed roots and adventitious roots of W. somnifera with further scale-up in bioreactors.
Somaclonal variants
Somaclonal variation can be either genetic or epigenetic in origin (Larkin and Scowcroft, 1981; Lee and Phillips, 1988). The occurrence of somaclonal variation is often associated with activation of transposable elements (Skirvin et al., 1994), point mutation, chromosomal rearrangement, recombination, DNA methylation, and altered sequence copy number. Somaclonal variation is influenced by explant type, culture medium, genotype, and the age of the donor plant, among other factors (Skirvin et al., 1994). It has been suggested that the frequency of somaclonal variation from cell culture is much higher than from field-grown plants because of a, higher rate of mutagenesis (Ahloowalia, 1986). Many plants regenerated via indirect organogenesis have shown somaclonal variation for a wide array of characteristics and this variation have been used to develop new varieties in some species, like tomato, sorghum, sugarcane, and chrysanthemum (Compton and Veilleux, 1991; Duncan et al., 1995; Jalaja et al., 2006; Miñano et al., 2009). Somaclonal variation could unleash the natural variability for withanolide production and accumulation, and could be exploited by breeders to develop W. somnifera varieties attracting commercial interest.
Rana et al. (2012) investigated and validated the applicability of an in vitro strategy to induce somaclonal variation in W. somnifera which manifested in the form of enhanced levels of 12-deoxywithastramonolide (WS-12D). Variations were examined in 54 regenerated plants obtained through indirect organogenesis from leaf explants. WS-R-1 somaclone displayed considerably elevated levels of WS-12D; 0.516% DWB in comparison to the explant donor mother plant (0.002% DWB). Somaclonal variations were investigated at cytological level, by investigating meiosis and mitosis in comparison to number of chromosome and structural organization. Chromosome phenotypes, somatic chromosome count, or meiotic behavior showed no alterations. Further, several genetic polymorphisms between explant donor mother plant and WS-12D over-producing somaclone was examined by random amplification of polymorphic DNA (RAPD) study. WS-R-1 somaclone was evaluated for 2 years to confirm genetic and chemical stability. This study supports the feasibility of an in vitro strategy for chemotypic variability induction to develop high-yielding clones considering the molecular instability displayed by W. somnifera. It also widens the genetic resource base for manipulative hybridization for quantitative chemotypic novelty in W. somnifera (Rana et al., 2012).
Future prospects
W. somnifera has enjoyed a long and important history in traditional medicine system wherein withanolides are attributed with significant remedying properties. Nevertheless, withanolide biosynthesis is still in its infancy with regard to being understood in entirety that enormously hampers the exploitation of its full biotechnological potential. Though, investigations at molecular and in vitro levels have begun, but we are still a long way from understanding how diverse withanolides are synthesized and regulated in W. somnifera. However, the gene elucidation data, omics resource and in vitro study inferences generated so far offers significant promise for enormous increase in correct annotation, functional characterization of enzymes and for comprehending the assorted interactions amongst sophisticated biosynthetic and regulatory mechanisms crucial for successful implementation of withanolide metabolic engineering strategies. Furthermore, advancing metabolic engineering technologies for transgenics, precursor feeding, gene overexpression and inhibition and mutant selection in W. somnifera still awaits investigation. There is much to be learned about the chemical ecology of withanolides to answer an important question about their evolution in the form of sophisticated and diverse structures and types. Though, anticipation about withanolides acting as growth regulators owing to their partial coinciding biosynthetic route with brassinosteroids do exist, but it further demands in-depth examination to build a framework for elaborate pathway modulation strategies.
Author contributions
SL, RV conceived and designed the review. ND and SL wrote the manuscript. ND, S Razdan, S Rana, WB, SL, and RV have contributed in the original studies published earlier vis-à-vis W. somnifera. ND, S Razdan, S Rana, and WB have also collated the up-to-date literature. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the financial grant from Council of Scientific and Industrial Research, Government of India, New Delhi under Network Projects NWP-0008 and BSC-0108. This manuscript represents institutional communication number IIIM/1793/2015.
References
- Abe I., Abe T., Lou W., Masuoka T., Noguchi H. (2007). Site-directed mutagenesis of conserved aromatic residues in rat squalene epoxidase. Biochem. Biophys. Res. Commun. 352, 259–263. 10.1016/j.bbrc.2006.11.014 [DOI] [PubMed] [Google Scholar]
- Abe I., Rohmer M., Prestwich G. D. (1993). Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 93, 2189–2206. 10.1021/cr00022a009 [DOI] [Google Scholar]
- Ahloowalia B. (1986). Limitations to the use of somaclonal variation in crop improvement, in Somoclonal Variations and Crop Improvement (Carlow: Springer; ), 14–27. 10.1007/978-94-015-7733-5_3 [DOI] [Google Scholar]
- Akhtar N., Gupta P., Sangwan N. S., Sangwan R. S., Trivedi P. K. (2013). Cloning and functional characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Withania somnifera: an important medicinal plant. Protoplasma 250, 613–622. 10.1007/s00709-012-0450-2 [DOI] [PubMed] [Google Scholar]
- al-Hindawi M. K., al-Khafaji S. H., Abdul-Nabi M. H. (1992). Anti-granuloma activity of Iraqi Withania somnifera. J. Ethnopharmacol. 37, 113–116. 10.1016/0378-8741(92)90069-4 [DOI] [PubMed] [Google Scholar]
- Anabalagan K., Sadique J. (1981). Influence of an Indian medicine (Ashwagandha) on acute-phase reactants in inflammation. Indian J. Exp. Biol. 19, 245–249. [PubMed] [Google Scholar]
- Arshad Jamal S., Qureshi S., Noor Ali S., Choudhary M. I., Atta Ur R. (1995). Bioactivities and structural studies of withanolides from Withania somnifera. Chem. Heterocycl. Compd. 31, 1047–1059. 10.1007/BF01165050 [DOI] [Google Scholar]
- Attaurrahman Arshad Jamal, S., Choudhary M. I., Asif E. (1991). Two withanolides from Withania somnifera. Phytochemistry 30, 3824–3826. 10.1016/0031-9422(91)80125-K [DOI] [Google Scholar]
- Bai M., Prestwich G. D. (1992). Inhibition and activation of porcine squalene epoxidase. Arch. Biochem. Biophys. 293, 305–313. 10.1016/0003-9861(92)90400-Q [DOI] [PubMed] [Google Scholar]
- Bak S., Beisson F., Bishop G., Hamberger B., Höfer R., Paquette S., et al. (2011). Cytochromes P450. Arabidopsis Book 9:e0144. 10.1199/tab.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Naqvi A., Mandal S., Ahuja P. (1994). Transformation of Withania somnifera (L) Dunal by Agrobacterium rhizogenes: infectivity and phytochemical studies. Phytother. Res. 8, 452–455. 10.1002/ptr.2650080803 [DOI] [Google Scholar]
- Benveniste P. (2002). Sterol metabolism. Arabidopsis Book 1:e0004. 10.1199/tab.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat W. W., Lattoo S. K., Razdan S., Dhar N., Rana S., Dhar R. S., et al. (2012). Molecular cloning, bacterial expression and promoter analysis of squalene synthase from Withania somnifera (L.) Dunal. Gene 499, 25–36. 10.1016/j.gene.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. K., Satyan K. S., Ghosal S. (1997). Antioxidant activity of glycowithanolides from Withania somnifera. Indian J. Exp. Biol. 35, 236–239. [PubMed] [Google Scholar]
- Budhiraja S. D., Sudhir S. (1987). Review of biological activity of withanolides. J. Sci. Ind. Res. 46, 488–491. [Google Scholar]
- Campbell J. A., Davies G. J., Bulone V., Henrissat B. (1997). A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326, 929. 10.1042/bj3260929u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Cervantes M., Gallagher C. E., Zhu C., Wurtzel E. T. (2006). Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol. 141, 220–231. 10.1104/pp.106.077008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasiya N. D., Gupta V. K., Sangwan R. S. (2007). Leaf ontogenic phase-related dynamics of withaferin a and withanone biogenesis in ashwagandha (Withania somnifera Dunal.)-an important medicinal herb. J. Plant Biol. 50, 508–513. 10.1007/BF03030691 [DOI] [Google Scholar]
- Chaurasiya N., Sangwan N., Sabir F., Misra L., Sangwan R. (2012). Withanolide biosynthesis recruits both mevalonate and DOXP pathways of isoprenogenesis in Ashwagandha Withania somnifera L. (Dunal). Plant Cell Rep. 31, 1889–1897. 10.1007/s00299-012-1302-4 [DOI] [PubMed] [Google Scholar]
- Chao C. H., Chou K. J., Wen Z. H., Wang G. H., Wu Y. C., Dai C. F., et al. (2011). Paraminabeolides A-F, cytotoxic and anti-inflammatory marine withanolides from the soft coral Paraminabea acronocephala. J. Nat. Prod. 74, 1132–1141. 10.1021/np2000705 [DOI] [PubMed] [Google Scholar]
- Choe S., Dilkes B. P., Gregory B. D., Ross A. S., Yuan H., Noguchi T., et al. (1999). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–908. 10.1104/pp.119.3.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. Y., Jung E., Jung D. H., Pandey B. P., Yun H., Park H. Y., et al. (2012). Cloning, expression and characterization of CYP102D1, a self-sufficient P450 monooxygenase from Streptomyces avermitilis. FEBS J. 279, 1650–1662. 10.1111/j.1742-4658.2011.08462.x [DOI] [PubMed] [Google Scholar]
- Choudhary M. I., Hussain S., Yousuf S., Dar A., Mudassar Atta-ur-Rahman. (2010). Chlorinated and diepoxy withanolides from Withania somnifera and their cytotoxic effects against human lung cancer cell line. Phytochemistry 71, 2205–2209. 10.1016/j.phytochem.2010.08.019 [DOI] [PubMed] [Google Scholar]
- Choudhary M. I., Nawaz S. A., Lodhi M. A., Ghayur M. N., Jalil S., Riaz N., et al. (2005). Withanolides, a new class of natural cholinesterase inhibitors with calcium antagonistic properties. Biochem. Biophys. Res. Commun. 334, 276–287. 10.1016/j.bbrc.2005.06.086 [DOI] [PubMed] [Google Scholar]
- Closa M., Vranová E., Bortolotti C., Bigler L., Arró M., Ferrer A., et al. (2010). The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J. 63, 512–525. 10.1111/j.1365-313X.2010.04253.x [DOI] [PubMed] [Google Scholar]
- Compton M. E., Veilleux R. E. (1991). Variation for genetic recombination among tomato plants regenerated from three tissue culture systems. Genome 34, 810–817. 10.1139/g91-125 [DOI] [Google Scholar]
- Coon M. J. (2005). Cytochrome P450: nature's most versatile biological catalyst. Annu. Rev. Pharmacol. Toxicol. 45, 1–25. 10.1146/annurev.pharmtox.45.120403.100030 [DOI] [PubMed] [Google Scholar]
- Cordoba E., Salmi M., León P. (2009). Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60, 2933–2943. 10.1093/jxb/erp190 [DOI] [PubMed] [Google Scholar]
- Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317. 10.1016/S0022-2836(03)00307-3 [DOI] [PubMed] [Google Scholar]
- Croteau R., Kutchan T. M., Lewis N. G. (2000). Natural products (secondary metabolites), in Biochemistry and Molecular Biology of Plant, eds Buchannan B. B., Gruissem W., Jones R. L. (Rockville, MD: ), 1250–1318. [Google Scholar]
- Davis L., Kuttan G. (1999). Effect of Withania somnifera on cytokine production in normal and cyclophosphamide treated mice. Immunopharmacol. Immunotoxicol. 21, 695–703. 10.3109/08923979909007135 [DOI] [PubMed] [Google Scholar]
- Davis L., Kuttan G. (2000). Immunomodulatory activity of Withania somnifera. J. Ethnopharmacol. 71, 193–200. 10.1016/S0378-8741(99)00206-8 [DOI] [PubMed] [Google Scholar]
- Dhar N., Rana S., Bhat W. W., Razdan S., Pandith S. A., Khan S., et al. (2013). Dynamics of withanolide biosynthesis in relation to temporal expression pattern of metabolic genes in Withania somnifera (L.) Dunal: a comparative study in two morpho-chemovariants. Mol. Biol. Rep. 40, 7007–7016. 10.1007/s11033-013-2820-z [DOI] [PubMed] [Google Scholar]
- Dhar N., Rana S., Razdan S., Bhat W. W., Hussain A., Dhar R. S., et al. (2014). Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) dunal. J. Biol. Chem. 289, 17249–17267. 10.1074/jbc.M114.571919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R. S., Gupta S. B., Singh P. P., Razdan S., Bhat W. W., Rana S., et al. (2012). Identification and characterization of protein composition in Withania somnifera—an Indian ginseng. J. Plant. Biochem. Biotechnol. 21, 77–87. 10.1007/s13562-011-0083-0 [DOI] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Duncan R., Waskom R., Nabors M. (1995). In vitro screening and field evaluation of tissue-culture-regenerated sorghum (Sorghum bicolor (L.) Moench) for soil stress tolerance, in The Methodology of Plant Genetic Manipulation: Criteria for Decision Making, eds Cassells A. C., Jones P. W. (Griffin, GA: Springer; ), 373–380. [Google Scholar]
- Elsakka M., Grigorescu E., Stanescu U., Dorneanu V. (1990). New data referring to chemistry of Withania somnifera species. Rev. Med. Chir. Soc. Med. Nat. Iasi. 94, 385–387. [PubMed] [Google Scholar]
- Estévez J. M., Cantero A., Reindl A., Reichler S., León P. (2001). 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276, 22901–22909. 10.1074/jbc.M100854200 [DOI] [PubMed] [Google Scholar]
- Evans D. A., Sharp W. R. (1986). Applications of somaclonal variation. Nat. Biotechnol. 4, 528–532. 10.1038/nbt0686-528 [DOI] [Google Scholar]
- Feldmann K. A. (2001). Cytochrome P450s as genes for crop improvement. Curr. Opin. Plant Biol. 4, 162–167. 10.1016/S1369-5266(00)00154-0 [DOI] [PubMed] [Google Scholar]
- Furmanowa M., Gajdzis-Kuls D., Ruszkowska J., Czarnocki Z., Obidoska G., Sadowska A., et al. (2001). In vitro propagation of Withania somnifera and isolation of withanolides with immunosuppressive activity. Planta Med. 67, 146–149. 10.1055/s-2001-11494 [DOI] [PubMed] [Google Scholar]
- Gauttam V. K., Kalia A. N. (2013). Development of polyherbal antidiabetic formulation encapsulated in the phospholipids vesicle system. J. Adv. Pharm. Technol. Res. 4, 108–117. 10.4103/2231-4040.111527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri A., Narasu M. L. (2000). Transgenic hairy roots: recent trends and applications. Biotechnol. Adv. 18, 1–22. 10.1016/S0734-9750(99)00016-6 [DOI] [PubMed] [Google Scholar]
- Glotter E. (1991). Withanolides and related ergostane-type steroids. Nat. Prod. Rep. 8, 415–440. 10.1039/np9910800415 [DOI] [PubMed] [Google Scholar]
- Guengerich F. (2001a). Uncommon P450-catalyzed reactions. Curr. Drug Metab. 2, 93–115. 10.2174/1389200013338694 [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. (2001b). Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 14, 611–650. 10.1021/tx0002583 [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. (2002). Cytochrome P450 enzymes in the generation of commercial products. Nat. Rev. Drug Discov. 1, 359–366. 10.1038/nrd792 [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. (2011). P450 Catalysis Mechanisms Introduction. NEW york, NY: Elsevier; Science Inc. Special Issue. [Google Scholar]
- Guevara-García A., San Román C., Arroyo A., Cortés M. E., de la Luz Gutiérrez-Nava M., León P. (2005). Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell. 17, 628–643. 10.1105/tpc.104.028860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Agarwal A. V., Akhtar N., Sangwan R. S., Singh S. P., Trivedi P. K. (2013a). Cloning and characterization of 2-C-methyl-D-erythritol-4-phosphate pathway genes for isoprenoid biosynthesis from Indian ginseng, Withania somnifera. Protoplasma 250, 285–295. 10.1007/s00709-012-0410-x [DOI] [PubMed] [Google Scholar]
- Gupta P., Akhtar N., Tewari S. K., Sangwan R. S., Trivedi P. K. (2011). Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regul. 65, 93–100. 10.1007/s10725-011-9578-x [DOI] [Google Scholar]
- Gupta P., Goel R., Pathak S., Srivastava A., Singh S. P., Sangwan R. S., et al. (2013b). De novo assembly, functional annotation and comparative analysis of Withania somnifera leaf and root transcriptomes to identify putative genes involved in the withanolides biosynthesis. PLoS ONE 8:e62714. 10.1371/journal.pone.0062714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm E. R., Moura M. B., Kelley E. E., Van Houten B., Shiva S., Singh S. V. (2011). Withaferin A-induced apoptosis in human breast cancer cells is mediated by reactive oxygen species. PLoS ONE 6:e23354. 10.1371/journal.pone.0023354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.-Y., In J.-G., Kwon Y.-S., Choi Y.-E. (2010). Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry 71, 36–46. 10.1016/j.phytochem.2009.09.031 [DOI] [PubMed] [Google Scholar]
- He F., Zhu Y., He M., Zhang Y. (2008). Molecular cloning and characterization of the gene encoding squalene epoxidase in Panax notoginseng. DNA Seq. 19, 270–273. 10.1080/10425170701575026 [DOI] [PubMed] [Google Scholar]
- Himmeldirk K., Kennedy I. A., Hill R. E., Sayer B. G., Spenser I. D. (1996). Biosynthesis of vitamins B1 and B6 in Escherichia coli: concurrent incorporation of 1-deoxy-D-xylulose into thiamin (B1) and pyridoxol (B6). Chem. Commun. 10, 1187–1188. 10.1039/cc9960001187 [DOI] [Google Scholar]
- Hrycay E. G., Bandiera S. M. (2012). The monooxygenase, peroxidase, and peroxygenase properties of cytochrome P450. Arch. Biochem. Biophys. 522, 71–89. 10.1016/j.abb.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Hu Z. B., Du M. (2006). Hairy root and its application in plant genetic engineering. J. Integr. Plant Biol. 48, 121–127. 10.1111/j.1744-7909.2006.00121.x [DOI] [Google Scholar]
- Hughes J., Hughes M. A. (1994). Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. Mitochondrial DNA 5, 41–49. 10.3109/10425179409039703 [DOI] [PubMed] [Google Scholar]
- Hunter W. N. (2007). The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 282, 21573–21577. 10.1074/jbc.R700005200 [DOI] [PubMed] [Google Scholar]
- Istvan E. S., Deisenhofer J. (2001). Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292, 1160–1164. 10.1126/science.1059344 [DOI] [PubMed] [Google Scholar]
- Jalaja N., Sreenivasan T., Pawar S., Bhoi P., Garker R. (2006). Co 94012—a new sugarcane variety through somaclonal variation. Sugar Tech 8, 132–136. 10.1007/BF02943647 [DOI] [Google Scholar]
- Jayaprakasam B., Zhang Y., Seeram N. P., Nair M. G. (2003). Growth inhibition of human tumor cell lines by withanolides from Withania somnifera leaves. Life Sci. 74, 125–132. 10.1016/j.lfs.2003.07.007 [DOI] [PubMed] [Google Scholar]
- Jones P., Vogt T. (2001). Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213, 164–174. 10.1007/s004250000492 [DOI] [PubMed] [Google Scholar]
- Julliard J.-H. (1992). Biosynthesis of the pyridoxal ring (vitamin B6) in higher plant chloroplasts and its relationship with the biosynthesis of the thiazole ring (vitamin B1). C. R. Acad. Sci. III, Sci. Vie 314, 285–290. [Google Scholar]
- Julliard J.-H., Douce R. (1991). Biosynthesis of the thiazole moiety of thiamin (vitamin B1) in higher plant chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 88, 2042–2045. 10.1073/pnas.88.6.2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaileh M., Vanden Berghe W., Heyerick A., Horion J., Piette J., Libert C., et al. (2007). Withaferin A strongly elicits I kappa B kinase beta hyperphosphorylation concomitant with potent inhibition of its kinase activity. J. Biol. Chem. 282, 4253–4264. 10.1074/jbc.M606728200 [DOI] [PubMed] [Google Scholar]
- Kim K. H., Choi S. U., Choi S. Z., Son M. W., Lee K. R. (2011). Withanolides from the rhizomes of Dioscorea japonica and their cytotoxicity. J. Agric. Food Chem. 59, 6980–6984. 10.1021/jf2006535 [DOI] [PubMed] [Google Scholar]
- Kim O. T., Kim S. H., Ohyama K., Muranaka T., Choi Y. E., Lee H. Y., et al. (2010). Upregulation of phytosterol and triterpene biosynthesis in Centella asiatica hairy roots overexpressed ginseng farnesyl diphosphate synthase. Plant Cell Rep. 29, 403–411. 10.1007/s00299-010-0831-y [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., Zhang D., Yang D.-C. (2015). Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 33, 717–735. 10.1016/j.biotechadv.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Kirson I., Glotter E., Lavie D., Abraham A. (1971). Constitutents of Withania somnifera Dun. Part XII. The withanolides of an indian chemotype. J. Chem. Soc. 1971, 2032–2044. 10.1039/j39710002032 [DOI] [Google Scholar]
- Klahre U., Noguchi T., Fujioka S., Takatsuto S., Yokota T., Nomura T., et al. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690. 10.1105/tpc.10.10.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koduru S., Kumar R., Srinivasan S., Evers M. B., Damodaran C. (2010). Notch-1 inhibition by withaferin-A: a therapeutic target against colon carcinogenesis. Mol. Cancer Ther. 9, 202–210. 10.1158/1535-7163.MCT-09-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboyama T., Tohda C., Komatsu K. (2005). Neuritic regeneration and synaptic reconstruction induced by withanolide A. Br. J. Clin. Pharmacol. 144, 961–971. 10.1038/sj.bjp.0706122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboyama T., Tohda C., Komatsu K. (2006). Withanoside IV and its active metabolite, sominone, attenuate A beta(25-35)-induced neurodegeneration. Eur. J. Neurosci. 23, 1417–1426. 10.1111/j.1460-9568.2006.04664.x [DOI] [PubMed] [Google Scholar]
- Kulkarni S. K., George B. (1996). Anticonvulsant action of Withania somnifera (Aswaganda) root extract against pentylenetetrazol-induced kindling in mice. Phytother. Res. 10, 447–449. [Google Scholar]
- Kurapati V. K. R., Atluri V. S. R., Samikkannu T., Yndart A. A., Nair M. P. N. (2013). Ashwagandha (Withania somnifera) reverses B-amyloid induced neuronal toxicity: implications in HAND. J. Neuroimmune Pharmacol. 8, 422–422. 10.1371/journal.pone.0077624 [DOI] [Google Scholar]
- Kuzuyama T. (2002). Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 66, 1619–1627. 10.1271/bbb.66.1619 [DOI] [PubMed] [Google Scholar]
- Lal P., Misra L., Sangwan R. S., Tuli R. (2006). New withanolides from fresh berries of Withania somnifera. Z. Naturforsch. B 61, 1143–1147. 10.1515/znb-2006-0914 [DOI] [Google Scholar]
- Larkin P. J., Scowcroft W. (1981). Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 60, 197–214. 10.1007/BF02342540 [DOI] [PubMed] [Google Scholar]
- Lattoo S. K., Dhar R. S., Khan S., Bamotra S., Dhar A. K. (2007). Temporal sexual maturation and incremental staminal movement encourages mixed mating in Withania somnifera - an insurance for reproductive success. Curr. Sci. 92, 1390–1399. [Google Scholar]
- Leber R., Landl K., Zinser E., Ahorn H., Spök A., Kohlwein S. D., et al. (1998). Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell 9, 375–386. 10.1091/mbc.9.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Jeong J.-H., Seo J.-W., Shin C.-G., Kim Y.-S., In J.-G., et al. (2004). Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 45, 976–984. 10.1093/pcp/pch126 [DOI] [PubMed] [Google Scholar]
- Lee M., Phillips R. L. (1988). The chromosomal basis of somaclonal variation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 413–437. 10.1146/annurev.pp.39.060188.002213 [DOI] [Google Scholar]
- Le Flem-Bonhomme V., Laurain-Mattar D., Fliniaux M. (2004). Hairy root induction of Papaver somniferum var. album, a difficult-to-transform plant, by A. rhizogenes LBA 9402. Planta 218, 890–893. 10.1007/s00425-003-1196-z [DOI] [PubMed] [Google Scholar]
- Li J. W., Vederas J. C. (2011). [Drug discovery and natural products: end of era or an endless frontier?]. Biomed. Khim. 57, 148–160. 10.18097/pbmc20115702148 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 47–65. 10.1146/annurev.arplant.50.1.47 [DOI] [PubMed] [Google Scholar]
- Mander L., Liu H.-W. (eds.). (2010). Comprehensive Natural Products II: Chemistry and Biology. Austin, TX: Elsevier Science. [Google Scholar]
- Mathur S., Kaur P., Sharma M., Katyal A., Singh B., Tiwari M., et al. (2004). The treatment of skin carcinoma, induced by UVB radiation, using 1-oxo-5 beta,6 beta-epoxy-witha-2-enolide, isolated from the roots of Withania somnifera, in a rat model. Phytomedicine 11, 452–460. 10.1016/j.phymed.2003.05.004 [DOI] [PubMed] [Google Scholar]
- Matsuda H., Murakami T., Kishi A., Yoshikawa M. (2001). Structures of withanosides I, II, III, IV, V, VI, and VII, new withanolide glycosides, from the roots of Indian Withania somnifera DUNAL. and inhibitory activity for tachyphylaxis to clonidine in isolated guinea-pig ileum. Bioorg. Med. Chem. 9, 1499–1507. 10.1016/S0968-0896(01)00024-4 [DOI] [PubMed] [Google Scholar]
- Matsushita Y., Kang W., Charlwood B. V. (1996). Cloning and analysis of a cDNA encoding farnesyl diphosphate synthase from Artemisia annua. Gene 172, 207–209. 10.1016/0378-1119(96)00054-6 [DOI] [PubMed] [Google Scholar]
- Miñano H. S., González-Benito M. E., Martín C. (2009). Molecular characterization and analysis of somaclonal variation in chrysanthemum cultivars using RAPD markers. Sci. Hortic. 122, 238–243. 10.1016/j.scienta.2009.05.001 [DOI] [Google Scholar]
- Miners J. O. (2002). Evolution of drug metabolism: hitchhiking the technology bandwagon. Clin. Exp. Pharmacol. Physiol. 29, 1040–1044. 10.1046/j.1440-1681.2002.03768.x [DOI] [PubMed] [Google Scholar]
- Mirjalili M. H., Moyano E., Bonfil M., Cusido R. M., Palazón J. (2009). Steroidal lactones from Withania somnifera, an ancient plant for novel medicine. Molecules 14, 2373–2393. 10.3390/molecules14072373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra L. C., Singh B. B., Dagenais S. (2000). Scientific basis for the therapeutic use of Withania somnifera (ashwagandha): a review. Altern. Med. Rev. 5, 334–346. [PubMed] [Google Scholar]
- Misico R. I., Nicotra V. E., Oberti J. C., Barboza G., Gil R. R., Burton G. (2011). Withanolides and related steroids. Prog. Chem. Org. Nat. Prod. 94, 127–229. 10.1007/978-3-7091-0748-5_3 [DOI] [PubMed] [Google Scholar]
- Misra L., Lal P., Sangwan R. S., Sangwan N. S., Uniyal G. C., Tuli R. (2005). Unusually sulfated and oxygenated steroids from Withania somnifera. Phytochemistry 66, 2702–2707. 10.1016/j.phytochem.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Misra L., Mishra P., Pandey A., Sangwan R. S., Sangwan N. S., Tuli R. (2008). Withanolides from Withania somnifera roots. Phytochemistry 69, 1000–1004. 10.1016/j.phytochem.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Mizutani M., Ohta D. (2010). Diversification of P450 genes during land plant evolution. Annu. Rev. Plant 61, 291–315. 10.1146/annurev-arplant-042809-112305 [DOI] [PubMed] [Google Scholar]
- Mohan R., Hammers H., Bargagna-Mohan P., Zhan X., Herbstritt C., Ruiz A., et al. (2004). Withaferin A is a potent inhibitor of angiogenesis. Angiogenesis 7, 115–122. 10.1007/s10456-004-1026-3 [DOI] [PubMed] [Google Scholar]
- Mondal S., Mandal C., Sangwan R., Chandra S. (2010). Withanolide D induces apoptosis in leukemia by targeting the activation of neutral sphingomyelinase-ceramide cascade mediated by synergistic activation of c-Jun N-terminal kinase and p38 mitogen-activated protein kinase. Mol. Cancer 9:239. 10.1186/1476-4598-9-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulabagal V., Subbaraju G. V., Rao C. V., Sivaramakrishna C., DeWitt D. L., Holmes D., et al. (2009). Withanolide sulfoxide from aswagandha roots inhibits nuclear transcription factor-kappa-B, cyclooxygenase and tumor cell proliferation. Phytother. Res. 23, 987–992. 10.1002/ptr.2736 [DOI] [PubMed] [Google Scholar]
- Murthy H. N., Dijkstra C., Anthony P., White D. A., Davey M. R., Power J. B., et al. (2008). Establishment of Withania somnifera hairy root cultures for the production of withanolide A. J. Integr. Plant Biol. 50, 975–981. 10.1111/j.1744-7909.2008.00680.x [DOI] [PubMed] [Google Scholar]
- Nagegowda D. A., Bach T. J., Chye M. L. (2004). Brassica juncea 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase 1: expression and characterization of recombinant wild-type and mutant enzymes. Biochem. J. 383, 517–527. 10.1042/BJ20040721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagella P., Murthy H. N. (2010). Establishment of cell suspension cultures of Withania somnifera for the production of withanolide A. Bioresour. Technol. 101, 6735–6739. 10.1016/j.biortech.2010.03.078 [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Schuler M. A., Paquette S. M., Werck-Reichhart D., Bak S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772. 10.1104/pp.104.039826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann K. H., Kumar A., Sopory S. K. (2008). Recent Advances in Plant Biotechnology and Its Applications: Prof. Dr. Karl-Hermann Neumann Commemorative, Vol. I. K Jaipur: International Publishing House. [Google Scholar]
- Newman J. D., Chappell J. (1999). Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. Crit. Rev. Biochem. Mol. Biol. 34, 95–106. 10.1080/10409239991209228 [DOI] [PubMed] [Google Scholar]
- Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405. 10.1038/sj.emboj.7600970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S., Narita K., Nakazawa T., Ishida C., Takeuchi Y., Ohto C., et al. (1996). A role of the amino acid residue located on the fifth position before the first aspartate-rich motif of farnesyl diphosphate synthase on determination of the final product. J. Biol. Chem. 271, 30748–30754. 10.1074/jbc.271.48.30748 [DOI] [PubMed] [Google Scholar]
- Owais M., Sharad K. S., Shehbaz A., Saleemuddin M. (2005). Antibacterial efficacy of Withania somnifera (ashwagandha) an indigenous medicinal plant against experimental murine salmonellosis. Phytomedicine 12, 229–235. 10.1016/j.phymed.2003.07.012 [DOI] [PubMed] [Google Scholar]
- Panda S., Kar A. (1997). Effects of root extract of Ashwagandha, Withania somnifera, on function of thyroid in cockerel. Indian J. Anim. Sci. 67, 575–576. [Google Scholar]
- Pandey K. B., Rizvi S. I. (2009). Plant polyphenols as dietary antioxidants in human health and disease. Oxid. Med. Cell. Longev. 2, 270–278. 10.4161/oxim.2.5.9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. R., Rasbery J. M., Bartel B., Matsuda S. P. (2006). Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314. 10.1016/j.pbi.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Rana S., Bhat W. W., Dhar N., Pandith S. A., Razdan S., Vishwakarma R., et al. (2014). Molecular characterization of two A-type P450s, WsCYP98A and WsCYP76A from Withania somnifera (L.) Dunal: expression analysis and withanolide accumulation in response to exogenous elicitations. BMC Biotechnol. 14:89. 10.1186/s12896-014-0089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S., Dhar N., Bhat W. W., Razdan S., Khan S., Dhar R. S., et al. (2012). A 12-deoxywithastramonolide-rich somaclonal variant in Withania somnifera (L.) Dunal—molecular cytogenetic analysis and significance as a chemotypic resource. In Vitro Cell. Dev. Biol. Plant. 48, 546–554. 10.1007/s11627-012-9458-8 [DOI] [Google Scholar]
- Rana S., Lattoo S. K., Dhar N., Razdan S., Bhat W. W., Dhar R. S., et al. (2013). NADPH-cytochrome P450 reductase: molecular cloning and functional characterization of two paralogs from Withania somnifera (L.) dunal. PLoS ONE 8:e57068. 10.1371/journal.pone.0057068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. R., Ravishankar G. A. (2002). Plant cell cultures: chemical factories of secondary metabolites. Biotechnol. Adv. 20, 101–153. 10.1016/S0734-9750(02)00007-1 [DOI] [PubMed] [Google Scholar]
- Rasool M., Varalakshmi P. (2006). Immunomodulatory role of Withania somnifera root powder on experimental induced inflammation: an in vivo and in vitro study. Vascul. Pharmacol. 44, 406–410. 10.1016/j.vph.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Ray A. B., Gupta M. (1994). Withasteroids, a growing group of naturally occurring steroidal lactones, in Fortschritte der Chemie organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products, eds Herz W., Kirby G. W., Moore R. E., Steglich W., Tamm C. (Vienna: Springer; ), 1–106. [DOI] [PubMed] [Google Scholar]
- Ray S., Ghosh B., Sen S., Jha S. (1996). Withanolide production by root cultures of Withania somnifera transformed with Agrobacterium rhizogenes. Planta Med. 62:571–573. 10.1055/s-2006-957977 [DOI] [PubMed] [Google Scholar]
- Ray S., Jha S. (1999). Withanolide synthesis in cultures of Withania somnifera transformed with Agrobacterium tumefaciens. Plant Sci. 146:1–7. 10.1016/S0168-9452(99)00077-1 [DOI] [PubMed] [Google Scholar]
- Razdan S., Bhat W. W., Rana S., Dhar N., Lattoo S. K., Dhar R. S., et al. (2013). Molecular characterization and promoter analysis of squalene epoxidase gene from Withania somnifera (L.) Dunal. Mol. Biol. Rep. 40, 905–916. 10.1007/s11033-012-2131-9 [DOI] [PubMed] [Google Scholar]
- Rees H. H., Goad L. J., Goodwin T. W. (1969). 2,3-Oxidosqualene cycloartenol cyclasefrom Ochromonas malhamensis. Biochim. Biophys. Acta 176, 892–894. 10.1016/0005-2760(69)90274-4 [DOI] [PubMed] [Google Scholar]
- Ro D.-K., Ehlting J., Douglas C. J. (2002). Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol. 130, 1837–1851. 10.1104/pp.008011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. W., Tsay Y. H., Kienzle B. K., Smith-Monroy C. A., Bishop R. W. (1993). Conservation between human and fungal squalene synthetases: similarities in structure, function, and regulation. Mol. Cell. Biol. 13, 2706–2717. 10.1128/MCB.13.5.2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepción M., Forés O., Martinez-Garcia J. F., González V., Phillips M. A., Ferrer A., et al. (2004). Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16, 144–156. 10.1105/tpc.016204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roja G., Heble M., Sipahimalani A. (1991). Tissue cultures of Withania somnifera: morphogenesis and withanolide synthesis. Phytother. Res. 5, 185–187. 10.1002/ptr.2650050411 [DOI] [Google Scholar]
- Roy R. V., Suman S., Das T. P., Luevano J. E., Damodaran C. (2013). Withaferin A, a steroidal lactone from Withania somnifera, induces mitotic catastrophe and growth arrest in prostate cancer cells. J. Nat. Prod. 76, 1909–1915. 10.1021/np400441f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder N. (1992). Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol. 126, 2–7. 10.1111/j.1365-2133.1992.tb00001.x [DOI] [PubMed] [Google Scholar]
- Sabir F., Sangwan R. S., Singh J., Misra L. N., Pathak N., Sangwan N. S. (2011). Biotransformation of withanolides by cell suspension cultures of Withania somnifera (Dunal). Plant Biotechnol. Rep. 5, 127–134. 10.1007/s11816-011-0165-4 [DOI] [Google Scholar]
- Sangwan R., Chaurasiya N., Misra L., Lal P., Uniyal G., Sharma R., et al. (2004). Phytochemical variability in commercial herbal products and preparations of Withania somnifera (Ashwagandha). Curr. Sci. 86, 461–464. [Google Scholar]
- Sangwan R. S., Chaurasiya N. D., Lal P., Misra L., Uniyal G. C., Tuli R., et al. (2007). Withanolide A biogeneration in in vitro shoot cultures of Ashwagandha (Withania somnifera Dunal), a main medicinal plant in Ayurveda. Chem. Pharm. Bull. 55, 1371–1375. 10.1248/cpb.55.1371 [DOI] [PubMed] [Google Scholar]
- Sangwan R. S., Das Chaurasiya N., Lal P., Misra L., Tuli R., Sangwan NS. (2008). Withanolide A is inherently de novo biosynthesized in roots of the medicinal plant Ashwagandha (Withania somnifera). Physiol. Plant. 133, 278–287. 10.1111/j.1399-3054.2008.01076.x [DOI] [PubMed] [Google Scholar]
- Santos P. A., Figueiredo A. C., Oliveira M. M., Barroso J. G., Pedro L. G., Deans S. G., et al. (2005). Growth and essential oil composition of hairy root cultures of Levisticum officinale WDJ Koch (lovage). Plant Sci. 168, 1089–1096. 10.1016/j.plantsci.2004.12.009 [DOI] [Google Scholar]
- Sehgal N., Gupta A., Valli R. K., Joshi S. D., Mills J. T., Hamel E., et al. (2012). Withania somnifera reverses Alzheimer's disease pathology by enhancing low-density lipoprotein receptor-related protein in liver. Proc. Natl. Acad. Sci. U.S.A. 109, 3510–3515. 10.1073/pnas.1112209109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil K., Karunanithi N., Kim G. S., Nagappan A., Sundareswaran S., Natesan S., et al. (2013). Proteome analysis of in vitro and in vivo root tissue of Withania somnifera. Afr. J. Biotechnol. 10, 16866–16874. 10.5897/AJB11.1244 [DOI] [Google Scholar]
- Seo J.-W., Jeong J.-H., Shin C.-G., Lo S.-C., Han S.-S., Yu K.-W., et al. (2005). Overexpression of squalene synthase in Eleutherococcus senticosus increases phytosterol and triterpene accumulation. Phytochemistry 66, 869–877. 10.1016/j.phytochem.2005.02.016 [DOI] [PubMed] [Google Scholar]
- Shahid M., Shahzad A., Malik A., Sahai A. (2013). Recent Trends in Biotechnology and Therapeutic Applications of Medicinal Plants. New Delhi: Springer. [Google Scholar]
- Sharma L. K., Madina B. R., Chaturvedi P., Sangwan R. S., Tuli R. (2007). Molecular cloning and characterization of one member of 3beta-hydroxy sterol glucosyltransferase gene family in Withania somnifera. Arch. Biochem. Biophys. 460, 48–55. 10.1016/j.abb.2007.01.024 [DOI] [PubMed] [Google Scholar]
- Simmons D. L., Lalley P. A., Kasper C. B. (1985). Chromosomal assignments of genes coding for components of the mixed-function oxidase system in mice. Genetic localization of the cytochrome P-450PCN and P-450PB gene families and the nadph-cytochrome P-450 oxidoreductase and epoxide hydratase genes. J. Biol. Chem. 260, 515–521. [PubMed] [Google Scholar]
- Singh B., Saxena A. K., Chandan B. K., Gupta D. K., Bhutani K. K., Anand K. K. (2001). Adaptogenic activity of a novel, withanolide-free aqueous fraction from the roots of Withania somnifera Dun. Phytother. Res. 15, 311–318. 10.1002/ptr.858 [DOI] [PubMed] [Google Scholar]
- Singh N., Bhalla M., Jager P. D., Gilca M. (2011). An Overview on Ashwagandha: a Rasayana (Rejuvenator) of Ayurveda. Afr. J. Tradit. Complement Altern. Med. 8, 208–213. 10.4314/ajtcam.v8i5S.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Guleri R., Singh V., Kaur G., Kataria H., Singh B., et al. (2015). Biotechnological interventions in Withania somnifera (L.) Dunal. Biotechnol. Genet. Eng. Rev. 19, 1–20. 10.1080/02648725.2015.1020467 [DOI] [PubMed] [Google Scholar]
- Sivanandhan G., Arun M., Mayavan S., Rajesh M., Jeyaraj M., Dev G. K., et al. (2012). Optimization of elicitation conditions with methyl jasmonate and salicylic acid to improve the productivity of withanolides in the adventitious root culture of Withania somnifera (L.) Dunal. Appl. Biochem. Biotechnol. 168, 681–696. 10.1007/s12010-012-9809-2 [DOI] [PubMed] [Google Scholar]
- Sivanandhan G., Rajesh M., Arun M., Jeyaraj M., Dev G. K., Arjunan A., et al. (2013). Effect of culture conditions, cytokinins, methyl jasmonate and salicylic acid on the biomass accumulation and production of withanolides in multiple shoot culture of Withania somnifera (L.) Dunal using liquid culture. Acta Physiol. Plant. 35, 715–728. 10.1007/s11738-012-1112-x [DOI] [Google Scholar]
- Skirvin R. M., McPheeters K. D., Norton M. (1994). Sources and frequency of somaclonal variation. HortScience 29, 1232–1237. [Google Scholar]
- Sono M., Roach M. P., Coulter E. D., Dawson J. H. (1996). Heme-containing oxygenases. Chem. Rev. 96, 2841–2888. 10.1021/cr9500500 [DOI] [PubMed] [Google Scholar]
- Sprenger G. A., Schörken U., Wiegert T., Grolle S., de Graaf A. A., Taylor S. V., et al. (1997). Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. U.S.A. 94, 12857–12862. 10.1073/pnas.94.24.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Sangwan R. S., Tripathi S., Mishra B., Narnoliya L. K., Misra L. N., et al. (2015). Light and auxin responsive cytochrome P450s from Withania somnifera Dunal: cloning, expression and molecular modelling of two pairs of homologue genes with differential regulation. Protoplasma. 252, 1421–1437. 10.1007/s00709-015-0766-9 [DOI] [PubMed] [Google Scholar]
- Sukanya D. H., Lokesha A. N., Datta G., Himabindu K. (2010). Phytochemical diversity in ashwagandha (Withania somnifera). Open Access J. Med. Aromat. Plants 1, 47. [Google Scholar]
- Takaya A., Zhang Y.-W., Asawatreratanakul K., Wititsuwannakul D., Wititsuwannakul R., Takahashi S., et al. (2003). Cloning, expression and characterization of a functional cDNA clone encoding geranylgeranyl diphosphate synthase of Hevea brasiliensis. Biochim. Biophys. Acta 1625, 214–220. 10.1016/S0167-4781(02)00602-4 [DOI] [PubMed] [Google Scholar]
- Takemura T., Chow Y.-L., Todokoro T., Okamoto T., Sato F. (2010). Over-expression of rate-limiting enzymes to improve alkaloid productivity, in Plant Secondary Metabolism Engineering, ed Fett-Neto A. G. (Kyoto: Springer; ), 95–109. 10.1007/978-1-60761-723-5_7 [DOI] [PubMed] [Google Scholar]
- Tohda C., Kuboyama T., Komatsu K. (2005). Search for natural products related to regeneration of the neuronal network. Neurosignals 14, 34–45. 10.1159/000085384 [DOI] [PubMed] [Google Scholar]
- Tuli R., Sangwan R., Kumar S., Bhattacharya S., Misra L., Trivedi P., et al. (2009). Ashwagandha (Withania somnifera)—A Model Indian Medicinal Plant. New Delhi: Council of Scientific and Industrial Research (CSIR). [Google Scholar]
- Vitali G., Conte L., Nicoletti M. (1996). Withanolide composition and in vitro culture of Italian Withania somnifera. Planta Med. 62, 287–288. 10.1055/s-2006-957884 [DOI] [PubMed] [Google Scholar]
- Vogt T., Jones P. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 380–386. 10.1016/S1360-1385(00)01720-9 [DOI] [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. 10.1146/annurev-arplant-050312-120116 [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D., Hehn A., Didierjean L. (2000). Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 5, 116–123. 10.1016/S1360-1385(00)01567-3 [DOI] [PubMed] [Google Scholar]
- Winters M. (2006). Ancient medicine, modern use: Withania somnifera and its potential role in integrative oncology. Altern. Med. Rev. 11, 269–277. [PubMed] [Google Scholar]
- Xu R., Fazio G. C., Matsuda S. P. (2004). On the origins of triterpenoid skeletal diversity. Phytochemistry 65, 261–291. 10.1016/j.phytochem.2003.11.014 [DOI] [PubMed] [Google Scholar]
- Yu Y., Hamza A., Zhang T., Gu M., Zou P., Newman B., et al. (2010). Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochem. Pharmacol. 79, 542–551. 10.1016/j.bcp.2009.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Fu C., Chen Y., Ma F. (2004). Transformation of Saussurea medusa for hairy roots and jaceosidin production. Plant Cell Rep. 23, 468–474. 10.1007/s00299-004-0840-9 [DOI] [PubMed] [Google Scholar]
- Zhao Z. J., Song Y. G., Liu Y. L., Qiao M., Zhai X. L., Xiang F. N. (2013). The effect of elicitors on oleanolic acid accumulation and expression of triterpenoid synthesis genes in Gentiana straminea. Biol. Plant. 57, 139–143. [Google Scholar]