Abstract
The small size of dendritic spines belies the elaborate role they play in excitatory synaptic transmission and ultimately complex behaviors. The cytoskeletal architecture of the spine is predominately composed of actin filaments. These filaments, which at first glance might appear simple, are also surprisingly complex. They dynamically assemble into different structures and serve as a platform for orchestrating the elaborate responses of the spine during spinogenesis and experience-dependent plasticity. Multiple mutations associated with human neurodevelopmental and psychiatric disorders involve genes that encode regulators of the synaptic cytoskeleton. A major, unresolved question is how the disruption of specific actin filament structures leads to the onset and progression of complex synaptic and behavioral phenotypes. This review will cover established and emerging mechanisms of actin cytoskeletal remodeling and how this influences specific aspects of spine biology that are implicated in disease.
Keywords: actin, adhesion, Arp2/3 complex, cofilin, dendritic spine, formin, Rho (Rho GTPase), synapse, synaptic plasticity, WASH
Introduction
Dendritic spines are morphologically diverse, actin-rich protrusions emerging from dendritic shafts that serve as the receiving sites for the majority of excitatory synaptic transmission in the brain. First described over a century ago by Ramón y Cajal, dendritic spines typically consist of a spine head, which can range in size from 0.5 to 2 μm in length, and thin spine neck (∼0.2 μm thick) (Fig. 1). Their essential function is to compartmentalize biochemical and electrical signals in response to synaptic activation (1). Dendritic spines are supported by an underlying cytoskeleton that is almost exclusively composed of actin filaments, which can be up to ∼200 nm long (2). Remodeling of this densely packed actin governs most, if not all, dendritic spine physiology. This includes spine formation and maintenance, synaptic adhesion, receptor endocytosis and exocytosis, and synaptic plasticity (Fig. 1). Further, the actin cytoskeleton drives dendritic spine turnover and morphological changes that occur in vivo in response to experience (3). Finally, aberrations in dendritic spine morphology and density are linked to a variety of neurological disorders such as schizophrenia (SZ)2 and intellectual disability (ID) (4). Recent studies link de novo mutations associated with increased risk of complex psychiatric disorders such as SZ to genes encoding regulators of the post-synaptic actin cytoskeleton (5) (see Fig. 3). Together these findings strongly imply that proper maintenance of the spine actin cytoskeleton is critical for spine functionality and neuronal connectivity. This review will focus on the nuts and bolts of actin dynamics in spines as well as recent developments in the modulation of the synaptic cytoskeleton in two crucial dendritic spine processes whose disturbances are linked with synapse pathologies: synaptic adhesion and synaptic plasticity.
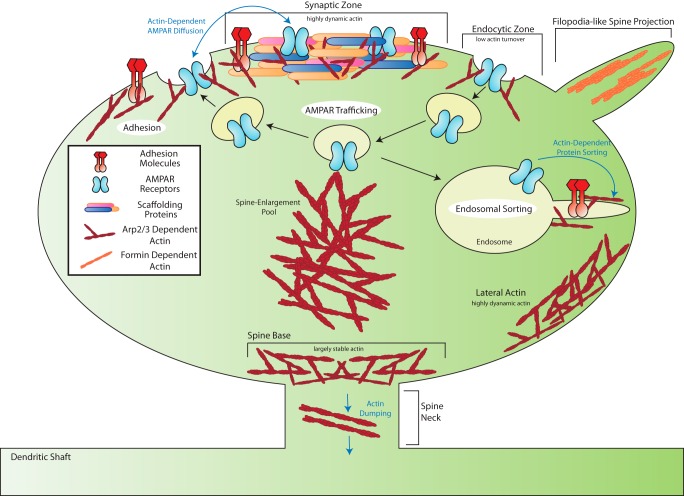
FIGURE 1.
Organization of distinct actin pools in dendritic spines. Shown is a schematic depicting the spatial organization of actin dynamics within different regions of the spine. Two types of actin, Arp2/3-dependent (red) and formin-based actin (orange), are delineated. Regions of the spine referenced in the text, including the synaptic zone, spine neck, and filopodial-like projections, are also labeled.
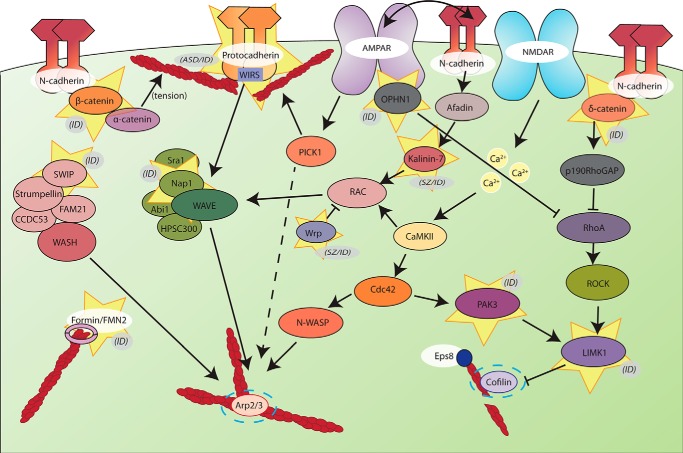
FIGURE 3.
Actin signaling pathways and their association with brain disorders. Shown is a schematic of proteins involved in regulating the spine actin cytoskeleton. The synaptic actin cytoskeleton is a common pathway in which mutations are associated with increased risk for brain disorders. Each protein highlighted by yellow stars has a genetic mutation associated with ID, SZ, or ASD, each of which are indicated in parentheses. Finally, the dashed lines surrounding the Arp2/3 complex and cofilin indicate the final points of signaling output for actin remodeling, both of which have been studied extensively through knock-out mouse studies. Loss of Arp2/3 complex activity in the mouse forebrain mimics aspects of psychiatric conditions such as schizophrenia-related disorders, whereas loss of cofilin is associated with decreased anxiety in mice. ROCK, Rho-associated kinase.
Dendritic Spine Actin Regulators
The cytoskeleton of the spine is a highly branched meshwork of filamentous actin (F-actin) that is assembled from a pool of monomeric actin (G-actin) (6). Before considering how actin modifies many of the key functions of the dendritic spine, it is important to first consider the essential actin regulatory proteins that facilitate actin dynamics within the spine (Fig. 2). The activity of these regulators is tightly modulated by Rho family GTPases such as Rho, Rac, and Cdc42. These GTPases are themselves regulated by a host of spine-enriched activators (GEFs) and inhibitors (GAPs) that tune GTPase activity within the spine in response to synaptic cues (reviewed in Ref. 7). Mutations in spine-enriched GEFs (for example, kalirin-7 (8)) and GAPs (for example, WAVE-associated Rac GAP (WRP) (9, 10)) are associated with SZ and ID, implicating the local control of Rho family GTPase activity and actin remodeling in essential spine functions (Fig. 3). In order for dendritic spines to maintain a highly active cytoskeleton that is capable of responding to adhesion molecules or neurotransmitter release, the actin cytoskeleton undergoes constant “treadmilling,” growing at the “barbed end” of actin filaments and disassembling at the “pointed end” of actin filaments (11) (Fig. 2). Actin regulators can facilitate actin polymerization, promote their disassembly, or stabilize filaments as part of this process.
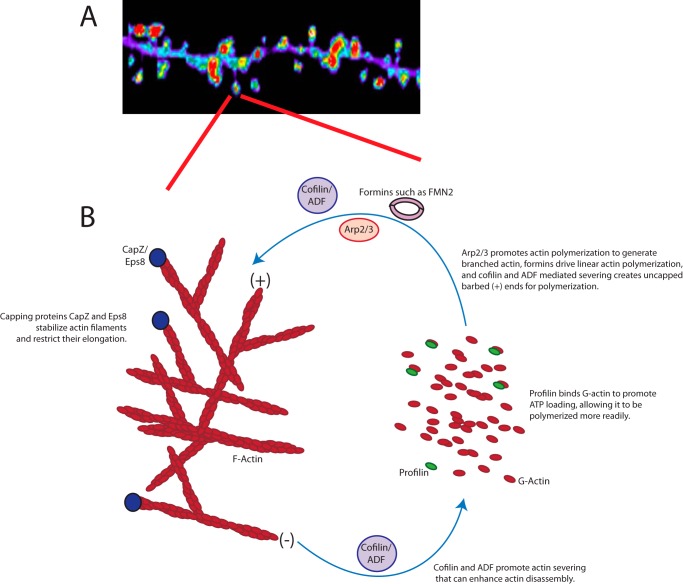
FIGURE 2.
The basics of actin dynamics in the dendritic spine. A, representative image of a dendritic section from a hippocampal neuron expressing GFP-actin. Fluorescent signal is pseudo-colored for relative intensity ranging from low (blue) to high (red). Note that actin is highly enriched in spines protruding from the dendritic shaft. B, schematic depicting actin turnover in dendritic spines and the proteins directly involved in its remodeling. Treadmilling of actin between polymerized F-actin to monomeric G-actin at the barbed (+) versus pointed (−) end is indicated.
Actin Filament Assembly
Elegant super-resolution microscopy of actin within spines at basal states has shown that the majority of foci for actin polymerization occurs at the tip and lateral regions (2). Three factors within the spine promote actin filament assembly: the actin-related proteins 2 and 3 complex (Arp2/3), formins, and profilin.
Perhaps the most prominent of these is Arp2/3, a seven-subunit protein complex that binds to the side of an existing actin filament and drives nucleation of a new actin filament at a 70° angle from the mother filament (12). Arp2/3 generates a dense network of branched actin within dendritic spines (Fig. 2), but to stimulate branched actin polymerization, the Arp2/3 complex must be activated by one of its nucleation-promoting factors (NPFs) such as N-WASP (neuronal Wiskott–Aldrich syndrome protein), WAVE1, or WASH (12) (Fig. 3). Ablating the expression of Arp2/3 activators such as WAVE1 leads to aberrations in spine morphology as well as behavioral abnormalities (13, 14). The Arp2/3 complex is essential for dendritic spine maintenance and function such as promoting the treadmilling of the actin cytoskeleton, morphological alterations in response to synaptic activity, and behavior (15, 16).
A second class of actin assembly proteins important in dendritic spines is the formin family, which is composed of 15 mammalian proteins (17). In contrast to the branched actin created by the Arp2/3 complex, formins facilitate the polymerization of linear actin filaments. Recent super-resolution imaging reveals that spines are not smooth bulbous structures, as they often appear in conventional light microscopy, but rather they contain fine, finger-like projections from the spine head (18) (Fig. 1). These spine projections are enriched in the formin family member, formin-like protein-2 (FMNL2), suggesting that it may be important for these structures. Further work will be required to determine the function of these spine protrusions and FMNL2 to spine physiology.
Formins are also thought to play an important role in dendritic spine development by driving the formation of spine precursors such as dendritic filopodia. These filopodia are understudied, highly transient structures that emerge from dendrites before spinogenesis (around day 9 in vitro in cultured hippocampal neurons) (9). They lack the rounded spine head and are thought to serve as the first contact sites between nascent axonal boutons and dendrites during the development of the synapse. Much (although not all) of the F-actin within dendritic filopodia is unbranched (6), and studies knocking down the formin mDia2 demonstrate that it is important for the actin-dependent emergence of filopodia during this initial stage of spine formation (19). The formin FMN2 may also be important for either spine formation or maintenance. In mice, loss of FMN2 leads to a 32% reduction in spines (20) and an age-related learning/memory deficit (21). This is particularly relevant as homozygous truncation of FMN2 is associated with profound human ID (20).
Finally, the assembly of actin by nucleators such as Arp2/3 or formins requires a local pool of available G-actin (Fig. 2). Profilin is a G-actin-binding protein that facilitates nucleotide exchange (ADP to ATP), a switch that allows actin to polymerize more readily at the barbed end of the growing filament (17) and is central to both dendritic spine development as well as maintenance. Profilin also binds to both WAVE1 and formins to enhance the local supply of actin during polymerization (22, 23). The likely importance of profilin is highlighted by findings that it is rapidly recruited to dendritic spines in an activity-dependent fashion where it may facilitate their stabilization (24). The recruitment of profilin to spines has also been observed following behaviorally induced activity such as fear conditioning (25).
Actin Filament Disassembly
Counterbalancing polymerization are the actin-depolymerizing factors (ADF)/cofilins (26), which sever actin filaments (Fig. 2). This severing can lead to the creation of new barbed ends for additional filament growth in addition to disassembling F-actin. Cofilin-1 (also termed n-cofilin) is found in the vertebrate brain and localizes to the post-synaptic density (PSD, the protein-rich compartment within spines where neurotransmitters are received from the pre-synapse) of dendritic spines (27). Because cofilin is enriched at the tip of the spine, it may be particularly relevant for the high rate of actin dynamics within this region (2). Cofilin is tightly regulated by phosphorylation at serine 3, which causes inactivation of cofilin by inhibiting its ability to bind F-actin (28). LIM kinase 1 (LIMK-1) phosphorylates cofilin downstream of either Rac or Cdc42 (Fig. 3), and loss of LIMK-1 is associated with cognitive deficiencies in Williams syndrome (29). Loss of LIMK-1 in mice leads to a reduced phosphorylation of cofilin, altered dendritic spine morphology, and enhanced long-term potentiation (LTP) (30), suggesting that neuron-specific regulation of cofilin is critical for actin homeostasis within dendritic spines. Further, a recent genetic study showed that ADF/cofilin genes are associated with anxiety (31). Specific roles of cofilin in dendritic spine plasticity will be further explored below.
Actin Filament Stabilization
Capping protein (CapZ) is a heterodimer of α and β subunits that binds to the barbed ends of actin filaments to stabilize and restrict their elongation (32) (Fig. 2). Capping protein is localized in dendritic spines (6) and the β2 subunit has been shown to be important for dendritic spine development and synapse formation through shRNA knockdown (33). A second type of actin capping protein, Eps8, further highlights the critical role of capping proteins in dendritic spine plasticity. Eps8 (Fig. 3) localizes to dendritic spines during chemical LTP (LTP induced pharmacologically), and loss of Eps8 leads to reduced spine enlargement and structural plasticity (34). Further, Eps8 expression may be reduced in patients with autism spectrum disorder (ASD) (34).
The Actin Cytoskeleton and Synaptic Adhesion
Trans-synaptic adhesion molecules play a critical role in synapse formation, adaptation, and maintenance through their regulation of the actin cytoskeleton (Fig. 1). The large diversity of adhesion molecules that mediate these contacts is generated through multiple gene families and extensive alternative splicing. The combinatorial effect of this diversity is thought to generate a sophisticated “chemo-affinity” code that specifies the fine wiring of synaptic contacts. Here we will restrict our review to the cadherins and how they modulate actin remodeling based on recent studies and their potential association with brain disorders in humans.
The Cadherin-Catenin Complex
Classical cadherins are homotypic transmembrane proteins that are composed of an extracellular domain containing five cadherin repeat sequences, a single transmembrane domain, and an intracellular tail that anchors cadherins to the actin cytoskeleton through catenins (α, β, and p120ctn family members). Classical cadherins bind β-catenin, which localizes to synapses similarly to N-cadherin and plays an important role in dendritic spine formation (35) and excitatory synaptic transmission (36) (Fig. 3). β-Catenins interact with the actin-binding α-catenins and couple cadherins directly to F-actin under tension (37) (Fig. 3). α-Catenin is thought to relay activity-dependent signals during structural plasticity to actin cytoskeleton reorganization (38). p120 catenin binds to the juxtamembrane region of N-cadherin, and loss of p120 catenin leads to reduced dendritic spine density, loss of N-cadherin expression, and reduced Rac1 activity with increased RhoA activity (39). The loss of dendritic spine density is rescued by inhibition of RhoA, suggesting that modulation of Rho family proteins by p120 catenin is sufficient for normal spine density. Finally, δ-catenin is also found almost exclusively in neurons, and it regulates the actin cytoskeleton through interactions with cortactin (40) and p190RhoGAP (41) (Fig. 3). Although the cadherin-catenin complex is important for coupling adhesion to actin under tension, the roles of actin tension in spine physiology are unclear. Future imaging using newly developed FRET probes (42) will be an exciting area to better understand the spatio-temporal coupling of cadherin adhesion with tension during synaptogenesis and synaptic plasticity. Mutations in both β-catenin and δ-catenin underlie forms of syndromic ID (Fig. 3), suggesting that dysregulation of cadherin adhesion and potentially spine actin tension have severe neurodevelopmental consequences relevant to human health (43, 44).
N-cadherin
Perhaps the best-characterized cadherin at synapses is N-cadherin, which is localized at perisynaptic sites (the area around the PSD) flanking the area of transmitter release (45). Dominant negative N-cadherin expression in neurons, which blocks multiple classic cadherin adhesion molecules (N-cadherin, cadherin-8, and canherin-11), impairs spine morphogenesis and synapse formation (46, 47). N-cadherin engagement during the transition between dendritic filopodia and spine morphogenesis may serve to stabilize the actin cytoskeleton in these nascent spines via myosin engagement and tension (48). Interestingly, N-cadherin also interacts directly with the extracellular portion of GluA2 (Fig. 3), which is recruited during spine maturation. This interaction may be important for regulating spine morphology in response to synaptic activity through Rac1 and cofilin-mediated remodeling of the actin cytoskeleton (46, 47, 49). N-cadherin can also couple to the actin cytoskeleton through the scaffolding protein afadin (Fig. 3). Afadin is an actin-binding protein that colocalizes in synapses with the GEF kalirin-7 (50, 51) and is critical for dendritic spine structure and function (52).
Protocadherins
Perhaps the most compelling link between cadherins and brain disorders, however, are the protocadherins (Fig. 3). Protocadherins are the largest subfamily of the cadherin superfamily and are also heavily implicated in neurodevelopmental disorders such as ID and ASD (53). The signaling mechanisms linking the diverse protocadherins to the actin cytoskeleton in neurons are still being worked out. Recent studies, however, show that the protocadherins PCDH10 and PCDH19 directly recruit the WAVE1 complex (54, 55). This interaction is mediated by a WAVE complex-interacting receptor sequence (WIRS) in the intracellular tail (56). Remarkably, the WIRS motif appears to be present in at least 115 proteins, many of which are adhesion molecules, including 24 protocadherins. Interaction between the WIRS motif and the WAVE1 complex may enhance the Rac1-mediated activation of Arp2/3, particularly in the case of the protocadherins PCDH10, PCDH12, and PCDH19.
Actin's Role in Synaptic Plasticity Mechanisms
Once synapses have been formed through the assistance of trans-synaptic adhesion receptors and signaling, dendritic spines undergo rapid remodeling in response to patterns of input from the pre-synapse (Fig. 1). Here we will focus on two of these post-synaptic processes implicated in NMDA-type glutamate receptor (NMDAR)-dependent hippocampal learning and memory: LTP and long-term depression (LTD) (57). LTP is a process in which periods of high frequency synaptic activity lead to a long-lasting increase in the strength of a synapse. LTD occurs after periods of low frequency synaptic activity lead to a decrease in synaptic strength. Although LTP and LTD involve many long-lasting changes at the receptor, signaling, and gene expression levels, we will focus here on two of these alterations that are modulated by actin remodeling and that occur in CA1 hippocampal dendritic spines. These are 1) electrophysiological changes largely resulting from alterations in synaptic AMPA-type glutamate receptor (AMPAR) density, and 2) morphological changes in dendritic spine size or structural plasticity.
AMPAR Trafficking and the Actin Cytoskeleton
AMPARs are ionotropic, excitatory glutamate receptors in dendritic spines that underlie the majority of current alterations during LTP and LTD in hippocampal CA1 synapses. The first evidence to suggest that the actin cytoskeleton was important for AMPAR localization came from studies in cultured hippocampal neurons that were exposed to actin-depolymerizing drugs such as latrunculin A, which sequesters monomeric actin. Loss of filamentous actin led to reduced AMPAR receptors in dendritic spines (58), inhibition of LTP (59), and an increase in AMPAR internalization (60). Although there is an extensive body of research on this topic, for the sake of space, we will focus our discussion on select actin regulatory proteins recently implicated in AMPAR trafficking: cofilin, PICK1, WASH/retromer, and oligophrenin-1.
Cofilin
As mentioned above, cofilin is a critical actin regulator, which severs f-actin, simultaneously breaking it down while creating barbed ends for polymerization. The current literature suggests two roles for cofilin in regulating AMPARs: 1) regulation of AMPAR diffusion/stabilization into the dendritic spine synaptic zone (the portion of the spine immediately apposing pre-synaptic transmitter release sites) and 2) exocytosis into the perisynaptic zone of spines (61) (Fig. 1). Work in cultured hippocampal neurons shows that activation of cofilin through dephosphorylation is required for AMPAR surface recruitment, suggesting that cofilin's severing of the actin cytoskeleton is critical for insertion of AMPARs into the synaptic membrane (62). Further in vivo work using cofilin conditional knock-out mice indicated that diffusion of AMPARs was diminished in the extra-synaptic domains, whereas the overall membrane fluidity remained unaffected (63). This suggests that cofilin has a receptor-specific effect on AMPAR diffusion. Recent work examining AMPAR clustering using fluorescence recovery after photo-bleaching of surface GluA1 at the spine found that AMPARs are highly dynamic within the PSD, and the dynamics of AMPARs are a result of constantly cycling actin (64). Interestingly, pharmacological loss of actin by administration of latrunculin A did not lead to rapid diffusion of AMPARs between the extra-synaptic and PSD compartments, suggesting that AMPARs are not stabilized in the PSD by actin filaments contrary to previous belief (64). Taken together, these data suggest that constant actin treadmilling facilitated by cofilin is critical for recruitment of AMPARs to the extra-synaptic zone and PSD, but that AMPARs are not anchored in the PSD by actin.
PICK1
Human protein interacting with C kinase 1 (PICK1) is a scaffold protein widely expressed in cells, including neurons (65). PICK1 was first identified as a binding partner for PKC (66) and was subsequently shown to associate with the AMPAR receptor subunits GluA2 and GluA3 (65) as well as to interact directly with actin and the Arp2/3 complex (67). Thus, PICK1 provides a potential link between AMPARs and the actin cytoskeleton (68).
Three domains of PICK1 are believed to be critical for binding these different factors: an N-terminal PSD-95/DlgA/ZO-1 (PDZ) domain, a central Bin/amphiphysin/RVS (BAR) domain, and an acidic C-terminal tail. The PDZ domain binds to PKC (69), as well as several neuron-specific proteins such as GluA2 (70). The BAR domain of PICK1 binds actin, and the acidic C-terminal tail domain binds to and inhibits the Arp2/3 complex in vitro (Fig. 3). Structure-function studies of PICK1 suggest that inhibition of the Arp2/3 complex is important for AMPAR internalization in cultured neurons downstream of NMDA receptor activation (67). Although this work strongly supports a mechanistic link between PICK1, AMPAR trafficking, and actin cytoskeleton remodeling, recent conflicting in vitro analysis of PICK1 and Arp2/3 suggests that PICK1 does not regulate Arp2/3 activity (71). Thus, although PICK1 clearly appears to be important for AMPAR trafficking, additional work is required to resolve conflicting data regarding the relationship between PICK1 and the Arp2/3 complex in the context of AMPAR trafficking in spines.
The WASH-Retromer Complex
Endosomal protein sorting is a process by which endocytosed proteins are sent to one of three possible destinations: the lysosome for degradation, the cell surface for recycling, or the trans-Golgi network. Because of the rapid turnover of AMPARs in the dendritic spine, this process is a particularly important feature, and endosomes have been thought to play a critical role in synaptic responsiveness for many years (57). Recently, branched actin, the Arp2/3 complex, and its endosomal regulator, the WASH complex, have been linked to AMPAR trafficking.
WASH is a recently discovered endocytic compartment NPF (72) that binds to lipids and is ubiquitously expressed. There are five proteins in the WASH complex: SWIP (also known as KIAA1033), strumpellin, FAM21, WASH1, and CCDC53 (Fig. 3). As proteins are sorted, the WASH complex is thought to provide endosomal membrane domains through the polymerization of actin into which different proteins can be organized (73) (Fig. 1). WASH complex is recruited to endosomes through a second protein complex, retromer (74). VPS35, a component of the retromer complex, localizes to dendritic spines, and expression of a VPS35 loss-of-function mutation leads to altered AMPAR surface expression and synaptic recycling and is also linked to familial parkinsonism (75). In another recent study of the role of retromer in neurons, it was found that retromer-associated endosomes are found throughout dendrites, that they contain receptors such as AMPARs and NMDARs, and that the endosomes provide a local source of shaft-directed receptor insertion (76). Mutations within the WASH complex member SWIP cause a form of ID (77), further suggesting that the neural function of this complex may be important for synaptic plasticity. However, further work is needed to functionally test the consequences of this mutation in SWIP.
Oligophrenin-1 (OPHN1)
AMPAR trafficking is also modulated by the Rho-GAP OPHN1, which inactivates RhoA (78) (Fig. 3). OPHN1 forms a complex with GluA1/2, and its knockdown leads to a significant reduction in LTP. This effect on LTP is likely due to altered AMPAR trafficking as overexpression of OPHN1 stabilizes the synaptic surface levels of GluA1/2. Furthermore, blocking GluA1/2 internalization occludes the effect of OPHN1 knockdown on AMPAR-mediated synaptic transmission. The regulation of AMPAR by OPHN1 is dependent on the Rho-GAP domain of OPHN1, and inhibition of the RhoA effector, Rho-associated kinase (ROCK), affected surface GluA1/2 stability similar to OPHN1 overexpression. OPHN1 is recruited to spines during NMDAR-dependent LTP, suggesting that it likely forms a positive feedback loop by inhibiting Rho-ROCK signaling to stabilize surface AMPAR levels. Interestingly, the effect of OPHN1 is also likely dependent on its ability to interact with the scaffolding protein Homer1b/c (79). Disruption of this interaction reduces the positioning of the endocytic zone within spines and impairs the recycling of AMPARs important for basal transmission and LTP. Together, these data suggest that OPHN1 operates in a multifaceted manner to coordinate actin cytoskeletal remodeling and endocytosis mechanisms in spines. The ability of OPHN1 to influence glutamate receptor trafficking may be one important clue into the mechanisms underlying how the loss of OPHN1 leads to ID in humans (80).
Structural Plasticity of Spines Associated with LTP and LTD
The first link between structure and function of dendritic spines was made when Ramón y Cajal hypothesized that changes in dendritic structure could be the mechanism behind information storage. Studies in the mid-1970s showed that synaptic activation increases spine volume and that these morphological changes persisted long-term (81). This change in spine size is associated with an increase in spine AMPAR number (82), synaptic strength (83), and PSD area (84). Spine structural plasticity is an actin-dependent process and is linked to functional changes associated with LTP and LTD in spines (85). It is well established that NMDAR activation triggers calmodulin kinase II (CaMKII) activation through calcium influx (Fig. 3). CaMKII activity then leads to downstream activation of Rho-GTPases such as Cdc42 and Rac via GEFs, triggering activation of NPFs and cytoskeleton reorganization that results in morphological changes of the dendritic spine (86).
Actin Dynamics during Structural Plasticity
The F-actin of the dendritic spine is divided into three subpopulations based on rate of turnover: the tip of the PSD has highly dynamic actin filaments, the base of the spine has largely stable actin filaments, and the central “spine-enlargement” pool has actin filaments whose dynamics are altered by synaptic activity (87) (Fig. 1). Further studies show that activation of F-actin contained within the central enlargement pool is dependent on CaMKII activity and enhanced during LTP (88). Interestingly, this rate of turnover is also subdomain-specific: low at perisynaptic endocytic zones and faster at filaments near the PSD (2). Regardless of zone, actin filaments treadmill constitutively, with filament lifetimes lasting less than 17 min (87). Interestingly, although actin remodeling constantly occurs under basal conditions, F-actin content in spines resulting from synaptic plasticity can be long-lasting. Increased actin density in dendritic spines is seen in vivo for up to 5 weeks following electrical LTP induction (89).
Arp2/3 Complex and the Rho-GTPases
As discussed above, the Arp2/3 complex is the essential factor that creates branched actin filaments in spines. Conditional knock-out of the Arp2/3 complex shows that its activity is critical for the turnover of actin in spines, dendritic spine maintenance, and spine structural plasticity (15). Furthermore, mice lacking Arp2/3 activity in forebrain exhibit classic endophenotypes associated with neuropsychiatric disorders such as SZ-related disorders (16) (Fig. 3). The Arp2/3 complex is activated downstream of two Rho-GTPases that are present in the dendritic spine: Cdc42 and Rac (90) (Fig. 3). A recent study of Cdc42 using conditional knock-out mice showed that Cdc42 is activated in the spine during the induction of structural LTP and that it is essential for structural plasticity and remote memory recall (91). Directly downstream of Cdc42 and Rac are the NPFs N-WASP and WAVE1, respectively (90) (Fig. 3). Although these NPFs are the established direct activators of the Arp2/3 complex, their temporal and spatial dynamics have yet to be fully explored during structural plasticity. Loss of WAVE1, however, enhances LTP, impairs LTD, and alters the morphology of synapses and a wide range of behaviors, including learning and memory (92, 93).
Cofilin
Actin severing by cofilin is another important mediator of structural LTP and LTD. There are three highly conserved cofilin genes with differing expression patterns: m-cofilin (muscle), n-cofilin (non-muscle), and ADF. Early in vitro studies linked ADF/cofilin phosphorylation to LTP and spine enlargement (94), whereas ADF/cofilin dephosphorylation was connected with LTD and spine shrinkage (95) through actin cytoskeletal remodeling. Further, genetic loss of n-cofilin leads to an increase and enlargement of spines in vivo as well as impairment of reward-induced and fear-conditioned learning (63). Together, these results suggest a model where spine expansion is mediated by reduced cofilin activity and spine volume loss is facilitated by increased cofilin activity. This role of cofilin in structural LTD or spine shrinkage was recently evaluated in a pharmacological study. The authors found that cofilin activity is required for dendritic spine maintenance and that cofilin is inhibited from severing actin during LTD (96), contradicting the current model. The role of cofilin during LTP has also been recently evaluated by another study (97) where the authors demonstrate that cofilin is recruited early in LTP, and the rate of cofilin turnover decreases as cofilin interacts with F-actin. This study provides a new regulatory pathway for cofilin during structural LTP, which suggests that instead of cofilin activity being down-regulated, cofilin in fact may facilitate structural LTP. Together, these studies indicate that cofilin activity is important for both LTP and LTD and that more work is needed to clearly understand cofilin recruitment and regulation of activity mechanisms during these processes.
Spine Neck Plasticity
Although most studies of spine structural plasticity have focused on the spine head, super-resolution imaging also suggests that morphometric changes, which are associated with LTP, occur in the spine neck (98). Following chemical LTP, spine neck lengths decreased by an average of 25% and neck widths increased by 30%. These structural changes in spine necks occurred concurrently with spine head expansion. Because the neck links the spine with the rest of the neuron, this form of plasticity may provide a novel mechanism for morphological regulation of both biochemical diffusion and electrical filtering. Moreover, spine neck plasticity may be important for regulating actin within the spine. For example, actin fibers are reported to be released from the spine head through the neck into the dendritic shaft following LTP, particularly in spines that fail to maintain spine head enlargement (87) (Fig. 1). As super-resolution microscopy is adopted more widely, it will be important to better understand the regulatory factors that control spine neck dynamics. Although actin may play a role, other cytoskeletal components such as septins could also influence neck dynamics (99, 100).
Conclusion
The signaling pathways operant to the remodeling of the dendritic spine actin cytoskeleton within the context of synaptic physiology are beginning to emerge. Furthermore, recent discoveries such as the WASH complex, direct links between WAVE/Arp2/3 to adhesion via the WIRS motif, and spine neck plasticity are likely to drive the field in new directions, providing a more complete picture of how spine actin signaling modulates spine physiology. This review has also highlighted multiple links between neurodevelopmental and psychiatric disorders and the regulation of spine actin (Fig. 3). The emerging consensus is that signaling to the actin cytoskeleton in dendritic spines is a commonly disrupted pathway, whose dysfunction greatly increases the risk of these disorders. Future work is needed to bridge our knowledge gaps between gene mutations relevant to the synaptic cytoskeleton, how they impact synaptic development and physiology, and the resultant neural circuit abnormalities driving disorder-relevant endophenotypes. Advances in mouse genetics (CRISPR (clustered regularly interspaced short palindromic repeats) genome editing), imaging (particularly single spine fluorescence lifetime imaging (FLIM)/FRET), synaptic proteomics (quantitative rather than qualitative), and circuit level manipulations (viral tracing, optogenetics) promise to reveal the interplay between mutations affecting the synaptic cytoskeleton and behavioral endophenotypes.
This work was supported by National Institutes of Health Grants MH103374 and NS059957 (to S. H. S.). This is the third article in the Thematic Minireview series “Molecular Mechanisms of Synaptic Plasticity.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
- SZ
- schizophrenia
- ID
- intellectual disability
- ASD
- autism spectrum disorder(s)
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- NPF
- nucleation-promoting factor
- WASH
- Wiskott–Aldrich syndrome protein and SCAR homologue
- WIRS
- WAVE complex-interacting receptor sequence
- WAVE
- Wiskott-Aldrich syndrome family, verprolin homologous protein
- ADF
- actin-depolymerizing factor(s)
- NMDAR
- NMDA-type glutamate receptor
- AMPAR
- AMPA-type glutamate receptor
- CaMKII
- calmodulin kinase II
- n-cofilin
- non-muscle cofilin
- SWIP
- strumpellin And WASH-interacting protein
- LTP
- long-term potentiation
- LTD
- long-term depression
- PSD
- post-synaptic density.
References
- 1.Yuste R. (2013) Electrical compartmentalization in dendritic spines. Annu. Rev. Neurosci. 36, 429–449 [DOI] [PubMed] [Google Scholar]
- 2.Frost N. A., Shroff H., Kong H., Betzig E., and Blanpied T. A. (2010) Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron 67, 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holtmaat A., and Svoboda K. (2009) Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10, 647–658 [DOI] [PubMed] [Google Scholar]
- 4.van Spronsen M., and Hoogenraad C. C. (2010) Synapse pathology in psychiatric and neurologic disease. Curr. Neurol. Neurosci. Rep. 10, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fromer M., Pocklington A. J., Kavanagh D. H., Williams H. J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D. M., Carrera N., Humphreys I., Johnson J. S., Roussos P., Barker D. D., Banks E., Milanova V., Grant S. G., Hannon E., Rose S. A., Chambert K., Mahajan M., Scolnick E. M., Moran J. L., Kirov G., Palotie A., McCarroll S. A., Holmans P., Sklar P., Owen M. J., Purcell S. M., and O'Donovan M. C. (2014) De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korobova F., and Svitkina T. (2010) Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell 21, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolias K. F., Duman J. G., and Um K. (2011) Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog. Neurobiol. 94, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell T. A., Blizinsky K. D., Cobia D. J., Cahill M. E., Xie Z., Sweet R. A., Duan J., Gejman P. V., Wang L., Csernansky J. G., and Penzes P. (2014) A sequence variant in human KALRN impairs protein function and coincides with reduced cortical thickness. Nat. Commun. 5, 4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson B. R., Lloyd K. E., Kruszewski A., Kim I. H., Rodriguiz R. M., Heindel C., Faytell M., Dudek S. M., Wetsel W. C., and Soderling S. H. (2011) WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 31, 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endris V., Wogatzky B., Leimer U., Bartsch D., Zatyka M., Latif F., Maher E. R., Tariverdian G., Kirsch S., Karch D., and Rappold G. A. (2002) The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc. Natl. Acad. Sci. U.S.A. 99, 11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bugyi B., and Carlier M. F. (2010) Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys. 39, 449–470 [DOI] [PubMed] [Google Scholar]
- 12.Rotty J. D., Wu C., and Bear J. E. (2013) New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12 [DOI] [PubMed] [Google Scholar]
- 13.Kim Y., Sung J. Y., Ceglia I., Lee K. W., Ahn J. H., Halford J. M., Kim A. M., Kwak S. P., Park J. B., Ho Ryu S., Schenck A., Bardoni B., Scott J. D., Nairn A. C., and Greengard P. (2006) Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature 442, 814–817 [DOI] [PubMed] [Google Scholar]
- 14.Soderling S. H., Guire E. S., Kaech S., White J., Zhang F., Schutz K., Langeberg L. K., Banker G., Raber J., and Scott J. D. (2007) A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 27, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I. H., Racz B., Wang H., Burianek L., Weinberg R., Yasuda R., Wetsel W. C., and Soderling S. H. (2013) Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J. Neurosci. 33, 6081–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim I. H., Rossi M. A., Aryal D. K., Racz B., Kim N., Uezu A., Wang F., Wetsel W. C., Weinberg R. J., Yin H., and Soderling S. H. (2015) Spine pruning drives antipsychotic-sensitive locomotion via circuit control of striatal dopamine. Nat. Neurosci. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris E. S., and Higgs H. N. (2004) Actin cytoskeleton: formins lead the way. Curr. Biol. 14, R520–R522 [DOI] [PubMed] [Google Scholar]
- 18.Chazeau A., Mehidi A., Nair D., Gautier J. J., Leduc C., Chamma I., Kage F., Kechkar A., Thoumine O., Rottner K., Choquet D., Gautreau A., Sibarita J. B., and Giannone G. (2014) Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 33, 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotulainen P., Llano O., Smirnov S., Tanhuanpaa K., Faix J., Rivera C., and Lappalainen P. (2009) Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J. Cell Biol. 185, 323–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law R., Dixon-Salazar T., Jerber J., Cai N., Abbasi A. A., Zaki M. S., Mittal K., Gabriel S. B., Rafiq M. A., Khan V., Nguyen M., Ali G., Copeland B., Scott E., Vasli N., Mikhailov A., Khan M. N., Andrade D. M., Ayaz M., Ansar M., Ayub M., Vincent J. B., and Gleeson J. G. (2014) Biallelic truncating mutations in FMN2, encoding the actin-regulatory protein Formin 2, cause nonsyndromic autosomal-recessive intellectual disability. Am. J. Hum. Genet. 95, 721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R. C., Cota P., Wittnam J. L., Gogol-Doering A., Opitz L., Salinas-Riester G., Dettenhofer M., Kang H., Farinelli L., Chen W., and Fischer A. (2010) Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 [DOI] [PubMed] [Google Scholar]
- 22.Miki H., Suetsugu S., and Takenawa T. (1998) WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17, 6932–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., and Carlier M. F. (2004) Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119, 419–429 [DOI] [PubMed] [Google Scholar]
- 24.Ackermann M., and Matus A. (2003) Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat. Neurosci. 6, 1194–1200 [DOI] [PubMed] [Google Scholar]
- 25.Lamprecht R., Farb C. R., Rodrigues S. M., and LeDoux J. E. (2006) Fear conditioning drives profilin into amygdala dendritic spines. Nat. Neurosci. 9, 481–483 [DOI] [PubMed] [Google Scholar]
- 26.Bernstein B. W., and Bamburg J. R. (2010) ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racz B., and Weinberg R. J. (2006) Spatial organization of cofilin in dendritic spines. Neuroscience 138, 447–456 [DOI] [PubMed] [Google Scholar]
- 28.Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., and Mizuno K. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809–812 [DOI] [PubMed] [Google Scholar]
- 29.Gray V., Karmiloff-Smith A., Funnell E., and Tassabehji M. (2006) In-depth analysis of spatial cognition in Williams syndrome: a critical assessment of the role of the LIMK1 gene. Neuropsychologia 44, 679–685 [DOI] [PubMed] [Google Scholar]
- 30.Meng Y., Zhang Y., Tregoubov V., Janus C., Cruz L., Jackson M., Lu W. Y., MacDonald J. F., Wang J. Y., Falls D. L., and Jia Z. (2002) Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 35, 121–133 [DOI] [PubMed] [Google Scholar]
- 31.Goodson M., Rust M. B., Witke W., Bannerman D., Mott R., Ponting C. P., and Flint J. (2012) Cofilin-1: a modulator of anxiety in mice. PLoS Genet. 8, e1002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards M., Zwolak A., Schafer D. A., Sept D., Dominguez R., and Cooper J. A. (2014) Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15, 677–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Y., Tang X., Vitriol E., Chen G., and Zheng J. Q. (2011) Actin capping protein is required for dendritic spine development and synapse formation. J. Neurosci. 31, 10228–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menna E., Zambetti S., Morini R., Donzelli A., Disanza A., Calvigioni D., Braida D., Nicolini C., Orlando M., Fossati G., Cristina Regondi M., Pattini L., Frassoni C., Francolini M., Scita G., Sala M., Fahnestock M., and Matteoli M. (2013) Eps8 controls dendritic spine density and synaptic plasticity through its actin-capping activity. EMBO J. 32, 1730–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X., and Malenka R. C. (2004) Multiple functions for the cadherin/catenin complex during neuronal development. Neuropharmacology 47, 779–786 [DOI] [PubMed] [Google Scholar]
- 36.Okuda T., Yu L. M., Cingolani L. A., Kemler R., and Goda Y. (2007) β-Catenin regulates excitatory postsynaptic strength at hippocampal synapses. Proc. Natl. Acad. Sci. U.S.A. 104, 13479–13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M. (2010) α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 [DOI] [PubMed] [Google Scholar]
- 38.Abe K., Chisaka O., Van Roy F., and Takeichi M. (2004) Stability of dendritic spines and synaptic contacts is controlled by α N-catenin. Nat. Neurosci. 7, 357–363 [DOI] [PubMed] [Google Scholar]
- 39.Elia L. P., Yamamoto M., Zang K., and Reichardt L. F. (2006) p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron 51, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez M. C., Ochiishi T., Majewski M., and Kosik K. S. (2003) Dual regulation of neuronal morphogenesis by a δ-catenin-cortactin complex and Rho. J. Cell Biol. 162, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H., Han J. R., Park J., Oh M., James S. E., Chang S., Lu Q., Lee K. Y., Ki H., Song W. J., and Kim K. (2008) δ-Catenin-induced dendritic morphogenesis: an essential role of p190RhoGEF interaction through Akt1-mediated phosphorylation. J. Biol. Chem. 283, 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim T. J., Zheng S., Sun J., Muhamed I., Wu J., Lei L., Kong X., Leckband D. E., and Wang Y. (2015) Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr. Biol. 25, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tucci V., Kleefstra T., Hardy A., Heise I., Maggi S., Willemsen M. H., Hilton H., Esapa C., Simon M., Buenavista M. T., McGuffin L. J., Vizor L., Dodero L., Tsaftaris S., Romero R., Nillesen W. N., Vissers L. E., Kempers M. J., Vulto-van Silfhout A. T., Iqbal Z., Orlando M., Maccione A., Lassi G., Farisello P., Contestabile A., Tinarelli F., Nieus T., Raimondi A., Greco B., Cantatore D., Gasparini L., Berdondini L., Bifone A., Gozzi A., Wells S., and Nolan P. M. (2014) Dominant β-catenin mutations cause intellectual disability with recognizable syndromic features. J. Clin. Invest. 124, 1468–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medina M., Marinescu R. C., Overhauser J., and Kosik K. S. (2000) Hemizygosity of δ-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics 63, 157–164 [DOI] [PubMed] [Google Scholar]
- 45.Uchida N., Honjo Y., Johnson K. R., Wheelock M. J., and Takeichi M. (1996) The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 135, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Togashi H., Abe K., Mizoguchi A., Takaoka K., Chisaka O., and Takeichi M. (2002) Cadherin regulates dendritic spine morphogenesis. Neuron 35, 77–89 [DOI] [PubMed] [Google Scholar]
- 47.Saglietti L., Dequidt C., Kamieniarz K., Rousset M. C., Valnegri P., Thoumine O., Beretta F., Fagni L., Choquet D., Sala C., Sheng M., and Passafaro M. (2007) Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 54, 461–477 [DOI] [PubMed] [Google Scholar]
- 48.Chazeau A., Garcia M., Czondor K., Perrais D., Tessier B., Giannone G., and Thoumine O. (2015) Mechanical coupling between transsynaptic N-cadherin adhesions and actin flow stabilizes dendritic spines. Mol. Biol. Cell 26, 859–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Z., Hu J., Passafaro M., Xie W., and Jia Z. (2011) GluA2 (GluR2) regulates metabotropic glutamate receptor-dependent long-term depression through N-cadherin-dependent and cofilin-mediated actin reorganization. J. Neurosci. 31, 819–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Z., Photowala H., Cahill M. E., Srivastava D. P., Woolfrey K. M., Shum C. Y., Huganir R. L., and Penzes P. (2008) Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J. Neurosci. 28, 6079–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tachibana K., Nakanishi H., Mandai K., Ozaki K., Ikeda W., Yamamoto Y., Nagafuchi A., Tsukita S., and Takai Y. (2000) Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J. Cell Biol. 150, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava D. P., Copits B. A., Xie Z., Huda R., Jones K. A., Mukherji S., Cahill M. E., VanLeeuwen J. E., Woolfrey K. M., Rafalovich I., Swanson G. T., and Penzes P. (2012) Afadin is required for maintenance of dendritic structure and excitatory tone. J. Biol. Chem. 287, 35964–35974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirabayashi T., and Yagi T. (2014) Protocadherins in neurological diseases. Adv. Neurobiol. 8, 293–314 [DOI] [PubMed] [Google Scholar]
- 54.Nakao S., Platek A., Hirano S., and Takeichi M. (2008) Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J. Cell Biol. 182, 395–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tai K., Kubota M., Shiono K., Tokutsu H., and Suzuki S. T. (2010) Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 1344, 13–24 [DOI] [PubMed] [Google Scholar]
- 56.Chen B., Brinkmann K., Chen Z., Pak C. W., Liao Y., Shi S., Henry L., Grishin N. V., Bogdan S., and Rosen M. K. (2014) The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huganir R. L., and Nicoll R. A. (2013) AMPARs and synaptic plasticity: the last 25 years. Neuron 80, 704–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allison D. W., Gelfand V. I., Spector I., and Craig A. M. (1998) Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J. Neurosci. 18, 2423–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim C. H., and Lisman J. E. (1999) A role of actin filament in synaptic transmission and long-term potentiation. J. Neurosci. 19, 4314–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Q., Xiao M., and Nicoll R. A. (2001) Contribution of cytoskeleton to the internalization of AMPA receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Opazo P., and Choquet D. (2011) A three-step model for the synaptic recruitment of AMPA receptors. Mol. Cell. Neurosci. 46, 1–8 [DOI] [PubMed] [Google Scholar]
- 62.Gu J., Lee C. W., Fan Y., Komlos D., Tang X., Sun C., Yu K., Hartzell H. C., Chen G., Bamburg J. R., and Zheng J. Q. (2010) ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat. Neurosci. 13, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rust M. B., Gurniak C. B., Renner M., Vara H., Morando L., Gorlich A., Sassoe-Pognetto M., Banchaabouchi M. A., Giustetto M., Triller A., Choquet D., and Witke W. (2010) Learning, AMPA receptor mobility and synaptic plasticity depend on n-cofilin-mediated actin dynamics. EMBO J. 29, 1889–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kerr J. M., and Blanpied T. A. (2012) Subsynaptic AMPA receptor distribution is acutely regulated by actin-driven reorganization of the postsynaptic density. J. Neurosci. 32, 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia J., Zhang X., Staudinger J., and Huganir R. L. (1999) Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron 22, 179–187 [DOI] [PubMed] [Google Scholar]
- 66.Staudinger J., Zhou J., Burgess R., Elledge S. J., and Olson E. N. (1995) PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 128, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rocca D. L., Martin S., Jenkins E. L., and Hanley J. G. (2008) Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat. Cell Biol. 10, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanley J. G. (2014) Actin-dependent mechanisms in AMPA receptor trafficking. Front. Cell. Neurosci. 8, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staudinger J., Lu J., and Olson E. N. (1997) Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-α. J. Biol. Chem. 272, 32019–32024 [DOI] [PubMed] [Google Scholar]
- 70.Erlendsson S., Rathje M., Heidarsson P. O., Poulsen F. M., Madsen K. L., Teilum K., and Gether U. (2014) Protein interacting with C-kinase 1 (PICK1) binding promiscuity relies on unconventional PSD-95/discs-large/ZO-1 homology (PDZ) binding modes for nonclass II PDZ ligands. J. Biol. Chem. 289, 25327–25340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Madasu Y., Yang C., Boczkowska M., Bethoney K. A., Zwolak A., Rebowski G., Svitkina T., and Dominguez R. (2015) PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner. Mol. Biol. Cell 26, 1308–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., and Gautreau A. (2009) The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 [DOI] [PubMed] [Google Scholar]
- 73.Puthenveedu M. A., Lauffer B., Temkin P., Vistein R., Carlton P., Thorn K., Taunton J., Weiner O. D., Parton R. G., and von Zastrow M. (2010) Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell 143, 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harbour M. E., Breusegem S. Y., Antrobus R., Freeman C., Reid E., and Seaman M. N. (2010) The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci. 123, 3703–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munsie L. N., Milnerwood A. J., Seibler P., Beccano-Kelly D. A., Tatarnikov I., Khinda J., Volta M., Kadgien C., Cao L. P., Tapia L., Klein C., and Farrer M. J. (2015) Retromer-dependent neurotransmitter receptor trafficking to synapses is altered by the Parkinson's disease VPS35 mutation p.D620N. Hum. Mol. Genet. 24, 1691–1703 [DOI] [PubMed] [Google Scholar]
- 76.Choy R. W., Park M., Temkin P., Herring B. E., Marley A., Nicoll R. A., and von Zastrow M. (2014) Retromer mediates a discrete route of local membrane delivery to dendrites. Neuron 82, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ropers F., Derivery E., Hu H., Garshasbi M., Karbasiyan M., Herold M., Nurnberg G., Ullmann R., Gautreau A., Sperling K., Varon R., and Rajab A. (2011) Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum. Mol. Genet. 20, 2585–2590 [DOI] [PubMed] [Google Scholar]
- 78.Nadif Kasri N., Nakano-Kobayashi A., Malinow R., Li B., and Van Aelst L. (2009) The Rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev. 23, 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakano-Kobayashi A., Tai Y., Nadif Kasri N., and Van Aelst L. (2014) The X-linked mental retardation protein OPHN1 interacts with Homer1b/c to control spine endocytic zone positioning and expression of synaptic potentiation. J. Neurosci. 34, 8665–8671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Billuart P., Bienvenu T., Ronce N., des Portes V., Vinet M. C., Zemni R., Roest Crollius H., Carrie A., Fauchereau F., Cherry M., Briault S., Hamel B., Fryns J. P., Beldjord C., Kahn A., Moraine C., and Chelly J. (1998) Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature 392, 923–926 [DOI] [PubMed] [Google Scholar]
- 81.Van Harreveld A., and Fifkova E. (1975) Swelling of dendritic spines in the fascia dentata after stimulation of the perforant fibers as a mechanism of post-tetanic potentiation. Exp. Neurol. 49, 736–749 [DOI] [PubMed] [Google Scholar]
- 82.Nusser Z., Lujan R., Laube G., Roberts J. D., Molnar E., and Somogyi P. (1998) Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron 21, 545–559 [DOI] [PubMed] [Google Scholar]
- 83.Matsuzaki M., Ellis-Davies G. C., Nemoto T., Miyashita Y., Iino M., and Kasai H. (2001) Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4, 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kasai H., Matsuzaki M., Noguchi J., Yasumatsu N., and Nakahara H. (2003) Structure-stability-function relationships of dendritic spines. Trends Neurosci. 26, 360–368 [DOI] [PubMed] [Google Scholar]
- 85.Matsuzaki M., Honkura N., Ellis-Davies G. C., and Kasai H. (2004) Structural basis of long-term potentiation in single dendritic spines. Nature 429, 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murakoshi H., Wang H., and Yasuda R. (2011) Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature 472, 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Honkura N., Matsuzaki M., Noguchi J., Ellis-Davies G. C., and Kasai H. (2008) The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron 57, 719–729 [DOI] [PubMed] [Google Scholar]
- 88.Yamagata Y., Kobayashi S., Umeda T., Inoue A., Sakagami H., Fukaya M., Watanabe M., Hatanaka N., Totsuka M., Yagi T., Obata K., Imoto K., Yanagawa Y., Manabe T., and Okabe S. (2009) Kinase-dead knock-in mouse reveals an essential role of kinase activity of Ca2+/calmodulin-dependent protein kinase IIα in dendritic spine enlargement, long-term potentiation, and learning. J. Neurosci. 29, 7607–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukazawa Y., Saitoh Y., Ozawa F., Ohta Y., Mizuno K., and Inokuchi K. (2003) Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 38, 447–460 [DOI] [PubMed] [Google Scholar]
- 90.Burianek L. E., and Soderling S. H. (2013) Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin. Cell Dev. Biol. 24, 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim I. H., Wang H., Soderling S. H., and Yasuda R. (2014) Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. eLife 3, e02839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soderling S. H., Langeberg L. K., Soderling J. A., Davee S. M., Simerly R., Raber J., and Scott J. D. (2003) Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hazai D., Szudoczki R., Ding J., Soderling S. H., Weinberg R. J., Sótonyi P., and Rácz B. (2013) Ultrastructural abnormalities in CA1 hippocampus caused by deletion of the actin regulator WAVE-1. PLoS One 8, e75248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen L. Y., Rex C. S., Casale M. S., Gall C. M., and Lynch G. (2007) Changes in synaptic morphology accompany actin signaling during LTP. J. Neurosci. 27, 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou Q., Homma K. J., and Poo M. M. (2004) Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron 44, 749–757 [DOI] [PubMed] [Google Scholar]
- 96.Calabrese B., Saffin J. M., and Halpain S. (2014) Activity-dependent dendritic spine shrinkage and growth involve downregulation of cofilin via distinct mechanisms. PLoS One 9, e94787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bosch M., Castro J., Saneyoshi T., Matsuno H., Sur M., and Hayashi Y. (2014) Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82, 444–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tønnesen J., Katona G., Rózsa B., and Nägerl U. V. (2014) Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17, 678–685 [DOI] [PubMed] [Google Scholar]
- 99.Xie Y., Vessey J. P., Konecna A., Dahm R., Macchi P., and Kiebler M. A. (2007) The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 17, 1746–1751 [DOI] [PubMed] [Google Scholar]
- 100.Ewers H., Tada T., Petersen J. D., Racz B., Sheng M., and Choquet D. (2014) A septin-dependent diffusion barrier at dendritic spine necks. PLoS One 9, e113916. [DOI] [PMC free article] [PubMed] [Google Scholar]