Background: Protein kinase G (PKG) is selectively activated by cGMP, but the mechanism of cGMP-versus-cAMP selectivity is not fully understood.
Results: cAMP-bound PKG exists in a dynamic exchange between three states, inactive, active, and a partially autoinhibited state, which results in partial agonism.
Conclusion: Partial agonism contributes to cGMP-versus-cAMP selectivity.
Significance: The cGMP-versus-cAMP selectivity controls the cross-talk between cGMP- and cAMP-dependent signaling pathways.
Keywords: allosteric regulation, cell signaling, cyclic AMP (cAMP), cyclic GMP (cGMP), molecular dynamics, nuclear magnetic resonance (NMR), protein kinase G (PKG), agonism
Abstract
Protein kinase G (PKG) is a major receptor of cGMP and controls signaling pathways often distinct from those regulated by cAMP. Hence, the selective activation of PKG by cGMP versus cAMP is critical. However, the mechanism of cGMP-versus-cAMP selectivity is only limitedly understood. Although the C-terminal cyclic nucleotide-binding domain B of PKG binds cGMP with higher affinity than cAMP, the intracellular concentrations of cAMP are typically higher than those of cGMP, suggesting that the cGMP-versus-cAMP selectivity of PKG is not controlled uniquely through affinities. Here, we show that cAMP is a partial agonist for PKG, and we elucidate the mechanism for cAMP partial agonism through the comparative NMR analysis of the apo, cGMP-, and cAMP-bound forms of the PKG cyclic nucleotide-binding domain B. We show that although cGMP activation is adequately explained by a two-state conformational selection model, the partial agonism of cAMP arises from the sampling of a third, partially autoinhibited state.
Introduction
Protein kinase G (PKG) is a major receptor of the cGMP second messenger. By binding to PKG, cGMP regulates intracellular signaling pathways that control a wide range of intracellular processes, such as cell differentiation, platelet activation, memory formation, and vasodilation (1–5). The PKG signaling pathways are often distinct from those regulated by cAMP-dependent proteins such as protein kinase A (PKA) (6–12). Thus, a key element of the cross-talk between PKG and PKA signaling pathways is the selective activation of PKG by cGMP rather than by cAMP. However, the mechanism of cGMP-versus-cAMP selectivity in PKG is currently only limitedly understood.
One possible determinant of cyclic nucleotide selectivity is the lower binding affinity of PKG for cAMP than for cGMP (13–16). Indeed, the C-terminal cyclic nucleotide-binding domain (CNB-B) of PKG Iβ was previously found to exhibit greater binding affinity for cGMP, which has been attributed to specific interactions between the base-binding region (BBR)5 of CNB-B and the cGMP base moiety, as observed in previously solved crystallographic x-ray structures (Fig. 1e) (17, 18). However, the intracellular concentration of cAMP is typically significantly greater than that of cGMP (19–23), thus potentially allowing cAMP to bind to PKG despite its lower binding affinity and suggesting that other means of cyclic nucleotide selectivity must be present in PKG.
FIGURE 1.
Introduction to PKG Iβ domain and structural architecture, and to cGMP versus cAMP binding and activation. a, schematic overview of the domain organization of PKG Iβ. The regulatory and catalytic regions of PKG are indicated, as are the major underlying structural domains, as follows: the N-terminal dimerization domain (D/D); the autoinhibitory linker region (AI); the cyclic nucleotide-binding domains (CNB-A and CNB-B); the switch helix region (SW); and the two lobes of the catalytic region (N-lobe and C-lobe). Residue numbers for the regulatory region domain boundaries are indicated, and the monomer fragment examined in the current study (referred to herein as PKG Iβ(219–369)) is highlighted in gray. b–d, ribbon structure overlays of the previously solved apo (red ribbon), cAMP-bound (blue ribbon), and cGMP-bound (green ribbon) structures of PKG Iβ(219–369) (17, 18). Bound cAMP and cGMP are shown as blue and green sticks, respectively, and the following key structural elements are indicated: the N-terminal α-helix bundle (N3A), the β-barrel (β-core); the BBR element involved in cGMP binding; and the C-terminal αB-helix (αB) and switch helix region. To clearly demonstrate the differences in structure, the structures are overlaid at their β-barrels. e, zoomed-in view of the cGMP-binding site of the cGMP-bound structure. Key cGMP-interacting residues (17) are indicated as sticks, hydrogen bonds as black dotted lines, and cGMP functional groups that are different/absent in cAMP are highlighted with black circles. f, activation of PKG Iβ by cGMP and cAMP, highlighting the difference in maximal activity levels in the presence of saturating amounts of cAMP versus cGMP. The phosphotransferase activity of PKG Iβ was monitored at various concentrations of cAMP (blue plot) and cGMP (red plot) by a spectrophotometric kinase assay. Here, 1 unit is equal to a substrate conversion rate of 1 μmol per min. Each data point was measured at least in duplicate, and the data sets were fitted using a sigmoidal dose-response algorithm.
Another key difference in the response of PKG Iβ to cAMP versus cGMP is that in contrast to cGMP, cAMP is only a partial agonist of PKG Iβ, as shown by Cook photometric kinase assays of PKG Iβ activity (Fig. 1f). Such partial agonism of cAMP is not fully explained by previously solved structures of PKG Iβ, as they reveal very similar structural rearrangements of both CNB domains in response to either cAMP or cGMP binding (Fig. 1, b–d) (2, 17, 18) that are analogous to structural rearrangements observed for other CNB domains (8, 12, 24–29). Hence, we hypothesize that the partial agonist response of PKG Iβ to cAMP may reflect differences in the dynamics of cAMP versus cGMP-bound PKG Iβ. For example, in the context of the two-state conformational selection mechanism, the partial agonism of cAMP could simply arise from a reduced inactive-to-active shift of the dynamic conformational equilibrium underlying PKG activation (30–33). Indeed, such conformational dynamics have been shown to play a role in the varying responses of other CNB domains to cyclic nucleotides (13, 34). Alternatively, in the presence of cAMP, the regulatory region of PKG may dynamically sample a third conformation, which is distinct from both the apo-inactive and cGMP-bound active structures (30) and is at least partially auto-inhibitory of the kinase function of PKG.
As a first step toward testing our hypotheses and differentiating between the two-state versus three-state hypothetical models for the partial agonism of cAMP, here we report the comparative NMR analyses of the apo, cAMP-, and cGMP-bound forms of the PKG Iβ CNB-B domain (i.e. “PKG Iβ(219–369)”; see Fig. 1a). This domain is adjacent to the catalytic domain (Fig. 1a), and it has been previously identified as a critical control unit for auto-inhibition and cGMP selectivity in PKG Iβ (17, 18). In addition, we examined the CNB-B domain because although the CNB-A domain contributes to the cyclic nucleotide modulation of PKG I function (17, 35–37), the CNB-A domain undergoes only minor structural changes upon cyclic nucleotide binding (2, 17). We found that the partial agonism of cAMP cannot be explained by a simple reversal of the two-state inactive/active conformational equilibrium that rationalizes cGMP activation (2, 17). Rather, our data show that the partial agonism exhibited by cAMP is the result of cAMP-bound CNB-B sampling a distinct third conformational state. In this third state, the lid region that caps the cyclic nucleotide base in the fully active state is disengaged, and kinase inhibition is only partially effective. By populating this third conformation, and to a lesser degree the inactive conformation, cAMP reduces the overall level of PKG activity compared to cGMP, thereby providing an explanation for the observed partial agonism of cAMP.
Experimental Procedures
Preparation of the PKG Iβ(219–369) Construct
The PKG Iβ(219–369) construct was expressed with an N-terminal poly-l-histidine tag in Escherichia coli strain BL-21(DE3). The E. coli cells were grown at 37 °C in isotopically enriched minimal media supplemented with trace metals, d-biotin, and thiamine-HCl. Expression was induced with 0.4 mm isopropyl β-d-1-thiogalactopyranoside at an optical density of 1.0 (at λ = 600 nm), and the cells were further incubated for 16 h at 20 °C before being harvested by centrifugation.
The harvested cells were lysed using a cell disruptor, and cell debris was subsequently removed by centrifugation (20,000 × g for 1 h). For this process, the cells were resuspended in 50 mm Tris buffer (pH 7.6) with 150 mm NaCl, 1 mm β-mercaptoethanol, and 0.2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride. An initial purification of the PKG construct was then performed using a Ni2+-Sepharose resin in a 5-ml gravity column. After passing the cell lysate through the column, the column was rinsed with more cell resuspension buffer, followed by 50 mm Tris buffer (pH 7.6) with 500 mm NaCl, 20 mm imidazole, and 1 mm β-mercaptoethanol. The protein was then eluted from the column using 50 mm Tris buffer (pH 7.6) with 50 mm NaCl, 300 mm imidazole, and 1 mm β-mercaptoethanol.
The collected gravity column eluant was dialyzed in 50 mm Tris buffer (pH 7.6) with 100 mm NaCl and 1 mm β-mercaptoethanol, and cleaved with tobacco etch virus protease for 48 h to remove the N-terminal poly-l-histidine tag from the PKG Iβ(219–369) construct. The cleaved poly-l-histidine tag and tobacco etch virus protease were removed from the dialyzed eluant by passing the eluant through a Ni2+-Sepharose gravity column, followed by a rinse with dialysis buffer. All column flow-through and rinse were collected, and the PKG Iβ(219–369) construct was further purified by size-exclusion FPLC (HiLoad Superdex 120-ml column). The FPLC was performed using a 50 mm Tris running buffer (pH 7.0) with 100 mm NaCl, 1 mm DTT, and 0.02% (w/v) NaN3, which also served as the NMR sample buffer for most of the NMR experiments (except for H/D hydrogen exchange, which was performed in a pH 7.0 2H2O-based phosphate buffer).
Spectrophotometric Kinase Activity Assay
Kinase activity was measured using the coupled spectrophotometric assay as described by Cook et al. (38). The phosphotransferase activity of full-length PKG Iβ purified from Sf9 cells was determined in 100-μl reaction volumes containing 35 nm PKG Iβ, 1 mm Kemptide (LRRASLG), varying concentrations of cGMP or cAMP, respectively, and the assay mix. The depletion of NADH directly correlating with the conversion of ATP was monitored using a spectrophotometer (SPECORD® 205; Analytik Jena). The buffer used for the spectrophotometric assay consisted of 100 mm MOPS (pH 7), 10 mm MgCl2, 1 mm phosphoenolpyruvate, 1 mm ATP, 150 units of lactate dehydrogenase, 84 units of pyruvate kinase, 220 μm NADH, and 5 mm β-mercaptoethanol. Phosphotransferase activity was plotted against the logarithmic cyclic nucleotide concentration, and data points were fitted using a sigmoidal dose-response fit in GraphPad Prism 6.01.
NMR Data Acquisition and Analysis
All NMR spectra for the PKG Iβ(219–369) construct were acquired at 306 K with a Bruker Avance 700-MHz NMR spectrometer equipped with a 5-mm TCI cryoprobe. For the chemical shift-based analyses of PKG Iβ(219–369), two-dimensional {1H,15N} HSQC NMR spectra were acquired for apo, cAMP-, and cGMP-bound samples of PKG Iβ(219–369). A PKG Iβ(219–369) concentration of 20 μm was used for all HSQC samples, and 80 μm 15N-labeled N-acetylglycine was added to the samples as an internal reference for subsequent alignment of the HSQC spectra with one another. The experiments for cAMP- and cGMP-bound PKG Iβ(219–369) were performed with a cyclic nucleotide concentration of 2 mm to achieve saturated ligand binding, thus minimizing the influence of differing ligand binding affinities on the results of the comparative analyses. The spectra were processed with NMRPipe (39) and analyzed using Sparky (40). Peak assignments for the HSQC spectra were obtained from standard three-dimensional triple-resonance NMR spectra (i.e. HNCACB, CBCA(CO)NH, HNCA, and HN(CO)CA), using the automated PINE-NMR server (41) to facilitate the initial assignment process. The assigned HSQC spectra were aligned using Sparky, and the resulting (1H,15N) chemical shift data for apo, cAMP-, and cGMP-bound PKG Iβ(219–369) were utilized to compute (1H,15N) combined chemical shifts, perform chemical shift projection analyses (CHESPA), and build chemical shift correlation plots, as described previously (13, 42–45). The CHESPA “outlier” residues are defined as those deviating significantly from the two-state equilibrium between conformations resembling the apo and cGMP-bound PKG Iβ(219–369) structures (i.e. residues with projection angle cosine values less than 0.8). The chemical shift correlation plots were confined to residues with CHESPA fractional activation values less than 1.2 and projection angle cosine values greater than 0.95.
To further examine PKG Iβ(219–369) dynamics, backbone 15N relaxation (R1, R2, HN-NOE) and hydrogen exchange (H/D and H/H) NMR data for the apo, cAMP-, and cGMP-bound samples of PKG Iβ(219–369) were acquired and analyzed using a protocol similar to that described previously (13, 46, 47), with the T1 and T2 data sets acquired as pseudo-three-dimensional matrices (48). Reduced spectral densities (RSDs) were then computed from the 15N relaxation rates as described previously (13, 46, 47). Considering the rigidity typically exhibited by the inner β-strands (i.e. β3, β4, β7, and β8) of the β-barrel of CNB domains (27, 46), R1R2 and/or J(0) values significantly greater than the inner-β-strand average were considered indicative of the presence of millisecond-microsecond backbone dynamics, whereas R1R2, J(0), and/or HN-NOE values significantly less than the inner-β-strand average, and/or J(ωH + ωN) values significantly greater than the inner-β-strand average, are indicative of the presence of picosecond-nanosecond backbone dynamics. Differences in the R1R2 and J(0) values between different samples of PKG Iβ(219–369) were deemed significant if the error bars of the compared data points did not overlap one another. Meanwhile, because of the larger errors affecting the HN-NOE and J(ωH + ωN) values, differences in these values were deemed significant if the error bar of one of the compared data points did not overlap the other data point, and vice versa. Finally, significant differences in hydrogen exchange rates were indicated by non-overlapping protection factor (PF) value error bars or transitions between different time scales of hydrogen exchange. The latter are defined, from fast to slow exchange, as follows: H/H-time-scale exchange, H/D-dead-time exchange, quantifiable exchange (i.e. measurable PF), and finally, exchange that is too slow for reliable PF quantification.
Determination of State Populations in the Three-state Model
The state populations were estimated based on the fractional activations measured through CHESPA. Specifically, the state populations were determined based on the average fractional activations observed for the “pre-lid” region, which is reflective of the population of the state in which this region adopts an active conformation (i.e. the active state), and for the CNB-B domain region preceding residue 340, which is reflective of the total population of states in which this region is active-like (i.e. the intermediate and active states). Thus, the population of the intermediate state is reflected by the difference between the two average fractional activations, whereas the population of the inactive state is the percentage not accounted for by the other two states.
Results
Initial Assessment of cAMP-Versus-cGMP Differences in PKG Iβ(219–369) through Compounded Chemical Shift Changes
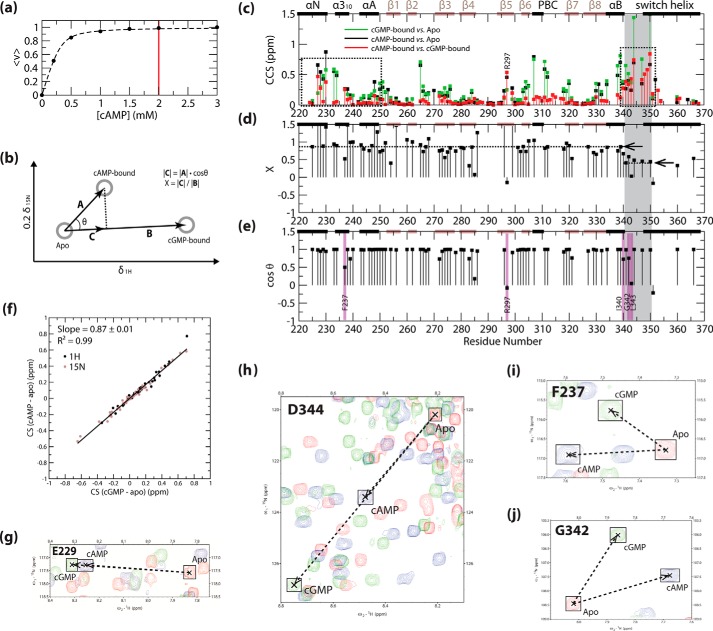
As an initial assessment of the differences between cAMP- and cGMP-bound PKG Iβ(219–369), we examined the pairwise compounded chemical shift changes between apo, cAMP-, and cGMP-bound PKG Iβ(219–369) (Fig. 2c). Chemical shifts from the apo state revealed that binding of both cAMP and cGMP results in major widespread structural changes within the CNB-B domain, including part of the switch helix region (Fig. 2c, black and green points), in agreement with the available x-ray structures (17, 18). In addition, as expected, the cAMP-versus-cGMP chemical shifts detected differences for residues at/near the base-binding pocket, including residues in the β5-strand of the BBR, and the cGMP-capping residue from the switch helix region (i.e. residue Tyr-351; Fig. 2c, red points). However, Fig. 2c also shows that the cAMP- and cGMP-bound states exhibit differences that are not apparent from previous structural comparisons. For example, unanticipated cAMP-versus-cGMP chemical shift differences were observed for residues outside the binding site, including the residues spanning the 340–350 region, and the N-terminal N3A motif spanning residues 220–250 (Fig. 2c, red points marked with dotted boxes). To better understand the origin of these long range differences, we analyzed the cAMP-bound chemical shifts using CHESPA (Fig. 2, b, d, and e).
FIGURE 2.
Effects of cAMP binding on PKG Iβ(219–369), as observed from CHESPA analysis. a, ligand binding isotherm for cAMP-bound PKG Iβ(219–369), as computed from the combined chemical shifts obtained by {1H,15N} HSQC. The fitted curve is shown as a black dashed line, and the cAMP concentration selected to achieve saturated binding is indicated by a vertical red line. This concentration was selected based on the lack of further shift in HSQC peak positions during ligand titration, as determined by HSQC overlays. b, schematic illustration of the CHESPA calculation methodology, using {1H,15N} HSQC chemical shifts as input (44). The 1H and 15N chemical shift values for the reference (i.e. apo and cGMP-bound) and perturbation (i.e. cAMP-bound) states are plotted as gray circles, and key vectors and parameters used in the analysis are indicated. Positive values of X and cosθ indicate a shift in favor of activation in the perturbation state, and negative values indicate a shift in favor of inactivation. c–e, results of the CHESPA analysis of cAMP-bound PKG Iβ(219–369), with apo and cGMP-bound PKG Iβ(219–369) as reference states, as shown in b. The secondary structural elements from the apo-state x-ray structure are indicated across the top of each graph as follows: black bars = α-helices; brown bars = β-strands. c, compounded chemical shifts of cAMP-bound (black points) and cGMP-bound (green points) PKG Iβ(219–369) from apo PKG Iβ(219–369), and of cAMP-bound from cGMP-bound PKG Iβ(219–369) (red points). Black dotted boxes indicate notable cAMP-versus-cGMP chemical shift differences observed for residues outside the cGMP-binding site. d and e, fractional shifts toward activation (“X”; d) and projection angle cosines (“cosθ”; e) achieved by cAMP binding, relative to apo and cGMP-bound PKG Iβ(219–369). The average fractional shift toward activation computed for the residue 340–350 region (average of X values from d) and the overall fractional shift toward activation computed for residues N-terminal to residue 340 (computed slope in f) are marked by arrows and black dotted lines. The residue 340–350 region is indicated in all plots by gray highlighting, and notable low-cosθ-value residues (outliers, as defined under “Experimental Procedures”) are indicated by purple highlights. f, chemical shift correlation plot of 1H (black points) and scaled 15N (brown points) chemical shifts of cAMP-bound versus cGMP-bound PKG Iβ(219–369) (both relative to the apo state). The black line delineates the line of best fit as determined from linear regression, and the corresponding slope and correlation coefficient are indicated. g–j, expansions of the overlaid {1H,15N} HSQC spectra of apo (red contours), cAMP-bound (blue contours), and cGMP-bound (green contours) PKG Iβ(219–369), illustrating the cAMP-associated perturbations of representative high-cosθ-value residues from the residue 340–350 region (h) and N-terminal to residue 340 (g), as well as representative low-cosθ-value (outlier) residues from the N3A (i) and residue 340–350 region (j).
CHESPA of cAMP-bound PKG Iβ(219–369) Reveals Three Major Types of Long Range Perturbations Caused by the Replacement of cAMP with cGMP
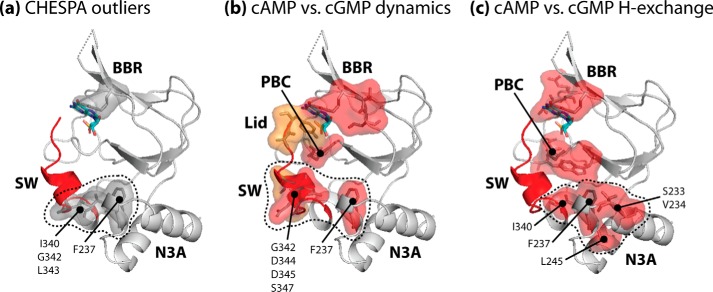
The CHESPA analysis was performed for cAMP-bound PKG Iβ(219–369), utilizing apo and cGMP-bound PKG Iβ(219–369) as reference states (Fig. 2b). The resulting computed fractional activations (Fig. 2d) and projection angle cosine values (Fig. 2e) were complemented by a cAMP-apo versus cGMP-apo chemical shift correlation plot analysis, whose slope provides an alternative estimation of the relative cAMP-versus-cGMP fractional activation (Fig. 2f). Examination of the CHESPA (Fig. 2, d and e) and chemical shift correlation results (Fig. 2f) revealed three groups of non-binding site residues demonstrating notable cAMP-versus-cGMP differences. The first group includes residues 340–350 (referred to herein as the “pre-lid” region), which link the αB-helix to the C-terminal capping residue (i.e. Tyr-351; a residue referred to herein as the “lid”). The residues in this first group exhibited an average fractional activation of only ∼40% (Fig. 2d), as was also evident from partial HSQC peak shifts for these residues (Fig. 2h), suggesting that in the presence of cAMP the pre-lid region is subject to a partial disengagement from its active-state structural arrangement. The second group of residues is composed of the residues N-terminal to residue 340, which exhibited an average fractional activation of ∼87% in the presence of cAMP (Fig. 2, d, f, and g), reflecting a markedly different degree of activation compared with the pre-lid region. The third notable group spans multiple “outlier” residues, which deviate from a two-state conformational equilibrium in the presence of cAMP, as seen through low projection angle cosine values (Fig. 2e, purple highlights) and non-linear arrangements of apo, cAMP-, and cGMP-bound state HSQC cross-peaks (Fig. 2, i and j). The identified outlier residues include cGMP-binding residue Arg-297 (from the β5-strand), as well as multiple non-binding-site residues in the pre-lid region N terminus and the N3A motif (Fig. 2e, purple highlights). When mapped on the structure of cGMP-bound PKG Iβ(219–369), the non-binding-site outlier residues form a cluster spanning the interface between the pre-lid region and the N3A motif (Fig. 3a, gray surface), indicating that the partial disengagement of the pre-lid region caused by the replacement of cGMP with cAMP may in turn influence the N3A motif as well. In addition, the low degree of fractional activation observed for the pre-lid region (Fig. 2d) points to a loss of structure in this segment, suggesting that dynamics are involved in the observed perturbations caused by cAMP. To test this hypothesis, we examined PKG Iβ(219–369) dynamics more closely through 15N relaxation NMR measurements.
FIGURE 3.
Three-dimensional maps of cAMP-versus-cGMP differential dynamics. The maps shown illustrate residues exhibiting low cosθ values from CHESPA (“outlier” residues) (a), differences in cAMP versus cGMP-bound dynamics (b), and differences in cAMP versus cGMP-bound hydrogen exchange rates (c), as identified in Figs. 2 and 4–6. All residue sets are illustrated in the context of the cGMP-bound structure of PKG Iβ(219–369), with bound cGMP shown as colored sticks, and the following structural elements are indicated: the N3A region; the BBR and PBC elements involved in cGMP binding; and the C-terminal switch helix region (SW) and cGMP-capping lid (both highlighted as red ribbon). The identified residues are shown as sticks and surfaces, and the surfaces in b and c follow the same color codes as the residue highlights in Figs. 4–6. The cluster of non-binding site residues identified in each case is delineated by a black dotted line, and the identities of the constituent residues are indicated.
Assessment of Apo-Versus-cGMP Differential Dynamics through Backbone 15N Relaxation Rates and Reduced Spectral Densities
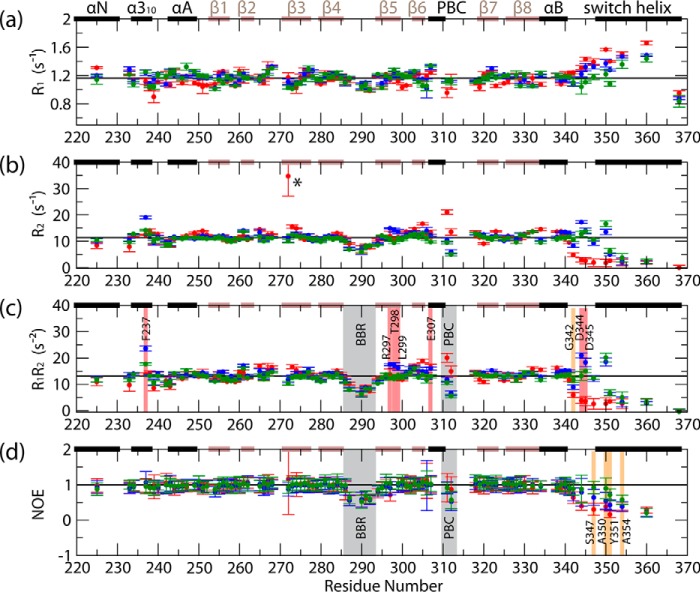
As a first assessment of PKG Iβ(219–369) dynamics, we measured backbone 15N relaxation rates and reduced spectral densities (RSDs) for the apo, cAMP-, and cGMP-bound states (Figs. 4 and 5). The apo versus cGMP-bound state comparison revealed several unanticipated changes in dynamics. First, the β4-β5 loop in the base binding region (BBR) exhibited consistent picosecond-nanosecond dynamics in both the apo and cGMP-bound states (Figs. 4, c and d, and 5a and c, gray highlights). In addition, the phosphate-binding cassette (PBC) exhibited a shift from millisecond-microsecond to picosecond-nanosecond dynamics upon cGMP binding (Figs. 4, c and d, and 5a and c, gray highlights), suggesting a retention of residual PBC dynamics. This is in marked contrast to the more quantitative quenching of PBC dynamics observed previously for the bound states of other CNB domains, which unlike PKG include a proline residue in the PBC (8, 12, 27, 46, 49, 50). In addition, our data indicated that the region C-terminal to the αB-helix (i.e. C-terminal to residue 340) is subject to picosecond-nanosecond dynamics in the apo state (Figs. 4, c and d, and 5, a and c) (17). The binding of cGMP produces a dramatic quench of picosecond-nanosecond dynamics in the pre-lid region residues (i.e. residues 340–350) that bridge the αB-helix to the lid residue Tyr-351 (Figs. 4, c and d, and 5, a and c). However, despite this dramatic quenching of dynamics, Tyr-351 retains partial picosecond-nanosecond flexibility (Figs. 4, c and d, and 5, a and c).
FIGURE 4.
Backbone 15N relaxation rates for apo (red points), cAMP-bound (blue points), and cGMP-bound (green points) PKG Iβ(219–369). a, R1 relaxation rates; b, R2 relaxation rates; c, product of R1 and R2; d, (1H,15N) NOE values, computed as Isat/Inonsat. Black horizontal lines denote the average values computed for the inner β-strands of the β-barrel, which, due to the rigidity of this region of the protein, are assumed to represent the overall tumbling motion of the protein in solution. Residues exhibiting notable enhancements of millisecond-microsecond/picosecond-nanosecond dynamics in cAMP versus cGMP-bound PKG Iβ(219–369) are indicated by red/orange highlights, respectively, while other regions exhibiting notable dynamics are indicated by gray highlights, and an apo-state residue for which the R2 rate could not be properly quantified (due to a poor signal-to-noise ratio) is marked with an asterisk. The secondary structure elements from the apo-state x-ray structure are indicated across the top of each graph (black bars = α-helices, brown bars = β-strands), and residues for which no data are shown are prolines or were not successfully assigned in the relaxation NMR spectra for one or more states.
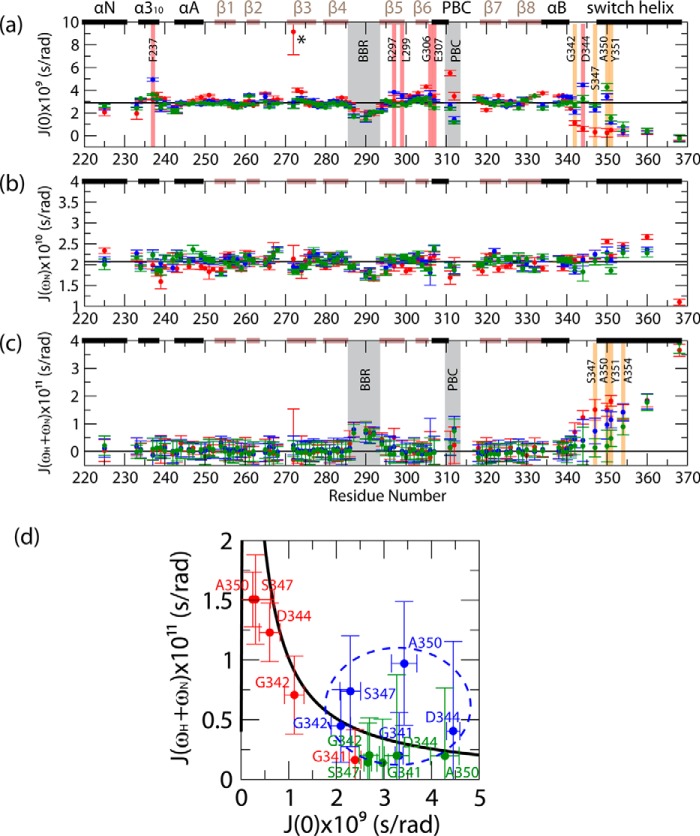
FIGURE 5.
RSDs for apo (red points), cAMP-bound (blue points), and cGMP-bound (green points) PKG Iβ(219–369). a, J(0) values; b, J(ωN) values; c, J(ωH + ωN) values. The RSDs were computed as described previously (13, 46, 47) based on the relaxation rates reported in Fig. 4. Black horizontal lines denote the average values computed for the inner β-strands of the β-barrel, which, due to the rigidity of this region of the protein, are assumed to represent the overall tumbling motion of the protein in solution. All color codes illustrated are as in Fig. 4. d, two-dimensional J(ωH + ωN) versus J(0) plots for residues 341–350. The values expected for a rigid rotor in the absence of millisecond-microsecond dynamics are indicated by the black plot, and the cAMP-bound state data are indicated by a blue dashed oval.
Assessment of cAMP-Versus-cGMP Differential Dynamics through Backbone 15N Relaxation Rates and Reduced Spectral Densities
The region C-terminal to the αB-helix is also a site of previously unanticipated cAMP-versus-cGMP differences in dynamics. In particular, partially enhanced picosecond-nanosecond dynamics were observed in the cAMP-bound state throughout the region C-terminal to residue 340, including the lid residue Tyr-351 (Figs. 4, c and d, and 5, a and c, orange highlights). Furthermore, several other residues exhibited enhanced millisecond-microsecond dynamics in the cAMP-bound state (Figs. 4c and 5a, red highlights). The latter enhancements include binding site residues from the β5-strand and the PBC N terminus, as well as non-binding-site residues such as those in the N3A motif and the pre-lid region N terminus (Figs. 4c and 5a, red highlights). These cAMP-versus-cGMP differences cannot be explained by variations in diffusional anisotropy, given the similarity of the cAMP- and cGMP-bound structures of the CNB-B domain (Fig. 1, b–d). Therefore, the pre-lid region emerges as a site affected by cAMP-versus-cGMP enhancements in both picosecond-nanosecond and millisecond-microsecond dynamics. This conclusion is further supported by the correlated changes in high- and low-frequency spectral densities for this region (i.e. J(ωH + ωN) versus J(0); Fig. 5d).
Fig. 5d shows that the cAMP-versus-cGMP changes deviate from the simple rigid rotor model, suggesting the simultaneous presence of both picosecond-nanosecond and millisecond-microsecond flexibility in the pre-lid region of the cAMP-bound state. These observations corroborate the partial disengagement of the lid, as suggested by the CHESPA analysis (Fig. 2d). Furthermore, the map of non-binding site loci subject to enhanced millisecond-microsecond dynamics in the presence of cAMP coincides quite well with that of the outlier residues identified from CHESPA (Fig. 3b, red surfaces versus Fig. 3a, gray surfaces). Overall, the data of Figs. 4 and 5 confirm that cGMP replacement with cAMP leads to increased structural dynamics not only at binding site residues but also at selected allosteric sites of PKG Iβ(219–369) (e.g. the pre-lid and N3A loci; Fig. 3b). Unlike the cAMP-versus-cGMP changes, the apo-versus-cGMP variations in the correlated J(ωH + ωN) versus J(0) spectral densities follow, within error, the values expected for a rigid rotor in the absence of millisecond-microsecond dynamics (Fig. 5d, black line), further confirming that cGMP binding quenches picosecond-nanosecond dynamics in the pre-lid region.
Further Assessment of Dynamics through Hydrogen Exchange Rates
Next, we measured the backbone hydrogen exchange rates for apo, cAMP-, and cGMP-bound PKG Iβ(219–369) (Fig. 6). The comparison of the apo and cGMP-bound states revealed unanticipated changes in dynamics. For example, multiple residues of both the αB-helix and the N3A motif showed significantly reduced hydrogen exchange rates, pointing to a reduced access to partially unfolded and solvent-exposed excited states upon cGMP binding (Fig. 6). However, when cGMP is replaced by cAMP, a partial recovery of solvent exposure is observed, as several residues exhibited greater hydrogen exchange rates in the cAMP-bound state than in the cGMP-bound state (Fig. 6). The affected areas include residues in the vicinity of the binding site (e.g. the β5- and β6-strands, the β2–3 loop adjacent to the PBC, and the PBC N and C termini), as well as non-binding-site residues, such as the N terminus of the pre-lid region and the N3A motif (Fig. 6, red highlights). Overall, the identified non-binding site residues affected by cAMP overlap quite well with the outlier residues identified from CHESPA (Fig. 3c, red surfaces with dotted outline versus Fig. 3a, gray surfaces with dotted outline), suggesting that in agreement with the 15N relaxation and RSD results, the cAMP-versus-cGMP perturbations observed for PKG Iβ(219–369) involve an increase in dynamics at the interface between the α-helical regions flanking the β-barrel (Fig. 3). Furthermore, the pre-lid and lid regions exhibited high solvent exposure in all three states, as suggested by fast hydrogen exchange (Fig. 6), thus confirming the finding from 15N relaxation and RSDs that the lid retains significant dynamics even in the cGMP-bound state.
FIGURE 6.
Backbone hydrogen exchange results for apo (red points), cAMP-bound (blue points), and cGMP-bound (green points) PKG Iβ(219–369). Residues are grouped into four types as follows: residues that undergo hydrogen exchange on the 100–102-ms time scale (as identified by H/H NMR) are denoted by H/H; other residues that undergo hydrogen exchange within the dead time of the H/D NMR experiment, and thus exhibit hydrogen exchange on a seconds-to-minutes time scale, are denoted by D.T.; residues with hydrogen exchange rates quantifiable by H/D NMR are plotted as the logarithms of PFs computed from the H/D NMR data, whereby higher values indicate slower hydrogen exchange; and residues whose hydrogen exchange rates were too slow to be properly quantified are denoted by S.E. and triangles. Residues exhibiting notable enhancements of hydrogen exchange rates in cAMP versus cGMP-bound PKG Iβ(219–369) are indicated by red highlights, and the secondary structure elements from the apo-state x-ray structure are indicated across the top of each graph: black bars = α-helices; brown bars = β-strands. Residues for which no data are shown are prolines or were not successfully assigned in the NMR spectra.
Discussion
The comparative NMR analysis of the CNB-B construct (i.e. PKG Iβ(219–369)) in its apo, cGMP-, and cAMP-bound states revealed that the cAMP-versus-cGMP differences are more extensive than previously anticipated, reaching well beyond the cGMP contact sites to crucial allosteric structural elements. In particular, the chemical shift, 15N relaxation, and hydrogen-exchange data (Figs. 2 and 4–6) consistently show that cAMP binding results in only a partial recruitment of the pre-lid region to its active-state conformation, which in turn leads to perturbations of the adjacent N3A region (Fig. 3). Furthermore, when bound to the PKG Iβ CNB-B domain, cAMP exhibits a higher propensity for an anti base orientation compared to cGMP, which mainly adopts a syn conformation (17, 18). A possible explanation for the differing syn/anti conformational propensities of bound cAMP versus cGMP is the intrinsic preference of free cAMP for the anti orientation together with a lack of stable interactions between the 6-NH2 moiety of cAMP, which is replaced by a less bulky hydrogen-bond-accepting 6-oxo moiety in cGMP, and the residues of the cyclic nucleotide-binding site responsible for cGMP selectivity, including residue Arg-297 (in β5), which interacts with the 6-oxo moiety of cGMP (Fig. 1e). The anti versus syn difference may reduce the efficacy with which the lid residue (i.e. Tyr-351) is recruited to its active-state capping interaction with the bound cyclic nucleotide, thus providing an explanation for the partial pre-lid region disengagement observed in the cAMP-bound state. Indeed, the analogous capping residue of the cAMP-bound PKA CNB-B domain (i.e. Tyr-371) adopts a structural arrangement similar to that of Tyr-351 in the cGMP-bound PKG CNB-B domain, and the bound cyclic nucleotides in both of these domains adopt a syn conformation (17), suggesting that replacement of cGMP by cAMP without a syn-to-anti transition is not sufficient to achieve the observed lid disengagement.
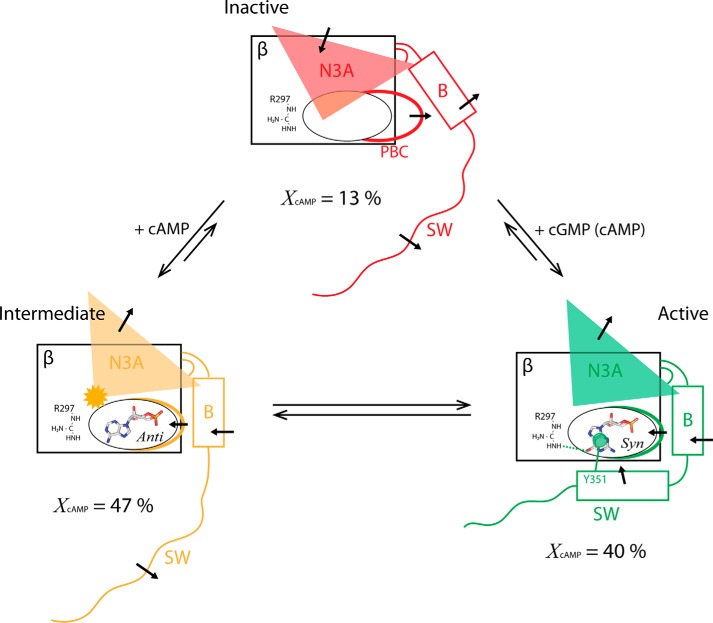
The inability of cAMP to recruit the pre-lid region into the active state to the same extent as observed for the majority of the CNB-B domain residues preceding residue 340 (Fig. 2d) suggests that the partial agonism of cAMP cannot be explained by a simple two-state active-inactive model of conformational selection. A purely two-state model dictates that all of the CNB-B domain residues should exhibit a similar fractional activation, which is not the case here (Fig. 2d). Hence, the dual pattern of fractional activations observed in Fig. 2d (horizontal arrows) implies the presence of a third state in which kinase inhibition is only partially effective. The deviation from a two-state model is also independently confirmed by the observation for multiple non-binding-site residues of clear deviations from a linear apo-cAMP-cGMP CHESPA pattern (Fig. 2, e, i, and j). These results consistently suggest that the partial agonist response of PKG Iβ to cAMP (Fig. 1f) cannot be explained by a simple reversal of the two-state inactive/active conformational equilibrium (Fig. 7, red and green structures). Instead, our results point to a dynamic exchange of the cAMP-bound CNB-B domain with a third, “intermediate,” conformational state (Fig. 7, orange structure), leading to a three-state model (Fig. 7).
FIGURE 7.
Schematic summary of the proposed CNB-B model for cAMP partial agonism, illustrating the “inactive” (red) and “active” (green) conformational states explored in the cGMP-modulated two-state activation equilibrium, and the additional intermediate state (orange) stabilized in the presence of cAMP. The structural shifts of the α-helical subdomain elements in each conformational state are indicated by black arrows, and the N3A perturbation unique to the intermediate state is indicated by an orange starburst. The bound ligands and their conformations are shown as labeled stick structures, and the key active-state cGMP interactions with residues Arg-297 (cGMP-selective hydrogen bond) and Tyr-351 (cGMP-capping lid interaction) are schematically indicated. The percentages given below each state identify the populations of each state in the presence of cAMP, as calculated from the average fractional shift toward activation computed for the residue 340–350 region (average of X values from Fig. 2d), and the overall fractional shift toward activation computed for residues N-terminal to residue 340 (computed slope in Fig. 2f).
In the proposed three-state model (Fig. 7), the population of the intermediate state is ∼47%, whereas that of the apo-like inactive state is ∼13%, as assessed based on the fractional activations measured through CHESPA (Fig. 2d). The intermediate state sampled in the presence of cAMP maintains an active-like conformation with the exception of the pre-lid region (i.e. residues 340–350). Upon cAMP binding, the pre-lid region becomes disengaged and more dynamic in the picosecond-nanosecond and millisecond-microsecond time scales (Fig. 7, orange structure). Notably, the disengagement of the pre-lid region would alter the relative positions of the PKG regulatory and catalytic regions, which are connected to one another by the CNB-B domain/switch helix region (Fig. 1a) (2, 4, 17, 18). Such repositioning is likely to affect the access of substrates to the PKG catalytic site (6), explaining why the intermediate state retains at least partial kinase inhibition. Hence, by promoting population of the intermediate conformation, and to a lesser degree the inactive conformation (Fig. 7), cAMP reduces the overall level of PKG activity compared to cGMP, explaining the partial agonism observed for cAMP (Fig. 1f). Such partial agonism represents a key determinant of the cGMP-versus-cAMP selectivity of PKG, and together with differing binding affinities for cAMP versus cGMP (17, 18), it contributes to the minimization of cross-talk between cGMP- and cAMP-controlled signaling pathways.
Finally, it is worth noting that while the partial agonism of cAMP contributes to cyclic nucleotide selectivity for cGMP-controlled signaling pathways, previous studies have suggested that selected signaling responses are influenced by cAMP via modulation of PKG activity, rather than cAMP-dependent proteins such as PKA (5, 51). A possible explanation for this phenomenon is that as the intracellular cAMP concentration rises, the minimal cGMP concentration necessary for full PKG activation increases, and the shape of the cGMP-dependent activation profile of PKG may also change. Such modulation of PKG activity by cAMP represents a notable contribution to the control of PKG, and may thus have relevant physiological implications for cGMP-dependent intracellular signaling.
Author Contributions
B. V. S., R. S., and G. M. conceived and designed the study. B. V. S., R. S., and R. G. performed and analyzed the NMR experiments. R. L. and F. W. H. performed and analyzed the kinase assay. C. K. designed and constructed vectors for expression. B. V. S. and G. M. wrote the paper. All authors reviewed the results and the manuscript.
Acknowledgments
We thank Professors Susan S. Taylor (University of California at San Diego) and W. Dostmann (University of Vermont) as well as M. Akimoto, S. Boulton, and K. Moleschi (McMaster University) for helpful discussions.
This work was supported by a grant from the Canadian Institutes of Health Research (to G. M.) and by the Natural Sciences and Engineering Research Council of Canada. The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.
- BBR
- base-binding region
- CHESPA
- chemical shift projection analysis
- CNB
- cyclic nucleotide-binding domain
- HSQC
- heteronuclear single-quantum coherence
- N3A
- N-terminal α-helix bundle
- PBC
- phosphate-binding cassette
- PF
- hydrogen exchange protection factor
- RSD
- reduced spectral density
- H/D
- hydrogen/deuterium.
References
- 1.Francis S. H., Busch J. L., Corbin J. D., and Sibley D. (2010) cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol. Rev. 62, 525–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J. J., Casteel D. E., Huang G., Kwon T. H., Ren R. K., Zwart P., Headd J. J., Brown N. G., Chow D. C., Palzkill T., and Kim C. (2011) Co-crystal structures of PKG Iβ(92–227) with cGMP and cAMP reveal the molecular details of cyclic-nucleotide binding. PLoS ONE 6, e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann F., Bernhard D., Lukowski R., and Weinmeister P. (2009) in Handbook of Experimental Pharmacology (Rosenthal W., ed) pp. 137–162, Springer-Verlag, Berlin: [DOI] [PubMed] [Google Scholar]
- 4.Osborne B. W., Wu J., McFarland C. J., Nickl C. K., Sankaran B., Casteel D. E., Woods V. L. Jr., Kornev A. P., Taylor S. S., and Dostmann W. R. (2011) Crystal structure of cGMP-dependent protein kinase reveals novel site of interchain communication. Structure 19, 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis S. H., and Corbin J. D. (1999) Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit. Rev. Clin. Lab. Sci. 36, 275–328 [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal D. K., Copps J., Smith-Nguyen E. V., Zhang P., Heller W. T., and Taylor S. S. (2014) The roles of the RIIβ linker and N-terminal cyclic nucleotide-binding domain in determining the unique structures of the type IIβ protein kinase A: a small angle x-ray and neutron scattering study. J. Biol. Chem. 289, 28505–28512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng C. Y., Yang J., Taylor S. S., and Blumenthal D. K. (2009) Sensing domain dynamics in protein kinase A-Iα complexes by solution x-ray scattering. J. Biol. Chem. 284, 35916–35925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim C., Cheng C. Y., Saldanha S. A., and Taylor S. S. (2007) PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 9.Kim J., Masterson L. R., Cembran A., Verardi R., Shi L., Gao J., Taylor S. S., and Veglia G. (2015) Dysfunctional conformational dynamics of protein kinase A induced by a lethal mutant of phospholamban hinder phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 112, 3716–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masterson L. R., Yu T., Shi L., Wang Y., Gustavsson M., Mueller M. M., and Veglia G. (2011) cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J. Mol. Biol. 412, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava A. K., McDonald L. R., Cembran A., Kim J., Masterson L. R., McClendon C. L., Taylor S. S., and Veglia G. (2014) Synchronous opening and closing motions are essential for cAMP-dependent protein kinase A signaling. Structure 22, 1735–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y., Dostmann W. R., Herberg F. W., Durick K., Xuong N. H., Ten Eyck L., Taylor S. S., and Varughese K. I. (1995) Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science 269, 807–813 [DOI] [PubMed] [Google Scholar]
- 13.Das R., Chowdhury S., Mazhab-Jafari M. T., Sildas S., Selvaratnam R., and Melacini G. (2009) Dynamically driven ligand selectivity in cyclic nucleotide binding domains. J. Biol. Chem. 284, 23682–23696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De S., Greenwood A. I., Rogals M. J., Kovrigin E. L., Lu K. P., and Nicholson L. K. (2012) Complete thermodynamic and kinetic characterization of the isomer-specific interaction between Pin1-WW domain and the amyloid precursor protein cytoplasmic tail phosphorylated at Thr668. Biochemistry 51, 8583–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenwood A. I., Kwon J., and Nicholson L. K. (2014) Isomerase-catalyzed binding of interleukin-1 receptor-associated kinase 1 to the EVH1 domain of vasodilator-stimulated phosphoprotein. Biochemistry 53, 3593–3607 [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman B., and Nicholson L. K. (2007) Thermodynamic dissection of the ezrin FERM/CERMAD interface. Biochemistry 46, 12174–12189 [DOI] [PubMed] [Google Scholar]
- 17.Huang G. Y., Kim J. J., Reger A. S., Lorenz R., Moon E. W., Zhao C., Casteel D. E., Bertinetti D., Vanschouwen B., Selvaratnam R., Pflugrath J. W., Sankaran B., Melacini G., Herberg F. W., and Kim C. (2014) Structural basis for cyclic-nucleotide selectivity and cGMP-selective activation of PKG I. Structure 22, 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang G. Y., Gerlits O. O., Blakeley M. P., Sankaran B., Kovalevsky A. Y., and Kim C. (2014) Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: insights for cGMP-dependent protein kinase agonist design. Biochemistry 53, 6725–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beavo J. A., and Brunton L. L. (2002) Cyclic nucleotide research–still expanding after half a century. Nat. Rev. Mol. Cell Biol. 3, 710–718 [DOI] [PubMed] [Google Scholar]
- 20.Börner S., Schwede F., Schlipp A., Berisha F., Calebiro D., Lohse M. J., and Nikolaev V. O. (2011) FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells. Nat. Protoc. 6, 427–438 [DOI] [PubMed] [Google Scholar]
- 21.Wolter S., Golombek M., and Seifert R. (2011) Differential activation of cAMP- and cGMP-dependent protein kinases by cyclic purine and pyrimidine nucleotides. Biochem. Biophys. Res. Commun. 415, 563–566 [DOI] [PubMed] [Google Scholar]
- 22.Zaccolo M., De Giorgi F., Cho C. Y., Feng L., Knapp T., Negulescu P. A., Taylor S. S., Tsien R. Y., and Pozzan T. (2000) A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat. Cell Biol. 2, 25–29 [DOI] [PubMed] [Google Scholar]
- 23.Zaccolo M., and Pozzan T. (2002) Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science 295, 1711–1715 [DOI] [PubMed] [Google Scholar]
- 24.Akimoto M., Selvaratnam R., McNicholl E. T., Verma G., Taylor S. S., and Melacini G. (2013) Signaling through dynamic linkers as revealed by PKA. Proc. Natl. Acad. Sci. U.S.A. 110, 14231–14236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akimoto M., Zhang Z., Boulton S., Selvaratnam R., VanSchouwen B., Gloyd M., Accili E. A., Lange O. F., and Melacini G. (2014) A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP. J. Biol. Chem. 289, 22205–22220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das R., Esposito V., Abu-Abed M., Anand G. S., Taylor S. S., and Melacini G. (2007) cAMP activation of PKA defines an ancient signaling mechanism. Proc. Natl. Acad. Sci. U.S.A. 104, 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das R., and Melacini G. (2007) A model for agonism and antagonism in an ancient and ubiquitous cAMP-binding domain. J. Biol. Chem. 282, 581–593 [DOI] [PubMed] [Google Scholar]
- 28.VanSchouwen B., Selvaratnam R., Fogolari F., and Melacini G. (2011) Role of dynamics in the auto-inhibition and activation of the exchange protein directly activated by cyclic AMP (EPAC). J. Biol. Chem. 286, 42655–42669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanSchouwen B., Akimoto M., Sayadi M., Fogolari F., and Melacini G. (2015) Role of dynamics in the auto-inhibition and activation of the hyperpolarization-activated cyclic nucleotide-modulated (HCN) ion channels. J. Biol. Chem. 290, 17642–17654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boehr D. D., Nussinov R., and Wright P. E. (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nussinov R., and Tsai C. J. (2014) Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci. 35, 256–264 [DOI] [PubMed] [Google Scholar]
- 32.Nussinov R., Ma B., and Tsai C. (2014) Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys. Chem. 186, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussinov R., Tsai C. J., and Liu J. (2014) Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 136, 17692–17701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulton S., Akimoto M., VanSchouwen B., Moleschi K., Selvaratnam R., Giri R., and Melacini G. (2014) Tapping the translation potential of cAMP signalling: molecular basis for selectivity in cAMP agonism and antagonism as revealed by NMR. Biochem. Soc. Trans. 42, 302–307 [DOI] [PubMed] [Google Scholar]
- 35.Alverdi V., Mazon H., Versluis C., Hemrika W., Esposito G., van den Heuvel R., Scholten A., and Heck A. J. (2008) cGMP-binding prepares PKG for substrate binding by disclosing the C-terminal domain. J. Mol. Biol. 375, 1380–1393 [DOI] [PubMed] [Google Scholar]
- 36.Smith J. A., Reed R. B., Francis S. H., Grimes K., and Corbin J. D. (2000) Distinguishing the roles of the two different cGMP-binding sites for modulating phosphorylation of exogenous substrate (heterophosphorylation) and autophosphorylation of cGMP-dependent protein kinase. J. Biol. Chem. 275, 154–158 [DOI] [PubMed] [Google Scholar]
- 37.Wall M. E., Francis S. H., Corbin J. D., Grimes K., Richie-Jannetta R., Kotera J., Macdonald B. A., Gibson R. R., and Trewhella J. (2003) Mechanisms associated with cGMP binding and activation of cGMP-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 100, 2380–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook P. F., Neville M. E. Jr., Vrana K. E., Hartl F. T., and Roskoski R. Jr. (1982) Adenosine cyclic 3′,5′-monophosphate-dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry 21, 5794–5799 [DOI] [PubMed] [Google Scholar]
- 39.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 40.Goddard T. D., and Kneller D. G. (2006) SPARKY, NMR Assignment and Integration Software, Version 3 University of California at San Francisco [Google Scholar]
- 41.Bahrami A., Assadi A. H., Markley J. L., and Eghbalnia H. R. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 5, e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulton S., Akimoto M., Selvaratnam R., Bashiri A., and Melacini G. (2014) A tool set to map allosteric networks through the NMR chemical shift covariance analysis. Sci. Rep. 4, 7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selvaratnam R., Chowdhury S., VanSchouwen B., and Melacini G. (2011) Mapping allostery through the covariance analysis of NMR chemical shifts. Proc. Natl. Acad. Sci. U.S.A. 108, 6133–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvaratnam R., VanSchouwen B., Fogolari F., Mazhab-Jafari M. T., Das R., and Melacini G. (2012) The projection analysis of NMR chemical shifts reveals extended EPAC autoinhibition determinants. Biophys. J. 102, 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selvaratnam R., Mazhab-Jafari M. T., Das R., and Melacini G. (2012) The auto-inhibitory role of the EPAC hinge helix as mapped by NMR. PLoS ONE 7, e48707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das R., Mazhab-Jafari M. T., Chowdhury S., SilDas S., Selvaratnam R., and Melacini G. (2008) Entropy-driven cAMP-dependent allosteric control of inhibitory interactions in exchange proteins directly activated by cAMP. J. Biol. Chem. 283, 19691–19703 [DOI] [PubMed] [Google Scholar]
- 47.Mazhab-Jafari M. T., Das R., Fotheringham S. A., SilDas S., Chowdhury S., and Melacini G. (2007) Understanding cAMP-dependent allostery by NMR spectroscopy: comparative analysis of the EPAC1 cAMP-binding domain in its apo and cAMP-bound states. J. Am. Chem. Soc. 129, 14482–14492 [DOI] [PubMed] [Google Scholar]
- 48.Mulder F. A., Schipper D., Bott R., and Boelens R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
- 49.Rehmann H., Das J., Knipscheer P., Wittinghofer A., and Bos J. L. (2006) Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439, 625–628 [DOI] [PubMed] [Google Scholar]
- 50.Rehmann H., Arias-Palomo E., Hadders M. A., Schwede F., Llorca O., and Bos J. L. (2008) Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 455, 124–127 [DOI] [PubMed] [Google Scholar]
- 51.White R. E., Kryman J. P., El-Mowafy A. M., Han G., and Carrier G. O. (2000) cAMP-dependent vasodilators cross-activate the cGMP-dependent protein kinase to stimulate BKCa channel activity in coronary artery smooth muscle cells. Circ. Res. 86, 897–905 [DOI] [PubMed] [Google Scholar]