Background: Hereditary mutations (D244G and M87T) of skeletal calsequestrin have been associated with skeletal myopathies.
Results: The D244G mutation loses Ca2+, resulting in structural instability. M87T inhibits polymerization of calsequestrin by altering the Casq1 dimer interface.
Conclusion: D244G is largely dysfunctional, whereas the Ca2+ binding capacity of M87T is mildly reduced.
Significance: Altered characteristics of Casq1 mutants are congruent with their associated disease phenotypes.
Keywords: calcium, calcium-binding protein, malignant hyperthermia, sarcoplasmic reticulum (SR), skeletal muscle, calsequestrin, vacuolar aggregate myopathy
Abstract
Calsequestrin 1 is the principal Ca2+ storage protein of the sarcoplasmic reticulum of skeletal muscle. Its inheritable D244G mutation causes a myopathy with vacuolar aggregates, whereas its M87T “variant” is weakly associated with malignant hyperthermia. We characterized the consequences of these mutations with studies of the human proteins in vitro. Equilibrium dialysis and turbidity measurements showed that D244G and, to a lesser extent, M87T partially lose Ca2+ binding exhibited by wild type calsequestrin 1 at high Ca2+ concentrations. D244G aggregates abruptly and abnormally, a property that fully explains the protein inclusions that characterize its phenotype. D244G crystallized in low Ca2+ concentrations lacks two Ca2+ ions normally present in wild type that weakens the hydrophobic core of Domain II. D244G crystallized in high Ca2+ concentrations regains its missing ions and Domain II order but shows a novel dimeric interaction. The M87T mutation causes a major shift of the α-helix bearing the mutated residue, significantly weakening the back-to-back interface essential for tetramerization. D244G exhibited the more severe structural and biophysical property changes, which matches the different pathophysiological impacts of these mutations.
Introduction
To activate contraction of skeletal muscle fibers, up to 200 μmol/liter Ca2+ are released into the cytosol after an action potential. Ca2+ is released from the sarcoplasmic reticulum (SR),2 an organelle of small volume where the ion is mostly bound to acidic proteins (for a review, see Ref. 1). Calsequestrin (2) (protein and genes denoted here as Casq) is the most abundant and capacious Ca2+-binding protein within the SR of both skeletal and cardiac muscle where tissue-specific isoforms Casq1 and Casq2, respectively, are expressed. In skeletal muscle, ions bound to Casq1 constitute ∼75% of the Ca2+ released for activation of contraction (3).
Ca2+ titrations of Casq in vitro feature multiple stages associated with progressive Casq polymerization (4, 5). We have interpreted this observation, together with crystallographic studies, as evidence that Casq binds Ca2+ cooperatively, which explains both its greater oligomerization and greater Ca2+ binding capacity as Ca2+ concentrations increase. In vivo, inside the SR, Casq exists in linear ramified polymers, forming a dense network that fills the SR terminal cisternae (6, 7). The structure of this network is entirely consistent with the lattice interactions observed in vitro (8). Polymerized in this way, Casq in living muscle appears to be in a state consistent with maximum storage capacity.
Other structural details suggest roles for Casq in addition to Ca2+ storage (for a review, see Ref. 9). The polymeric network inside the SR ends with slender pillars or tendrils that lead to the junctional membrane near the mouth of the RyR channels (7). Casq1 binds between 40 and 60 mol of Ca2+/mol, but in past crystallographic studies only up to 17 Ca2+ ions were identified, the majority of which appeared to be loosely bound and consequently diffusible (8). These features suggest that Casq polymers facilitate Ca2+ diffusion toward the open Ca2+ release channels both by increasing the local concentration of diffusible Ca2+ ions and by adding one-dimensional directionality down the polymer like a “calcium wire” (10, 11). This mechanism, a form of diffusion enhancement by reduction of dimensionality (12), remains hypothetical.
Finally, Casq appears to have a gating role, specifically operating in the termination of Ca2+ release. This termination, which is essential for rapid contractile relaxation, requires fast closing of SR Ca2+ release (RyR) channels. Some evidence indicates that Casq is required for adequate channel closure in skeletal muscle (13, 14), but the issue remains controversial (15).
Much of the interest in the properties of this protein stems from observations of linkage between its mutations and human disease (special cases of “couplonopathies” or diseases of the couplon (19)). This association is especially clear in the heart where at least 15 Casq2 mutations have been linked with a disease known as catecholaminergic polymorphic ventricular tachycardia (for a review, see Ref. 16).
As for Casq1, its association with muscle disease rests on three observations. (i) A syndrome similar to malignant hyperthermia (MH) has been reported for Casq1-null mice (17, 18). The causation of MH by Casq1 absence is consistent with the observations in cardiac muscle as both are mechanistically similar diseases of enhancement and loss of control of Ca2+ release (19), and polymorphic ventricular tachycardia is, in most cases, associated with severe deficits in the amount of protein present (16). (ii) A missense mutation, D244G, is linked to a myopathy characterized by the presence of vacuoles containing SR protein inclusions (20). The patients experience muscle weakness, and their cells show altered Ca2+ dynamics. (iii) A second missense mutation, M87T, was found in 16 of 205 MH probands (21). M87T could not be formally linked to MH (it is clinically called a variant, rather than a mutation, and that is the term we use here), but a mild association of mutation and disease was found (with an odds ratio of ∼2). Additionally, this mutation occurred in patients also carrying a MH-causative mutation of RyR1 at an unexpectedly high frequency. Although the M87T mutation appears to cause mild malfunction per se, it might contribute to the disease phenotype when associated to RyR mutations. M87T also has relevance in the context of its polymerization-dependent Ca2+ storage functions as replacing the highly conserved Met-87 by Thr at the dimer interface should hamper dimerization (21). Here, we report the first structural and biophysical characterization of D244G and M87T hCasq1, compare them with wild type hCasq1, and propose mechanisms for how these mutations may contribute to the associated disease phenotypes.
Materials and Methods
Site-directed Mutagenesis
The D244G and M87T hCasq1 genes were generated by site-directed mutagenesis of the wild type hCasq1 gene (GenBank accession number AB277764.1) in pET30a and transformed into Rosetta(DE3)pLysS cells for protein expression.
Protein Expression and Purification
Rosetta(DE3)pLysS Escherichia coli cells containing the Casq1 vectors were grown in LB media at 37 °C, and overexpression was induced by 0.5 mm isopropyl β-d-1-thiogalactopyranoside after reaching an A600 of 0.6. Following induction, the cells were harvested; suspended in 20 mm Tris, 0.05 g/100 ml NaN3, pH 7.5; and sonicated at 8000 rpm using a 450 Sonifier® (Branson Ultrasonics) until they reached apparent homogeneity. The resulting lysate was clarified by centrifugation at 20,000 × g and loaded onto a Toyopearl DEAE-650 M (Tosoh Biosciences) column equilibrated with DEAE wash buffer (20 mm Tris, 0.05 g/100 ml NaN3, pH 7.5). hCasq1 eluted between a linear gradient of 12.5 and 25% DEAE elution buffer (20 mm Tris, 2 m NaCl, 0.05 g/100 ml NaN3, pH 7.5). DEAE fractions containing hCasq1 were then purified using a CHT ceramic hydroxyapatite column (Bio-Rad) equilibrated with hydroxyapatite wash buffer (5 mm sodium phosphate, 0.05 g/100 ml NaN3, pH 6.8). hCasq1 eluted between a gradient of 50 and 100% hydroxyapatite elution buffer (0.5 m sodium phosphate, 0.05 g/100 ml NaN3, pH 6.8). Finally, hydroxyapatite fractions containing purified hCasq1 were purified using a phenyl-Sepharose 6 Fast Flow high substitution column (GE Healthcare) equilibrated with phenyl-Sepharose wash buffer (20 mm MOPS, 0.5 m NaCl, 0.05 g/100 ml NaN3, 1 mm EGTA). After extensive washing, hCasq1 was eluted from the column using wash buffer containing 10 mm CaCl2. Fractions containing hCasq1 were then buffer-exchanged into Casq assay buffer (20 mm MOPS, 0.3 m KCl, 0.05 g/100 ml, pH 7.2). Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Thermo Scientific).
Crystallization
Wild type, D244G, and M87T hCasq1 (12.5 mg/ml in 20 mm HEPES, 0.5 m NaCl, 0.05 g/100 ml NaN3, pH 7.0) were crystallized using the hanging drop vapor diffusion method at 4 °C. For wild type and M87T, 1.5 μl of protein solution was mixed with 1.5 μl of crystallization buffer (0.1 m HEPES, 0.2 m NaCl, 27.5% (v/v) 2-methyl-2,4-pentanediol, pH 7.0), whereas for D244G, 1.25 μl of protein solution was mixed with 1.75 μl of crystallization buffer to obtain the low Ca2+ form. Crystals generally formed within 1 week. In the case of M87T, crystals grew from protein incubated with 5 mm CaCl2 prior to mixing with crystallization solution. High Ca2+ D244G crystals were obtained by slowly diffusing 10 mm CaCl2 across a semipermeable membrane to D244G over the course of 24 h.
Structure Determination
Crystallographic data were collected at the Advanced Light Source Beamline 8.2.1 and reduced and scaled using HKL2000 (22). The low Ca2+ wild type hCasq1 structure was solved by molecular replacement using PHENIX with native rabbit Casq1 (Protein Data Bank code 3TRQ) as an input model. The D244G and M87T structures were then solved by molecular replacement using the low Ca2+ wild type hCasq1 structure as the input model. The high Ca2+ forms were built manually as necessary. Iterative model adjustment and refinement were completed using Coot (23) and PHENIX. Crystallographic coordinates and structure factors for wild type hCasq1 (Protein Data Bank code 5CRD), low Ca2+ D244G hCasq1 (Protein Data Bank code 5CRE), high Ca2+ D244G hCasq1 (Protein Data Bank code 5CRG), and M87T hCasq1 (Protein Data Bank code 5CRH) have been deposited in the Protein Data Bank. Refinement statistics are listed in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
r.m.s., root mean square. Statistics for the highest resolution shell are shown in parentheses.
| Wild-type hCasq1 | D244G (low Ca2+) | D244G (high Ca2+) | M87T hCasq1 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2221 | P21212 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 59.170, 145.132, 110.242 | 66.106, 82.815, 89.269 | 91.179, 67.462, 158.062 | 65.681, 68.553, 99.262 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 96.485, 90.0 | 90.0, 92.845, 90.0 |
| Resolution (Å) | 43.89–2.08 (2.15–2.08) | 44.72–3.315 (3.43–3.32) | 45.30–1.97 (2.04–1.97) | 49.57–2.03 (2.10–2.03) |
| Rmerge | 7.4 (76.9) | 6.7 (20.7) | 8.6 (44.0) | 5.1 (33.0) |
| I/σI | 17.3 (2.6) | 31.4 (10.6) | 10.0 (2.3) | 19.0 (5.0) |
| Completeness (%) | 99.4 (98.8) | 99.7 (97.4) | 96.1 (90.5) | 99.6 (96.2) |
| Redundancy | 6.8 | 7.0 | 3.4 | 3.7 |
| Refinement | ||||
| Resolution | 43.89–2.08 | 45.274–3.32 | 45.311–1.97 | 49.57–2.03 |
| Unique reflections | 28,782 (2,822) | 7,652 (723) | 129,973 (12,177) | 57,095 (5,636) |
| Rwork/Rfree | 0.1893/0.2168 (0.2433/0.2653) | 0.2488/0.2904 (0.3003/0.2651) | 0.1754/0.1931 (0.2201/0.2703) | 0.1757/0.2074 (0.2199/0.2886) |
| Number of atoms | ||||
| Macromolecules | 2,825 | 2,682 | 11,425 | 5,693 |
| Ion | 4 | 2 | 82 | 27 |
| Ligand | 16 | 16 | 0 | 0 |
| Water molecules | 233 | 0 | 2,554 | 727 |
| B-factors | ||||
| Protein | 40.28 | 95.29 | 21.7 | 33.18 |
| Ligand/ion | 48.98 | 95.38 | 33.17 | 38.84 |
| Water | 44.66 | 32.41 | 37.49 | |
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.002 | 0.002 | 0.015 | 0.002 |
| Bond angles (°) | 0.510 | 0.510 | 0.764 | 0.600 |
| Ramachandran analysis | ||||
| Favored | 99.14 | 97.40 | 98.50 | 98.30 |
| Outliers | 0 | 0 | 0 | 0 |
| Clashscore | 2.19 | 3.32 | 2.93 | 3.09 |
Turbidity Assays
The turbidity of hCasq1 solutions (i.e. absorbance at 350 nm) as a function of Ca2+ concentration was monitored using a Genesys 10S UV-visible spectrophotometer (Thermo Scientific). Assays were performed using 1.0 ml of 2.0 mg/ml wild type, D244G, and M87T hCasq1 in Casq assay buffer. Concentrated Ca2+ solutions (0.10, 0.25, 0.50, and 1.0 m) were added in 1.0-μl aliquots to the 1.0-ml Casq1 solutions in a quartz cuvette to achieve the proper calcium concentration. Upon addition of each concentrated Ca2+ aliquot, the samples were mixed by aspiration and allowed to equilibrate (dA350/dt = 0) before addition of the next aliquot. Dilutions from adding Ca2+ aliquots were included in data analysis. For descriptive purposes, the A350 as a function of Ca2+ concentration was least square-fitted to the sum of two Boltzmann functions, namely Equation 1.
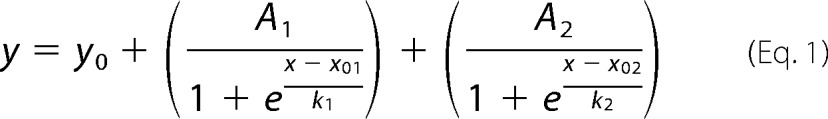 |
where y is A350, x is the Ca2+ concentration, y0 is the initial value, A1 and A2 are the spans of the absorbance changes, x01 and x02 are the centers of the first and second transitions, and k1 and k2 are the first and second slope factors, respectively.
Equilibrium Dialysis and Inductively Coupled Plasma Optical Emission Spectrometry
For equilibrium dialysis cells, two adjoined 1.4-ml acrylic half-cells separated by a 4.0-cm2 circular regenerated cellulose dialysis membrane (12–14-kDa cutoff) were used. For each cell, 0.5-ml aliquots of either 10 μm wild type, D244G, or M87T hCasq1 in Casq assay buffer were added to one half-cell, and solutions of varying CaCl2 concentrations in Casq assay buffer were added to the other. The two solutions within each cell equilibrated for 3 days at room temperature on a rocking shaker. After equilibration, the 317.933 nm Ca2+ emission intensity of each sample was measured using a PerkinElmer Life Sciences Optima 3200 RL inductively coupled plasma optical emission spectrometer. The difference between the Ca2+ contained within the protein-containing and protein-free sides of a given cell determined the bound Ca2+ (i.e. Ca2+(protein) − Ca2+(no protein) = ΔCa2+), which, when divided by its corresponding moles of protein, gave the fractional occupancy. The vertical bars at each point are standard deviations of three independent measurements. The fractional occupancies versus their corresponding free Ca2+ concentrations were least square-fitted to the sum of two Hill equations.
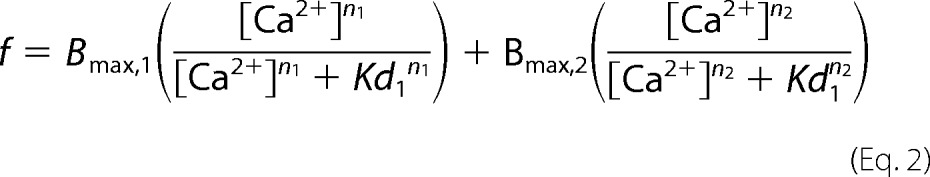 |
where f is fractional occupancy, Bmaxj (with j = 1 or 2) are the maxima of the two binding components, Kdj are their corresponding dissociation constants, and nj are their Hill coefficients.
Quantum Mechanics-Molecular Mechanics Optimization and Electrostatic Potential Surface Generation
The structures of wild type human, low Ca2+ D244G, and high Ca2+ D244G Casq1 were prepared for quantum mechanics/molecular mechanics calculations using the PDB Prep Wizard in Schrödinger Maestro (24). The high affinity site C Ca2+-binding sites in each structure were then optimized in Gaussian 09 using the ONIOM (quantum mechanics-molecular mechanics) method with AMBER used for the low layer (25). The B3LYP level of theory with double ζ correlation-consistent basis sets (cc-pVDZ for hydrogen and carbon, aug-cc-pVDZ for nitrogen and oxygen, and cc-pwCVDZ for Ca2+) was used for the high layer (26, 27). Single point calculations at the B3LYP level of theory with triple ζ basis sets (cc-pVTZ for hydrogen and carbon, aug-cc-pVTZ for nitrogen and oxygen, and cc-pwCVTZ for calcium) were then performed on each optimized structure. Self-consistent field total electron density and electrostatic potentials were generated from the single point calculation at 12 and six points per bohr, respectively. Electrostatic potential surfaces were generated by plotting the electrostatic potential on its corresponding electron density at an isovalue of 0.0200 electron/bohr3 in GaussView 3.09 (28).
Results
We compared the Ca2+-dependent polymerization properties of the wild type protein and two mutants by light scattering and turbidity measurements. We also compared their equilibrium Ca2+ binding properties as well as their crystal structures obtained with or without Ca2+ in the medium.
Ca2+-dependent Polymerization
The size exclusion chromatography profiles of wild type, D244G, and M87T hCasq1 (Fig. 1) show that the three proteins are all monomers in 0 mm Ca2+ buffer (Fig. 1A) and have roughly the same response to rising Ca2+ concentrations at 1.0 and 1.5 mm Ca2+ (Fig. 1, B and C). There appeared to be a small presence of wild type and D244G Casq1 tetramers at 1.5 mm Ca2+ (Fig. 1C, inset); because of the low absorbance of each peak (implying a very small amount of protein in this state), the significance of this observation is not clear.
FIGURE 1.
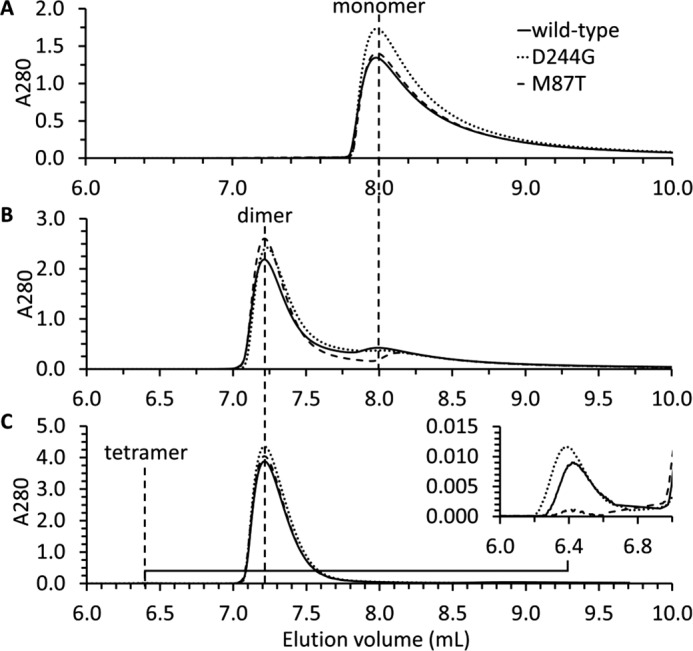
Multiangle static light scattering. Multiangle static light scattering profiles for wild type (solid line), D244G (dotted line), and M87T (dashed line) in 0 mm Ca2+ (A), 1.0 mm Ca2+ (B), and 1.5 mm Ca2+ (C) are shown. The inset in C is an expanded view that shows the low absorbance peaks at 1.5 mm Ca2+ between 5.5 and 7.5 ml that correspond to the tetramer.
Turbidity Assays
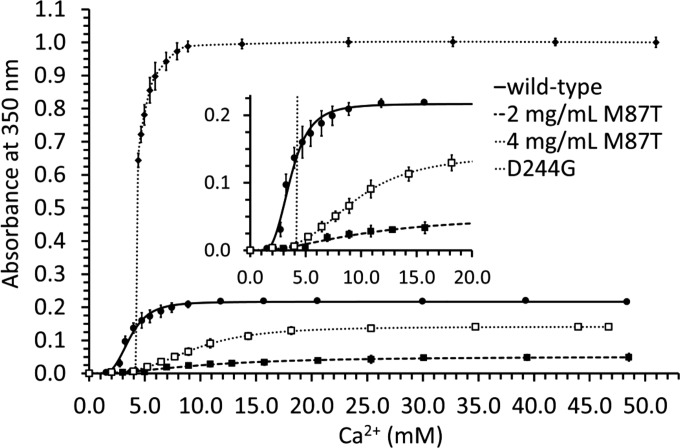
Above 1.5 mm Ca2+, monitoring Ca2+-dependent polymerization of Casq1 by multiangle light scattering was not possible because the higher order Casq1 polymers clogged the column matrix. To circumvent these limitations, Casq1 aggregation beyond 1.5 mm Ca2+ was monitored by turbidity assays. Wild type hCasq1 (Fig. 2, solid line) started a major transition at 1.4 mm Ca2+ and became saturated by 9 mm Ca2+ with an absorbance of 0.2. D244G (dotted line) underwent a large increase in turbidity that started at 1.4 mm Ca2+ and ended at 4.4 mm Ca2+ with a final absorbance of 1.0. The increase in absorbance for M87T (dashed line) was substantially lower than that of wild type and D244G. Expecting to enhance the response, M87T was studied at 2.0 and 4.0 mg/ml (Fig. 2, filled and open squares, respectively). The 4.0 mg/ml solution attained a maximum turbidity 2.8 times higher than the 2.0 mg/ml maxima, but both still required high Ca2+ concentrations to become turbid, and neither became as turbid as wild type.
FIGURE 2.
Ca2+-dependent turbidity. Ca2+-dependent turbidity curves for wild type hCasq1 (solid line, ●), D244G (dotted line, ♦), 2 mg/ml M87T (dashed line, ■), and 4 mg/ml M87T (dashed line, □) are shown. The inset is a magnified partial view of the same curves. Error bars represent S.D.
Equilibrium Ca2+ Binding Studies
Equilibrium Ca2+ binding curves calculated from equilibrium dialysis/inductively coupled plasma optical emission spectrometry data are shown in Fig. 3. The binding data were fit to the sum of two “Hill” functions (Equation 2). The best fit parameters are listed in Table 2.
FIGURE 3.

Equilibrium dialysis Ca2+ binding curve. Equilibrium dialysis-inductively coupled plasma optical emission spectrometry Ca2+ binding curve is plotted as fractional occupancy (mol of bound Ca2+/mol of hCasq1) versus free Ca2+ (mm) for wild type hCasq1 (solid line, ●), D244G (dotted line, ♦), and M87T hCasq1 (dashed line, ■). Error bars represent S.D. of three independent measurements.
TABLE 2.
Best fit parameters for Ca2+ binding curves fit to a “Hill function”
| Wild type | D244G | M87T | |
|---|---|---|---|
| Kd1 (mm)a | 0.56 | 1.01 | 2.50 |
| Kd2 (mm)a | 3.60 | 4.35 | 4.70 |
| Bmax,1b | 23.3 | 37.5 | 42.7 |
| Bmax,2b | 29.7 | 10.3 | 11.0 |
| n1c | 1.50 | 1.72 | 1.00 |
| n2c | 8.80 | 25.1 | 29.1 |
a First (Kd1) and second (Kd2) dissociation constants.
b Binding component maxima in terms of fractional occupancy (mol of Ca2+/mol of hCasq1).
c First (n1) and second (n2) Hill coefficients.
Each curve features two distinct binding stages with roughly sigmoidal Ca2+ concentration dependence. The mutants have lower Ca2+ binding capacity, and in both cases, the second binding stage occurs at a greater Ca2+ concentration than for wild type. In all cases, the second binding stage has a higher Kd than the first stage and, most notably, a very high Hill coefficient, which is suggestive of high cooperativity in this stage. Although each Casq1 features the two-stage binding property, the high Ca2+ high cooperativity component is most prominent in wild type and decays in size (i.e. Bmax2) for both mutants, especially D244G. Both mutants also have a greater Kd2 and a much greater Hill coefficient of the second component (the one with highest cooperativity). A final detail worth noting is that at 0 mm Ca2+ both wild type and M87T had an average fractional occupancy of 4 mol of Ca2+/mol of hCasq1, whereas D244G had an average fractional occupancy of only 2, corresponding to the number of Ca2+ ions observed per monomer in its low Ca2+ crystal structure (described below).
Crystal Structure of Wild Type hCasq1
Wild type hCasq1 crystallized in the absence of added Ca2+ in the C2221 space group with one molecule in the asymmetric unit, yielding 2.08-Å resolution. Like the published rabbit Casq1 (Protein Data Bank code 3TRQ) crystallized in the same condition, wild type hCasq1 contained Ca2+ ions at high affinity sites A, B, and C and a fourth Ca2+ bound by Thr-189 (Fig. 4A). Wild type hCasq1 and rabbit Casq1 had similar structures as demonstrated by the small root mean square deviation of their least square superposition (0.51 Å). In addition, the major lattice-packing interactions observed in both Casq1 crystals were mostly those involved in stabilizing a long linear polymer (Fig. 5A). The similarities of Casq1 between the two species, in terms of tertiary and polymeric structures, highlight the conserved nature of Casq1.
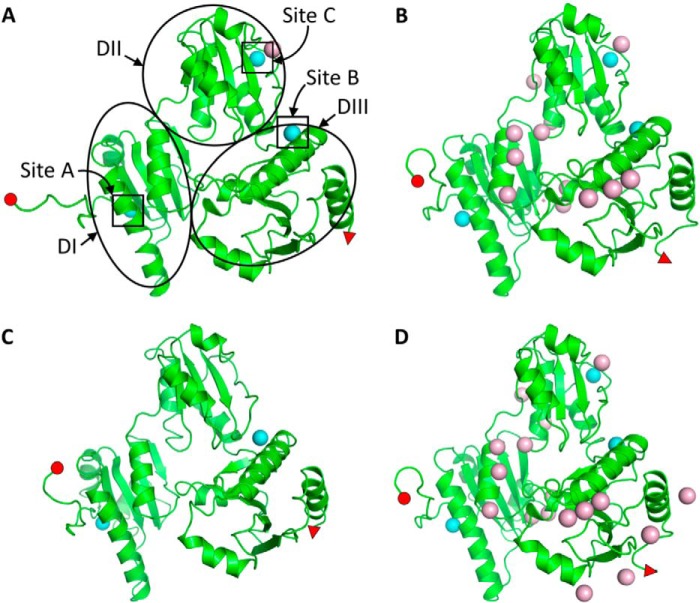
FIGURE 4.
hCasq1 crystal structures. Monomeric structures of low Ca2+ wild type hCasq1 (A), high Ca2+ M87T hCasq1 (B), low Ca2+ D244G hCasq1 (C), and high Ca2+ D244G hCasq1 (D) are shown. Cyan and pink spheres represent Ca2+ bound at high affinity and low affinity sites, respectively. Note the four Ca2+ ions in A, two in B, 21 in C, and 13 in D. The small red circles and red triangles located on each structure indicate the N and C termini, respectively, to the furthest identifiable extent. The three high affinity Ca2+ sites are boxed, and Domains I–III (DI–DIII) are circled in the wild type structure (A).
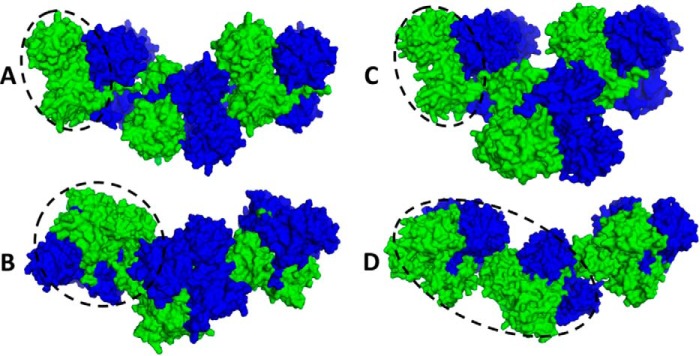
FIGURE 5.
Crystal packing. Crystal packing and polymeric structure of wild type hCasq1 (A), low Ca2+ D244G hCasq1 (B), high Ca2+ D244G hCasq1 (C), and M87T hCasq1 (D) are shown. Individual hCasq1 monomers are represented as their van der Waals surface and colored either blue or green. The asymmetric unit for each protein is circled.
Crystal Structures of D244G hCasq1
Diffraction quality crystals of D244G grew in both the absence of added Ca2+ and in the presence of 10 mm Ca2+, producing what we here call the low and high Ca2+ forms of D244G. Low Ca2+ D244G crystallized in the P21212 space group with one molecule in the asymmetric unit and two Ca2+ ions per Casq1 monomer and at a resolution of 3.32 Å (Fig. 4C). High Ca2+ D244G crystallized in the P21 space group with four molecules in the asymmetric unit and 21 Ca2+ ions per Casq1 monomer and at a resolution of 1.97 Å (Fig. 4D).
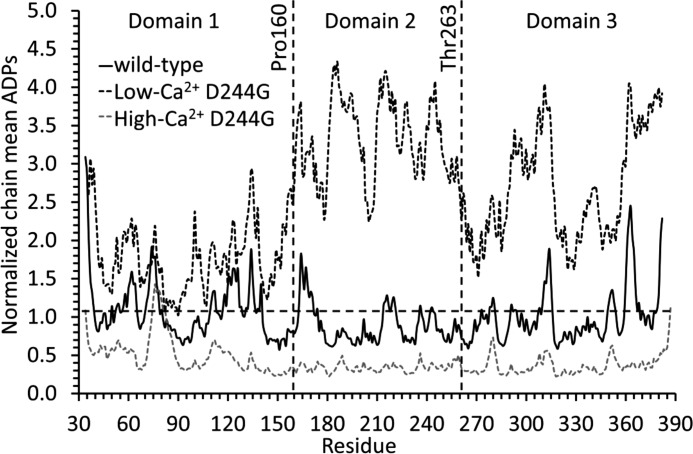
Low Ca2+ D244G had a vacant and highly disordered high affinity Ca2+-binding site C (Fig. 6A, right) presumably due to the mutation, which destabilized the hydrophobic core of Domain II (Pro-160 through Thr-263). Also affected were regions of Domain III connected to Domain II through high affinity site B, a site responsible for attaching Domain II to Domain III and located only a short stretch away from site C. The high Ca2+ D244G structure, however, had bound Ca2+ at site C and, as expected, a well ordered tertiary structure. Backbone atomic displacement parameter plots confirmed these observations (Fig. 7). In the low Ca2+ D244G structure, Domain I had similar order to wild type (i.e. similar atomic displacement parameters) but had high disorder in Domain II as well as in areas of Domain III connected to Domain II. In contrast, the high Ca2+ D244G displayed a highly ordered structure throughout.
FIGURE 6.
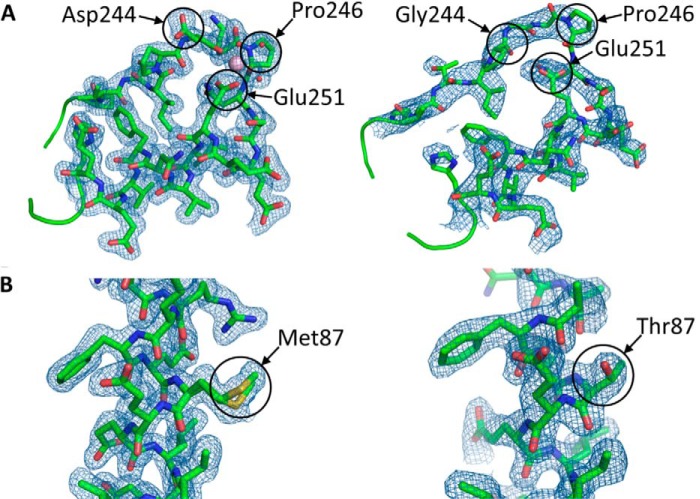
Mutation site electron densities. Crystallographic electron density maps of the D244G (A, right) and M87T mutation sites (B, right), corresponding to residues 240 through 259 and 84 through 98, respectively, compared with wild type hCasq1 (A, left and B, left) are shown. Each electron density map (blue mesh) is a feature-enhanced map generated in PHENIX at a contour of 1.5 σ. The locations of mutated residues and their corresponding wild type residues are indicated.
FIGURE 7.
Atomic displacement parameter plot. A plot of wild type (black solid line), low Ca2+ D244G (black dashed line), and high Ca2+ D244G (gray dashed line) main chain atomic displacement parameters (ADPs) normalized to the mean wild type chain value is shown. The regions corresponding to Domains I–III and their limits at Pro-160 and Thr-263 are marked with vertical dashed lines. Residue 34 marks the N terminus of Casq1 in its physiologically relevant form (i.e. after the signal peptide is removed).
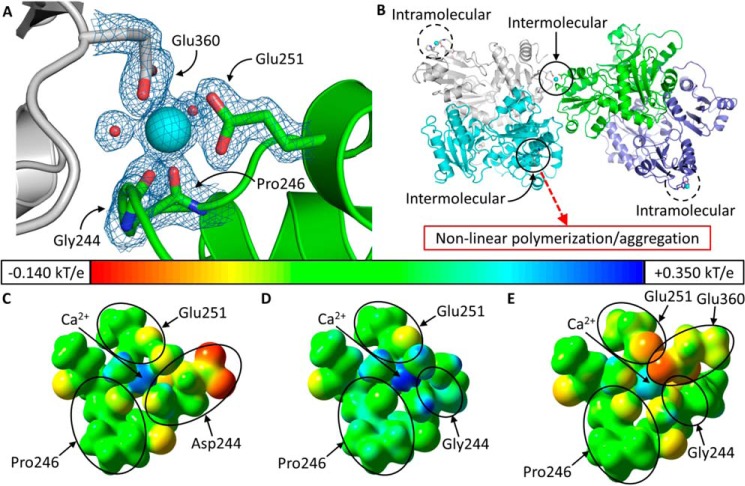
Each high Ca2+ D244G dimer features intramolecular and intermolecular Ca2+ coordination at site C with one monomer coordinating intramolecularly and the second coordinating intermolecularly. The intramolecular coordination is similar to that of site C in wild type hCasq1 except for an additional solvent molecule (for a total of four) that is seen coordinating Ca2+ in the high Ca2+ D244G structure Conversely, the intermolecular coordination at C is unique to D244G. As shown in Fig. 8A, the Ca2+ bound by site C from a monomer in one dimer is coordinated by Glu-360 from a monomer in a second dimer. This intermolecular coordination of site C forms non-canonical Casq1 tetramers (Fig. 8B). In each of the two dimers depicted, intramolecular coordination occurs on the Glu-360-donating monomer. An intermolecular site C appears to be present in each dimer and causes this intermolecular coordination to propagate through the crystal lattice, making this interaction the possible cause for the Ca2+-dependent crystalline aggregation of D244G seen in experiments.
FIGURE 8.
Ca2+ coordination in D244G. A, intermolecular Ca2+ coordination at high affinity site C of D244G. The Ca2+ ion at site C is shown as a large cyan sphere, and the small red spheres are coordinating water molecules. The green protein contains site C, and the gray protein contributes Glu-360 for intermolecular coordination. Blue and red segments in each protein represent nitrogen and oxygen, respectively. The electron density map is contoured at 1.5 σ. B, dimers of D244G have two asymmetrical high affinity site Cs; open circles mark intermolecularly coordinated site Cs, which support non-canonical tetramerization. Thus, site C in the green monomer (whose dimeric partner is in light purple) is complemented by Glu-360 from the gray monomer (whose dimeric partner is in light blue). The dashed circles indicate high affinity site Cs that are coordinated solely intramolecularly. Electrostatic potentials for wild type hCasq1 site C (C), high Ca2+ D244G intramolecular (D), and intermolecular site C (E) are shown. The potential surfaces are shown at an isovalue of 0.200 electron/bohr3. The color scale corresponds to a range of −0.140 (red) to +0.350 kT/e (blue) with green corresponding to a potential of 0.000 kT/e (or −0.200 for red to +0.500 in hartrees for blue where 1 hartree = 627.509 kcal/mol).
To rationalize the different coordination modes, we analyzed the electrostatic potential surfaces of high affinity site C from wild type human and high Ca2+ D244G. In wild type human Casq1 (Fig. 8C), the carboxylate side chain of Asp-244, due to its proximity to the Ca2+ ion in site C, neutralizes the +2 charge conferred by the Ca2+ ion bound at site C. In the high Ca2+ D244G structure, site C coordinated intramolecularly (Fig. 8D) is left with a +1 charge as a consequence of losing the Asp-244 carboxylate side chain to the mutation. As shown by the electrostatic potential surface, this +1 charge is distributed throughout the Ca2+-binding residues (here Gly-244, Pro-246, Glu-251, and four water molecules). In the case of the intermolecular coordination of site C by Glu-360 from a monomer in a second dimer (Fig. 8E), direct Ca2+ coordination by Glu-360 neutralizes the +1 charge on site C. Two coordinating water molecules are displaced in the process, giving a final coordination sphere constituted by Gly-244, Pro-246, Glu-251, Glu-360, and two water molecules. As shown in Fig. 8, the intermolecularly coordinated Ca2+ ion is more neutralized than in wild type probably because the intermolecular case has two carboxylates directly coordinating Ca2+ (Glu-251 and Glu-360), whereas wild type has one direct coordination (Glu-251) and one outer sphere anion (the Asp-244 side chain).
Crystal Structure of M87T hCasq1
M87T crystallized in the P21 space group with two molecules in the asymmetric unit. Crystals were obtained only in the presence of 2.5 mm Ca2+, and each monomer contained 13 Ca2+ ions. The crystal structure had a resolution of 2.03 Å, which allowed clear imaging of the mutated residue (Fig. 6B, right).
Least square superposition of the M87T structure onto the wild type hCasq1 structure, crystallized without adding Ca2+, gave a root mean square deviation of 1.6 Å. This high deviation could have originated from the mutation, Ca2+-dependent structural changes, or both. To separate these factors, we compared the bovine Casq1 crystal structure (Protein Data Bank code 4TLY), obtained previously at 2.5 mm Ca2+, with M87T hCasq1 (Fig. 9A). Their superposition had a lower root mean square deviation of 0.74 Å. The corresponding difference between the superposed structures, which was due to the mutation and not Ca2+, was an outward shift of α-helix 2 (α2) in Domain I where Met-87 is located (Fig. 9B).
FIGURE 9.
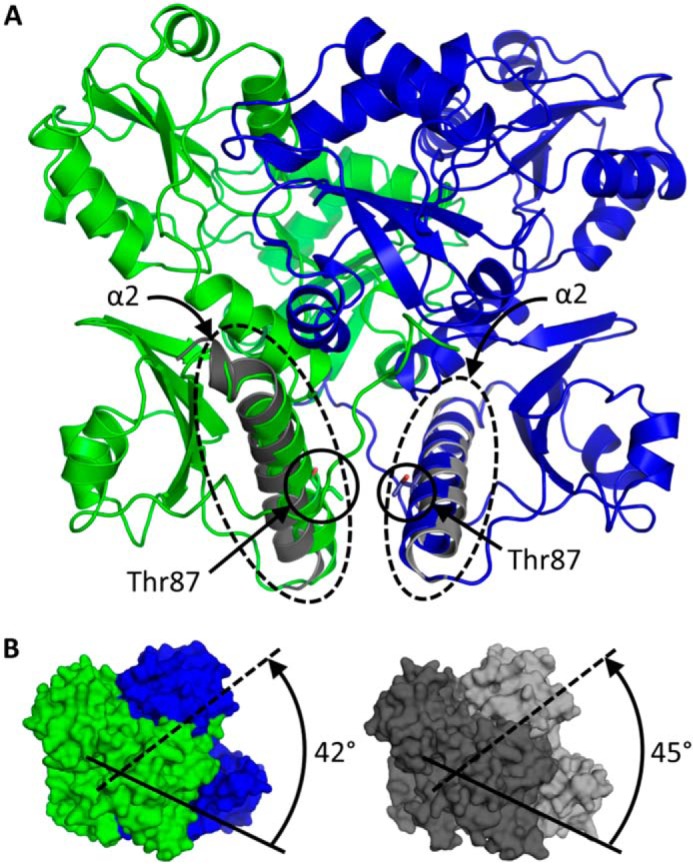
M87T dimeric interactions. A, M87T front-to-front dimer. The locations of α-helix 2 and Thr-87 from each monomer are indicated. The gray helices represent the least square-superposed α2 from high Ca2+ wild type Casq1. B, Ca2+-induced rotation of dimers around front-to-front interface of M87T (left) and wild type Casq1 (right). The solid and dashed lines represent the relative orientations of Domain I from each of the dimeric partners. The angles of rotation from the initial relative rotation of 180° across the front-to-front dimer interface are listed.
Discussion
We compared physicochemical properties of wild type hCasq1 and two naturally occurring mutants, D244G and M87T. D244G causes a myopathy associated with limited muscle function and characterized by vacuoles with inclusions consisting of aggregates of SR proteins. M87T has an ill defined nosologic status. Its allele frequency in a large group of patients diagnosed as MH-susceptible was greater than in control groups, but the difference was not significant (21). Despite this inconclusive result, there are multiple indications that the mutation alters muscle function to some degree. Indeed, its presence increases the odds of MH by a factor of 2. Furthermore, M87T was associated with causative RyR1 mutations in patients with the disease at a frequency higher than the expected random association value, which suggests that it enhances the disease phenotype.
Simultaneous Alteration of Ca2+-dependent Polymerization, Precipitation, and Ca2+ Binding Capacity
At the free in vivo SR Ca2+ concentration of ∼1 mm, Casq1 is known to be fully polymerized as shown by EM images of the junctional SR (6, 7). Although there are technical limitations to imitating an in vivo scenario where Casq1 concentrations approach 100 mg/ml and it exists together with a number of other cellular components and structural constraints, our turbidity data confirmed that, at least in vitro, D244G and M87T displayed different Ca2+-dependent aggregation, in both quality and quantity, than that observed in the wild type. Although all forms showed similar monomer-to-dimer oligomerization between zero (Fig. 1A) and 1.0 mm Ca2+ (Fig. 1B), wild type began to polymerize at lower Ca2+ concentrations than either D244G or M87T (Fig. 2). However, D244G overtook wild type and M87T by 4 mm, reaching its maximum turbidity and, presumably, maximum aggregation by 4.4 mm Ca2+ (Fig. 2). D244G precipitation was unique in that it produced by far the greatest increase in turbidity; furthermore, it settled rapidly at the bottom of the cuvette. The differences imply that the Ca2+-induced quaternary change induced in D244G is qualitatively different, leading to larger aggregates. As argued below, these aggregates are less able to bind Ca2+.
The M87T mutant responded less than the other Casq1 forms to rising Ca2+ concentrations. The increase in turbidity only started around 5 mm Ca2+ and reached a lower maximum even at double the protein concentration (Fig. 2).
For all three Casq1s, the Ca2+ binding data (Fig. 3) showed a binding process of roughly two successive stages. The first stage corresponds largely to the occupancy of sites on individual protein monomers, a process of low cooperativity, which occurs at low Ca2+ concentrations. Two aspects of the second binding stage, the requirement for higher Ca2+ concentrations (reflected in a higher Kd) and the high Hill coefficient (n2), are in agreement with the second stage consisting of Ca2+ binding to polymerized Casq1 as multiple Ca2+ ions are required to form the polymers. The second stage, at least in the wild type, has the greatest binding capacity, which explains the observed loss of Ca2+ buffering power in vivo as the SR is depleted (1–3).
Both mutations resulted in a reduction of the total Ca2+ binding capacity, mostly in the second stage (a reduction of Bmax2), an increased Kd2, and a large increase in n2. These changes suggest that the highly cooperative second stage, which appears to be associated with Casq polymerization, is both more difficult to reach for the mutants and less effective as a means to provide additional binding capacity.
The mutation-dependent changes to the Ca2+ binding properties of the two mutants were similar to the observed differences in the Ca2+ concentration-dependent increase in turbidity. The Ca2+-dependent increase in turbidity had the lowest steepness in M87T and required Ca2+ concentrations much higher than did wild type to reach its maximum turbidity. Both changes are consistent with the reduced Bmax2 and increased Kd2 of the mutant. For D244G as well, the Ca2+-dependent increase in turbidity shifted to higher Ca2+ concentrations in agreement with the change in Kd2.
Overall, the quantitative and qualitative differences between D244G and wild type are probably causative for the vacuolar inclusion phenotype of vacuolar aggregate myopathy. Impaired targeting might be an additional cause of the functional deficits as other important SR proteins were also detected in the vacuolar inclusions characteristic of this disease (21).
In agreement with the more overt disease phenotype linked to D244G, the physicochemical alterations appear greater in the D244G mutant than in the M87T variant. Other changes due to mutation, including the lower Ca2+ binding capacity of both mutants, could explain some of the functional impairment observed in patients who have the diseases associated with these Casq1 mutations.
Mutation-dependent Changes to Protein Structure and Oligomeric Interactions
In wild type Casq1, including Casq1 from humans, the backbone carbonyl oxygen atoms of Asp-244 and Pro-246, together with the carboxylate of Glu-251 and three water molecules, form a Ca2+ coordination site known as high affinity site C (Fig. 6A) (8). In the D244G mutant, the inherent flexibility of Gly-244 destabilized site C and left it vacant, which caused an order-to-disorder transition not only for the immediate neighbors of the mutated residue but also for the adjacent hydrophobic core of Domain II. These disordered regions regained order when Ca2+ concentrations rose sufficiently high enough for Ca2+ to bind into the site C of D244G (Fig. 7). These observations confirm the previously hypothesized structural role of high affinity site C.
The low Ca2+ D244G crystal lattice lacked the linear back-to-back interaction seen in wild type Casq1 (Fig. 5A), whereas the front-to-front interaction was relatively unchanged (Fig. 5B). Furthermore, the normal back-to-back interface did not reappear after D244G regained structural integrity under high Ca2+ conditions (Fig. 5C). Instead, high Ca2+ D244G displayed a unique dimer-dimer interaction mediated by intermolecular coordination of Ca2+ at high affinity site C. This intermolecular coordination would likely disrupt normal Ca2+-dependent polymerization in vivo. The disordered and inadequately polymerized low Ca2+ D244G structure, taken together with the intermolecular coordination at site C in the high Ca2+ structure, shows that on a molecular level the consequence of the D244G mutation is interference with the normal modes of action of high affinity site C.
The M87T mutation, at its core, is a disruption of the hydrophobic interactions that strengthen the front-to-front dimerization of Casq1. Met-87 is located in Domain I on α2, which associates with α2 of its dimeric partner in complementary fashion within the front-to-front dimer interface (Fig. 9A). The hydrophobic residues on and near α2 form a hydrophobic pocket that plays a key role in dimerization by associating Met-87 from the dimeric partner in a symmetric, reciprocal interaction. Because of the polar, hydrogen bonding nature of the substituted Thr residue, the M87T mutation inhibits closure of the hydrophobic pocket under low Ca2+ concentrations.
Upon exposure to rising Ca2+ concentrations, Domain I of Casq1 undergoes a large Ca2+-dependent conformational change that exposes the hydrophobic face of α2 to solvent with concomitant rotation of monomers around the front-to-front dimer interface. In M87T, the monomers rotated 3° less than wild type bovine Casq1 under the same Ca2+ conditions (Fig. 9B). It is most likely that Thr-87 can hydrogen bond with neighboring residues on α2 to form a more rigid structure, which may hamper Ca2+-dependent rotation around the front-to-front interface. Supporting these expectations, instead of the linear lattice packing observed in the wild type (Fig. 5A), the crystal lattice of M87T was established mainly through side-to-side interactions between tetramers (Fig. 5D).
The altered front-to-front interface of M87T renders the dimer unsuitable for the linear back-to-back tetramerization that occurs with the wild type. If these non-linear tetramers were present in vivo, they would hinder further Ca2+-dependent polymerization. A predictable consequence would be a reduced ability of the SR to store and release Ca2+ ions.
Conclusion
Overall, these missense mutations measurably alter the Ca2+-dependent properties of Casq1. The D244G Casq1 mutant aggregates quickly and abnormally in response to rising Ca2+ concentrations, which explains the main pathological feature of the associated disease. In contrast, the M87T Casq1 mutant features lower reactivity to rising Ca2+ concentrations, probably due to an inability to polymerize to its full physiologically relevant extent. Because these properties (Ca2+ buffering and polymerization) are necessary for Casq1 to fulfill its role in the SR Ca2+ release/reuptake cycle, the D244G and M87T variants should lead to functional impairment. A reduction of the advantage for Ca2+ diffusion that calsequestrin “wires” putatively provide would also be expected. That they are most severe in the case of D244G, the most pathogenic mutation, indicates that its identified alterations are causative in the pathogenic process of vacuolar aggregate myopathy.
Author Contributions
C. K. and E. R. conceived and coordinated this study. K. M. L. performed the biochemical/biophysical work, computational chemistry, and structure determination. L. A. R. assisted with biochemical/biophysical work and structure determination. All authors contributed to data interpretation and the writing of the manuscript and approved of all content contained herein.
This work was supported by National Institutes of Health Grant 1R01GM11125401 (to E. R and C. K.) and the M. J. Murdock Charitable Trust (to C. K.). The authors declare that they have no conflicts of interest with the content of this article.
The atomic coordinates and structure factors (codes 5CRD, 5CRE, 5CRG, and 5CRH) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SR
- sarcoplasmic reticulum
- Casq
- calsequestrin
- hCasq
- human Casq
- Casq1
- skeletal calsequestrin
- Casq2
- cardiac calsequestrin
- D244G
- Asp-244 to Gly-244 mutation
- M87T
- Met-87 to Thr-87 mutation
- MH
- malignant hyperthermia
- RyR
- ryanodine receptor (calcium release channel)
- α2
- α-helix 2.
References
- 1.Sztretye M., Yi J., Figueroa L., Zhou J., Royer L., and Ríos E. (2011) D4cpv-calsequestrin: a sensitive ratiometric biosensor accurately targeted to the calcium store of skeletal muscle. J. Gen. Physiol. 138, 211–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLennan D. H., and Wong P. T. (1971) Isolation of a calcium-sequestering protein from sarcoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 68, 1231–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manno C., Sztretye M., Figueroa L., Allen P. D., and Ríos E. (2013) Dynamic measurement of the calcium buffering properties of the sarcoplasmic reticulum in mouse skeletal muscle. J. Physiol. 591, 423–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park H., Park I. Y., Kim E., Youn B., Fields K., Dunker A. K., and Kang C. (2004) Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization. J. Biol. Chem. 279, 18026–18033 [DOI] [PubMed] [Google Scholar]
- 5.Park H., Wu S., Dunker A. K., and Kang C. (2003) Polymerization of calsequestrin. Implications for Ca2+ regulation. J. Biol. Chem. 278, 16176–16182 [DOI] [PubMed] [Google Scholar]
- 6.Perni S., Close M., and Franzini-Armstrong C. (2013) Novel details of calsequestrin gel conformation in situ. J. Biol. Chem. 288, 31358–31362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boncompagni S., Thomas M., Lopez J. R., Allen P. D., Yuan Q., Kranias E. G., Franzini-Armstrong C., and Perez C. F. (2012) Triadin/Junctin double null mouse reveals a differential role for Triadin and Junctin in anchoring CASQ to the jSR and regulating Ca2+ homeostasis. PLoS One 7, e39962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez E. J., Lewis K. M., Danna B. R., and Kang C. (2012) High-capacity Ca2+ binding of human skeletal calsequestrin. J. Biol. Chem. 287, 11592–11601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Royer L., and Ríos E. (2009) Deconstructing calsequestrin. Complex buffering in the calcium store of skeletal muscle. J. Physiol. 587, 3101–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacLennan D. H., and Reithmeier R. A. (1998) Ion tamers. Nat. Struct. Biol. 5, 409–411 [DOI] [PubMed] [Google Scholar]
- 11.Kang C., Trumble W. R., and Dunker A. K. (2002) Crystallization and structure-function of calsequestrin. Methods Mol. Biol. 172, 281–294 [DOI] [PubMed] [Google Scholar]
- 12.Adam G., and Delbrück M. (1968) in Structural Chemistry and Molecular Biology (Rich A., and Davidson N., eds) pp. 198–215, W. H. Freeman, San Francisco [Google Scholar]
- 13.Sztretye M., Yi J., Figueroa L., Zhou J., Royer L., Allen P., Brum G., and Ríos E. (2011) Measurement of RyR permeability reveals a role of calsequestrin in termination of SR Ca2+ release in skeletal muscle. J. Gen. Physiol. 138, 231–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J., Valle G., Nani A., Chen H., Ramos-Franco J., Nori A., Volpe P., and Fill M. (2009) Ryanodine receptor luminal Ca2+ regulation: swapping calsequestrin and channel isoforms. Biophys. J. 97, 1961–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fénelon K., Lamboley C. R., Carrier N., and Pape P. C. (2012) Calcium buffering properties of sarcoplasmic reticulum and calcium-induced Ca2+ release during the quasi-steady level of release in twitch fibers from frog skeletal muscle. J. Gen. Physiol. 140, 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faggioni M., Kryshtal D. O., and Knollmann B. C. (2012) Calsequestrin mutations and catecholaminergic polymorphic ventricular tachycardia. Pediatr. Cardiol. 33, 959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dainese M., Quarta M., Lyfenko A. D., Paolini C., Canato M., Reggiani C., Dirksen R. T., and Protasi F. (2009) Anesthetic- and heat-induced sudden death in calsequestrin-1-knockout mice. FASEB J. 23, 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomasi M., Canato M., Paolini C., Dainese M., Reggiani C., Volpe P., Protasi F., and Nori A. (2012) Calsequestrin (CASQ1) rescues function and structure of calcium release units in skeletal muscles of CASQ1-null mice. Am. J. Physiol. Cell Physiol. 302, C575–C586 [DOI] [PubMed] [Google Scholar]
- 19.Ríos E., Figueroa L., Manno C., Kraeva N., and Riazi S. (2015) The couplonopathies: a comparative approach to a class of diseases of skeletal and cardiac muscle. J. Gen. Physiol. 145, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi D., Vezzani B., Galli L., Paolini C., Toniolo L., Pierantozzi E., Spinozzi S., Barone V., Pegoraro E., Bello L., Cenacchi G., Vattemi G., Tomelleri G., Ricci G., Siciliano G., Protasi F., Reggiani C., and Sorrentino V. (2014) A mutation in the CASQ1 gene causes a vacuolar myopathy with accumulation of sarcoplasmic reticulum protein aggregates. Hum. Mutat. 35, 1163–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraeva N., Zvaritch E., Frodis W., Sizova O., Kraev A., MacLennan D. H., and Riazi S. (2013) CASQ1 gene is an unlikely candidate for malignant hyperthermia susceptibility in the North American population. Anesthesiology 118, 344–349 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Schrödinger, LLC (2015) Schrödinger Suite 2015-2 Protein Preparation Wizard; Maestro, Version 10.2, Schrödinger, LLC, New York [Google Scholar]
- 25.Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., and Fox D. J. (2009) Gaussian 09, Revision C.01, Gaussian, Inc., Wallingford, CT [Google Scholar]
- 26.Peterson K. A., and Dunning T. H. (2002) Accurate correlation consistent basis sets for molecular core-valence correlation effects: the second row atoms Al-Ar, and the first row atoms B-Ne revisited. J. Chem. Phys. 117, 10548–10560 [Google Scholar]
- 27.Koput J., and Peterson K. A. (2002) Ab initio potential energy surface and vibrational-rotational energy levels of X2Σ+ CaOH. J. Phys. Chem. A 106, 9595–9599 [Google Scholar]
- 28.Frisch M. (2004) GaussView, Version 3, Gaussian, Inc., Wallingford, CT [Google Scholar]