Background: PARP-1 has importance in the immune system.
Results: Poly(ADP-ribosyl)ation of FOXP3 mediated by PARP-1 destabilizes FOXP3 and negatively regulates the suppressive activity of Treg cells.
Conclusion: PARP-1 negatively regulates Treg function via FOXP3 poly(ADP-ribosyl)ation.
Significance: This study helps the development of PARP-1 inhibitors to prevent autoimmune diseases.
Keywords: ADP-ribosylation, forkhead box P3 (FOXP3), immunosuppression, inhibitor, posttranscriptional regulation, T cell, regulatory T cell
Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) is an ADP-ribosylating enzyme participating in diverse cellular functions. The roles of PARP-1 in the immune system, however, have not been well understood. Here we find that PARP-1 interacts with FOXP3 and induces its poly(ADP-ribosyl)ation. By using PARP-1 inhibitors, we show that reduced poly(ADP-ribosyl)ation of FOXP3 results in not only FOXP3 stabilization and increased FOXP3 downstream genes but also enhanced suppressive function of regulatory T cells. Our results suggest that PARP-1 negatively regulates the suppressive function of Treg cells at the posttranslational level via FOXP3 poly(ADP-ribosyl)ation. This finding has implications for developing PARP-1 inhibitors as potential agents for the prevention and treatment of autoimmune diseases.
Introduction
Poly(ADP-ribose) polymerases (PARP),3 composed of 18 members, play diverse roles in multiple cellular processes (1). Proteins in the PARP family transfer ADP-ribose from NAD on Lys/Glu/Asp amino acid residues of protein acceptors to generate a branched poly(ADP-ribose) (PAR) chain, a process named poly(ADP-ribosyl)ation or PARylation (2, 3). PARP-1, the most characterized member of the PARP family, has been shown to regulate the protein stabilization, protein-protein interaction, cellular localization, and transcriptional activity of its substrates through poly(ADP-ribosyl)ation (4). Studies have shown that activated PARP-1 regulates the chromatin structure and DNA repair through the poly(ADP-ribosyl)ation of histones and the recruitment of single-strand DNA repair enzymes (5–7). In addition, PARP-1 has also been reported to regulate gene transcription in tumor generation, metabolism, and immune responses (8–10). Because of its diverse and important roles, PARP-1 inhibitors have been used in the treatment of cancer and inflammation (11, 12).
Regulatory T (Treg) cells are a subpopulation of CD4+ T cells and induce and maintain peripheral tolerance and modulate immune responses. Dysregulation of the development and function of Treg cells has been shown to be associated with many immune-related diseases, including autoimmune diseases, chronic inflammation, acute and chronic infection, and transplant rejection (13). Forkhead box P3 (FOXP3), known as a key transcription factor of Treg cells, is required for their development, maintenance, and function (14, 15). FOXP3 abnormality leads to the severe autoimmune disease IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome) in humans and the fatal lymphoproliferative disorder in mice (16, 17). Studies have shown that the transcriptional activity, stability, and localization of FOXP3 are regulated at posttranslational levels, including acetylation, ubiquitination, and phosphorylation (18–20). Recently, it has been shown that mice deficient in PARP-1 display increased numbers of regulatory T cells, indicating that PARP-1 may affect Treg cell differentiation (21). However, how PARP-1 may regulate the suppressive function of Treg cells has not been clear (22).
Here we provide evidence demonstrating that PARP-1 binds to FOXP3 and induces poly(ADP-ribosyl)ation of FOXP3. More importantly, we find that, after treatment with PARP-1-specific inhibitors, decreased poly(ADP-ribosyl)ation of FOXP3 in Treg cells leads to an increased level of FOXP3 by preventing proteasome-mediated degradation, resulting in an increased suppressive function of Treg cells. Our results suggest that PARP-1 negatively regulates the suppressive function of Treg cells at the posttranslational level through FOXP3 poly(ADP-ribosyl)ation.
Experimental Procedures
Plasmids, Antibodies, and Reagents
pIP-HA/Myc/FLAG-tagged FOXP3, FOXP3 truncations, His-tagged Ub, and FLAG-tagged Stub1 were constructed as described previously (20). PARP-1 was amplified from human peripheral blood mononuclear cell (PBMC) cDNA with the primers 5′-ATCATGGCGGAGTCTTCGGATAAGC-3′ (forward) and 5′-CCGCTCGAGTTACCACAGGGAGGTCTTAAAATTG-3′ (reverse). The antibodies used were as follows: anti-FLAG (catalog no. M2, Sigma), anti-HA (catalog no. F-7, Santa Cruz Biotechnology), anti-Myc (Santa Cruz Biotechnology), anti-FOXP3 (catalog no. hFOXY, eBioscience), anti-PARP-1 (Sigma), anti-PAR (catalog no. 4335-AMC-050, Trevigen), anti-GAPDH (catalog no. 1C4, Sungene Biotech), anti-α-tubulin (catalog no. DM1A, Sigma), anti-ubiquitin (Santa Cruz Biotechnology), anti-Lamin B (Santa Cruz Biotechnology), anti-H3 (catalog no. 9715, Cell Signaling Technology), anti-β-actin (catalog no. 6G3, Sungene Biotech), anti-CD25-PE (catalog no. BC96, Biolegend), anti-CD4-FITC (catalog no. RPA-T4, Biolegend), and anti-CD127-PE-cy7 (catalog no. eBioRDR5, eBioscience). The PARP-1-specific inhibitor AG14361 (catalog no. S2178) and the PARP inhibitor 3-aminobenzamide (3-AB) were purchased from Selleck and Santa Cruz Biotechnology, respectively. MG132 was purchased from Merck (catalog no. 474790).
Cells and Transfection
HEK293T cells were cultured in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin. Human Jurkat T cells were grown in RPMI 1640 medium supplemented with 10% FCS, 1% penicillin/streptomycin, 1% sodium pyruvate, and 1% non-essential amino acids. All cells were maintained at 37 °C in a 5% CO2 incubator. HEK293T cells were transfected with the indicated plasmids using polyethylenimine (Polyscience, catalog no. 23966-2) reagent according to the instructions of the manufacturer.
Isolation of Human Treg Cells and Naive T Cells
Human PBMCs were isolated from the buffy coat of healthy donors (Shanghai Blood Center). Human CD4+CD25hiCD127lo Treg cells and CD4+CD127hiCD45RAhi naive T cells were sorted using a FACS ARIA II cell sorter (BD Biosciences). The purity of the isolated cells was 95–99%. In vitro expansion of Treg cells was performed in X-VIVO (Lonza) medium supplemented with 10% human AB serum, 1% GlutaMax (Gibco), 1% sodium pyruvate (Gibco), and 500 units/ml rIL-2 (R&D Systems) in the presence of anti-human CD3/CD28-conjugated Dynabeads (Invitrogen) at a bead-to-cell ratio of 4:1. The purified naive T cells were differentiated into iTreg (induced Treg) cells with 5 ng/ml TGF-β (R&D Systems), 100 units/ml rIL-2, and anti-human CD3/CD28-conjugated Dynabeads at a ratio of 1:1.
Quantitative Real-time PCR
Total RNA was isolated from whole PBMCs using TRIzol reagent (Invitrogen) and following the instructions of the manufacturer. RNA was quantified, and cDNA was reverse-transcribed with the PrimeScript RT reagent kit (TakaRa). The cDNA samples were used at 10 ng/well in a 384-well plate and run in triplicate. PCR reactions were set up in 10-μl volumes with SYBR Premix Ex Taq reagent (TakaRa) on an ABI 7900HT sequence detection system. Quantification of the target mRNA expression level was normalized to β-actin expression. The RT primers of human genes used were as follows: PARP-1 forward, 5′-AAGCCCTAAAGGCTCAGAACG-3′; PARP-1 reverse, 5′-ACCATGCCATCAGCTACTCGGT-3′; CD25 forward, 5′-GAGACGTCCATATTTACAACAG-3′; CD25 reverse, 5′-CCTTTGATTTCACTTGGGCTTC-3′; CTLA4 forward, 5′-CTTCTCTTCATCCCTGTCTTC-3′; CTLA4 reverse, 5′-AAGGTCAACTCATTCCCCATC-3′; FOXP3 forward, 5′-TCCCAGAGTTCCTCCACAAC-3′; FOXP3 reverse, 5′-ATTGAGTGTCCGCTGCTTCT-3′; Il10 forward, 5′-TGCAAAACCAAACCACAAGA-3′; Il10 reverse, 5′-TCTCGGAGATCTCGAAGCAT-3′; β-actin forward, 5′-GGACTTCGAGCAAGAGATGG-3′; and β-actin reverse, 5′-AGCACTGTGTTGGCGTACAG-3′.
Immunoprecipitation and Immunoblot Analysis
Cells were lysed in radioimmune precipitation assay buffer containing 50 mm Tris/HCl (pH 7.4), 0.5% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA with 1 mm PMSF, 1 mm Na3VO4, 1 mm NaF, and protease inhibitor (Sigma). The supernatants were immunoprecipitated with 1 μg of the indicated antibodies and 10 μl of protein A/G Plus-agarose (Santa Cruz Biotechnology), followed by separation in SDS/PAGE and analysis with Western blotting.
Ubiquitin Pulldown Assay
After 4-h treatment with 5 μm MG132, cells were washed with ice-cold 1× PBS and lysed in urea buffer (10 mm Tris/HCl (pH 8.0), 8 m urea, 100 mm Na2HPO4, 0.2% Triton X-100, 1 mm N-ethylmaleimide, and 10 mm imidazole) for 30 min. The lysates were incubated with nickel-nitrilotriacetic acid beads (catalog no. 30210, Qiagen) for 3 h at room temperature. The beads were washed three times with urea buffer (pH 8.0) before incubation. After 3 h of incubation, the beads were washed twice with urea buffer (pH 8.0) buffer, twice with urea buffer (pH 6.3) (10 mm Tris/HCl (pH 6.3), 8 m urea, 100 mm Na2HPO4, 0.2% Triton X-100, and 10 mm imidazole), and once with a wash buffer (20 mm Tris/HCl (pH 8.0), 100 mm NaCl, 20% glycerol, 1 mm dithiothreitol, and 10 mm imidazole). Ubiquitination levels were evaluated by Western blotting with specific antibodies as indicated.
Cytoplasm, Nucleus, and Chromatin Extraction
1 × 106 cells were harvested in 1 ml of ice-cold 1× PBS buffer and then suspended in 300 μl of cytoplasm buffer (10 mm Tris/HCl (pH 7.5), 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, and 0.5 mm PMSF). Nonidet P-40 was added to a final concentration of 0.6% after 15 min incubation on ice. Another 15 min later, the lysate was centrifuged, and the supernatant was cytoplasm fraction. The pellet was resuspended in 200 μl of nucleus extract buffer (20 mm Tris/HCl (pH 8.0), 400 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 1 mm Na3VO4, and 1 mm NaF) and then incubated on ice for another 30 min. After centrifugation, the supernatant was nucleus fraction, and the pellet was chromatin fraction.
In Vitro Poly(ADP-ribosyl)ation Assay
An in vitro poly(ADP-ribosyl)ation assay was performed as described previously (23). MBP-FOXP3 protein was expressed in Escherichia coli and purified with amylose resin as described previously (20). His-PARP-1 recombinant protein was bought from Sino. Purified MBP-FOXP3 (1 μg) and His-PARP-1 (100 ng) were incubated at 37 °C with NAD (50 μm), DTT (1 mm), MgCl2 (10 mm), Tris/HCl (pH 7.4) (100 mm), and 10 mg of braked salmon germ DNA for 1 h. The reaction was stopped with 2× loading buffer followed by Western blotting, and anti-PAR antibody was used to specifically detect the poly(ADP-ribosyl)ation level of MBP-FOXP3 protein.
In Vitro Suppression Assay
Before incubation, Treg cells were pretreated with PARP-1 inhibitors for 6 or 12 h. After this treatment, human PBMC cells were labeled with 5 μm CFSE (Invitrogen) for 10 min at 37 °C, followed by incubation with anti-CD3/CD28 beads and pretreated Treg cells at a different ratio in a U-bottom 96-well plate. After 4 days, cells were harvested to stain with viability dye (fixable viability dye, eFluor® 780, eBioscience) and anti-CD8-APC antibody (eBioscience) and then analyzed on a Fortessa cytometer (BD Biosciences).
Statistics
Two-paired Student's t tests were used for the calculation of p values.
Results
PARP-1 Interacts with and Poly(ADP-ribosyl)ates FOXP3
The role of PARP-1 in regulating Treg cell function has been controversial (21, 22). A previous study has found that PARP-1 knockout mice displayed an increased frequency of Treg cells but normal suppression function, indicating that PARP-1 might affect Treg cell differentiation but not function. However, another study has shown that PARP-1 knockout Treg cells exhibited stronger suppressive activity than the WT. Therefore, whether and how PARP-1 regulates the suppressive function of Treg cells is still not clear.
Studies have shown that PARP-1, as a ADP-ribose transferase, poly(ADP-ribosyl)ates not only histones but also several transcription factors, including Smad2/3 and NFAT (10, 23). Therefore, we first set out to examine whether FOXP3 could be a substrate of PARP-1 in Treg cells. In a co-immunoprecipitation assay in which Myc-tagged PARP-1 and FLAG-tagged FOXP3 were co-transfected into HEK293T cells, we found that PARP-1 interacted with FOXP3 in reciprocal immunoprecipitation (Fig. 1A). Such an interaction of PARP-1 and FOXP3 was also confirmed in Jurkat cells stably expressing HA-tagged FOXP3 and in human primary iTreg cells (Fig. 1, B and C).
FIGURE 1.
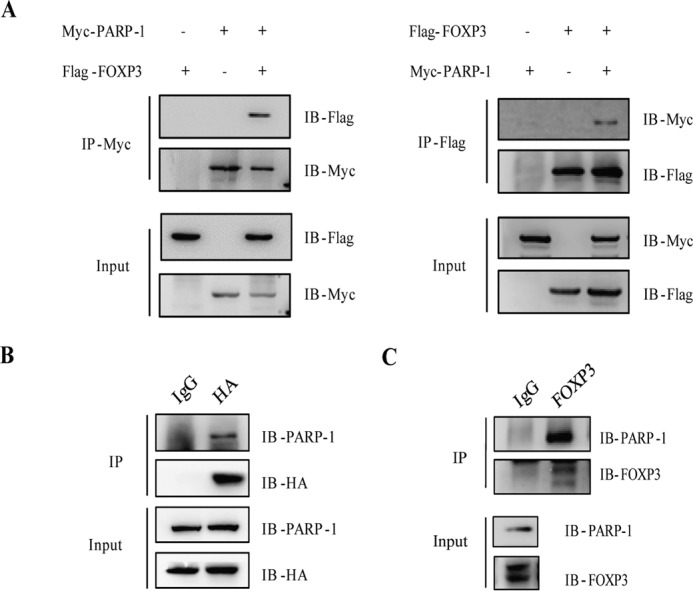
PARP-1 interacts with FOXP3. A, reciprocal immunoprecipitation (IP) of PARP-1 and FOXP3. HEK 293T cells were co-transfected with plasmids encoding for PARP-1 and FOXP3. Cell lysates were immunoprecipitated with anti-Myc or anti-FLAG antibodies, followed by immunoblotting (IB) with different antibodies as indicated. B, cell extracts prepared from Jurkat cells stably expressing HA-FOXP3 were immunoprecipitated with anti-HA antibodies, followed by immunoblotting with anti-PARP-1 antibodies. C, iTreg cells were immunoprecipitated with anti-FOXP3 (FOXY) antibody and analyzed by Western blotting. Data represent at least two independent experiments.
To further determine which domains in PARP-1 and FOXP3 were responsible for the interaction between these two proteins, we generated a panel of PARP-1 and FOXP3 truncation expression constructs. Human PARP-1 has been shown to contain a DNA-binding domain, a BRCA1 C terminus, a tryptophan-glycine-arginine domain (WGR), and a catalytic domain (CA). The FOXP3 protein is composed of several functional domains, including proline-rich, zinc finger, Leu zipper, coiled-coil (CC), and forkhead domains (FK). In the co-transfection experiments in HEK293T cells followed by immunoprecipitation, we found that the zinc finger and Leu zipper domains of FOXP3 and the DNA-binding domain and BRCA1 C terminus of PARP-1 were crucial for the interaction between PARP-1 and FOXP3 (Fig. 2, A and B).
FIGURE 2.
The zinc finger/Leu zipper domains of FOXP3 interact with the DNA-binding domain/BRCA1 C terminus of PARP-1. A, schematic of the FOXP3 protein. HEK293T cells were transfected with plasmids expressing various truncated FOXP3 proteins, followed by immunoprecipitation (IP) and Western blotting. IB, immunoblot; CC, coiled-coil domain; FK, forkhead domain. B, different domains of PARP-1 were co-expressed with FOXP3 in HEK293T cells, and the interaction of the PARP-1 domain with FOXP3 was examined by immunoprecipitation and immunoblotting. Data represent at least two independent experiments. DBD, DNA-binding domain; BRCT, BRCA1 C terminus; WGR, tryptophan-glycine-arginine domain; CA, catalytic domain.
To examine whether PARP-1 promoted the poly(ADP-ribosyl)ation of FOXP3, we used anti-PAR-specific antibody to detect the poly(ADP-ribosyl)ation level of FOXP3 in immunoprecipitates. We found that this modification of FOXP3 was increased with anti-PAR-specific antibody when FOXP3 and PARP-1 were co-transfected into HEK293T cells (Fig. 3A). Furthermore, in an in vitro poly(ADP-ribosyl)ation assay, we also examined the poly(ADP-ribosyl)ation of FOXP3 mediated by recombinant His-tagged PARP-1 protein (Fig. 3B).
FIGURE 3.
PARP-1 poly(ADP)-ribosylates FOXP3. A, immunoprecipitation (IP) of exogenous Myc-FOXP3 from HEK293 T cells that were co-transfected with Myc-FOXP3 and FLAG-PARP-1 plasmids. PARylation was detected by anti-PAR antibody. IB, immunoblot. B, poly(ADP-ribosyl)ation assay in vitro. Recombinant His-tagged PARP-1 was incubated with MBP-tagged FOXP3 in poly(ADP-ribosyl)ation reaction buffer, and anti-PAR antibody was used to detect PARylated FOXP3. C, HEK293T cells were co-transfected with PARP-1, PARG, and FOXP3. After harvesting, immunoprecipitation and Western blotting were performed. D, HEK293T cells were co-transfected with plasmids as indicated, and then 10 mm 3-AB, a PARP inhibitor, was added to cell culture and incubated for 6 h. Cell lysates were immunoprecipitated with anti-FLAG antibody, and PARylated FOXP3 proteins were detected by anti-PAR antibody. Data represent at least two independent experiments.
Studies have shown that poly(ADP-ribose) glycohydrase (PARG) rapidly degrades the poly(ADP) ribose chain (24). Therefore, we wanted to test whether PARG would inhibit the poly(ADP-ribosyl)ation of FOXP3 by PARP-1. HEK293T cells were simultaneously transfected with HA-tagged PARP-1, FLAG-tagged FOXP3, and Myc-tagged PARG. In the subsequent immunoprecipitation assays, we found that PARG did not affect the association between PARP-1 and FOXP3 but, rather, the opposite. The presence of PARG seemed to promote this interaction (Fig. 3C). Nevertheless, by using anti-PAR antibody, we found that the presence of PARG resulted in decreased poly(ADP-ribosyl)ation of FOXP3 (Fig. 3D). All of these results suggest that PARP-1 poly(ADP-ribosyl)ates FOXP3 and that FOXP3 is a substrate of PARP-1.
Reduced Poly(ADP-ribosyl)ation of FOXP3 Results in FOXP3 Stabilization in Treg Cells
3-AB is a widely used PARP-1 inhibitor, and AG14361 is a potent PARP-1-specific inhibitor, and they have been shown to effectively block PARP-1 catalytic activity (24). To test whether PARP-1 inhibitors affect the poly(ADP-ribosyl)ation of FOXP3, we firstly co-transfected FLAG-tagged FOXP3 and HA-tagged PARP-1 into HEK293T cells and treated the cells with 3-AB for 6 h. In the subsequent immunoprecipitation assay using anti-PAR antibody, we found decreased poly(ADP-ribosyl)ation of FOXP3, suggesting that 3-AB inhibited FOXP3 poly(ADP-ribosyl)ation mediated by PARP-1 (Fig. 3D). Such a reduced poly(ADP-ribosyl)ation of endogenous FOXP3 was also confirmed in 3-AB- or AG14361-treated Treg cells (Fig. 4A).
FIGURE 4.
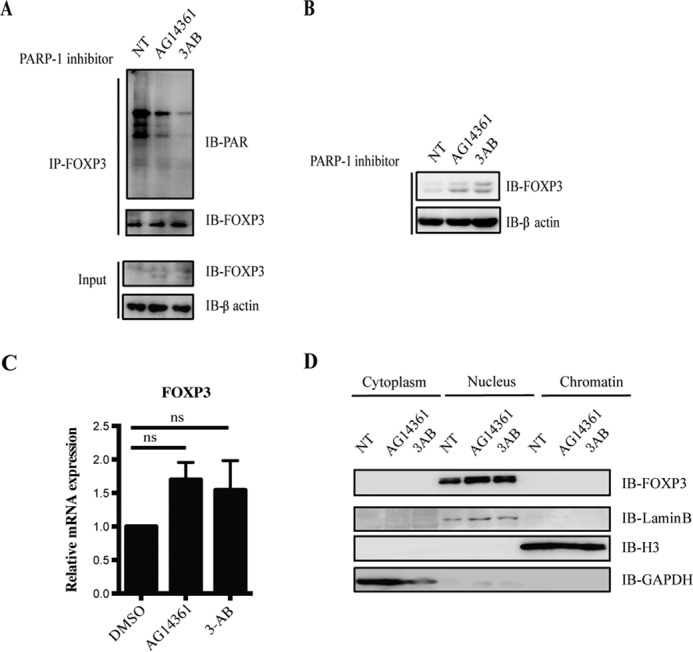
PARP-1 inhibitors stabilize the FOXP3 protein level but not cellular localization in human Treg cells. A, human primary Treg cells were cultured with the PARP-1 inhibitors 3-AB (10 mm) or AG14361 (10 μm) for 12 h. Cell lysates were immunoprecipitated (IP) with anti-FOXP3 antibody, and PARylated FOXP3 proteins were detected by anti-PAR antibody. IB, immunoblot; NT, no treatment. B and C, human primary Treg cells were cultured with the PARP-1 inhibitors 3-AB (10 mm) or AG14361 (10 μm) for 12 h. The protein level of FOXP3 was detected by Western blotting, and mRNA level of FOXP3 was examined by RT-PCR. DMSO, dimethyl sulfoxide; ns, no significance (two-paired Student's t test). Data represent at least three independent experiments. D, human primary Treg cells were cultured with the PARP-1 inhibitors 3-AB (10 mm) or AG14361 (10 μm) for 12 h. Cytoplasm, nucleus, and chromatin fractions were extracted followed the protocol detailed under “Experimental Procedures” and detected by Western blotting. Data represent at least two independent experiments.
Poly(ADP-ribosyl)ation, as a crucial posttranslational modification, has been reported to regulate protein stabilization, cellular localization, and transcriptional activity (4). To further investigate how PARP-1 inhibitors regulate FOXP3 function by inhibiting its poly(ADP-ribosyl)ation, we first set out to examine FOXP3 protein and mRNA levels in Treg cells treated with 3-AB or AG14361. We found that, 12 h after human primary Treg cells had been treated with 3-AB or AG14361, the protein level of FOXP3 was up-regulated, but the mRNA level did not change (Fig. 4, B and C), indicating that PARP-1 inhibitors stabilized FOXP3 protein level in Treg cells through inhibiting poly(ADP-ribosyl)ation of FOXP3.
Also, we detected the cellular localization of FOXP3 in PARP-1 inhibitor-treated Treg cells and found increased FOXP3 after PARP-1 inhibitor treatment. However, FOXP3 localization still did not change (Fig. 4D), suggesting that the suppression of PARP-1 enzymatic activity stabilized FOXP3 but not localization in human Treg cells.
PARP-1 Promotes the Poly-ubiquitination and Degradation of FOXP3
The E3 ubiquitin ligase Stub1 has already been identified to negatively regulate the suppressive function of Treg cells by promoting the poly-ubiquitination and degradation of FOXP3 (20). Given the fact that the inhibition of PARP-1 enzymatic activity not only inhibited the poly(ADP-ribosyl)ation of FOXP3 but also stabilized FOXP3 in Treg cells, we decided to further examine whether PARP-1 promoted the poly-ubiquitination and degradation of FOXP3. First, His-tagged Ub, HA-tagged FOXP3, Myc-tagged PARP-1, and FLAG-tagged Stub1 were co-transfected into HEK293T cells. As expected, the FOXP3 level was reduced with addition of Stub1, and, importantly, even less FOXP3 was found in the Stub1, PARP-1, and FOXP3 co-expressed group, suggesting that PARP-1 may promote FOXP3 degradation mediated by Stub1 (Fig. 5A). To further examine whether PARP-1 affected FOXP3 poly-ubiquitination to promote FOXP3 degradation, a His pulldown assay was used to detect the endogenous poly-ubiquitination level of FOXP3 after 4 h of MG132 treatment. The addition of the proteasome inhibitor MG132 prevented FOXP3 loss, suggesting that this process was proteasome-dependent. Moreover, the poly-ubiquitination level of FOXP3 was up-regulated significantly by addition of PARP-1, indicating that PARP-1 promoted FOXP3 poly-ubiquitination and degradation (Fig. 5B).
FIGURE 5.

PARP-1 promotes the poly-ubiquitination and degradation of FOXP3. A, HEK 293T cells were co-transfected with plasmids encoding for PARP-1, FOXP3, Ub, and Stub1. Cell lysates were examined by immunoblotting (IB) with different antibodies as indicated. B, HEK 293T cells were co-transfected with plasmids encoding for PARP-1, FOXP3, Ub, and Stub1. Before harvesting, these cells were treated with 5 μm MG132 for 4 h. The ubiquitin pulldown assay for His-tagged Ub was performed as described under “Experimental Procedures.” C, human primary Treg cells were immunoprecipitated (IP) with anti-Ub antibody and analyzed by Western blotting. Data represent at least two independent experiments.
To determine whether the enzymatic activity of PARP-1 affected FOXP3 poly-ubiquitination, we detected the endogenous poly-ubiquitination level of FOXP3 in PARP-1 inhibitor-treated Treg cells. After 12 h of treatment, we found decreased FOXP3 in Ub immunoprecipitates after PARP-1 inhibitor treatment (Fig. 5C), suggesting that the inhibition of PARP-1 enzymatic activity suppressed the poly-ubiquitination and degradation of FOXP3.
PARP-1 Inhibitors Enhanced the Suppressive Function of Treg Cells through PARylated FOXP3
To further investigate whether PARP-1 inhibitors regulated Treg cell function by inhibiting FOXP3 poly(ADP-ribosyl)ation, we examined some of the FOXP3-associated genes in Treg cells treated with 3-AB. We found that, 6 h after human primary Treg cells had been treated with 3-AB, the expression levels of CD25, CTLA4, and Il10 in the cells were up-regulated (Fig. 6A), suggesting that PARP-1 inhibitor affected FOXP3 downstream genes. Given the fact that the inhibition of PARP-1 enzymatic activity not only inhibited the poly(ADP-ribosyl)ation of FOXP3 but also regulated FOXP3 stabilization and its downstream genes, we decided to further examine the suppressive function of Treg cells in the standard Treg suppression cell co-culture system in vitro. For some of the cell culture groups, Treg cells were pretreated with the PARP-1 inhibitor 3-AB for 6 h before the suppression assay. Human PBMCs from healthy donors were labeled with CFSE before being cultured with human primary Treg cells at different ratios and stimulated with anti-CD3/CD28 beads for 4 days. The results showed that 3-AB-pretreated Treg cells exhibited a much stronger suppressive effect than untreated Treg cells on the proliferation of PBMCs (Fig. 6B), suggesting that the poly(ADP-ribosyl)ation of FOXP3 mediated by PARP-1 negatively regulates the suppressive function of Treg cells. We also used another PARP-1-specific inhibitor, AG14361, and found that AG14361 pretreated Treg cells had an increased suppressive function in vitro as well (Fig. 6C). All of these data strongly suggest that PARP-1 inhibitors promote the suppressive function of Treg cells through the inhibition of FOXP3 poly(ADP-ribosyl)ation.
FIGURE 6.
PARP-1 inhibitors regulate Treg cell suppressive function through inhibiting PARylated FOXP3. A, human primary Treg cells were cultured with the PARP-1 inhibitor 3-AB for 6 h. The mRNA levels of FOXP3 downstream genes in selected cells were examined by RT-PCR. *, p < 0.05; **, p < 0.02; ns, not significant (two-paired Student's t test); NT, no treatment. Data represent at least three independent experiments. B, in vitro suppression assay. Human primary Treg cells were treated with or without 10 mm PARP inhibitor 3-AB for 6 h prior to the suppression assay. The Treg cells were then cultured with CFSE-labeled human PBMCs and anti-CD3/CD28 beads at the indicated ratio for 4 days. Proliferation of CD8+ T effector cells was examined by flow cytometry. C, human primary Treg cells were treated with or without 5 μm PARP-1 inhibitor AG14361 for 12 h. Then Treg cells were cultured with CFSE-labeled human PBMCs and anti-CD3/CD28 beads at the indicated ratio for 4 days in the in vitro suppression assay. The proliferation of CD8+ T effector cells was examined by flow cytometry. Data represent at least two independent experiments.
Discussion
In this study, we demonstrated that PARP-1 interacted with human FOXP3 and promoted its poly(ADP-ribosyl)ation. We identified the zinc finger/Leu zipper domains of FOXP3 and the DNA-binding domain/BRCA1 C terminus of PARP-1 as crucial domains for the direct interaction between these two proteins. We also showed that the poly(ADP-ribosyl)ation of FOXP3 mediated by PARP-1 was inhibited by PARG or PARP-1 inhibitors. Most importantly, we found that reduced poly(ADP-ribosyl)ation of FOXP3 by PARP-1 inhibitor stabilized FOXP3, up-regulated the expression levels of FOXP3 downstream genes in Treg cells, and enhanced the suppressive function of Treg cells. Our results suggest that the poly(ADP-ribosyl)ation of FOXP3 mediated by PARP-1 negatively regulates the suppressive function of Treg cells.
Studies have reported that PARylation does not act alone but in concert with other posttranslational modifications (4). We found PARP-1 promoted FOXP3 poly-ubiquitination and degradation mediated by Stub1. In addition, the inhibition of PARP-1 enzymatic activity suppressed the poly-ubiquitination and degradation of FOXP3, suggesting that poly(ADP-ribosyl)ation of FOXP3 mediated by PARP-1 affected FOXP3 poly-ubiquitination and degradation. However, the relationship and mechanism between these two important posttranslational modifications still needs to be investigated further.
PARP-1 is an abundant nuclear protein with low enzyme activity that can be activated by DNA break, reactive oxygen species, and inflammation. The activation of PARP-1 results in the poly(ADP-ribosyl)ation of its target proteins, which leads to the modulation of target protein transcriptional activity, localization, and the creation of new protein interaction scaffolds (25). Our data show that PARP-1 regulates the suppressive function of Treg cells through FOXP3 poly(ADP-ribosyl)ation. However, the regulation of PARP-1 activity and the signal induces FOXP3 poly(ADP-ribosyl)ation in Treg cells are still unclear. Besides, PARP-1 takes part in several cellular processes, including cell death, transcriptional regulation, and inflammation. Additional studies will be needed to figure out how these diverse functions of PARP-1 are integrated and controlled in different cells.
Because of its diverse and important roles, a number of PARP-1 inhibitors are under clinical development for the treatment of cancer, such as iniparib (BSI-201), Olaparib (AZD-2281, oral) and veliparib (ABT-888, oral) (11, 12). Our study not only has implications in the developing PARP-1 inhibitors as potential agents for the treatment of autoimmune diseases but also shows the potential risk for cancer treatment.
In summary, here we uncovered the previously unrecognized molecular mechanism that PARP-1 regulated the suppressive function of Treg cells at a posttranslational modification level through poly(ADP-ribosyl)ation of FOXP3. Furthermore, more specific PARP-1 inhibitors will be required both as tools and therapeutics of autoimmune diseases on the basis of the role of PARP-1 in the immune system.
Author Contributions
X. L. designed, performed, and analyzed the experiments shown in Figs. 1–3 and 6. J. N. and S. W. designed, performed, and analyzed the experiments shown in Figs. 4 and 5. Z. C. purified the MBP-FOXP3 protein and constructed vectors for FOXP3 truncations. W. J. C., D. L., and H. H. provided technical assistance and helped with the preparation of the manuscript. B. L. conceived and coordinated the study, analyzed and interpreted data, and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by Chinese National Program on Key Basic Research Project Grants 2014CB541800, 2014CB541900, NSFC 81330072, 31300711, 31370863, 31170825, 31200646, and 31200647; Shanghai Key Grant 14JC1406100; and Shanghai Three-year Plan on Promoting TCM Development Grant ZY3-LCPT-2-1003. The authors declare that they have no conflicts of interest with the contents of this article.
- PARP
- poly(ADP-ribose) polymerase
- PAR
- poly(ADP-ribose)
- Treg cell
- regulatory T cell
- Ub
- ubiquitin
- PBMC
- peripheral blood mononuclear cell
- 3-AB
- 3-aminobenzamide
- PARG
- poly(ADP-ribose) glycohydrase
- MBP
- maltose binding protein
- CFSE
- carboxyfluorescein succinimidyl ester
- NFAT
- nuclear factor of activated T-cells
- WGR
- tryptophan-glycine-arginine.
References
- 1.Amé J. C., Spenlehauer C., and de Murcia G. (2004) The PARP superfamily. BioEssays 26, 882–893 [DOI] [PubMed] [Google Scholar]
- 2.Kiehlbauch C. C., Aboul-Ela N., Jacobson E. L., Ringer D. P., and Jacobson M. K. (1993) High resolution fractionation and characterization of ADP-ribose polymers. Anal. Biochem. 208, 26–34 [DOI] [PubMed] [Google Scholar]
- 3.Ruf A., Rolli V., de Murcia G., and Schulz G. E. (1998) The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol. 278, 57–65 [DOI] [PubMed] [Google Scholar]
- 4.Kraus W. L., and Lis J. T. (2003) PARP goes transcription. Cell 113, 677–683 [DOI] [PubMed] [Google Scholar]
- 5.D'Amours D., Desnoyers S., D'Silva I., and Poirier G. G. (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M. Y., Mauro S., Gévry N., Lis J. T., and Kraus W. L. (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 7.Kraus W. L. (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell. Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldinucci A., Gerlini G., Fossati S., Cipriani G., Ballerini C., Biagioli T., Pimpinelli N., Borgognoni L., Massacesi L., Moroni F., and Chiarugi A. (2007) A key role for poly(ADP-ribose) polymerase-1 activity during human dendritic cell maturation. J. Immunol. 179, 305–312 [DOI] [PubMed] [Google Scholar]
- 9.Hauschildt S., Scheipers P., Bessler W., Schwarz K., Ullmer A., Flad H. D., and Heine H. (1997) Role of ADP-ribosylation in activated monocytes/macrophages. Adv. Exp. Med. Biol. 419, 249–252 [DOI] [PubMed] [Google Scholar]
- 10.Valdor R., Schreiber V., Saenz L., Martínez T., Muñoz-Suano A., Dominguez-Villar M., Ramírez P., Parrilla P., Aguado E., García-Cózar F., and Yélamos J. (2008) Regulation of NFAT by poly(ADP-ribose) polymerase activity in T cells. Mol. Immunol. 45, 1863–1871 [DOI] [PubMed] [Google Scholar]
- 11.Curtin N. J. (2005) PARP inhibitors for cancer therapy. Expert. Rev. Mol. Med. 7, 1–20 [DOI] [PubMed] [Google Scholar]
- 12.Mabley J. G., Jagtap P., Perretti M., Getting S. J., Salzman A. L., Virág L., Szabó E., Soriano F. G., Liaudet L., Abdelkarim G. E., Haskó G., Marton A., Southan G. J., and Szabó C. (2001) Anti-inflammatory effects of a novel, potent inhibitor of poly (ADP-ribose) polymerase. Inflamm. Res. 50, 561–569 [DOI] [PubMed] [Google Scholar]
- 13.Sakaguchi S., Yamaguchi T., Nomura T., and Ono M. (2008) Regulatory T cells and immune tolerance. Cell 133, 775–787 [DOI] [PubMed] [Google Scholar]
- 14.Hori S., Nomura T., and Sakaguchi S. (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 [DOI] [PubMed] [Google Scholar]
- 15.Fontenot J. D., Gavin M. A., and Rudensky A. Y. (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 [DOI] [PubMed] [Google Scholar]
- 16.Bennett C. L., Christie J., Ramsdell F., Brunkow M. E., Ferguson P. J., Whitesell L., Kelly T. E., Saulsbury F. T., Chance P. F., and Ochs H. D. (2001) The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 [DOI] [PubMed] [Google Scholar]
- 17.Brunkow M. E., Jeffery E. W., Hjerrild K. A., Paeper B., Clark L. B., Yasayko S. A., Wilkinson J. E., Galas D., Ziegler S. F., and Ramsdell F. (2001) Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Lin F., Zhuo C., Deng G., Chen Z., Yin S., Gao Z., Piccioni M., Tsun A., Cai S., Zheng S. G., Zhang Y., and Li B. (2014) PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J. Biol. Chem. 289, 26872–26881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Samanta A., Song X., Iacono K. T., Bembas K., Tao R., Basu S., Riley J. L., Hancock W. W., Shen Y., Saouaf S. J., and Greene M. I. (2007) FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. U.S.A. 104, 4571–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Barbi J., Bu S., Yang H. Y., Li Z., Gao Y., Jinasena D., Fu J., Lin F., Chen C., Zhang J., Yu N., Li X., Shan Z., Nie J., Gao Z., Tian H., Li Y., Yao Z., Zheng Y., Park B. V., Pan Z., Zhang J., Dang E., Li Z., Wang H., Luo W., Li L., Semenza G. L., Zheng S. G., Loser K., Tsun A., Greene M. I., Pardoll D. M., Pan F., and Li B. (2013) The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 39, 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasta F., Laudisi F., Sambucci M., Rosado M. M., and Pioli C. (2010) Increased Foxp3+ regulatory T cells in poly(ADP-Ribose) polymerase-1 deficiency. J. Immunol. 184, 3470–3477 [DOI] [PubMed] [Google Scholar]
- 22.Zhang P., Maruyama T., Konkel J. E., Abbatiello B., Zamarron B., Wang Z. Q., and Chen W. (2013) PARP-1 controls immunosuppressive function of regulatory T cells by destabilizing Foxp3. PLoS ONE 8, e71590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lönn P., van der Heide L. P., Dahl M., Hellman U., Heldin C. H., and Moustakas A. (2010) PARP-1 attenuates Smad-mediated transcription. Mol. Cell. 40, 521–532 [DOI] [PubMed] [Google Scholar]
- 24.Gibson B. A., and Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell. Biol. 13, 411–424 [DOI] [PubMed] [Google Scholar]
- 25.Schreiber V., Dantzer F., Ame J. C., and de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell. Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]





