Background: The Jhd2 PHD finger is required for H3K4 demethylation, but how it contributes to chromatin binding is not known.
Results: Mutating two H2A residues impacts chromatin association and H3K4 demethylation by Jhd2.
Conclusion: The PHD finger-H2A interaction controls Jhd2 functions.
Significance: Histone H2A is a novel recognition target for the PHD finger, and it contributes to the chromatin binding dynamics, enzymatic activities, and transcriptional regulatory functions of Jhd2.
Keywords: gene transcription, histone, histone demethylase, histone methylation, histone modification, epigenetics, nucleosome, PHD finger, protein-protein interaction, transcription, H3K4 methylation
Abstract
Histone H3 lysine 4 (H3K4) methylation is a dynamic modification. In budding yeast, H3K4 methylation is catalyzed by the Set1-COMPASS methyltransferase complex and is removed by Jhd2, a JMJC domain family demethylase. The catalytic JmjC and JmjN domains of Jhd2 have the ability to remove all three degrees (mono-, di-, and tri-) of H3K4 methylation. Jhd2 also contains a plant homeodomain (PHD) finger required for its chromatin association and H3K4 demethylase functions. The Jhd2 PHD finger associates with chromatin independent of H3K4 methylation and the H3 N-terminal tail. Therefore, how Jhd2 associates with chromatin to perform H3K4 demethylation has remained unknown. We report a novel interaction between the Jhd2 PHD finger and histone H2A. Two residues in H2A (Phe-26 and Glu-57) serve as a binding site for Jhd2 in vitro and mediate its chromatin association and H3K4 demethylase functions in vivo. Using RNA sequencing, we have identified the functional target genes for Jhd2 and the H2A Phe-26 and Glu-57 residues. We demonstrate that H2A Phe-26 and Glu-57 residues control chromatin association and H3K4 demethylase functions of Jhd2 during positive or negative regulation of transcription at target genes. Importantly, we show that H2B Lys-123 ubiquitination blocks Jhd2 from accessing its binding site on chromatin, and thereby, we have uncovered a second mechanism by which H2B ubiquitination contributes to the trans-histone regulation of H3K4 methylation. Overall, our study provides novel insights into the chromatin binding dynamics and H3K4 demethylase functions of Jhd2.
Introduction
Histone H3 lysine 4 (H3K4)3 methylation plays an important role in transcriptional regulation (1, 2) and other cellular processes (3–7). H3K4 methylation occurs in three forms or degrees because the lysine ϵ-amino group can be mono-, di-, or tri-methylated (indicated as H3K4me1, H3K4me2, and H3K4me3, respectively). A distinct role for each of these three degrees of H3K4 methylation was demonstrated or proposed from genetic and/or genomics-based studies (8–11). H3K4me3 is enriched at transcriptional start sites from yeast to humans and is generally considered a mark of active or activated state of transcription (8, 12). Although the role for H3K4me1 in Saccharomyces cerevisiae or the budding yeast is not yet known, it is present at “poised” developmental enhancers in embryonic stem cells (13) and implicated in gene repression in mammals (14). H3K4me2 and H3K4me3 are found predominantly at transcriptionally active loci (15–17), but recent studies in budding yeast have also linked these marks to transcriptional repression (9, 18, 19). H3K4 methylation is a dynamic modification. In budding yeast, H3K4 methylation is catalyzed by the Set1 methyltransferase (20, 21), which is present in a multisubunit protein complex called COMPASS (22). The H3K4 methyl marks are removed by Jhd2, a JARID family JMJC domain-containing demethylase in budding yeast (23–26). Although the regulation and functions of Set1-COMPASS have been studied extensively (27), regulation of Jhd2 and its contributions to transcription have remained far from fully understood.
Jhd2 is not present in a stable protein complex in vivo (23). Hence, elements or components that regulate its functions might exist intrinsically within Jhd2 itself. Our study and those of others have shown that Jhd2 is regulated at its protein level by proteasomal degradation following polyubiquitination by the E3 ligase Not4 (26, 28). Furthermore, we showed that Not4 monitors the structural integrity of Jhd2, which is maintained by the interdomain interactions between the JmjN and JmjC catalytic domains (26). Jhd2 also contains a plant homeodomain (PHD) finger present between its catalytic JmjN and JmjC domains (Fig. 1A). We previously demonstrated that this PHD finger is required for the association of Jhd2 with chromatin (26). The PHD finger is an evolutionarily conserved domain present in many proteins involved in regulating chromatin structure and dynamics (29, 30). The PHD fingers in mammalian ING2 and BPTF were the first to be shown to bind the H3K4me3 mark and mediate chromatin association (31–33). Subsequently, other PHDs were reported to “read” or bind to unmodified or modified H3 or to acetylated H4 (29, 34–37). We previously also showed that the association of Jhd2 with chromatin is not dependent on its substrate (H3K4 methylation) or the H3 N-terminal tail (amino acids 2–28) (26). Therefore, how Jhd2 via its PHD finger binds to chromatin was not understood. Whereas H3K4 methylation is well connected to gene transcription, how Jhd2 contributes to this process is also not fully understood. Madhani and co-workers (38) have demonstrated a role for Jhd2 in regulating H3K4 methylation and transcription during yeast gametogenesis. They showed that during spore differentiation, Jhd2 activates protein-coding gene transcription by countering the transcriptional shut-off that normally occurs at this developmental stage. Apart from this study, target genes of Jhd2 and contributions of Jhd2 to their regulation have remained poorly defined.
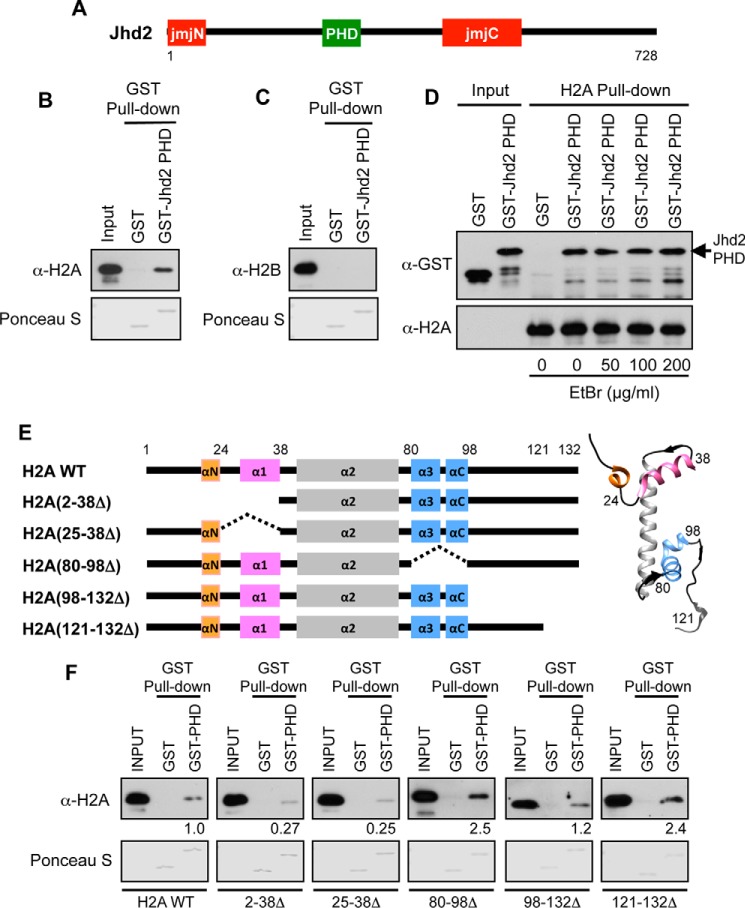
FIGURE 1.
The Jhd2 PHD finger interacts with H2A in vitro. A, schematic representation of Jhd2 with the catalytic JmjN and JmjC domains and the PHD is shown. Numbers denote the first and the last amino acid positions. B and C, recombinant GST alone or GST-tagged Jhd2 PHD finger was incubated with E. coli lysate containing recombinant histones H2A (B) or H2B (C) and subjected to pull-down using glutathione-conjugated beads. After extensive washing, the Jhd2 PHD finger-bound H2A or H2B was eluted in 1× Laemmli sample buffer. E. coli lysates (1%, Input) and eluates (40%) were subjected to Western blotting using α-H2A or α-H2B antibody. The top part of the blot was stained with Ponceau S to show equal loading of GST and GST-tagged PHD finger. D, the α-H2A antibody and protein A-conjugated beads were added to capture the recombinant H2A present in E. coli lysates. After extensive washing, the H2A-bound beads were incubated with the recombinant GST-tagged Jhd2 PHD finger in the absence or in the presence of the indicated concentrations of EtBr. As a negative control, GST alone was incubated with H2A-bound beads. The H2A-bound proteins were eluted from beads using Laemmli sample buffer. Purified GST or GST-Jhd2 PHD finger (1%, Input) and eluates (40%) were subjected to Western blotting using α-GST and α-H2A antibodies. E, schematic representations of histone H2A and its deletion mutants. Helices in H2A (αN, α1, α2, α3, and αC) are also shown. Right, crystal structure of yeast H2A (Protein Data Bank entry 1ID3) with helices colored as in the schematic on the left. F, recombinant GST alone or GST-tagged Jhd2 PHD finger was incubated with the E. coli lysates containing H2A or the deletion mutants, subjected to pull-down using glutathione-conjugated beads, and examined by Western blotting as described for A. Quantitations of the GST-Jhd2 PHD finger interaction with wild-type H2A and with deletion mutants are shown. Blots were quantified by densitometry using ImageJ. The signal for bead-bound histone was normalized to the signal for input control. The signal obtained for a truncated H2A is shown relative to the signal for H2A, which was set as 1.
In this study, we demonstrate an interaction between the Jhd2 PHD finger and histone H2A. Using mutational analyses, we have identified two residues in H2A, Phe-26 and Glu-57, as being important for the ability of Jhd2 to bind histone H2A in vitro and also for the chromatin association and H3K4 demethylase functions of Jhd2 in vivo. Using RNA-seq for gene expression analysis, we have identified common functional target genes for Jhd2 and the two H2A residues. We show that the H2A Phe-26 and Glu-57 residues mediate proper chromatin association and H3K4 demethylase functions of Jhd2 at target loci in vivo. Importantly, we also describe how access to these sites on H2A by Jhd2 is regulated by H2BK123 monoubiquitination, providing another mode by which H2BK123 monoubiquitination acts in the trans-histone regulation of H3K4 methylation via the control of both H3K4 demethylase functions as well as the H3K4 methyltransferase activity. Collectively, our study provides insights into the mechanism by which the Jhd2 H3K4 demethylase associates with chromatin and functions during transcriptional regulation.
Experimental Procedures
DNA Constructs
For recombinant H2A production in bacteria, mutations or deletions were introduced into HTA1 using PCR and mobilized as an NdeI-BamHI fragment to replace the wild-type sequence in H2A-pET11a (39). The H2A variant HTZ1 was PCR-amplified and mobilized into pET11a. The plasmid to express Myc epitope-tagged Jhd2 in yeast was described previously (26). Alanine substitution mutants of H2A created in HTA1-HTB1-pRS313 for expression in yeast were described previously (66). For the UBP8 γ-knock-out construct, ∼400-bp regions upstream of the UBP8 start codon and downstream of the stop codon were PCR-amplified. An overlap PCR strategy was used to combine these two UBP8 fragments and to include an intervening SacII site. The resulting 800-bp PCR product was cloned into the XbaI-SacI sites in YIPlac211. All constructs created using PCR-amplified products were verified by DNA sequencing.
Yeast Strains and Media
Yeast cells were grown in YPAD broth (1% yeast extract, 2% peptone, 2% dextrose, and 0.004% adenine hemisulfate) or in synthetic dropout media. Liquid broth supplemented with 2% agar was used to prepare solid media. Gene deletion strains were created by transforming PCR products containing the target locus and an inserted KanMX6 selection cassette amplified using genomic DNA isolated from the respective deletion mutant strains present in the Open Biosystem yeast gene knock-out collection. Epitope tagging of Jhd2 with 12 copies of V5 at its C terminus was performed by PCR amplification using pFA6a-12Pk-kanMX6 (40) as the template and subsequent transformation of the PCR product into FY406 strain. Histone mutations were mobilized and expressed in yeast strains derived from parental Y131 or FY406 strains using a standard plasmid shuffle approach. UBP10 was deleted in YMC85 by transforming a PCR product containing 500-bp regions upstream and downstream of the UBP10 ORF replaced with a NatMX4 selection module. This PCR product was amplified using genomic DNA from the UCC6398 strain (55). The UBP8 γ-knock-out construct described above was linearized with SacII and transformed into YMC88 to create the ubp8Δubp10Δ double mutant. Detailed genotypes of the yeast strains used in this study are listed in Table 1.
TABLE 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| FY406 | Mat a hta1-htb1Δ hta2-htb2Δ lys2-128δhis3Δ200 ura3-52 HTA1-HTB1 (CEN URA3) | Ref. 64 |
| HTA1WT | HTA1-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| HTA1L24A | hta1-L24A-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| HTA1T25A | hta1-T25A-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| HTA1F26A | hta1-F26A-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| HTA1P27A | hta1-P27A-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| HTA1E57A | hta1-E57A-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| HTA1L59A | hta1-L59A-HTB1 (CEN HIS3); derived from FY406 | Ref. 66 |
| Y131 | Mat a hta1-htb1Δ::LEU2, hta2-htb2Δ, leu2-2,-112, ura3-1, trp1-1, his3-11,-15 ade2-1, can1-100, ssd1, HTA1-HTB1 (2μ URA3) | Ref. 52 |
| YZS315 | HTA1-FLAG-HTB1 (CEN HIS3); derived from Y131 | Ref. 72 |
| YZS606 | HTA1-FLAG-HTB1 (CEN HIS3) ubp8Δ::KanMX6 ubp10Δ::NatMX4; derived from Y131 | Ref. 72 |
| FY120 | Mat a leu2Δ1 ura3 his4-912∂ lys2-128∂ | Ref. 75 |
| YZS613 | ubp8Δ::KanMX6 ubp10Δ::NatMX4; derived from FY120 | This study |
| YMH171 | Mat a ura3-52 leu2-3,112 his3 trp1Δ | Ref. 76 |
| YZS513 | sdc1Δ::KanMX6; derived from YMH171 | Ref. 26 |
| YZS515 | set1Δ::KanMX6; derived from YMH171 | Ref. 26 |
| YZS524 | sdc1Δ::KanMX6; derived from Y131 | This study |
| YZS652 | jhd2Δ::KanMX6; derived from FY406 | This study |
| YZS653 | sdc1Δ::KanMX6 HTA1-HTB1 (CEN HIS3); derived from YZS524 | This study |
| YZS654 | sdc1Δ::KanMX6 hta1-L24A-HTB1 (CEN HIS3); derived from YZS653 | This study |
| YZS655 | sdc1Δ::KanMX6 hta1-F26A-HTB1 (CEN HIS3); derived from YZS653 | This study |
| YZS656 | sdc1Δ::KanMX6 hta1-E57A-HTB1 (CEN HIS3); derived from YZS653 | This study |
| YMC64 | set1Δ::KanMX6 HTA1-HTB1 (CEN HIS3); derived from HTA1WT | This study |
| YMC70 | jhd2Δ::KanMX6 HTA1-HTB1 (CEN HIS3); derived from YZS652 | This study |
| YMC84 | Jhd2–12V5::KanMX6; derived from FY406 | This study |
| YMC85 | HTA1-HTB1 (CEN HIS3) Jhd2–12V5::KanMX6; derived from YMC84 | This study |
| YMC86 | hta1-F26A-HTB1 (CEN HIS3) Jhd2–12V5::KanMX6; derived from YMC84 | This study |
| YMC87 | hta1-E57A-HTB1 (CEN HIS3) Jhd2–12V5::KanMX6; derived from YMC84 | This study |
| YMC88 | ubp10Δ::NatMX4 Jhd2–12V5::KanMX6; derived from YMC85 | This study |
| YMC90 | ubp8Δ::URA3 ubp10Δ::NatMX4 Jhd2–12V5::KanMX6; derived from YMC88 | This study |
Western Blot Analysis
Crude nuclear extracts were prepared to examine H3 modifications following the cell fractionation procedure as described (41). To examine changes in the H2Bub1 levels, whole cell extracts were prepared by boiling 3 × 107 cells in 100 μl of 1× Laemmli sample buffer (Bio-Rad) for 10 min. After centrifugation (16,100 × g for 5 min), equal volumes of whole cell extracts were subjected to Western blot analysis using α-H2B (1:5000; Active Motif) and α-Pgk1 (1:5000; 22C5, Molecular Probes) antibodies. The following antibodies were purchased from Millipore unless specified otherwise and used in Western blotting (dilutions used and source are indicated in parentheses): α-H3K4me1 (1:1000), α-H3K4me2 (1:10,000), α-H3K4me3 (1:5000), and α-H3K14ac (1:5000) (Active Motif). The histone H3 loading was monitored using α-H3 (1:10,000; Abcam), and Jhd2 levels were examined using α-Myc (1:2000, clone 9E10) or α-V5 (1:1000; Invitrogen). Quantitation of Western blot signals was performed by densitometry using ImageJ (National Institutes of Health).
In Vitro GST Pull-down Assay
Plasmid constructs pFH54 (26) (to express glutathione-S-transferase (GST)-tagged Jhd2 PHD finger) or pBG101 (to express GST alone) were transformed into Escherichia coli strain BL21(DE3)-RIL (Agilent Technologies). Cells were grown in Luria broth medium supplemented with 0.15 mm ZnSO4 to an A600 of 0.6 at 37 °C, induced with 1 mm isopropyl-β-d-thiogalactopyranoside, and grown overnight at 16 °C before harvesting. Cell pellets were resuspended in NET buffer (25 mm Tris-Cl, pH 8.0, 50 mm NaCl, 5% glycerol, 0.1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) and sonicated. The lysate was then incubated on ice for 30 min following the addition of Triton X-100 (1%) to solubilize proteins. Recombinant GST-Jhd2 PHD finger or GST alone was purified using glutathione-Sepharose 4B beads following the manufacturer's instructions (GE Healthcare). Plasmid constructs to express wild-type HTB1, HTZ1, or the wild-type or mutant HTA1 were also introduced into the E. coli strain BL21 (DE3)-RIL. Cells were grown as described above, except they were induced with 0.1 mm isopropyl-β-d-thiogalactopyranoside for 5 h at 37 °C. E. coli cells expressing recombinant H2B, H2A, or H2A mutants were lysed in lysis/binding buffer (20 mm Tris-Cl, pH 7.5, 150 mm NaCl, 5% glycerol, 0.01% Nonidet P-40, 1 mm DTT, 1 mm PMSF, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) by sonication. Following centrifugation (16,100 × g for 15 min), the supernatant (lysate) was collected. Purified GST-tagged PHD fingers (15 μm) were incubated with the E. coli lysates containing H2B, H2A, or H2A mutants (10 μg of total protein) in 240 μl of lysis/binding buffer for 1 h at 4 °C. Incubation was continued for another 1 h at 4 °C after the addition of 10 μl glutathione-Sepharose beads to pull down GST alone or the GST-tagged Jhd2 PHD finger. Following four washes with a wash buffer (lysis/binding buffer containing 0.1% Nonidet P-40), the beads were boiled in 1× Laemmli sample buffer (Bio-Rad) for 10 min. Following centrifugation (16,100 × g for 5 min), the supernatant was subjected to Western blotting using α-H2B (1:5000) or α-H2A (1:5000; Active Motif) antibodies. The amount of GST-tagged proteins present in the binding reaction was monitored by Ponceau S staining.
In Vitro H2A Pull-down Assay
Lysates containing recombinant histone H2A (600 μg of total protein) from E. coli were prepared as described above and incubated with 10 μl of α-H2A antibody for 1 h at 4 °C. After the addition of 50 μl of Dynabeads® protein A (Invitrogen), incubation was continued for another 1 h at 4 °C to pull down the recombinant H2A. The H2A-bound beads were washed four times with the wash buffer as described above and then incubated with purified GST-Jhd2 PHD finger (5 μg) in 200 μl of lysis/binding buffer with 0, 50, 100, or 200 μg/ml ethidium bromide (EtBr) for 1 h at 4 °C. After four washes with the wash buffer, the beads were boiled in 1× Laemmli sample buffer (Bio-Rad) for 10 min. Following centrifugation (16,100 × g for 5 min), the supernatant was subjected to Western blot analysis using α-GST (1:40,000; GE Healthcare) and α-H2A (1:5000) antibodies.
Chromatin Immunoprecipitation (ChIP) Assay
Soluble chromatin was prepared from formaldehyde-cross-linked cells for ChIP of H3 or H3K4 methylation and dimethyl adipimidate/formaldehyde-double-cross-linked cells for ChIP of V5-tagged Jhd2 as described previously (41). Soluble chromatin was prepared from three independent cultures each for the Jhd2-12V5-containing strains (YMC85, YMC86, and YMC87) or the HTA1WT strain without V5-tagged Jhd2 (“no tag” control) and used in immunoprecipitation with α-V5 antibody. For H3K4 methylation, soluble chromatin was prepared from three independent cultures each of HTA1WT, HTA1F26A, HTA1E57A, and YMC70 strains, in addition to YMC64 (a set1Δ strain) used for background subtraction. An aliquot of the soluble chromatin was used to isolate the input DNA control. The following antibodies were used in a ChIP assay (volume used and source are indicated in parentheses): α-H3K4me1 (4 μl; Active Motif), α-H3K4me2 (1 μl; Millipore), α-H3K4me3 (1 μl; Millipore), α-H3 (1 μl; Abcam), and α-V5 (2 μl; Invitrogen). ChIP was performed essentially as described (41) except for modifications in the wash steps for ChIP of Jhd2-12V5. After immunoprecipitation with α-V5 antibody, the chromatin-bound α-V5-tag-mab-magnetic beads (MBL International Corp.) were washed twice with 1 ml of FA140 buffer (50 mm HEPES, pH 7.6, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mm PMSF, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin); once with FA1000 (FA140 buffer with 1 m NaCl); once with 1 ml of FA500 buffer (FA140 buffer with 500 mm NaCl); once with 1 ml of LiCl/Nonidet P-40 buffer (10 mm Tris·Cl, pH 8.0, 250 mm LiCl, 1 mm EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm PMSF, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin); and once with TE (10 mm Tris·Cl, pH 8.0, 1 mm EDTA, pH 8.0). ChIP DNA was eluted using 450 μl of ChIP elution buffer (25 mm Tris·Cl, pH 8.0, 2 mm EDTA, pH 8.0, 200 mm NaCl, and 0.5% SDS). ChIP elution buffer was also added to soluble chromatin set aside for input DNA control. 2 μl of Proteinase K (Roche Applied Science) was added, and reverse cross-linking was performed at 65 °C for 5 h. Input and ChIP DNA were then isolated using Qiagen PCR Cleanup Kit following the manufacturer's protocol. 2 μl of ChIP DNA (undiluted or 1:5 diluted) or input DNA (1:20 diluted) was the template in quantitative PCR performed in triplicate using a Bio-Rad CFX96 real-time PCR detection system and iQTM SYBR Green Supermix (Bio-Rad), which yielded the threshold cycles (CT). Primers sequence used in this study are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| SER3-PF (−298) | CTTCTCGTTCCCACCTAATTTCGC |
| SER3-PR (−127) | TTATCCTCTGCTCCCTCCTTCC |
| MET6 PF (−242) | GATAGATGCACTAATTTAAGGGAAG |
| MET6 PR (−38) | TTTCTTAGAGTGTTGTCAATGCTC |
| CAR1 PF (−250) | CGAATAGTCTCTAGCTCTTGCCC |
| CAR1 PR (−92) | CTTAGATCTTGATAGAAAGTGGGCC |
| SPL2-PRO-FOR | AATGCATGGCAGTTATGGCAATTAC |
| SPL2-PRO-FOR | AATGCATGGCAGTTATGGCAATTAC |
| GFD2-FOR | GTGGGTACATCCGTGCACCATTTCTG |
| GFD2-REV | AGCACCGGTATTACCGCTTCTGGAACG |
| NAT4-FOR | GCGGTGCGCGCGCGCCATTAAAAG |
| NAT4-REV | GGTGGTGAAGTTACCGTTTCCATCTG |
| PHO84-antisense-FOR | TCACAACCTTGTTGATCCCAGAAACTAAGAG |
| PHO84-antisense-REV | GGATGGCAGAGAGATGTGAGGAAATAATG |
| PHO84-sense-FOR | ATGCAACCACATTGCACACCGGTAATG |
| PHO84-sense-REV | ACCACCTTCGGTAAGGTGTTCTTTATGAAG |
| SHB17-antisense-FOR | TTGCACCGCAAGCACCAGAGTGAGGG |
| SHB17-antisense-REV | GGTTCGTCAATGTTGTGGTGAGCGTATG |
| DUR1-antisense-FOR | GCTCCTAAATCGGGTAAGATTATCAAGATTTG |
| DUR1-antisense-REV | GTTCAGCAAATTGAGAACATAACAAACACACG |
| BUB3-antisense-FOR | ACACGGCTGGCTCTGATGGCATAATTTC |
| BUB3-antisense-REV | CACGAATCTTCACGAAGATATCAGTTCTC |
| HOL1-antisense-FOR | GTATTTGCAATCTGTCAATTTGAGAGACGG |
| HOL1-antisense-REV | TTTCTTGGTACTGCTTCAGTCTTAGGAATGG |
ChIP Data Analysis
Data from Jhd2-12V5 ChIP experiments were analyzed as follows. The CT value for ChIP DNA was normalized to the CT value for the input DNA using the 2−ΔΔCT method (42). The resulting normalized 2−ΔCT values for each strain from three independent ChIP experiments were then averaged. The average 2−ΔCT value obtained for the “no tag” strain was then subtracted from the values obtained for the V5-tagged Jhd2-expressing strains, and the resulting background subtracted value is ChIP enrichment and is defined as Jhd2 occupancy. In general, we obtained ∼3–4-fold enrichment for Jhd2-12V5 in the wild-type H2A strain relative to the “no tag” strain. Jhd2 occupancy in each mutant strain was then normalized to the Jhd2 occupancy in the control wild-type H2A strain (set as 1). The resulting value, -fold change in Jhd2 occupancy in a mutant relative to the control, was plotted using Graphpad Prism version 6.0 software. S.E. values were calculated using the normalized 2−ΔCT values for each strain and from three independent ChIP experiments. They were further normalized to the Jhd2 occupancy value obtained for the control wild-type H2A strain and used within the graphs. For H3K4 methylation ChIP assays, the CT value for ChIP DNA from an antibody recognizing one of the H3K4 methyl marks (H3K4me1, H3K4me2, or H3K4me3) was normalized to the CT value for ChIP DNA obtained using α-H3 antibody, employing the 2−ΔΔCT method. The normalized values for each strain from three independent ChIP experiments were then averaged. The average 2−ΔCT value obtained for the set1Δ strain was subtracted from the average 2−ΔCT values obtained for the test strains in order to obtain the final ChIP enrichment value, termed H3K4 methyl occupancy. On average, we obtained >3-fold, >30-fold, and >100-fold enrichment for H3K4me1, H3K4me2, and H3K4me3, respectively, in the wild-type H2A strain relative to the set1Δ strain. The H3K4me1, H3K4me2, or H3K4me3 occupancy in each mutant was normalized to its occupancy in the control wild-type H2A (set as 1), in order to obtain the -fold change in the occupancy of an H3K4 methyl mark in a mutant relative to the wild type. The -fold change and the normalized S.E. values were calculated as described above and were plotted using GraphPad Prism version 6.0. Statistical significance was calculated using the Student's t test option available in Microsoft Excel.
RNA-seq Analysis
Three independent cultures for HTA1WT, HTA1F26A, HTA1E57A, and YMC70 strains were grown in YPAD medium to mid-log phase. 3 × 108 cells/culture were harvested and washed once with nuclease-free water (Sigma). Total RNA was isolated using the RiboPureTM-yeast kit (Life Technologies). To remove any residual DNA contamination, total RNA was treated with 2 μl of DNase I (TURBO DNA-freeTM kit; Life Technologies) for 45 min at 37 °C. RNA was digested once more using DNase I (RNase-free DNase set; Qiagen) for 10 min at room temperature before purification using the Qiagen RNeasy kit. Prior to library construction for RNA-seq, the quality of RNA was ascertained using a Bioanalyzer (Agilent), and the RNA integrity number for the samples ranged from 6.8 to 8.1. The Ribo-ZeroTM Magnetic Gold Kit (Yeast) (Epicenter) was used to remove ribosomal RNA from total RNA. Library construction was performed using the Illumina TruSeq RNA sample preparation kit version 2 and sequenced using the Illumina Hiseq2000 sequencer. For bioinformatics analysis, an S. cerevisiae genome index was initially created. The UCSC sacCer3 reference sequence and transcript table were downloaded from yeastgenome.org (S288C_reference_genome_R64-1-1_20110203). Standard differential expression was then performed using the open source USeq and DESeq analysis packages. Briefly, each replica data set was aligned to the genome and splice junctions using Novoalign. After alignment, splice junction coordinates were converted to genomic coordinates, counts for each gene were collected, variance was normalized, and differentially expressed genes between a mutant and control were identified using a negative binomial model with DESeq. Genes passing two thresholds, a false discovery rate of <5% and absolute log2 ratio of 0.585 (i.e. 1.5-fold change), were considered differentially expressed and were used in subsequent analysis. Read depth-normalized coverage was used for visualization in a genome browser to confirm changes in expression. Total counts, gene lists, and other relevant metrics associated with the RNA-seq analysis are included in supplemental Tables 1 and 2. Gene ontology enrichment analysis was performed using the GO Term Finder (version 0.83) available at the Saccharomyces Genome Database (supplemental Table 3).
Results
The N-terminal Region of Histone H2A Is Required for Its Interaction with the Jhd2 PHD Finger in Vitro
We previously showed that the chromatin binding and in vivo demethylase activities of Jhd2 are dependent on its PHD finger (26). We also showed that Jhd2 binds chromatin in the absence of H3K4 methylation (the substrate) and in the absence of the H3 N-terminal tail region (26). This led us to hypothesize that the Jhd2 PHD finger might interact with a histone(s) other than H3 in order to associate with chromatin. Hence, we tested the binding of Jhd2 PHD finger to histones using in vitro GST pull-down assays. Purified recombinant Jhd2 PHD finger was expressed as a GST-tagged fusion protein in bacteria and incubated with a bacterial lysate containing either yeast H2A or H2B before binding to glutathione beads. Our pull-down assays showed that the GST-Jhd2 PHD finger binds H2A (Fig. 1B) but not H2B (Fig. 1C). Also, GST-Jhd2 PHD finger did not bind Htz1 (the sole H2A variant in yeast) in our in vitro binding assays (data not shown). To confirm that the interaction between the Jhd2 PHD finger and H2A is not mediated by DNA, we performed the binding reaction in the presence of increasing amounts of ethidium bromide, a DNA intercalating agent known to disrupt protein-DNA interactions (43). The Jhd2 PHD finger bound to H2A even in the presence of high ethidium bromide concentrations (≥100 μg/ml) (Fig. 1D). Taken together, our results revealed a DNA-independent direct interaction between the Jhd2 PHD finger and histone H2A. To identify the region(s) in H2A required for its interaction with the Jhd2 PHD finger, we generated truncation mutants of H2A as shown in Fig. 1E. The ability or inability of H2A mutants to bind the Jhd2 PHD finger was then evaluated using GST pull-down assays. Deleting either amino acids 2–38 or amino acids 25–38 near the N-terminal region of H2A resulted in a >70% reduction in the binding of H2A to the GST-Jhd2 PHD finger (Fig. 1F). Thus, we identified a region in H2A, comprised of residues in helix 1 (α1) and the linker region between α1 and the N-terminal helix (αN), as being important for the interaction between H2A and the Jhd2 PHD finger.
Histone H2A Residues Phe-26 and Glu-57 Control Demethylase Functions of Jhd2 in Vivo
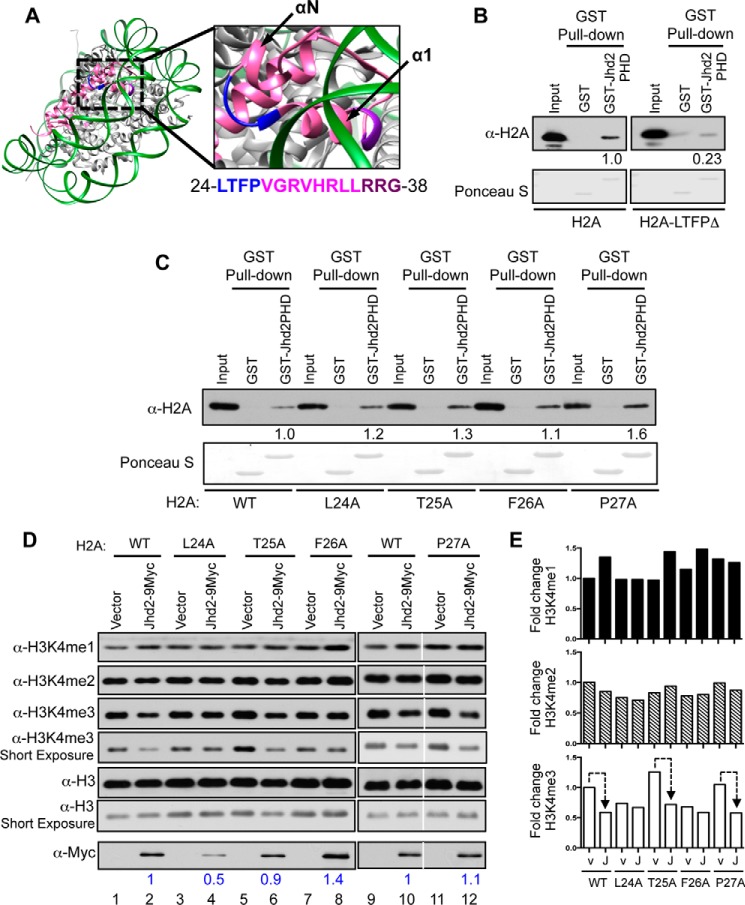
From the crystal structure of the yeast nucleosome (Fig. 2A), it appears that the H2A helix 1 (α1) is inaccessible for factor binding because it is located within the nucleosome. However, the linker region between helices α1 and αN is surface-accessible. We therefore asked whether this linker region mediates the H2A-Jhd2 PHD finger interaction. Hence, we deleted the four residues that make up this linker region (Leu-24, Thr-25, Phe-26, and Pro-27) and created the H2A-LTFPΔ mutant. We then tested the ability of this deletion mutant to interact with the Jhd2 PHD finger using a GST pull-down assay. The GST-Jhd2 PHD finger showed reduced binding to the H2A-LTFPΔ deletion mutant when compared with wild-type H2A (Fig. 2B). This finding confirmed the importance of the linker region to the in vitro interaction between histone H2A and the Jhd2 PHD finger.
FIGURE 2.
The Phe-26 residue in histone H2A is required for the removal of H3K4me3 marks by Jhd2 demethylase in vivo. A, crystal structure of yeast nucleosome (Protein Data Bank entry 1ID3) is shown together with an enlarged view of a region containing H2A residues involved in interacting with the Jhd2 PHD finger. H2A is shown in magenta; the surface-accessible loop region (residues 24LTFP27) is in blue; and the surface-inaccessible residues of the α1 helix (residues 36RRG38) are in purple. αN and α1 helices are indicated. B and C, recombinant GST alone or the GST-tagged PHD finger was incubated with E. coli lysate containing recombinant H2A or its truncation mutant lacking amino acids 24–27 (H2A-LTFPΔ) (B) or alanine substitution mutants (C) prior to pull-down using glutathione-conjugated beads. After extensive washing, the PHD finger-bound histone was eluted in sample buffer. E. coli lysates (1%, Input) and eluates were subjected to Western blotting using α-H2A antibody. The top part of the blot was stained with Ponceau S to show equal loading of GST and GST-tagged Jhd2 PHD finger. Densitometry signal for the bead-bound histones were normalized to the input control signals. Signal for H2A-LTFPΔ is shown relative to the signal for H2A, which was set as 1. D and E, Western blots for H3K4 methylation and Jhd2 were performed using nuclear extracts from yeast stains expressing either wild-type H2A or the indicated H2A mutant and transformed with an empty plasmid vector (pRS426; v) or a plasmid to overexpress Jhd2 C-terminally tagged with nine copies of the Myc epitope (Jhd2–9Myc; J). H3 served as a loading control. E, blots were quantified by densitometry using ImageJ. Signals for H3K4me1, H3K4me2, H3K4me3, and Jhd2–9Myc in each strain were normalized to the H3 signal. Graphs show -fold change in the normalized signal for an H3K4 methyl mark in various strains relative to that in the control H2AWT strain transformed with vector pRS426, which was arbitrarily set as 1. Dotted line, decrease in H3K4me3 levels upon Jhd2 overexpression. Normalized signals for Jhd2–9Myc in various H2A mutant strains are shown (in blue) relative to its signal in control H2A strain transformed with vector (set as 1).
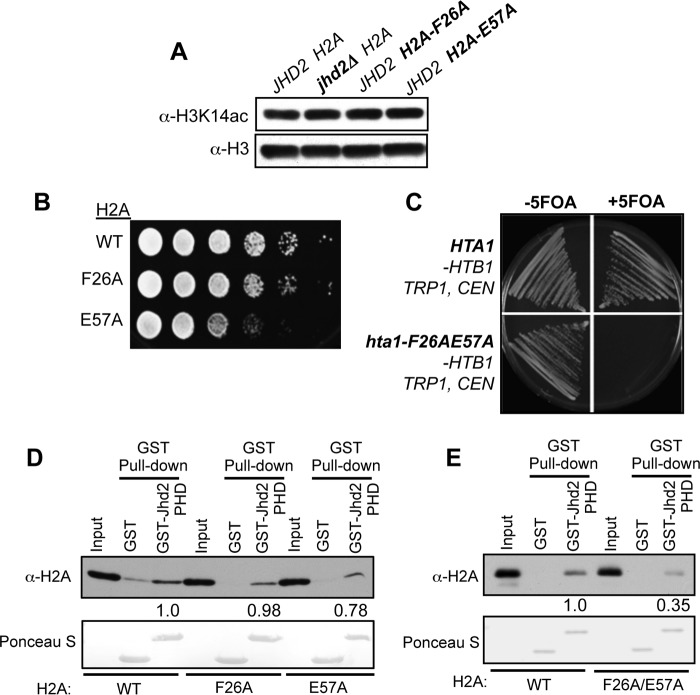
Next, we tested the contributions of individual linker region residues to the interaction between H2A and the Jhd2 PHD finger. Unlike the H2A-LTFPΔ deletion mutant, an alanine substitution mutation at each of the four linker region residues did not disrupt the histone H2A-Jhd2 PHD finger interaction (Fig. 2C). We then asked whether the linker region residues play any role in controlling the H3K4 demethylase functions of Jhd2 in vivo in yeast cells. Overexpression of Jhd2 in yeast cells leads to a decrease in global H3K4me3 levels (23–26). We used this decrease in bulk H3K4me3 levels following Jhd2 overexpression as an assay to measure the in vivo demethylase functions of Jhd2. To address our question, we overexpressed Jhd2 in yeast strains containing either the wild-type H2A or an H2A mutant with alanine substitution in one of the linker region residues. We also transformed wild-type H2A or H2A mutant strains with the plasmid vector as a control. We used Western blotting to then examine changes, if any, in bulk H3K4 methylation levels in extracts prepared from these strains. First, mutating any one of the four H2A linker region residues caused an alteration (i.e. increase or decrease) in the steady state levels of H3K4me3 when compared with the control wild-type H2A strain (Fig. 2, D and E, lanes labeled Vector), which suggested that the linker region residues are required for maintaining global H3K4me3 levels. Second, overexpression of Jhd2 in the H2A-T25A or H2A-P27A strain led to >50% reduction in bulk H3K4me3 levels when compared with these H2A mutant strains transformed with the plasmid vector alone (Fig. 2, D and E, lanes 5 and 6 and lanes 11 and 12). This is very similar to that observed upon Jhd2 overexpression in control strain with wild-type H2A (Fig. 2, D and E, lanes 1 and 2). Therefore, mutating either H2A Thr-25 or Pro-27 residue did not disrupt the H3K4 demethylase functions of Jhd2. On the other hand, overexpressing Jhd2 in the H2A-L24A or H2A-F26A strain showed no drastic reduction in bulk H3K4me3 levels, which is unlike what was observed upon overexpressing Jhd2 in the control wild-type H2A strain (Fig. 2D, compare lanes 3 and 4 and lanes 7 and 8 with lanes 1 and 2). Therefore, mutating H2A L24 or Phe-26 residue disrupted the ability of Jhd2 to remove H3K4me3 in vivo. To test whether these H2A mutations impair H3K4 demethylation by adversely affecting Jhd2 protein levels, we examined the levels of the Myc epitope-tagged Jhd2 in extracts prepared from yeast strains containing wild-type H2A or one of the H2A mutants. A 2-fold reduction in Jhd2–9Myc levels was observed in the H2A-L24A strain when compared with the control wild-type H2A strain (Fig. 2D), which explains the failure to observe a decrease in H3K4me3 upon Jhd2 overexpression in the H2A-L24A mutant. However, overexpressed Jhd2 protein levels were not reduced in the H2A-F26A strain (Fig. 2D). Therefore, the inability of overexpressed Jhd2 to decrease H3K4me3 levels in the H2A-F26A strain suggested that the H2A Phe-26 residue is required for the removal of the H3K4me3 marks by Jhd2 in vivo.
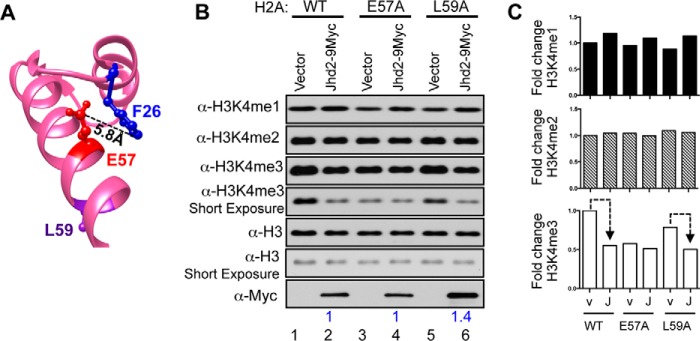
From the yeast nucleosome structure, it is apparent that the H2A Glu-57 residue is in the vicinity of the H2A Phe-26 residue (Fig. 3A). This spatial proximity prompted us to speculate that the H2A Glu-57 residue might also play a role in Jhd2-mediated H3K4 demethylation. Because the H2A Leu-59 residue resides on the opposite surface of the helix away from the H2A Glu-57 residue (Fig. 3A), we presumed that the H2A Leu-59 residue might not impact Jhd2 recruitment and could serve as a negative control. We then tested whether mutating the H2A Glu-57 residue could also adversely affect the demethylase functions of Jhd2 akin to the H2A-F26A mutation. Western blotting showed that the H2A-E57A and H2A-L59A mutations themselves led to a decrease in bulk H3K4me3 levels without overexpression of Jhd2 (Fig. 3, B and C, compare lanes 3 and 5 with lane 1). This result suggested that the H2A Glu-57 and Leu-59 residues also contribute to the maintenance of global H3K4me3 levels in addition to the H2A linker region residues (Fig. 2D). Overexpression of Jhd2 in the H2A-E57A strain did not reduce the global H3K4me3 level, which was otherwise reduced upon Jhd2 overexpression in both control wild-type H2A and the mutant H2A-L59A strains (Fig. 3, B and C). Therefore, mutating the H2A Glu-57 residue, but not the Leu-59 residue, adversely affected the removal of H3K4me3 by Jhd2. Moreover, global Jhd2 levels were not decreased in the H2A-E57A mutant (Fig. 3B). Taken together, these results showed that the H2A Glu-57 residue is also required for the removal of H3K4me3 marks by Jhd2 in vivo in addition to the H2A Phe-26 residue.
FIGURE 3.
Histone H2A Glu-57 residue is required for the removal of H3K4me3 marks by Jhd2 demethylase in vivo. A, ball-and-stick representations for H2A Phe-26, Glu-57, and Leu-59 residues are shown and colored in blue, red, and purple, respectively. The distance between the Phe-26 and Glu-57 residues calculated using the UCSF Chimera software is indicated. B and C, Western blot analysis of H3K4 methylation and Jhd2–9Myc (B) and quantitation (C) were performed as described in the legend to Fig. 2. Dotted line, decrease in H3K4me3 upon overexpressing Jhd2.
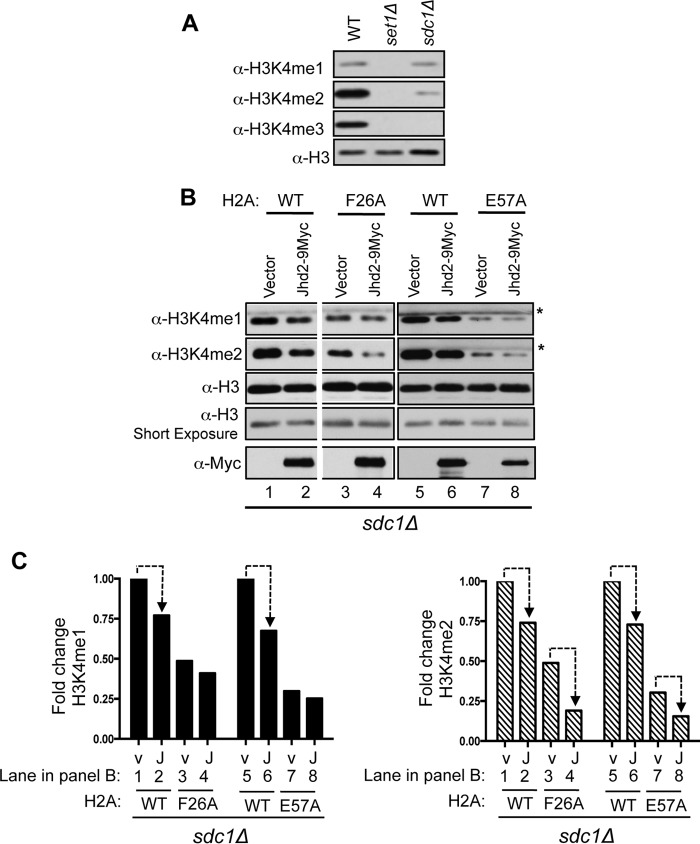
The absence of Sdc1 (a Set1-COMPASS regulatory subunit) results in cells lacking any detectable H3K4me3 and containing very low levels of H3K4me1 and H3K4me2 (Fig. 4A). As reported previously (26) and also shown in Fig. 4, B and C (lanes 1 and 2), a decrease in H3K4me1 and H3K4me2 levels is evident upon overexpressing Jhd2 in the strain lacking Sdc1 (sdc1Δ), indicating that Jhd2 also targets these two H3K4 methyl marks in addition to H3K4me3. We next asked whether the H2A Phe-26 and Glu-57 residues contribute to the removal of H3K4me1 and H3K4me2 marks by Jhd2 in vivo. To address this question, we transformed the sdc1Δ strain containing wild-type H2A or a mutant H2A (H2A-F26A or H2A-E57A) with either the plasmid vector alone or the plasmid to overexpress Jhd2. As shown in Fig. 4, B and C, H3K4me2 levels were reduced following Jhd2 overexpression in sdc1Δ strain containing either H2A-F26A or H2A-E57A mutant, similar to overexpression of Jhd2 in the control sdc1Δ/H2A strain (Fig. 4, B and C). On the other hand, H3K4me1 levels were not reduced in these strains, suggesting a lack of Jhd2 function (Fig. 4, B and C). Collectively, these findings revealed that the H2A Phe-26 and Glu-57 residues are required for the Jhd2-mediated removal of H3K4me1 but not H3K4me2. Taken together, our results showed that the H2A Phe-26 and Glu-57 residues are required for the enzymatic activities of Jhd2 toward H3K4me1 and H3K4me3 methyl marks in vivo.
FIGURE 4.
Histone H2A Phe-26 and Glu-57 residues contribute to the removal of H3K4me1 marks by Jhd2 demethylase in vivo. A, Western blot analysis of H3K4 methylation using nuclear extracts prepared from control wild-type, set1Δ, and sdc1Δ strains. B and C, Western blots showing H3K4me1, H3K4me2, and Jhd2 levels in crude nuclear extracts prepared from sdc1Δ strain expressing wild-type H2A or the H2A mutant and transformed with either an empty vector (pRS426) or a plasmid to overexpress Jhd2–9Myc. H3 serves as a loading control. *, a cross-reacting protein. C, blots were quantified by densitometry using ImageJ. Signals for H3K4me1 and H3K4me2 in each strain were normalized to the H3 signal. Graphs show -fold change in the normalized signals for H3K4me1 or H3K4me2 mark in various strains relative to the control sdc1Δ/H2AWT strain transformed with vector pRS426 (arbitrarily set as 1). Dotted line, decrease in H3K4me1 or H3K4me2 upon Jhd2 overexpression.
Histone H2A Residues Phe-26 and Glu-57 Mediate the in Vitro Interaction of Jhd2 with H2A
We then addressed how H2A residues Phe-26 and Glu-57 might control the enzymatic functions of Jhd2 in vivo. Acetylation of H3K14 (H3K14ac) was reported to negatively regulate the H3K4 demethylase activity of Jhd2 (44). However, bulk H3K14ac levels remained unchanged in the H2A-F26A and H2A-E57A mutants when compared with the control (Fig. 5A). This result suggested that the H2A-F26A and H2A-E57A mutations impair the in vivo demethylase functions of Jhd2 by some other mechanism and not via increasing H3K14ac. Whereas the H2A-F26A and H2A-E57A mutations conferred a subtle or a severe slow growth to yeast cells, respectively (Fig. 5B), the H2A-F26A/E57A double mutation rendered yeast cells inviable (Fig. 5C). Therefore, we were unable to examine H3K14ac levels in the H2A-F26A/E57A double mutant.
FIGURE 5.
Mutating H2A Phe-26 and/or Glu-57 residues alters yeast growth and the H2A-Jhd2 PHD finger interaction in vitro. A, crude nuclear extracts prepared from the indicated strains were analyzed by Western blotting using α-H3K14 acetyl antibody. H3 serves as the loading control. B, 10-fold serial dilutions of yeast cultures were spotted on YPAD medium and grown at 30 °C for 2 days. C, histone shuffle strain (Y131) harboring HTA1-HTB1, URA3 CEN was transformed with either HTA1-HTB1, TRP1 CEN or hta1-F26A/E57A-HTB1, TRP1 CEN plasmid and then streaked onto synthetic complete medium supplemented with or without 5-fluoro-orotic acid (5-FOA) for negative selection of the URA3-containing plasmid. D and E, in vitro GST pull-down assays were performed as described for Figs. 2 and 3, except the E. coli lysate contained wild-type H2A or the single (D) or double mutant (E) (H2A-F26A, H2A-E57A, and H2A-F26A/E57A). Densitometry signals for the bead-bound histones were normalized to the signals from the input control. -Fold change in the signal for H2A-F26A/E57A is shown relative to the signal for H2A (set as 1).
To test whether the Phe-26 and Glu-57 residues play any role in the H2A-Jhd2 PHD finger interaction, we returned to the GST pull-down assays. We found that the GST-Jhd2 PHD finger bound to the H2A-F26A mutant similarly to wild-type H2A but showed a slightly reduced binding to the H2A-E57A mutant (Fig. 5D). However, the GST-Jhd2 PHD finger interaction with the H2A-F26A/E57A double mutant was severely reduced (>60%) when compared with its interaction with wild-type H2A (Fig. 5E). Therefore, both H2A Phe-26 and Glu-57 residues are required to mediate the in vitro interaction between histone H2A and the Jhd2 PHD finger.
Deleting Jhd2 or Mutating the H2A Phe-26 or Glu-57 Residue Alters Transcription at a Common Set of Target Genes
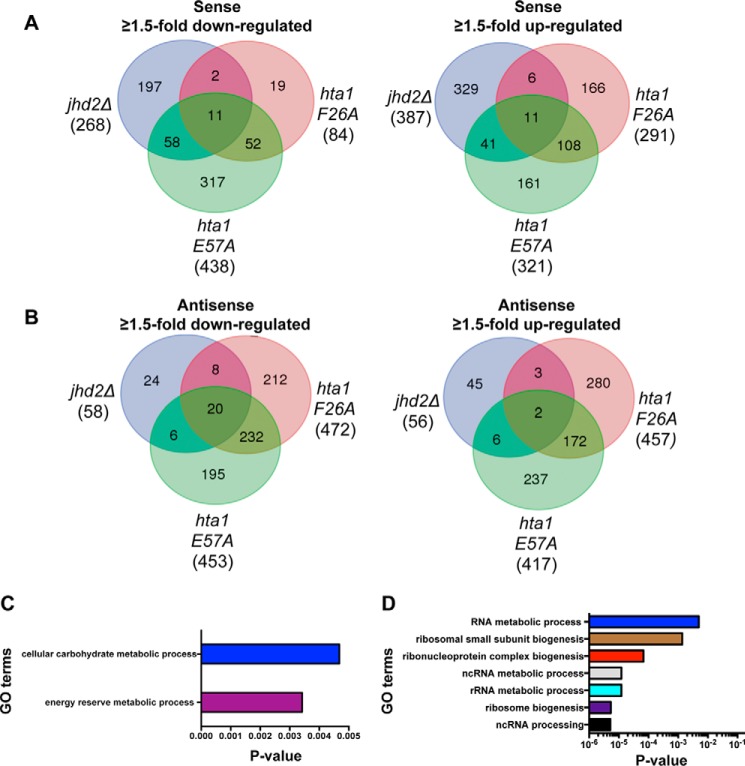
Regulation and functions of H3K4 methylation are well connected to gene transcription (6). It is therefore conceivable that the H2A Phe-26 or Glu-57 residue might control the chromatin association and H3K4 demethylase functions of Jhd2 during gene transcription. We hypothesized that mutating either the H2A Phe-26 or Glu-57 residue might impact genes whose transcriptional regulation is dependent on Jhd2. To test this possibility, we set out to identify the functional target genes of Jhd2 and the two H2A residues (Phe-26 or Glu-57). We performed a global differential gene expression analysis, where we sequenced in a strand-dependent fashion RNA isolated from three independent cultures for strains containing wild-type H2A (control), a mutant H2A allele (H2A-F26A or H2A-E57A), or the null allele for Jhd2 (jhd2Δ). By comparing with wild-type expression and using dual threshold ≥1.5-fold change (up or down) and a false discovery rate of <5%, we identified 769, 1304, and 1629 transcripts that were differentially expressed in jhd2Δ, H2A-F26A, and H2A-E57A strains, respectively (Fig. 6, A and B) (supplemental Tables 1 and 2). In addition to sense transcription, we also extended our analysis to the antisense transcription of annotated genes, which revealed that the expression of a greater number of sense transcripts was altered than antisense transcripts in jhd2Δ strain, suggesting that Jhd2 predominantly controls sense transcription in yeast (Fig. 6, A and B). Corresponding well with its adverse effects on yeast cell growth (Fig. 5B), a greater number of sense transcripts were impacted in the H2A-E57A mutant than in the H2A-F26A and jhd2Δ mutants (Fig. 6A). A greater number of antisense transcripts were affected in the histone mutants than in jhd2Δ (Fig. 6B), implicating a role for the H2A Phe-26 and Glu-57 residues in antisense transcription. Taken together, our gene expression analysis uncovered the functional roles for Jhd2 and histone H2A during the regulation of sense and antisense transcription in yeast.
FIGURE 6.
Transcriptional regulation at a common set of target genes is altered in the H2A-F26A, H2A-E57A, and jhd2Δ mutants. A and B, total RNA isolated from control wild-type H2A, H2A-F26A, H2A-E57A, and jhd2Δ strains was subjected to next generation sequencing. The total number of sense or antisense transcripts showing ≥1.5-fold change (up- or down-regulation at a false discovery rate of ≤5%) in the indicated mutant compared with the control H2A strain is shown in parentheses. The number of sense or antisense transcripts whose expression is increased or decreased in any one mutant or in any two mutants or in all three mutants is shown within intersections in the Venn diagrams. C, GO enrichment analysis of down-regulated sense transcripts common to the H2A-E57A and jhd2Δ mutants. D, GO enrichment analysis of up-regulated sense transcripts common to the H2A-E57A and jhd2Δ mutants.
To confirm whether the genes identified as differentially expressed were shared between the three mutants, we performed an intersection of the gene lists. We found that the affected genes in jhd2Δ were most commonly shared with the H2A-E57A mutant (Fig. 6, A and B). About 25% of sense transcripts and ∼50% of genic antisense transcripts were commonly down-regulated in jhd2Δ and H2A-E57A mutants. Furthermore, 13% of sense transcripts and 14% of antisense transcripts were commonly up-regulated in jhd2Δ and the H2A-E57A mutants. We performed a gene ontology (GO) analysis to identify the biological processes associated with these shared transcripts. The down-regulated sense transcripts common to jhd2Δ and H2A-E57A strains were enriched for genes governing energy reserve metabolic process (p = 0.003) and carbohydrate metabolic process (p = 0.004) (Fig. 6C). The up-regulated sense transcripts shared between jhd2Δ and H2A-E57A strains were significantly enriched for genes governing ribosome biogenesis (p < 10−5) (Fig. 6D). These findings therefore suggested that the transcriptional regulation at certain yeast genes, such as those controlling energy reserve metabolism or ribosome biogenesis, is highly dependent on Jhd2 and the H2A Glu-57 residue.
We then identified up or down-regulated transcripts that were shared between all three mutants. We found 11 sense transcripts with increased expression and another 11 sense transcripts with reduced expression in the jhd2Δ, H2A-F26A, and H2A-E57A strains when compared with the wild-type control (Fig. 6A). Additionally, antisense expression of 2 and 20 genes were increased and decreased, respectively, in all three mutants compared with the wild-type control (Fig. 6B). GO analysis of the up-regulated sense or antisense genes common to all three mutants showed no enrichment for any particular biological process. However, the shared down-regulated sense transcripts were enriched for genes involved in carbohydrate metabolic process (p = 0.03) and also contained genes involved in serine (SER3), methionine (MET6), or arginine (CAR1) metabolism (supplemental Table 3). GO analysis of the shared down-regulated antisense transcription showed that they were enriched for genes governing the fatty acid metabolic processes (p = 0.028) (supplemental Table 3). Collectively, our gene expression studies revealed that Jhd2 and the two H2A residues (Phe-26 and Glu-57) together contribute to either positive or negative regulation of transcription at a subset of yeast genes.
Mutating H2A Phe-26 or Glu-57 Residue Alters Jhd2 Occupancy on Chromatin at the Target Genes
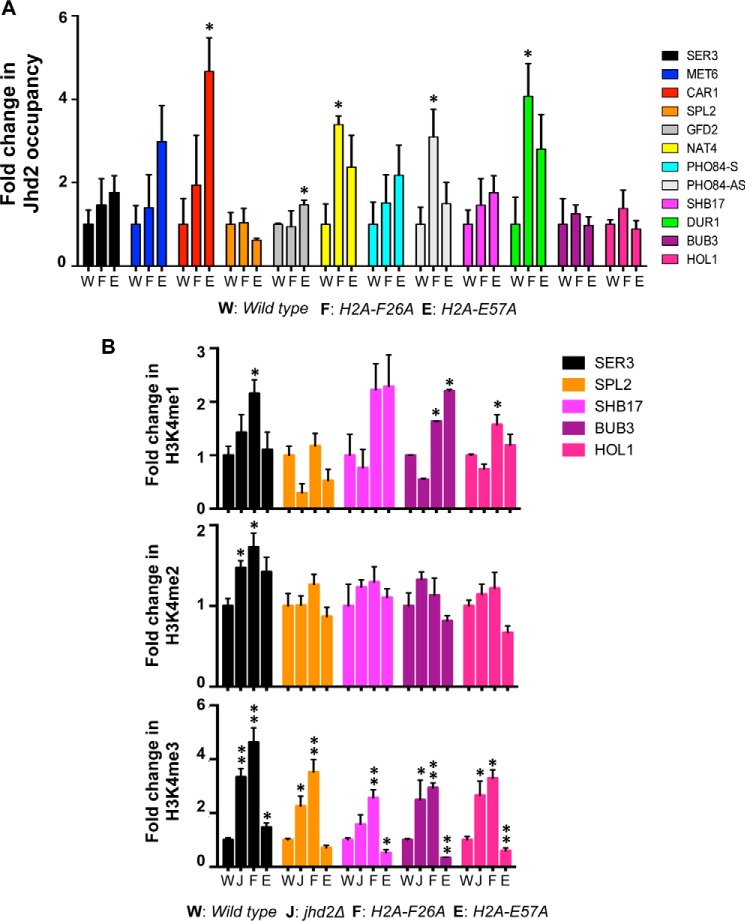
Next, we asked whether the H2A Phe-26 and Glu-57 residues control Jhd2 chromatin association at their shared functional target genes. While enrichment of Jhd2 on chromatin is not robust enough for genome-wide analysis, possibly due to the transitory nature of its interaction with nucleosomes, it is nevertheless sufficient for targeted ChIP-PCR analysis. We evaluated Jhd2 occupancy in control and H2A mutant strains at the following 11 candidate target loci, where transcript levels were either up- or down-regulated in jhd2Δ and the two H2A mutants: SER3, MET6, and CAR1 (sense transcription down-regulated); SPL2, GFD2, and NAT4 (sense transcription up-regulated); PHO84-AS, SHB17/YKR043C, and DUR1 (antisense transcription down-regulated); and BUB3 and HOL1 (antisense transcription up-regulated). Whereas PHO84 antisense transcripts were decreased in jhd2Δ and H2A-E57A strains, its sense transcript levels were increased in these two mutants (supplemental Table 1). Hence, we examined Jhd2 occupancy at the 5′-end of the PHO84 gene (PHO84-S) in addition to its 3′-end (PHO84-AS).
Our ChIP assays showed that Jhd2 occupancy was significantly increased at NAT4, PHO84-AS, and DUR1 genes, but remained mostly unchanged at other loci, in the H2A-F26A mutant when compared with the control strain (Fig. 7A). Jhd2 occupancy was significantly increased at the CAR1 and GFD2 genes but generally remained unchanged at many other target loci in the H2A-E57A mutant when compared with the control (Fig. 7A). Whereas the no change in chromatin-bound Jhd2 occupancy at target loci in the H2A-F26A mutant agrees well with the normal in vitro binding between the Jhd2 PHD finger and the H2A-F26A mutant (Fig. 5D), the no change or increased Jhd2 occupancy on chromatin in the H2A-E57A mutant differs from the decreased in vitro interaction between the Jhd2 PHD finger and H2A-E57A mutant. One explanation for this apparent difference is that the in vivo interaction of Jhd2 with chromatin is probably mediated by other factors that are not present in the in vitro binding assays. Another explanation is the molecular context of the substrate used in the in vitro and in vivo assays: free histone versus chromatin, which is similar to previous reports for other chromatin modifiers (45). Because the H2A-F26A/E57A double mutation severely reduces the in vitro H2A-Jhd2 PHD finger interaction, it is conceivable that the combined mutation of H2A Phe-26 and Glu-57 residues might drastically reduce Jhd2 occupancy on chromatin at target loci. However, we were unable to test this possibility, due to the lethality conferred by the H2A-F26A/E57A double mutation to yeast cells (Fig. 5C). Nevertheless, results from ChIP assays showed that the H2A Phe-26 and Glu-57 residues are required for the proper association of Jhd2 with chromatin at target genes.
FIGURE 7.
H2A-F26A and H2A-E57A mutations impact chromatin-bound occupancy and H3K4 demethylase functions of Jhd2 at target genes. A, Jhd2-12V5 occupancies at the indicated target genes in control wild-type strain or the two H2A mutants (H2A-F2A or H2A-E57A) were examined by a ChIP assay using α-V5 antibody. Following ChIP and quantitative PCR, the ChIP signal (i.e. V5 immunoprecipitation/input value) obtained from a control strain without V5-tagged Jhd2 (background) was subtracted from the ChIP signal obtained from strains expressing Jhd2-12V5, and the resulting difference was defined as Jhd2 occupancy. The graph shows -fold change in Jhd2 occupancy at a given locus in the H2A-F2A or H2A-E57A mutant relative to Jhd2 occupancy in the control strain, which was set as 1. Error bars, S.E. values obtained from three independent ChIP experiments. Statistical significance was calculated using Student's t test. *, p < 0.05. B, occupancies of H3K4me1, H3K4me2, and H3K4me3 at the indicated target loci in control wild-type H2A, jhd2Δ, H2A-F26A, and H2A-E57A strains were examined with ChIP assays using modification-specific antibodies. ChIP signals for α-H3K4me1, α-H3K4me2, or α-H3K4me3 antibodies obtained in set1Δ strain without any H3K4 methylation were subtracted from the ChIP signal obtained for these antibodies in the four test strains. Background-subtracted ChIP signals for the H3K4 methyl marks were further normalized to the ChIP signal obtained for histone H3, and the resulting value was defined as the H3K4 methyl mark occupancy. -Fold change in the normalized H3K4me1, H3K4me2, or H3K4me3 occupancy in a mutant is shown relative to its occupancy in the control wild-type strain (set as 1). Error bars, S.E. values obtained from three independent ChIP experiments. *, p < 0.05; **, p < 0.001.
H2A-F26A and H2A-E57A Mutations Alter H3K4 Methylation Levels at the Target Genes
We then asked whether H2A Phe-26 and Glu-57 residues control the H3K4 demethylase functions of Jhd2 at their shared functional target genes. Therefore, we used ChIP assays to measure the occupancies of H3K4me1, H3K4me2, and H3K4me3 at five candidate target genes in jhd2Δ, H2A-F26A, and H2A-E57A strains in addition to the control wild-type strain. Occupancies of H3K4me2 remained unchanged at most target loci in all three mutants when compared with the control (Fig. 7B), which is line with the observation that the H2A-F26A and H2A-E57A mutations do not alter the ability of Jhd2 to remove bulk H3K4me2 mark (Figs. 2D, 3B, 4B). However, at the SER3 gene, occupancy of H3K4me2 is increased in both H2A-F26A and H2A-E57A mutants similar to jhd2Δ when compared with its occupancy in the control strain (Fig. 7B). These results suggested that the H2A Phe-26 and Glu-57 residues in general do not impact Jhd2-mediated removal of H3K4me2, except at certain specific target loci.
Whereas overexpression of Jhd2 decreases global H3K4me3 and increases H3K4me1 (Fig. 2, D and E, lanes 1 and 2), deletion of JHD2 increases H3K4me3 but decreases bulk H3K4me1 (26). These results suggest that Jhd2 might preferentially target H3K4me3 and convert it to H3K4me1. However, we previously showed that the H3K4me1 is also targeted by Jhd2 (26). Our studies indicate a role for the H2A Phe-26 and Glu-57 residues in the Jhd2-mediated demethylation of the H3K4me1 mark, because mutating these H2A residues inhibits Jhd2 from removing H3K4me1 in vivo (Fig. 4B). Consistent with this finding, our ChIP assays showed a general increase in H3K4me1 occupancy at most target genes in the H2A-F26A mutant (Fig. 7B). Additionally, an increase in H3K4me1 occupancy at target genes, barring the SER3 and SPL2 genes, was also observed in the H2A-E57A mutant (Fig. 7B). Therefore, these results together showed that the H2A Phe-26 and Glu-57 residues contribute to the demethylase activity of Jhd2 toward the H3K4me1 mark during transcriptional regulation at target genes.
Agreeing well with the increase in global H3K4me3 in the absence of Jhd2, our ChIP assays showed an increase in H3K4me3 occupancy at target loci in jhd2Δ when compared with the control (Fig. 7B). Global H3K4me3 levels are reduced in the H2A-E57A mutant (Fig. 2E), and in line with this finding, occupancy of H3K4me3 is decreased at all target genes, except SER3, in the H2A-E57A mutant when compared with the control (Fig. 7B). Interestingly, H3K4me3 occupancy was increased at the SER3 gene in the H2A-E57A mutant, suggesting that the H2A Glu-57 residue controls the ability of Jhd2 to remove H3K4me3 at this particular gene. H3K4me3 occupancy was significantly increased at all target loci tested in the H2A-F26A mutant when compared with the control, and this increase is strikingly similar to that observed in the jhd2Δ strain (Fig. 7B). The increase in H3K4me3 occupancy is not due to any enhanced transcription, because transcript levels for SER3 and SHB17 are in fact attenuated in the jhd2Δ, H2A-F26A, and H2A-E57A mutants (supplemental Table 1). Collectively, our ChIP results demonstrated that the H2A Phe-26 and Glu-57 residues control the demethylase functions of Jhd2 in removing the H3K4me3 mark during transcriptional regulation.
In our ChIP assays, the H2A/F26A or H2A/E57A mutation resulted in an increase in Jhd2 occupancy at certain loci (Fig. 7A). It is possible that in the absence of one of the binding sites, Jhd2 perhaps associates very well with the other available binding site or with a binding site elsewhere on chromatin. Nevertheless, the increase in H3K4 methyl marks at several target loci in the H2A/F26A or H2A/E57A mutants clearly indicates that the H3K4 demethylase functions of Jhd2 are disrupted despite its ability to better associate with chromatin. Taken together, these findings suggest that the H2A Phe-26 and Glu-57 residues mediate the proper association of Jhd2 with chromatin and control its H3K4 demethylase functions during positive or negative regulation of transcription.
H2BK123 Ubiquitination Restricts Chromatin Association and H3K4 Demethylase Functions of Jhd2
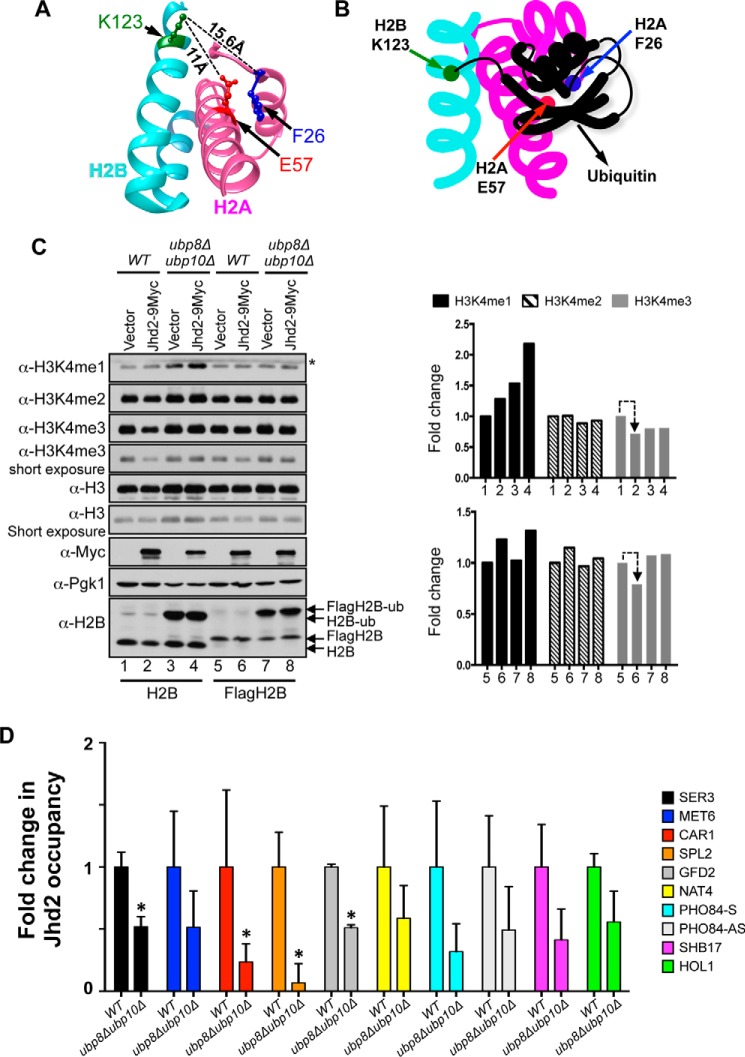
In a well studied trans-histone cross-talk, H2BK123 monoubiquitination (H2Bub1) is required for the Set1-COMPASS-mediated H3K4me2 and H3K4me3 and for the high levels of H3K4me1 (46–51). H2Bub1 is a dynamic histone modification; Rad6 and Bre1, the E2 conjugating enzyme and E3 ligase, respectively, conjugate ubiquitin onto the H2B Lys-123 residue (46, 52–54), and this conjugated ubiquitin is then removed by deubiquitinases, Ubp8 and Ubp10 (55–59). The two H2A residues (Phe-26 and Glu-57) that mediate the Jhd2 PHD finger-H2A interaction are present on the same surface-accessible phase of the nucleosome as the H2B Lys-123 residue (Fig. 8A). We therefore hypothesized that the bulky ubiquitin moiety conjugated to the H2B Lys-123 residue might sterically hinder Jhd2 from gaining access to the H2A Phe-26 and Glu-57 residues (Fig. 8B) and thereby impinge on its H3K4 demethylase functions.
FIGURE 8.
Ubp8 and Upb10, the H2B deubiquitinases, control chromatin association and H3K4 demethylase functions of Jhd2. A, structures of H2A (magenta) and H2B (cyan) in the yeast nucleosome are shown together with the H2B Lys-123 residue (green), the site ubiquitination, and the H2A Phe-26 (blue) and Glu-57 (red) residues, the binding sites for Jhd2. Distances between H2B Lys-123 and the two H2A residues calculated using the UCSF Chimera software are shown. B, a schematic model depicting the putative masking of the H2A Phe-26 and Glu-57 residues by H2BK123 ubiquitination. C, Western blots for H3K4 methylation, H2B ubiquitination and Jhd2–9Myc were examined in control wild-type strain or in strains lacking the two H2B deubiquitinases (ubp8Δubp10Δ) and transformed with an empty plasmid vector (pRS426) or a plasmid to overexpress Jhd2–9Myc. Wild-type and ubp8Δubp10Δ in two independent yeast backgrounds, namely FY120 or Y131 (labeled as H2B and FlagH2B, respectively), were used. Crude nuclear extracts were used for examining H3K4 methylation and Jhd2 levels. Pgk1 and H3 levels serve as loading controls. For H2B ubiquitination, cells were lysed by boiling in Laemmli sample buffer. *, a cross-reacting protein. D, Jhd2-12V5 occupancies at the indicated target genes in control wild-type and ubp8Δubp10Δ strains were assessed with ChIP assays using α-V5 antibody. Following ChIP and quantitative PCR, the ChIP signal (V5 immunoprecipitation/input value) obtained from a strain without V5-tagged Jhd2 (background) was subtracted from the ChIP signal obtained from strains expressing Jhd2-12V5, and the resulting difference was defined as Jhd2 occupancy. The graph shows -fold change in Jhd2 occupancy at a given target locus in the ubp8Δubp10Δ mutant relative to Jhd2 occupancy in the control wild-type strain (set as 1). Error bars, S.E. obtained from three independent ChIP experiments. Statistical significance was calculated using Student's t test. *, p < 0.05.
Absence of the two H2B deubiquitnases, Ubp8 and Ubp10, results in the accumulation of high levels of global H2Bub1 (>12-fold increase) and a concomitant strain-specific increase in H3K4 methylation (55, 60) (Fig. 8C). To test our hypothesis, we then examined the effect of increased H2Bub1 on H3K4 demethylation by overexpressing Jhd2 in control wild-type strain and a strain lacking the two H2B deubiquitinases (ubp8Δubp10Δ). Whereas overexpression of Jhd2 in wild-type strain led to a decrease in bulk H3K4me3 levels (Fig. 8C, lanes 1 and 2), overexpression of Jhd2 in the ubp8Δubp10Δ strain did not cause any reduction in H3K4me3 levels (Fig. 8C, lanes 3 and 4). Similar results were obtained for H3K4me3 levels upon overexpression of Jhd2 in control and ubp8Δubp10Δ strains in an independent yeast background (Y131) (Fig. 8C, lanes 5–8). Because global Jhd2 levels are not reduced in the ubp8Δubp10Δ strain compared with the wild-type control (Fig. 8C), our findings suggested that the high levels of H2Bub1 inhibit the removal of H3K4me3 by Jhd2. We then tested the occupancy of Jhd2 at the candidate target genes in control and ubp8Δubp10Δ strains using ChIP assays. Jhd2 occupancy was significantly reduced at the SER3, CAR1, SPL2, and GFD2 genes and generally decreased at many of the target loci in the ubp8Δubp10Δ strain compared with the control wild-type strain (Fig. 8D). This result showed that the absence of the deubiquitinases and the ensuing increase in H2Bub1 blocks the association of Jhd2 with chromatin. Collectively, our results have therefore uncovered a novel mechanism by which H2BK123 ubiquitination functions in the trans-histone regulation of H3K4 methylation via obstructing the Jhd2 H3K4 demethylase functions.
Discussion
A Binding Site on Histone H2A Controls Chromatin Association and H3K4 Demethylase Functions of Jhd2
Here, we have now demonstrated histone H2A as a novel recognition target for the PHD finger present in Jhd2 (Fig. 1B). Structural studies have previously shown that the PHD fingers bind with high specificity to unmodified or modified histone H3 or to acetylated H4 peptide (29). The PHD finger in ACF1 was shown to bind recombinant histones and nucleosomes (61). Our results have therefore for the first time shown that PHD fingers can bind to histones other than H3 or H4. Using mutational analysis, we have identified residues in H2A (Phe-26 and Glu-57) that mediate the in vitro interaction with the Jhd2 PHD finger (Fig. 5). We have further shown that Jhd2 and the two H2A residues Phe-26 and Glu-57 contribute to either positive or negative regulation of transcription at a shared set of functional target genes (Fig. 6). Whereas mutating both Phe-26 and Glu-57 disrupted the binding of H2A to the Jhd2 PHD finger in vitro, mutating either the Phe-26 or Glu-57 residue did not lead to any dramatic change in this interaction (Fig. 5). In agreement with this finding, Jhd2 occupancy on chromatin remained unaffected at certain target loci in the H2A/F26A and H2A/E57A mutants, but interestingly, Jhd2 occupancy on chromatin was increased at some other target genes in these two mutants (Fig. 6A). This result demonstrated that the H2A Phe-26 and Glu-57 residues contribute to the proper association of Jhd2 with chromatin. Although Jhd2 occupancy on chromatin is not reduced at target genes upon mutating either the H2A Phe-26 or Glu-57 residue, these H2A mutations disrupted the H3K4 demethylase functions of Jhd2 in vivo both globally and locally at the shared functional target genes (Figs. 2D, 3B, and 6B). In summary, we have uncovered the presence of a binding site for the Jhd2 PHD finger on histone H2A (composed of residues Phe-26 and Glu-57), which controls both the chromatin binding dynamics and H3K4 demethylase functions of Jhd2 during transcriptional regulation.
Among the JMJC family demethylases, those containing both JmjN and JmjC domains target either di- or trimethylated lysine or all three degrees of lysine methylation (62). The crystal structure of human JMJD2A shows extensive contacts between the β-strands present in its JmjN and JmjC catalytic core domains (63). In our previous work (26), we showed that mutating a highly conserved residue, lysine 37, to glutamine (K37Q) within the β-strand of the Jhd2 JmjN domain abolished the ability of Jhd2 to remove H3K4me1 and H3K4me3 marks but not the H3K4me2 mark. Therefore, the Jhd2 JmjN-JmjC interdomain interactions specify its catalytic functions toward removing a particular degree(s) of lysine methylation. Similar to the Jhd2 JmjN(K37Q), mutating the H2A Phe-26 or Glu-57 residue also disrupts the ability of Jhd2 to remove H3K4me1 and H3K4me3 marks (Figs. 2–4). Based on these findings and given the location of the Jhd2 PHD finger between the catalytic JmjN and JmjC domains (Fig. 1A), we propose a model wherein Jhd2 associates with chromatin via the PHD finger-histone H2A interaction, which then induces a conformational change within the JmjN and JmjC catalytic domains needed for the removal of H3K4me1 and H3K4me3 marks.
Histone H2A as a “Hub” for the Enzymes Involved in the trans-Histone Cross-talk
Histone H2A is an integral component of chromatin, and as such, it is expected to play a significant role in gene transcription. Indeed, mutations in various H2A residues were shown to impact gene regulation in yeast (64–66). Here, we have now uncovered a role for H2A Phe-26 and Glu-57 residues in regulating antisense transcription in addition to sense transcription in yeast (Fig. 6). Deleting the H2A N-terminal tail (residues 4–20) decreased the expression of certain Jhd2 target genes (e.g. SPL2 and CAR1) (65). Furthermore, residues 16–20 within the H2A N-terminal tail, which constitute the H2A repression domain (65), were shown to be important for H2B ubiquitination and H3K4 methylation (67). In a library screen of H2A mutants, alanine substitutions in certain H2A acidic patch residues reduced H3K4 methylation without affecting H2B ubiquitination (68). Therefore, histone H2A is linked to the control of H2B ubiquitination and H3K4 methylation. In this study, we have shown that alanine substitution at the H2A Glu-57 or Leu-59 residue, both of which occur close to the H2A acidic patch, decreases bulk H3K4 methylation (Fig. 3). Additionally, whereas alanine substitution at the H2A Thr-25 or Pro-27 residue increased H3K4me3, alanine substitution at the H2A Leu-24 or Phe-26 residue decreased H3K4me3 (Fig. 2D). Therefore, our results along with the published studies together suggest a role for a large region in histone H2A composed of the linker residues and helices α1 and α2 in mediating the H3K4 methyltransferase functions of Set1-COMPASS. Furthermore, we have now demonstrated that alanine substitution in two H2A residues (Phe-26 and Glu-57) adversely impacts the enzymatic functions of the Jhd2 H3K4 demethylase. The Jhd2 PHD finger does not bind Htz1 (the H2A variant) (data not shown), which agrees with the observation that Htz1 acts redundantly with H3K4 methylation in an anti-silencing mechanism (69). Collectively, our study and those of others together reveal that histone H2A controls H3K4 methylation either indirectly via controlling Rad6-Bre1-mediated H2B ubiquitination or directly through regulating the enzymatic functions of Set1-COMPASS methyltransferase and Jhd2 demethylase.
Blocking Jhd2-mediated H3K4 Demethylation, a Second Mechanism for the trans-Histone Regulation of H3K4 Methylation, by H2BK123 Ubiquitination
Kim et al. (70) demonstrated that the n-SET domain of Set1 methyltransferase and Spp1 (a Set1-COMPASS subunit) mediate the H2BK123 ubiquitination-dependent H3K4 methylation. Spp1 and the n-SET domain are also required for Set1 stability (71). We previously showed that the histone H2B residues Arg-119 and Thr-122 mediate the in vitro Spp1-H2B interaction, and mutations in these H2B residues disrupted chromatin association and complex stability of Set1-COMPASS in addition to its processive H3K4 methylation in an H2BK123 ubiquitination-independent manner (60). We as well as others have also shown that H2BK123 ubiquitination controls chromatin dynamics and nucleosome stability (72–74). Based on all of these studies, a model proposed for the regulation of H3K4 methylation by H2BK123 ubiquitination is that the addition of an ubiquitin moiety onto H2B stabilizes the nucleosome by preventing H2A-H2B eviction, which then leads to the retention of the binding sites for Spp1 in H2B on chromatin. Histone or nucleosome-bound Spp1 then stabilizes Set1-COMPASS and, acting along with other COMPASS subunits, stimulates the processive methylation by Set1 via the n-SET domain. In this study, we have now demonstrated that H2BK123 ubiquitination also controls the functions of Jhd2 H3K4 demethylase. The binding sites for Jhd2 on H2A (residues Phe-26 and Glu-57) are on the same nucleosomal surface as H2BK123, the site of ubiquitination (Fig. 8A), which prompted the hypothesis that the bulky ubiquitin moiety might mask the binding site for Jhd2 on chromatin (Fig. 8B). Indeed, a high amount of H2BK123 ubiquitination not only inhibits Jhd2-mediated removal of H3K4me3 marks but also decreases its association with chromatin (Fig. 8, C and D). Therefore, we have uncovered a novel mechanism by which H2B ubiquitination contributes to the trans-histone regulation of H3K4 methylation. In summary, we propose that H2B ubiquitination promotes H3K4 methylation in two ways: 1) it stimulates the Set1 H3K4 methyltransferase functions by stabilizing the nucleosome and retaining the binding sites for the Spp1 subunit on chromatin, and 2) it inhibits the Jhd2 H3K4 demethylase functions by occluding its binding site on chromatin and preventing the removal of H3K4 methyl marks.
Author Contributions
F. H. and M. B. C. designed the study and wrote the paper. F. H., S. R., S. P., and M. B. C. performed the experiments. C. P. and T. J. P. performed the bioinformatics analysis. S. L. C. and N. B. provided technical advice on protein purification. M. H. provided the histone alanine mutant library. M. M. K. provided recombinant histones. B. J. G., B. R. C., and S. B. provided valuable suggestions.
Supplementary Material
Acknowledgments
We are grateful to Fred Winston, Mary Ann Osley, and Dan Gottschling for kindly providing the yeast strains. We thank Gennie Parkman and Sowmiya Palani for technical assistance. We thank Alisha Schlichter, Cedric Clapier, and members of the Graves laboratory for suggestions. We also thank Brian Dalley at the Huntsman Cancer Institute's genomic analysis core facility for RNA-seq. M. B. C. is very grateful to Zu-Wen Sun and Dennis Shrieve for encouragement and support of this work.
This work was supported by funds provided by the Huntsman Cancer Institute, the Department of Radiation Oncology, and the Cancer Center Support Grant (Nuclear Control of Cell Growth and Differentiation program) (to M. B. C.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables 1–3.
- H3K4
- histone H3 Lys-4
- H2BK123
- histone H2B Lys-123
- H3K4me1
- H3K4me2, and H3K4me3, mono-, di-, or tri-methylated H3K4, respectively
- PHD
- plant homeodomain
- GO
- gene ontology
- RNA-seq
- RNA sequencing.
References
- 1.Ruthenburg A. J., Allis C. D., and Wysocka J. (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 [DOI] [PubMed] [Google Scholar]
- 2.Eissenberg J. C., and Shilatifard A. (2010) Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizzardi L. F., Dorn E. S., Strahl B. D., and Cook J. G. (2012) DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics 192, 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faucher D., and Wellinger R. J. (2010) Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 10.1371/journal.pgen.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel J. A., and Nussenzweig A. (2012) Roles for histone H3K4 methyltransferase activities during immunoglobulin class-switch recombination. Biochim. Biophys. Acta 1819, 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shilatifard A. (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter D., Matter A., and Fahrenkrog B. (2014) Loss of histone H3 methylation at lysine 4 triggers apoptosis in Saccharomyces cerevisiae. PLoS Genet. 10, e1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., and Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 9.Margaritis T., Oreal V., Brabers N., Maestroni L., Vitaliano-Prunier A., Benschop J. J., van Hooff S., van Leenen D., Dargemont C., Géli V., and Holstege F. C. (2012) Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 8, e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinskaya M., and Morillon A. (2009) Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity? Epigenetics 4, 302–306 [DOI] [PubMed] [Google Scholar]
- 11.Kim T., and Buratowski S. (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., and Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 13.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., and Wysocka J. (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng J., Blum R., Bowman C., Hu D., Shilatifard A., Shen S., and Dynlacht B. D. (2014) A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell 53, 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider R., Bannister A. J., Myers F. A., Thorne A. W., Crane-Robinson C., and Kouzarides T. (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6, 73–77 [DOI] [PubMed] [Google Scholar]
- 16.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., and Kouzarides T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 17.Bernstein B. E., Humphrey E. L., Erlich R. L., Schneider R., Bouman P., Liu J. S., Kouzarides T., and Schreiber S. L. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U.S.A. 99, 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinskaya M., Gourvennec S., and Morillon A. (2009) H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28, 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiner A., Chen H. V., Liu C. L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S., Pleiss J. A., Friedman N., and Rando O. J. (2012) Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 10, e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., and Allis C. D. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roguev A., Schaft D., Shevchenko A., Pijnappel W. W., Wilm M., Aasland R., and Stewart A. F. (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., Johnston M., and Shilatifard A. (2002) COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277, 10753–10755 [DOI] [PubMed] [Google Scholar]
- 23.Liang G., Klose R. J., Gardner K. E., and Zhang Y. (2007) Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 14, 243–245 [DOI] [PubMed] [Google Scholar]
- 24.Tu S., Bulloch E. M., Yang L., Ren C., Huang W. C., Hsu P. H., Chen C. H., Liao C. L., Yu H. M., Lo W. S., Freitas M. A., and Tsai M. D. (2007) Identification of histone demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 282, 14262–14271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seward D. J., Cubberley G., Kim S., Schonewald M., Zhang L., Tripet B., and Bentley D. L. (2007) Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 14, 240–242 [DOI] [PubMed] [Google Scholar]
- 26.Huang F., Chandrasekharan M. B., Chen Y. C., Bhaskara S., Hiebert S. W., and Sun Z. W. (2010) The JmjN domain of Jhd2 is important for its protein stability, and the plant homeodomain (PHD) finger mediates its chromatin association independent of H3K4 methylation. J. Biol. Chem. 285, 24548–24561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shilatifard A. (2012) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mersman D. P., Du H. N., Fingerman I. M., South P. F., and Briggs S. D. (2009) Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 23, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez R., and Zhou M. M. (2011) The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musselman C. A., and Kutateladze T. G. (2011) Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 39, 9061–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., and Gozani O. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., and Patel D. J. (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wysocka J., Swigut T., Xiao H., Milne T. A., Kwon S. Y., Landry J., Kauer M., Tackett A. J., Chait B. T., Badenhorst P., Wu C., and Allis C. D. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 [DOI] [PubMed] [Google Scholar]
- 34.Shi X., Kachirskaia I., Walter K. L., Kuo J. H., Lake A., Davrazou F., Chan S. M., Martin D. G., Fingerman I. M., Briggs S. D., Howe L., Utz P. J., Kutateladze T. G., Lugovskoy A. A., Bedford M. T., and Gozani O. (2007) Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews A. G., Kuo A. J., Ramón-Maiques S., Han S., Champagne K. S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D. N., Walter K. L., Utz P. J., Shi Y., Kutateladze T. G., Yang W., Gozani O., and Oettinger M. A. (2007) RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan F., Collins R. E., De Cegli R., Alpatov R., Horton J. R., Shi X., Gozani O., Cheng X., and Shi Y. (2007) Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448, 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H. H., Whetstine J. R., Bonni A., Roberts T. M., and Shi Y. (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 38.Xu M., Soloveychik M., Ranger M., Schertzberg M., Shah Z., Raisner R., Venkatasubrahmanyan S., Tsui K., Gebbia M., Hughes T., van Bakel H., Nislow C., Madhani H. D., and Meneghini M. D. (2012) Timing of transcriptional quiescence during gametogenesis is controlled by global histone H3K4 demethylation. Dev. Cell 23, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittmeyer J., Saha A., and Cairns B. (2004) DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 377, 322–343 [DOI] [PubMed] [Google Scholar]
- 40.Gadaleta M. C., Iwasaki O., Noguchi C., Noma K., and Noguchi E. (2013) New vectors for epitope tagging and gene disruption in Schizosaccharomyces pombe. BioTechniques 55, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandrasekharan M. B., Huang F., and Sun Z. W. (2011) Decoding the trans-histone crosstalk: methods to analyze H2B ubiquitination, H3 methylation and their regulatory factors. Methods 54, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaffl M. W. (2004) in A–Z of Quantitative PCR (Bustin S. A., ed.) pp. 87–112, Internation University Line, La Jolla, CA [Google Scholar]
- 43.Lai J. S., and Herr W. (1992) Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 89, 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maltby V. E., Martin B. J., Brind'Amour J., Chruscicki A. T., McBurney K. L., Schulze J. M., Johnson I. J., Hills M., Hentrich T., Kobor M. S., Lorincz M. C., and Howe L. J. (2012) Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 109, 18505–18510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lalonde M. E., Cheng X., and Côté J. (2014) Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev. 28, 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Z. W., and Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 47.Dehé P. M., Pamblanco M., Luciano P., Lebrun R., Moinier D., Sendra R., Verreault A., Tordera V., and Géli V. (2005) Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 353, 477–484 [DOI] [PubMed] [Google Scholar]
- 48.Shahbazian M. D., Zhang K., and Grunstein M. (2005) Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19, 271–277 [DOI] [PubMed] [Google Scholar]
- 49.Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., and Shilatifard A. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 50.Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., and Strahl B. D. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakanishi S., Lee J. S., Gardner K. E., Gardner J. M., Takahashi Y. H., Chandrasekharan M. B., Sun Z. W., Osley M. A., Strahl B. D., Jaspersen S. L., and Shilatifard A. (2009) Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 186, 371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robzyk K., Recht J., and Osley M. A. (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 53.Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., and Madhani H. D. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11, 261–266 [DOI] [PubMed] [Google Scholar]
- 54.Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., and Shilatifard A. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 55.Gardner R. G., Nelson Z. W., and Gottschling D. E. (2005) Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 25, 6123–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daniel J. A., Torok M. S., Sun Z. W., Schieltz D., Allis C. D., Yates J. R. 3rd, and Grant P. A. (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279, 1867–1871 [DOI] [PubMed] [Google Scholar]
- 57.Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., and Berger S. L. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emre N. C., Ingvarsdottir K., Wyce A., Wood A., Krogan N. J., Henry K. W., Li K., Marmorstein R., Greenblatt J. F., Shilatifard A., and Berger S. L. (2005) Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell 17, 585–594 [DOI] [PubMed] [Google Scholar]
- 59.Schulze J. M., Hentrich T., Nakanishi S., Gupta A., Emberly E., Shilatifard A., and Kobor M. S. (2011) Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 25, 2242–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chandrasekharan M. B., Huang F., Chen Y. C., and Sun Z. W. (2010) Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol. Cell. Biol. 30, 3216–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eberharter A., Vetter I., Ferreira R., and Becker P. B. (2004) ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 23, 4029–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Secombe J., and Eisenman R. N. (2007) The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle 6, 1324–1328 [DOI] [PubMed] [Google Scholar]
- 63.Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T. G., Simpson M., Mao Q., Pan C. H., Dai S., Hagman J., Hansen K., Shi Y., and Zhang G. (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702 [DOI] [PubMed] [Google Scholar]
- 64.Hirschhorn J. N., Bortvin A. L., Ricupero-Hovasse S. L., and Winston F. (1995) A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15, 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parra M. A., and Wyrick J. J. (2007) Regulation of gene transcription by the histone H2A N-terminal domain. Mol. Cell. Biol. 27, 7641–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsubara K., Sano N., Umehara T., and Horikoshi M. (2007) Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12, 13–33 [DOI] [PubMed] [Google Scholar]
- 67.Zheng S., Wyrick J. J., and Reese J. C. (2010) Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol. Cell. Biol. 30, 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakanishi S., Sanderson B. W., Delventhal K. M., Bradford W. D., Staehling-Hampton K., and Shilatifard A. (2008) A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venkatasubrahmanyam S., Hwang W. W., Meneghini M. D., Tong A. H., and Madhani H. D. (2007) Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. U.S.A. 104, 16609–16614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim J., Kim J. A., McGinty R. K., Nguyen U. T., Muir T. W., Allis C. D., and Roeder R. G. (2013) The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell 49, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thornton J. L., Westfield G. H., Takahashi Y. H., Cook M., Gao X., Woodfin A. R., Lee J. S., Morgan M. A., Jackson J., Smith E. R., Couture J. F., Skiniotis G., and Shilatifard A. (2014) Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 28, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandrasekharan M. B., Huang F., and Sun Z. W. (2009) Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. U.S.A. 106, 16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fleming A. B., Kao C. F., Hillyer C., Pikaart M., and Osley M. A. (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31, 57–66 [DOI] [PubMed] [Google Scholar]
- 74.Batta K., Zhang Z., Yen K., Goffman D. B., and Pugh B. F. (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartzog G. A., Wada T., Handa H., and Winston F. (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12, 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y., Sun Z. W., Iratni R., Erdjument-Bromage H., Tempst P., Hampsey M., and Reinberg D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1, 1021–1031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.