Background: In bacteria, type I signal peptidase is targeted to the membrane co-translationally.
Results: Plastidic type I signal peptidase 1 (Plsp1) could insert into the membrane in an incorrect orientation, which was prevented by a stroma chaperone and the cpSec1 machine.
Conclusion: Correct membrane insertion of Plsp1 is ensured by stromal components.
Significance: The results demonstrate an adaptive mechanism of protein transport during organelle evolution.
Keywords: chaperone, chloroplast, protein export, signal peptidase, transmembrane domain, Sec channel, thylakoid
Abstract
Type I signal peptidase (SPase I) is an integral membrane Ser/Lys protease with one or two transmembrane domains (TMDs), cleaving transport signals off translocated precursor proteins. The catalytic domain of SPase I folds to form a hydrophobic surface and inserts into the lipid bilayers at the trans-side of the membrane. In bacteria, SPase I is targeted co-translationally, and the catalytic domain remains unfolded until it reaches the periplasm. By contrast, SPases I in eukaryotes are targeted post-translationally, requiring an alternative strategy to prevent premature folding. Here we demonstrate that two distinct stromal components are involved in post-translational transport of plastidic SPase I 1 (Plsp1) from Arabidopsis thaliana, which contains a single TMD. During import into isolated chloroplasts, Plsp1 was targeted to the membrane via a soluble intermediate in an ATP hydrolysis-dependent manner. Insertion of Plsp1 into isolated chloroplast membranes, by contrast, was found to occur by two distinct mechanisms. The first mechanism requires ATP hydrolysis and the protein conducting channel cpSecY1 and was strongly enhanced by exogenously added cpSecA1. The second mechanism was independent of nucleoside triphosphates and proteinaceous components but with a high frequency of mis-orientation. This unassisted insertion was inhibited by urea and stroma extract. During import-chase assays using intact chloroplasts, Plsp1 was incorporated into a soluble 700-kDa complex that co-migrated with the Cpn60 complex before inserting into the membrane. The TMD within Plsp1 was required for the cpSecA1-dependent insertion but was dispensable for association with the 700-kDa complex and also for unassisted membrane insertion. These results indicate cooperation of Cpn60 and cpSecA1 for proper membrane insertion of Plsp1 by cpSecY1.
Introduction
Approximately half of the proteins in an average cell are transported into or across one membrane or more to reach their final destinations (1). Among the mechanisms that catalyze translocation of polypeptides across the membranes are Sec and twin-Arg translocation (Tat) pathways. Their substrates carry N-terminal transport signals, which consist of a positive n-domain followed by a hydrophobic core of 12–18 residues (h-domain) and a C-terminal domain usually ending with the Ala-Xaa-Ala motif (2). Notably, the Tat transport signals contain the twin Arg (Arg-Arg) motif in the n-domain. The transport signals are removed by a type I signal peptidase (SPase I)3 at the trans-side of the membrane. Escherichia coli SPase I, known as LepB, spans the membrane twice and faces its large C-terminal portion containing the catalytic site toward the periplasm (3). Signal cleavage is assumed to occur in or at the surface of the lipid bilayer because the h-domain of most transport signals is too short to span the membrane completely (2, 4). This model agrees with structural and biochemical properties of the periplasmic portion of LepB without the transmembrane domains (TMDs), which was found to form a large hydrophobic surface composed of multiple β-strands (5) and was also shown to insert into membrane lipids in vivo and in vitro (6). Membrane insertion of the full-length LepB itself occurs co-translationally by the signal recognition particle (SRP) and the membrane insertase YidC (7). Translocation of its large C terminus requires the SecA motor and the SecYEG channel (8, 9).
The chloroplast is an organelle found in photosynthetic eukaryotes and surrounded by a double-membrane envelope, housing photosynthetic electron transport in the internal membrane system known as the thylakoid. As an organelle of endosymbiotic origin, the chloroplast has its own genome. However, most proteins found in the chloroplast are encoded in the nucleus, synthesized on cytosolic ribosomes, and targeted to the organelle post-translationally (10). Specific chloroplast targeting is usually ensured by an N-terminal extension called transit peptide, which is cleaved in the stroma (11, 12). Four pathways are known to target proteins from the stroma to thylakoids: the three pathways homologous to bacterial Sec, Tat, and SRP pathways, respectively, and the chloroplast-specific unassisted pathway for a subset of proteins that spontaneously insert into the membrane (13, 14). All known chloroplast Sec1 (cpSec1) and Tat (cpTat) substrates, except for the Rieske iron/sulfur subunit of cytochrome b6f complexes (ISP), and all known unassisted pathway substrates with a single TMD carry an N-terminal transport signal. Removal of this signal, called cleavable thylakoid-transfer signal (cTTS), is catalyzed by a LepB homolog known as thylakoidal processing peptidase in the lumen (11, 15). ISP is an unusual cpTat substrate in that it carries an uncleavable TTS, which appears to act as a membrane anchor, and contains Lys-Arg, instead of Arg-Arg, in the n-domain (16, 17).
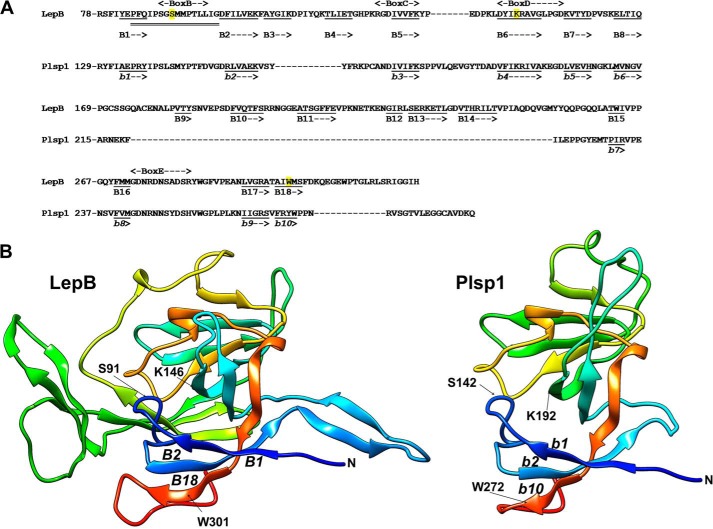
In land plants, thylakoidal processing peptidase is encoded in the nucleus by a small gene family called plastidic SPase I 1 and 2 (Plsp1 and 2) (18). Plsp1 is predicted to span the membrane once and face a large C terminus including the catalytic site into the thylakoid lumen (19). Its lumenal domain shows a high sequence similarity to the periplasmic domain of LepB (20) and is predicted to form a β-sheet that presents a hydrophobic surface (Fig. 1). Genetic and biochemical data have demonstrated that Plsp1 is the main thylakoidal processing peptidase isoform in the reference plant Arabidopsis thaliana and is required for proper thylakoid development (20–22). Interestingly, Plsp1 is present not only in thylakoids but also in the envelope membrane where it cleaves the envelope-targeting sequence off the precursor of an outer membrane protein Toc75 (19, 21). Localization of Plsp1 correlates with the abundance of membranes, shifting from the envelope to thylakoids as chloroplasts develop (19).
FIGURE 1.
Predicted structure of the lumenal portion of Plsp1. A, sequence alignment with the C-terminal periplasmic portion of LepB and the secondary structure prediction using Phyre2 (71) based on d1b12a (E. coli LepB; Protein Data Bank code 1b12) (5), which shows 41% sequence identity and 100% confidence for Plsp1. Shown in the diagram are the number of the most N-terminal residue in each line, conserved boxes B–E based on the work of Paetzel (2), residues in β-strands as underlined (B1–B18 for LepB and b1 to b10 for Plsp1), and the region (from Pro-84 to Gly-99) of LepB necessary for the TMD-independent membrane insertion with double line (=) (6). For LepB, the catalytic nucleophile Ser-91 and the general base Lys-146, as well as Trp-301, the residue predicted to help facilitate membrane insertion of the catalytic region (2), are highlighted in yellow. Note that the assignments to β-strands are based on the prediction and do not completely match with the ones show by Paetzel (2) and that not only the β-strands but also the region needed for membrane insertion is conserved in Plsp1. B, three-dimensional structural models of LepB78–323 (left panel) and Plsp1129–280 (right panel). The models were visualized with color by rainbow N to C terminus with the UCSF Chimera package (72), which is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS, National Institutes of Health Grant P41-GM103311). N indicates the N terminus of each structure. Indicated in LepB are the catalytic nucleophile Ser-91 (S91) and the general base Lys-146 (K146), as well as the three β-strands (B1, B2, and B18) including the essential Trp-301 (W301) (73). The corresponding catalytic residues, β-strands, and Trp in Plsp1 are indicated as S142, K192, b1, b2, b10, and W272, respectively.
Here we have used various assays to elucidate the mechanism of Plsp1 transport. Available systems have not allowed us to define the mechanism of envelope targeting. Nonetheless, our results demonstrate that Plsp1 is imported into the chloroplast stroma using an N-terminal extension and sorted to thylakoids by the cpSec1 pathway, even though it lacks a cTTS. Our data also show that the lumenal portion of Plsp1 can spontaneously insert into the membrane from the cis-side, but this nonspecific event is prevented by integration into a large complex in the stroma that co-migrates with a chaperonin-60 (Cpn60) complex. These results represent an evolutionary adaptation from the ribosome-dependent co-translational insertion to the chaperone-dependent post-translational transport of SPase I.
Experimental Procedures
DNA Constructs
Two sets of DNA constructs were prepared using oligonucleotide primers listed in Table 1. The first set was used for in vitro transcription/translation to generate radiolabeled precursors for transport assays. Coding sequences for prPlsp1 and Plsp1Δ2–67 (21), Citrine (23), and Plsp11–70-Citrine (24) were amplified from appropriate templates and then were subcloned using the Gateway® system into a unidirectional transcription vector called pIVTGW-SP6. pIVTGW-SP6 was generated by ligating a 4-kb fragment of pCR®II-TOPO® (Life Technologies, Inc.) and a 1.8-kb fragment of pMDC32 (25) after digesting each plasmid with KpnI and SacI, followed by insertion of an oligonucleotide linker into the AscI-KpnI site to remove an in-frame start codon upstream of the cloning site. Plasmids encoding Pisum sativum (pea) Tic22 (26), Arabidopsis Tic40 (27), and pea LHCP (28) were as described previously. Arabidopsis OE23 (U19899), OE33 (U13665), and PsbW (U12467) in pUNI51 were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The second set of DNA constructs was used for production of recombinant proteins in E. coli. Constructs for His10-Plsp1Δ2–67 and His10-Plsp1Δ1–134 were as described (20). The cDNA sequence encoding residues 63–1042 of cpSecA1 (At4g01800) was amplified by PCR using cDNA synthesized from total RNA isolated from 15-day-old Arabidopsis (Columbia) seedlings as a template. The obtained fragment was digested with NotI and ligated into a pET28a vector, yielding the cpSecA1 expression plasmid.
TABLE 1.
Oligonucleotide primers used to make constructs
The oligonucleotide primers are depicted from left to right as 5′ to 3′. F indicates forward, and R indicates reverse.
| For in vitro transcription | |
| AscI-KpnI linker | F: CGTGAGTGAGG |
| R: CGCGCCTCACTCACGGTAC | |
| Plsp1 | F: CAAAAAAAGCAGGCTCGCCCATGATGGTGATGATATCTC |
| R: GTACAAGAAAGCTGGGTCCTATTGCTTATCCACAGCAC | |
| Plsp12–67 | F: AAAAAGCAGGCTTGATGGATTCAAGTGAGACAACGAAG |
| R: the same as the one for Plsp1 | |
| Citrine | F: AAAAGCAGGCTCGCCCATGGTCTCCAAG |
| R: GAAAGCTGGGTTTACTACTTGTATAGCTCGTC | |
| Plsp11–70-Citrine | F: the same as the one for Plsp1 |
| R: the same as the one for Citrine | |
| For overexpression in E. coli | |
| cpSecA163−1042 | F: GGGCGGCCGCGAGAACCTGTACTTCCAGGGTTGCTCTCGGAAGAGAAGCACG |
| R: CCGCGGCCGCGGCGCTTGCAATTGAGGATGG | |
Preparation of Chloroplasts, Chloroplast Membranes, and Stroma Extracts
Intact chloroplasts were isolated from seedlings of pea (Little Marvel) and resuspended in import buffer (IB: 50 mm Hepes-KOH, pH 8.0, and 330 mm sorbitol) as described (29). Chloroplast membranes and stroma extract were prepared as described (30). Briefly, after precipitating by centrifugation at 1,000 × g, 4 °C for 6 min, intact chloroplasts were resuspended with a hypotonic lysis buffer (HM buffer: 10 mm Hepes-KOH, pH 8.0, and 10 mm MgCl2) to 1 mg/ml chlorophyll on ice in dark for 10 min. The lysed chloroplasts were centrifuged at 3,200 × g, 4 °C for 8 min, yielding the crude stroma and pellet fractions. The obtained pellet fraction was further resuspended in HM buffer and centrifuged at 3,200 × g, 4 °C for 8 min, yielding the crude membrane fraction. To prepare stroma extract (SE), the crude stroma fraction was diluted with an equal volume of 100 mm Hepes-KOH, pH 8.0, 660 mm sorbitol, and 10 mm MgCl2 and centrifuged at 42,000 × g, 4 °C for 30 min. An aliquot (95%) of the obtained supernatant was concentrated to 5-fold using an Amicon Ultra-4 30000 NMWL column (EMD Millipore, Billerica, MA). To prepare chloroplast membranes for transport assays, the crude membrane fraction was resuspended in HM buffer to ∼0.25 mg/ml chlorophyll and centrifuged at 3,200 × g, 4 °C for 8 min. The obtained pellet was resuspended to 1 mg/ml chlorophyll in IB containing 10 mm MgCl2 (IBM).
Import and Transport Assays in Vitro
Protein import assays using isolated chloroplasts was performed as described previously (29). After import, intact chloroplasts were reisolated through a 40% Percoll cushion and resuspended in IB. An aliquot was used to quantify the amount of chlorophyll as described (31). Other aliquots were centrifuged to yield chloroplast pellet, which was resuspended to 0.2 mg/ml chlorophyll of 1 × SDS-PAGE sample loading solution. For import time course experiments, the reactions with different incubation times were staggered so that all reactions would be terminated at the same time and processed together. For import-chase assays, after 10-min import in the light, the chloroplasts were pelleted at 900 × g and 4 °C for 3 min and resuspended with IB containing 0.5 mg/ml thermolysin and 10 mm CaCl2 and then incubated on ice in the dark for 30 min. The thermolysin activity was quenched with one volume of IB containing 20 mm EDTA. Intact chloroplasts were then reisolated through a 1 ml of 40% Percoll cushion in IB containing 50 mm EDTA, resuspended in IB containing 5 mm EDTA, and aliquoted into multiple tubes. Each sample was then centrifuged, and the resultant pellets were resuspended with IB containing one of the following five sets of chemicals: (i) 3 mm MgCl2 and 3 mm LiCl (blank), (ii) 3 mm MgATP and 3 mm LiCl (ATP), (iii) 3 mm Li-AMP-PNP and 3 mm MgCl2 (AMP-PNP), (iv) 10 mm NaN3, 3 mm MgCl2 and 3 mm LiCl (NaN3), and (v) 1.5 μm nigericin, 1.5 μm valinomycin, 3 mm MgCl2, and 3 mm LiCl (N/V). The reaction was further incubated under import conditions for the specified time, and then intact chloroplasts were reisolated through Percoll cushions and analyzed as above.
For analysis of the soluble mPlsp1 complex by BN-PAGE, radiolabeled prPlsp1 was imported into intact chloroplasts in the light with 3 mm MgATP for 10 min. Chloroplasts reisolated through Percoll cushions were lysed with HM buffer and incubated for 5 min in the dark on ice. The mixture was then centrifuged at 16,000 × g, 4 °C for 20 min. The obtained supernatant was centrifuged again, and the resultant soluble fraction was subjected to various treatments or directly added to 1 × BN-PAGE buffer and analyzed by BN-PAGE as described previously (24).
Protein transport into isolated chloroplast membranes was performed as described previously (30) with some modifications. Briefly, each reaction (72 μl) contained chloroplast membranes equivalent to 24 μg of chlorophyll and 5 mm MgATP in IBM. Where indicated, concentrated stroma (24 μl) was also included in the mixture. The reaction was performed at room temperature for 30 min in the dark or in the light, followed by centrifugation at 3,200 × g, 4 °C for 8 min. The obtained pellet was resuspended with 120 μl of IB, and 6 μl of 2 mg/ml thermolysin in IB containing 10 mm CaCl2 was added. After incubation on ice in the dark for 40 min, the thermolysin activity was quenched with one volume IB containing 14 mm EDTA and then centrifuged at 3,200 × g, 4 °C for 8 min. The pellet was washed with IB containing 5 mm EDTA and subjected to SDS-PAGE analysis.
Trypsin pretreatment of isolated chloroplast membranes was performed as previously described (32). Briefly, the membranes were resuspended to 50 μg/ml chlorophyll with HM buffer without or with trypsin (60 μg/ml) and incubated on ice in the dark for 10 min, followed by the addition of one volume of 0.6 mg/ml soybean trypsin inhibitor in HM buffer. The chloroplast membranes were then recovered as a pellet after centrifugation and washed with 0.12 mg/ml trypsin inhibitor in HM buffer. The resultant membranes were further washed with IBM, resuspended to 1 mg/ml chlorophyll with IBM, and used in transport experiments as above. Antibody inhibition experiments were performed as described (30). When the effect of cpSecA1 was tested, the control reaction was supplemented with 3 mm NaH2PO4, 19 mm NaCl, and 16 mm imidazole.
Quantification of Radioactive Signals and Statistical Analysis
Intensity of radioactive signals from bands on gels was quantified using ImageJ 1.48v (National Institutes of Health). For the import time course and import-chase assays, the signals for each repetition of the experiment were normalized to the mean of signals for all of the time points or treatments for the total chloroplast fraction. These normalized values were adjusted to the mean of the normalized signals at the 30-min time point for the import time course or the mean of all of the signals in the total chloroplast fractions for the import-chase experiments. For the membrane transport experiments, the signals were adjusted to the mean within each repetition for each substrate. The normalized signals were then adjusted to the mean signals of the untreated transport reaction with ATP and SE for the repetitions for each substrate, except for sets of transports including His10-Plsp1Δ1–134, where they were adjusted to the untreated transport reaction without SE. Statistical analyses were performed with R, version 3.1.2. The Tukey honest significant difference (HSD) groupings were found for the inhibitor treatment, the interaction between SE and inhibitor treatment, and the interaction between SE, inhibitors, and cpSecA1 using Tukey HSD test of the agricolae package, version 1.1–9 with α = 0.05.
Preparation of Recombinant Proteins
Production of His6-tagged cpSecA1 was performed using BL21(DE3)-CodonPlus-RIL cells (Agilent Technologies). An aliquot of overnight culture (15 ml) was added to 300 ml of lysogeny broth containing 25 mg/liter kanamycin and 12.5 mg/liter chloramphenicol and grown further at 37 °C and 220 rpm until it reached an A600 of ∼0.6. Isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1 mm, and the cultures were incubated at 28 °C and 220 rpm for 4 h and pelleted by centrifugation at 4,000 × g and 4 °C for 20 min. The recovered cells were resuspended with buffer containing 50 mm NaH2PO4, 200 mm NaCl, and 20 mm imidazole, pH 8.0, lysozyme added to 1 mg/liter and were further incubated on ice for 30 min. They were then sonicated and centrifuged at 11,000 × g and 4 °C for 20 min. Recombinant cpSecA1 was purified from the resultant supernatant using Ni-NTA-agarose and eluted with buffer containing 50 mm NaH2PO4, 200 mm NaCl, 250 mm imidazole, pH 8.0, under the native conditions according to the manufacturer's instructions (Qiagen). His10-tagged Plsp1 variants were prepared as described (17).
Antisera Used for Immunoblotting and Antibody Inhibition Assay
Antisera used for immunoblotting include those against a His tag (His probe, H-15; Santa Cruz Biotechnology), Plsp1276–291 (20), human Hsp60 (SPA807; Enzo Life Sciences, Farmingdale, NY), and pea Hsp70 (66). For the antibody inhibition assay, antisera against pea translocon components (Alb3, cpSecY1, and Hcf106) were used (30).
Results
Plsp1 Accumulates as a Stromal Intermediate before ATP Hydrolysis-dependent Membrane Association
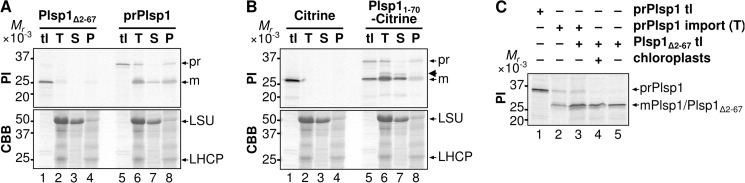
Plsp1 is encoded as a larger precursor (prPlsp1) in the nuclear genome (21). The antibody against its C terminus recognizes the endogenous protein (19), suggesting that prPlsp1 carries an N-terminal extension. Indeed, a neural network-based program ChloroP (33) predicts a transit peptide that is cleaved after residue 67. This idea was tested by in vitro targeting assays. As shown in Fig. 2, the N-terminal extension of Plsp1 was found to be necessary for import (compare lanes 2–4 and lanes 6–8 in A) and also sufficient for targeting a passenger protein, Citrine, to the chloroplast stroma (compare lanes 2–4 and lanes 7 and 8 in B). The mobility of imported Plsp1 on SDS-PAGE was comparable with that of Plsp1 lacking most of the predicted transit peptide (Plsp1Δ2–67) (Fig. 2C), confirming the prediction. In chloroplasts isolated from mature pea leaves, endogenous Plsp1 was detected only in the thylakoid fraction with the Nstroma-Clumen orientation (19). This proper orientation of Plsp1 is assessed by thermolysin treatment of lysed chloroplasts, which yields a 23-kDa fragment under the reducing conditions, lacking the stroma-exposed N terminus (named dp1 for degradation product 1 in Fig. 3A) (19). Interestingly, approximately one-fourth of mPlsp1 generated by in vitro import assay was found in the soluble fraction (Fig. 2A, lane 7), and the membrane-associated mPlsp1 was converted by thermolysin to a fragment of ∼21 kDa under the reducing conditions (named dp2) in addition to dp1 (Fig. 3A, lane 10).
FIGURE 2.
Import of Plsp1 and Plsp11–70-Citrine into isolated chloroplasts. A, in vitro chloroplast import of Plsp1 without or with the predicted transit peptide. After import of radiolabeled proteins listed on top with 3 mm ATP in the light for 30 min at room temperature, chloroplasts were reisolated and examined by SDS-PAGE directly as total (T) or after hypotonic lysis and separation by centrifugation into soluble (S) and pellet (P) fractions. Protein signals on the same gels were visualized using phosphorimaging (PI) or Coomassie Brilliant Blue staining (CBB). Each lane contained samples equivalent to 3 μg of total chlorophyll. tl lanes were loaded with 20% of the translation product equivalent to the amount used for the assay containing 3 μg of chlorophyll. The radioactive bands corresponding to prPlsp1 (pr) and mPlsp1 (m) and Coomassie Brilliant Blue-stained bands corresponding to the stroma marker large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase and the thylakoid marker light-harvesting chlorophyll a/b binding protein (LHCP) are indicated at right. B, in vitro chloroplast import of Citrine without or with the N-terminal extension of Plsp1. The import assay and analysis were done as described for A. Plsp11–70-Citrine and the protein derived from Plsp11–70-Citrine after import are indicated as pr and m, respectively. Note that the translation product of Plsp11–70-Citrine (lane 5) includes a band corresponding to mature Citrine, which is due to the in-frame initiation of AUG located at the 5′-end of the Citrine-coding sequence. This protein is most likely different from imported and matured Citrine (lanes 6–8) because Citrine by itself was not imported (lanes 2–4). A minor band slightly larger than mature Citrine, whose identity remains unknown, is indicated with an arrow. C, comparison of mPlsp1 derived from prPlsp1 after chloroplast import and Plsp1Δ2–67 in the mobility on SDS-PAGE. The radiolabeled proteins indicated on top were separated by SDS-PAGE and visualized using phosphorimaging. prPlsp1 import (T) is equivalent to the sample loaded on lane 6 in A.
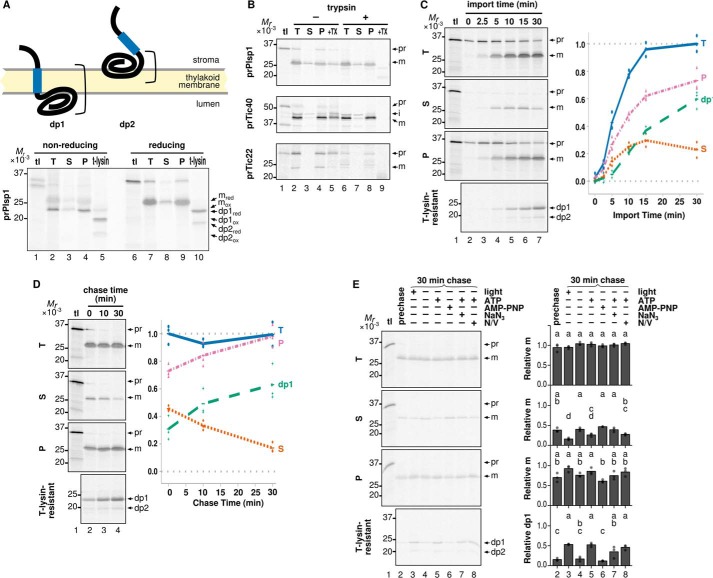
FIGURE 3.
Distribution of Plsp1 into soluble and membrane fractions after import into isolated chloroplasts. A, membrane topologies of imported Plsp1. Blue boxes denote the region corresponding to TMD (residues 111–128), and brackets indicate the portion protected from thermolysin. Radiolabeled prPlsp1 was imported into isolated chloroplasts and separated into total (T), soluble (S), pellet (P), and thermolysin-resistant (t-lysin) fractions as described in the legend to Fig. 2A. Samples were resuspended in sample loading buffer contained either no reducing agents (nonreducing) or 100 mm β-mercaptoethanol (reducing) and separated by SDS-PAGE, and protein signals were visualized using phosphorimaging. Reduced mPlsp1 (mred; detected in lanes 7–9), oxidized mPlsp1(mox; detected in lanes 2–4), reduced dp1 and dp2 (dp1red and dp2red; detected in lane 10), and oxidized dp1 and dp2 (dp1ox and dp2ox; detected in lane 5) are indicated to the right. B, in vitro chloroplast import followed by trypsin treatment and fractionation. After import of radiolabeled proteins indicated at left with 3 mm ATP in the light for 30 min at room temperature, chloroplasts were reisolated and incubated with buffer without (−) or with (+) trypsin at room temperature in the dark for 30 min. After the protease activity was quenched with trypsin inhibitor, chloroplasts were reisolated and separated into total (T), soluble (S), and pellet (P) fractions and analyzed as described above. Each lane contained samples equivalent to 3 μg of chlorophyll. As a control for trypsin activity, chloroplast samples were solubilized with 1% Triton X-100 before treatment (+TX). Bands corresponding to precursor (pr), intermediate (i; for Tic40 only), and mature (m) forms of each protein are indicated. C, import time course of Plsp1. Radiolabeled prPlsp1 was imported into isolated chloroplasts in the light with 3 mm ATP for the times indicated, followed by fractionation into total (T), soluble (S), and pellet (P) fractions and analyzed as with A. An aliquot of the total chloroplasts recovered after each time point was also lysed and incubated with thermolysin to yield dp1 and dp2. Shown at right is quantification of recovered mPlsp1 and dp1 in each fraction. The results of three repetitions and their means are shown as dots and lines, respectively. D, import-chase assay of Plsp1. Radiolabeled prPlsp1 was imported into isolated chloroplasts for 10 min in the light with 3 mm ATP, followed by treatment with 0.5 mg/ml thermolysin for 30 min on ice in the dark to remove unintegrated Plsp1 and quenching with 20 mm EDTA. Intact chloroplasts were then reisolated, incubated under light with 3 mm ATP for the times indicated, and then analyzed as described for A. Note that the EDTA treatment followed by wash with import buffer did not affect membrane insertion of Plsp1 (data not shown). E, energy requirement of membrane integration of soluble Plsp1 after import into isolated chloroplasts. Import-chase was performed and analyzed as in A under the conditions varying in the light, ATP (3 mm), AMP-PNP (3 mm), NaN3 (10 mm), and uncouplers (N/V, 1.5 μm each of nigericin and valinomycin). tl lanes were loaded as in Fig. 2. Prechase lane was loaded with the sample before the chase. Bands corresponding to prPlsp1 (pr), mPlsp1 (m), and the thermolysin degradation products (dp1 and dp2) are indicated at right. Quantification of three repetitions (circles) and their means (bars) is shown for each treatment. Means of treatments with the same letter do not differ significantly based on HSD for α = 0.05.
The soluble form of imported mPlsp1 may be located in the space between the outer and inner envelope membranes or in the stroma. It may also represent a mis-sorted end product or a productive intermediate en route to the membrane. These possibilities were tested by the following three assays. In the first assay, after import, intact chloroplasts were reisolated and treated with the protease trypsin, which can access the intermembrane space but not the stroma when chloroplasts remain intact (34). As shown in Fig. 3B, Tic40, whose large hydrophilic domain faces into the stroma (35), was resistant, whereas an intermembrane space control Tic22 (26) was digested (compare lanes 4 and 8). Under these conditions, imported Plsp1 in the soluble fraction remained intact (Fig. 3B, compare lanes 3 and 7), indicating its stroma localization. The second assay was an import time course (Fig. 3C). There, the level of mPlsp1 in total and membrane fractions, as well as that of dp1, increased over time, whereas that of the soluble form increased up to 15 min but leveled off and decreased at 30 min. The third assay was 30-min chase after 10-min import, in which the soluble form decreased, whereas the membrane-associated form and dp1 increased (Fig. 3D). The changes during the chase were not due to protein degradation because the amounts of Plsp1 in total chloroplasts after varying chase times were comparable (Fig. 3D). Taken together, these results indicate that soluble mPlsp1 represents a productive intermediate in the stroma en route to the membrane.
As for the thermolysin-resistant products, both dp1 and dp2 showed redox-dependent mobility shifts on SDS-PAGE (Fig. 3A, lanes 5 and 10), indicating that both contained the redox active pair of Cys residues at positions 166 and 286 (20). The size difference between dp1 and dp2 is ∼2 kDa, which is roughly equivalent to the size of TMD. During the import time course or chase assay, both dp1 and dp2 increased, and conversion of dp1 to dp2, or vice versa, was not detected (Fig. 3, C and D). These data suggest that dp1 and dp2 originated from distinct proteins and that the Plsp1 form that leads to dp2 after protease treatment is stably associated with the membrane with its TMD exposed at the stroma side. Structural predictions identified β-sheets but no α-helical domains long enough to span the membrane in the lumenal domain (Fig. 1). Thus, dp2 is most likely derived from a protein that inserts into but does not cross the membrane (Fig. 3A). This prediction is consistent with previous reports showing tight folding of multiple β-strands, such as a soluble β-barrel protein (36), and results of targeting assays described below.
During the import-chase, the correct membrane integration of the soluble intermediate was observed in the light or in the dark with ATP, but not without ATP or in the presence of the slowly hydrolyzed ATP analog AMP-PNP (Fig. 3E, lanes 3–6). Sodium azide, which is known to inhibit the cpSec1 motor cpSecA1 (37), also abolished the decrease of soluble Plsp1 during the chase (Fig. 3E, lane 7). An effect of the ionophores nigericin and valinomycin, which inhibit the cpTat pathway by collapsing the proton and charge gradients across the membranes, respectively (28), was not observed for membrane integration of Plsp1 (Fig. 3E, lane 8).
Membrane Insertion of Plsp1 Occurs with or without Stroma Extract
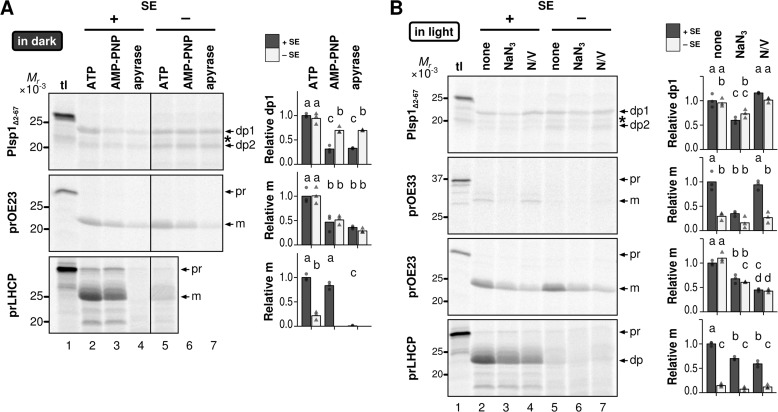
To gain further insight into the membrane transport of Plsp1, we tested whether Plsp1Δ2–67, which lacks the predicted transit peptide and thus mimics the stroma intermediate (Fig. 2), could properly insert into thylakoid-enriched membranes isolated from mature chloroplasts (19). ATP requirements were tested in the dark to avoid the effect of photosynthetic phosphorylation and thus strictly control the endogenous level of ATP (Fig. 4A). Effects of other inhibitors were tested under light (Fig. 4B). As reported previously (38), transport of the cpSec1 substrate OE33 was much lower in the dark than in the light (data not shown). Under light, OE33 was properly transported, but it required SE (compare lanes 2 and 5 in Fig. 4B). SE-dependent OE33 transport was inhibited by NaN3 to a degree comparable with that seen in the absence of SE (compare lanes 2, 3, and 5 in Fig. 4B) as reported previously (30). Transport of the cpTat substrate OE23, by contrast, could occur independently of light and SE (Figs. 4, A, lanes 2 and 5, and B, lanes 2 and 5) and was inhibited by AMP-PNP and apyrase, either of which prevents ATP hydrolysis-dependent formation of proton motive force across the thylakoid membrane in the dark (Fig. 4A). Sodium azide and ionophores also inhibited OE23 transport in the light regardless of the presence or absence of SE (Fig. 4B). These results are consistent with proton motive force-dependent transport of OE23 (28). Transport of the chloroplast SRP (cpSRP) substrate LHCP occurred in the dark in the presence of ATP and SE (Fig. 4A, lanes 2 and 5); it was not affected by AMP-PNP (lane 3) but was inhibited by apyrase (lane 4), which removes all NTPs including GTP as reported previously (38). LHCP transport in the light also depended on SE, which was inhibited by ionophores as reported previously (39) and also by NaN3 (Fig. 4B). Under these conditions, radiolabeled Plsp1Δ2–67 correctly inserted into the membrane in the dark when ATP was included in the assay, either in the presence or absence of SE (Fig. 4A, lanes 2 and 5), and was inhibited by AMP-PNP by ∼70% in the presence of SE (compare lanes 2 and 3 and lanes 2 and 4) or by ∼26% in the absence of SE (compare lanes 5–7). Transport of Plsp1 in the light was inhibited by NaN3 by 40% (Fig. 4B, compare lanes 2 and 3 and lanes 5 and 6) but not by ionophores (compare lanes 2 and 4 and lanes 5 and 7), regardless of the presence or absence of SE. Notably, after transport in the dark, the ratio of dp2 to dp1 was higher in the absence of SE (∼0.9) than in the presence of SE (∼0.6) (Fig. 4A, lanes 2 and 5). Furthermore, in the presence of SE, ATP depletion by apyrase led to an increase of the dp2 to dp1 ratio to ∼1.0 (Fig. 4A, lane 4).
FIGURE 4.
Energy requirement of integration of Plsp1 into isolated chloroplast membranes. A, effects of ATP and ATP depletion. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes for 30 min in the dark with (+) or without (−) SE. The transport reactions also contained 5 mm ATP, 5 mm AMP-PNP, or apyrase (1 unit/150 μl). The resultant chloroplast membranes were treated with thermolysin, and the products were separated by SDS-PAGE and analyzed using phosphorimaging. The lanes labeled tl were loaded with 10% of translation products used for the assay. Shown at right are quantifications of transport for three replications of the experiment (circles and triangles) and their means (bars). The results of experiments with SE are depicted with circles and dark gray bars, whereas those without SE are shown with triangles and light gray bars. B, effects of NaN3 and uncouplers. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes for 30 min in the light with 5 mm ATP with (+) or without (−) SE. The transport reactions also contained 5 mm NaN3 or 1.5 μm each of nigericin and valinomycin (N/V). The resultant chloroplasts were treated with thermolysin, and the products were analyzed as A. A band between dp1 and dp2 is indicated with an asterisk. It appeared occasionally after thermolysin treatment; when it appeared, its intensity did not vary between treatments. Thus, we did not include it in the analysis.
Together, these data suggest that Plsp1 inserts into the chloroplast membrane by at least two mechanisms. The first mechanism depends on ATP hydrolysis to achieve a functional orientation, similar to the cpSec1-dependent targeting of OE33. This is observed by the import assay using isolated chloroplasts and the membrane transport assay in the presence of SE. The second mechanism occurs in the absence of SE, NTP, and proton motive force and increases the frequency of mis-sorting as shown by the high dp2 to dp1 ratio. A population of Plsp1 may insert into the membrane via the second mechanism even in the presence of SE, especially when ATP is depleted. Given the lack of Arg-Arg or Lys-Arg motif in the region N-terminal to TMD, it was not a surprise that Plsp1 transport occurs independently of the cpTat pathway. However, its similarity to OE33 in the presence of SE was unexpected, because Plsp1 lacks cTTS.
Membrane Insertion of Plsp1 Occurs by the cpSecY1 Channel in the Presence of the Stroma or Spontaneously in the Absence of Stroma
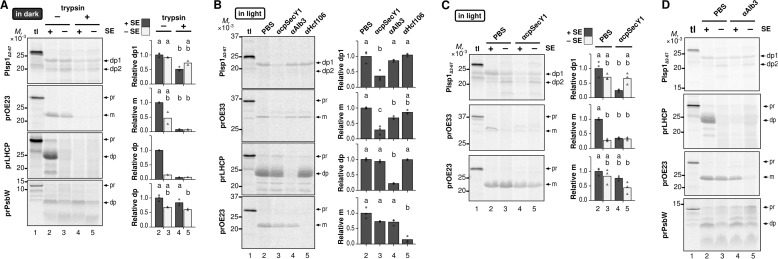
To test involvement of proteinaceous components, chloroplast membranes were first treated with trypsin and used for transport assay in the dark. As shown in Fig. 5A, OE23 transport was almost completely abolished regardless of the presence or absence of SE (compare lanes 2 and 4 and lanes 3 and 5, respectively), and so was the SE-dependent LHCP transport (compare lanes 2 and 4). By contrast, transport of PsbW, which is known to occur spontaneously in the dark (40), was not affected in all the conditions used (Fig. 5A). Under these conditions, Plsp1 transport in the dark was decreased by 50% in the presence of SE (Fig. 5A, compare lanes 2 and 4) but not affected in the absence of SE (Fig. 5A, compare lanes 3 and 5).
FIGURE 5.
Requirements of proteinaceous components for integration of Plsp1 into isolated chloroplast membranes. A, effects of pretransport protease treatment. Radiolabeled proteins indicated at left were incubated with chloroplast membranes pretreated without (−) or with (+) trypsin. The reactions were performed with 5 mm ATP in the dark for 30 min with (+) or without (−) SE, followed by thermolysin treatment. The products were then analyzed as in Fig. 4. The tl lanes were loaded with 10% of translation products used for the assay. Shown at right are quantifications of each of two replications (without SE, circles) or three (with SE, triangles) and their means (bars). Samples without trypsin pretreatment are indicated with circles and dark bars, and those pretreated with trypsin are shown with triangles and light bars. Only the two repetitions of the experiment with all treatments were used for statistical analysis except for prLHCP, which had only one repetition; thus statistical analysis could not be performed. B, effects of antibodies against thylakoid translocons in the presence of SE. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes pretreated with PBS or antibodies against cpSecY1 (αcpSecY1), Alb3 (αAlb3), or cpHcf106 (αcpHcf106) in the presence of SE and 5 mm ATP under light for 30 min. The reactions were analyzed as in A. C and D, effects of the antibody against cpSecY1 (C) or Alb3 (D) in the presence or the absence of SE. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes pretreated with PBS or the antibody indicated in the light with 5 mm ATP for 30 min with (+) or without (−) SE. The reactions were analyzed in A, with circles and dark bars for PBS, and triangles and light bars for the anti-cpSecY1 antibody pretreatment. Bands corresponding to the expected sizes of the precursor (pr), mature (m), and degradation products (dp for LHCP and PsbW; dp1 and dp2 for Plsp1) are indicated to the right of each gel. For A–C, treatments with the same letter do not have significantly different means based on Tukey HSD for α = 0.05.
To further define proteinaceous components required for Plsp1 insertion, an antibody inhibition assay was done by pretreating the isolated chloroplast membranes with antisera against integral components of the three transport pathways as demonstrated previously (30). To visualize the effect on transport of the control cpSec1 substrate OE33, the assay was done in the light. As shown in Fig. 5B, in the presence of SE, Plsp1 insertion was significantly lowered only when the chloroplast membranes were pretreated with the anti-cpSecY1 antibody (64% inhibition). This pattern was similar to that of OE33 (71% inhibition), but not to those of LHCP and OE23, which are cpSRP and cpTat substrates and were inhibited specifically by anti-Alb3 and anti-Hcf106 antibodies, respectively (Fig. 5B). In the absence of SE, by contrast, Plsp1 insertion was not affected by antibodies against cpSecY1 (Fig. 5C, compare lanes 3 and 5) or Alb3 (Fig. 5D).
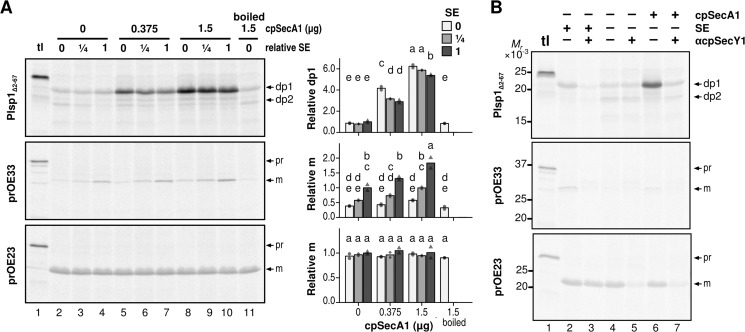
Proper Integration of Plsp1 Is Enhanced by cpSecA1
Our data suggest that SE-dependent proper integration of Plsp1 uses the cpSec1 pathway, which involves the ATP-consuming motor component cpSecA1, whereas SE-independent transport of Plsp1 occurs spontaneously. To confirm these ideas, we tested the effect of bacterially produced cpSecA1 on Plsp1 transport as demonstrated previously with soluble cpSec1 substrates (41). As shown in Fig. 6A, exogenously added cpSecA1 enhanced transport of the cpSec1 substrate OE33 and proper insertion of Plsp1 in a dose-dependent manner regardless of the presence or absence of SE, whereas it did not affect cpTat transport (OE23). The correct Plsp1 integration in the absence of SE was greatly enhanced by the presence of cpSecA1 as judged by a change in the dp2 to dp1 ratio from 1.0 to 0.2 (Fig. 6B, compare lanes 4 and 6). The enhancing effect by cpSecA1 was inhibited by preincubation of the chloroplast membranes with the anti-cpSecY1 antibody (Fig. 6B, compare lanes 6 and 7). These data indicate that cpSecA1 prevents Plsp1 from spontaneous mistargeting and ensures the correct insertion by the cpSecY channel.
FIGURE 6.
Effects of recombinant cpSecA1 on transport of Plsp1 into isolated chloroplast membranes. A, effects of cpSecA1 and SE. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes in the light with 5 mm ATP and varying amounts of SE and recombinant cpSecA1 for 30 min. Analyses of the products were done as described in Fig. 4B. tl lanes were loaded with 10% of translation products used for the assay. To the right are graphs showing quantifications of two repetitions (circles, diamonds, and triangles) and their means (bars) for each treatment. Circles with light gray bars, diamonds with medium gray bars, and triangles with dark gray bars denote specific amounts of SE included in the assay. Letters indicate groupings of treatment with means that do not significantly differ from each other based on Tukey HSD with α = 0.05. Quantifications are relative to transport in the presence of undiluted stroma extract without cpSecA1. B, effects of cpSecA1. SE and the antibody against cpSecY1. Chloroplast membranes preincubated with PBS (αcpSecY1−) or anti-cpSecY1 (αcpSeY1+) were used for the transport assay including radiolabeled proteins indicated at left, 5 mm ATP and without (−) or with (+) SE and cpSecA1 in the light. The resultant products were analyzed as described in the legend to A. Precursor (pr) and mature (m) forms of OE23 and OE33, and thermolysin-resistant fragments of Plsp1 (dp1 and dp2) are indicated to the right.
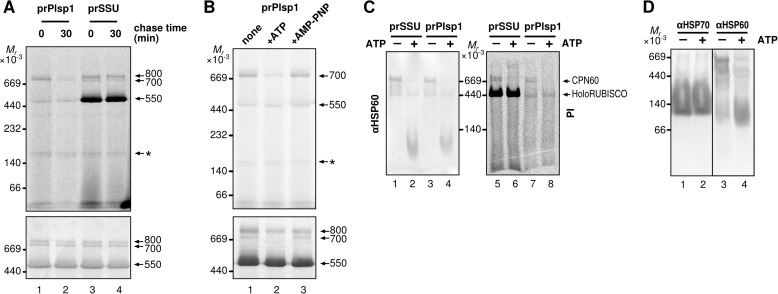
Stromal Plsp1 Is Present in a 700-kDa Complex That Co-migrates with the Cpn60 Chaperone Complex
Interestingly, cpSecA1-dependent enhancement of Plsp1 transport, but not that of OE33, appeared to be inhibited by SE (Fig. 6A, compare lanes 5–7 and 8–10, respectively). To gain a molecular base for the effect of SE, we examined the oligomeric status of imported mPlsp1 in the stroma using BN-PAGE, which has been used to separate membrane and soluble protein complexes by apparent molecular mass (42). As a control, radiolabeled ribulose-1,5-bisphosphate carboxylase/oxygenase (RUBISCO) small subunit was also imported and examined. The newly imported small subunit of RUBISCO is known to be integrated into the Cpn60 complex as the folding intermediate in addition to the holoenzyme (43, 44). As shown in Fig. 7A, the radiolabeled small subunit of RUBISCO was found mainly in three bands, all of which co-migrated with protein complexes visible by Coomassie Brilliant Blue staining: the 550-kDa band corresponding to the holoenzyme showed the strongest signal intensity, followed by two bands at 800 and 700 kDa, respectively. Radiolabeled Plsp1 recovered in the stroma after import co-migrated with the 700-kDa band, whose level decreased after a 30-min chase (Fig. 7A, lanes 1 and 2). Decline of the level of this radiolabeled band was also observed when the stroma fraction was incubated with ATP but not with AMP-PNP (Fig. 7B, lanes 2 and 3). To gain insight into the identity of the 700-kDa band, the stroma proteins including the imported radiolabeled proteins separated by BN-PAGE were transferred from the gel to a PVDF membrane and examined by autoradiography or immunoblotting with the antibody against the Cpn60 homolog HSP60. Compared with the gel (Fig. 7, A and B), separation of the 800- and 700-kDa bands was less clear on the blot (Fig. 7C). Nonetheless, these bands appear to co-migrate with the immunoreactive band, which was dissociated from the large complex when ATP was added (Fig. 7C) as reported previously for Cpn60 (45, 46). Finally, the presence of Hsp70 in the 700-kDa complex was under the detection limit of immunoblotting (Fig. 7D).
FIGURE 7.
Incorporation of soluble import intermediates into a large oligomeric complex. A, separation of soluble intermediate by BN-PAGE after import-chase. After 10-min import of radiolabeled proteins indicated on top, chloroplasts were treated with thermolysin, reisolated, and subjected to 0- or 30-min chase as described in the legend to Fig. 3D. Chloroplasts were then lysed hypotonically and fractionated by centrifugation. The resultant soluble fraction was separated by BN-PAGE and proteins on the same gel visualized using phosphorimaging (top panel) or by Coomassie Brilliant Blue staining (bottom panel). Three complexes of 800, 700, and 550 kDa are indicated at right with the numbers. An asterisk indicates a nonspecific band. Note that the lane loaded with imported Plsp1 contains a 550-kDa band. This corresponds to the RUBISCO holoenzyme, either as a shadow of the endogenous one, which is abundant, or the one incorporating its large subunit translated from the chloroplast ribosomes with residual radiolabeled amino acids derived from in vitro translation. B, mobility of soluble import intermediate on BN-PAGE after 10-min import followed by various treatments. After 10-min import of radiolabeled prPlsp1, chloroplasts were reisolated, lysed hypotonically, and fractionated by centrifugation. The resultant soluble fraction was further incubated on ice for 10 min with buffer (none), 10 mm ATP or 10 mm AMP-PNP, then separated, and analyzed as described in the legend to A. The 700-kDa complex and the band corresponding to the RUBISCO holoenzyme are indicated at right. C, co-migration of soluble import intermediates with Cpn60 on BN-PAGE. After 10-min import of radiolabeled proteins indicated on top into intact chloroplasts, the soluble fraction was recovered as in A and treated without (−) or with 10 mm ATP (+) for 10 min, separated by BN-PAGE, transferred to a PVDF membrane, and examined by immunoblotting with the antibody against Hsp60 (αHSP60, left panel) or using phosphorimaging (PI, right panel). Cpn60 complex and RUBISCO holoenzyme (HoloRUBISCO) are indicated at right. D, migration of Cpn60 and Hsp70 complexes on BN-PAGE. Soluble chloroplast fractions prepared as in A without import of radiolabeled proteins were incubated without (−) and with (+) 10 mm ATP, separated by SDS-PAGE, transferred to a PVDF membrane, and examined by immunoblotting with antibodies against Hsp70 (αHSP70) or Hsp60 (αHSP60).
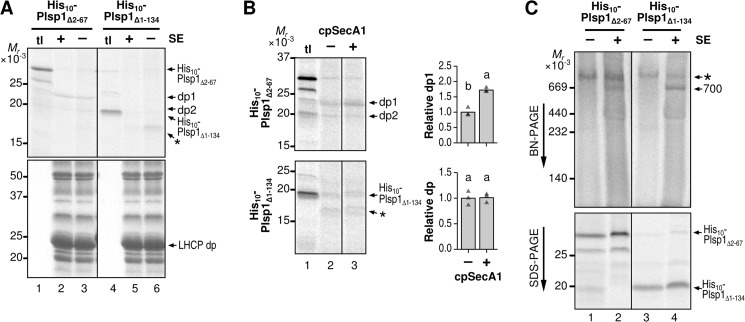
The Transmembrane Domain Is Dispensable for Spontaneous Membrane Insertion and Integration into the 700-kDa Complex
We wondered whether spontaneous membrane insertion was due to the presence of the TMD within Plsp1 or whether it occurred independently of the TMD as was shown for LepB (6). To test these ideas, we used the construct encoding the C-terminal lumenal portion of Plsp with an N-terminal decahistidine tag (His10-Plsp1Δ1–134) (20) for in vitro transcription and translation and subjected the resultant radiolabeled 19-kDa protein to the transport assay. As shown in Fig. 8A, after transport, thermolysin treatment yielded a fragment of ∼16 kDa in the absence (lane 6) but not in the presence of SE (lane 5). Notably, SE-independent insertion of His10-Plsp1Δ1–134 was not enhanced by cpSecA1, unlike insertion of His10-Plsp1Δ2–67 (Fig. 8B).
FIGURE 8.
Requirements of TMD within Plsp1 for spontaneous membrane insertion, cpSecA1-dependent transport, and association with the stroma complex. A, effects of SE on transport of His-tagged Plsp1 variants. Radiolabeled proteins indicated on top were incubated with isolated chloroplast membranes with 5 mm ATP in the light for 30 min. After treatment with thermolysin, products were separated by SDS-PAGE. Proteins on the same gel were visualized by phosphorimaging (top panel) or Coomassie Brilliant Blue staining (bottom panel). tl lanes were loaded with 10% of translation products used for the assay. B, effects of cpSecA1 on transport of His-tagged Plsp1 variants. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes in the light in the absence of SE, without (−) or with (+) 0.375 μg of recombinant cpSecA1, followed by thermolysin treatment. Products were separated by SDS-PAGE and visualized using phosphorimaging. Shown at right are quantifications of data from three repetitions (triangles) and their means (bars) with letters indicating Tukey HSD groupings above for α = 0.05. C, incorporation of His-tagged Plsp1 variants into the 700-kDa complex in SE. Radiolabeled proteins indicated on top were incubated without (−) or with (+) SE for 30 min at room temperature, followed by transfer to ice and addition of cold BN-PAGE sample buffer. Samples were then separated by BN-PAGE (top) or SDS-PAGE (bottom) and visualized using phosphorimaging. The 700-kDa complex incorporating the radiolabeled Plsp1 variants (700) and a larger complex present in the translation product mixture (*) are indicated to the right of the BN-PAGE gel. The bands corresponding to each of the two Plsp1 variants are indicated at the right of the SDS-PAGE gel.
Because integration into the large stroma complex appeared to compete with the spontaneous insertion of Plsp1, we tested whether radiolabeled His10-Plsp1Δ1–134 could associate with the complex. Because the protein lacks the chloroplast import signal, the assay was done by incubating the radiolabeled protein with SE. As shown in Fig. 8C, the TMD-less protein showed a stronger association with the 700-kDa complex than the one with TMD (His10-Plsp1Δ2–67) (compare lanes 2 and 4 in top panel). Together, the data indicate that the Plsp1 TMD is needed for cpSecA1-dependent transport but is dispensable for association with the 700-kDa complex and spontaneous membrane insertion.
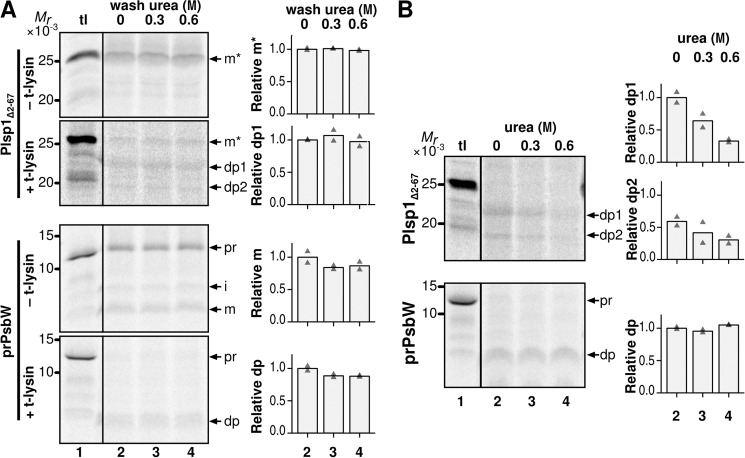
Urea Inhibits SE-independent Insertion of Plsp1
The predicted structure (Fig. 1) and results of the biochemical assays (Fig. 8A) suggest that the lumenal domain of Plsp1 may fold to form a hydrophobic surface and insert into the membrane spontaneously, similar to the case with the periplasmic domain of LepB (6). To test this idea, we examined the effect of the denaturing agent urea (47) on SE-independent insertion of Plsp1Δ2–67. As controls, we tested effects of urea on the stability of the inserted proteins and also transport of PsbW, which depends on its TMD and occurs spontaneously (48). As shown in Fig. 9A, when urea of up to 0.6 m was added after the insertion, the amounts of thermolysin-protected forms (dp1 and dp2 for Plsp1Δ2–67 and dp for PsbW) were not affected (compare lanes 2–4), indicating that these concentrations of urea did not affect stability of the inserted protein. By contrast, when urea was included during the SE-independent insertion assay, membrane insertion of Plsp1Δ2–67, as judged by the production of dp1 and dp2 but not that of PsbW, was lowered by 67 and 49%, respectively (Fig. 9B, compare dp1 and dp2 in lanes 2 and 4). PsbW inserts into the membrane via α-helices (49), which are known to be more resistant to urea than β-sheets (50). These data indicate that spontaneous insertion of Plsp1, but not stability of the membrane-inserted form, was disturbed by urea, which most likely affects premature folding of the β-sheet.
FIGURE 9.
The effect of urea on membrane association and transport of Plsp1. A, effects of post-transport treatment of urea on membrane-associated Plsp1. After the assay as described in Fig. 4A, the recovered chloroplast membranes were resuspended with buffer containing the indicated concentrations of urea and incubated at room temperature in the dark for 10 min. The membranes were recovered by centrifugation and treated with the same buffer again. The resultant membrane fractions were resuspended in import buffer and treated without (−) or with (+) thermolysin (t-lysin) before analysis as described in Fig. 2A. The degradation products of Plsp1 (dp1 and dp2) and precursor (pr), intermediate (i), mature (m), and degradation product (dp) of PsbW are indicated. B, effects of urea on SE-independent insertion of Plsp1 into isolated chloroplast membranes. Radiolabeled proteins indicated at left were incubated with isolated chloroplast membranes in the light in the absence of SE with the indicated concentrations of urea. The reactions were incubated and analyzed as described for Fig. 4.
Discussion
Genetic and immunolocalization studies have established the presence of Plsp1 in both the envelope and thylakoid membranes (19, 21), although the localization mechanism to one or both remained unknown. The present study used two distinct systems to address part of this issue. The first system used intact chloroplasts, which has helped identify the presence of the stroma intermediate and the requirement of ATP hydrolysis for membrane insertion. The second assay used the membrane fraction of chloroplasts from mature pea leaves, which is enriched with thylakoids (19). The result demonstrates that proper membrane insertion of Plsp1 is assisted by Cpn60 and the cpSec1 pathway. One of the key results is the inhibition by the antibody against cpSecY1, which was found only in thylakoids (51). Thus, our results explain the mechanism of Plsp1 targeting to the thylakoid. Whether Cpn60 also assists the targeting of Plsp1 to the envelope, which may or may not require another translocon, remains to be investigated. A robust assay to quantify protein distribution to the envelope and thylakoids, which is currently missing, needs to be established to elucidate the envelope-targeting mechanism.
The Sec translocon transports unfolded proteins into and across the membranes in a wide range of organisms. In bacteria, the Sec system transports large periplasmic portions of membrane proteins co-translationally or exports proteins destined to the periplasm, outer membrane, or extracellular space, post-translationally (52). Export signals of post-translational substrates are cleaved upon transport, allowing protein release from the membrane (52), whereas those of co-translational Sec substrates often remain in the mature protein as a membrane anchor (53). In chloroplasts, the cpSec1 system has been known to catalyze thylakoid transport of both soluble (lumenal) and membrane proteins, all of which carry a signal sequence that is cleaved upon transport (13). Our results demonstrate that a single-pass membrane protein Plsp1 is a cpSec1 substrate, which does not carry a cleavable signal, although it is targeted post-translationally. TMD of Plsp1 is necessary for the cpSecA1-dependent transport; thus it may act as an uncleavable TTS. This is reminiscent of the unusual cpTat substrate ISP, which appears to use its TMD as an uncleavable TTS (17). However, the detailed transport mechanism of ISP has not been examined because it cannot insert into isolated thylakoids. Further studies should use Plsp1 as a model protein to define the mechanism of cTTS-less post-translational protein transport and also to test whether there are cTTS-less cpSec1 substrates other than Plsp1. Their outcomes should advance our mechanistic understanding of evolutionarily conserved Sec transport.
Our results also show that Plsp1 can spontaneously insert into the chloroplast membranes but with a high frequency of mis-sorting. Integration into a 700-kDa complex that co-migrates with Cpn60 in the stroma appears to prevent premature folding and thus mistargeting. Cpn60 is related to a bacterial chaperonin known as GroEL (54). GroEL binds to various client proteins at a hydrophobic surface and uses ATP hydrolysis to catalyze protein folding and unfolding (55). It is also known to assist Sec- and Tat-dependent protein export from the cytoplasm in bacteria (56). In chloroplasts, Cpn60 has been shown to mediate folding and assembly of the RUBISCO holoenzyme (57) and the catalytic subunit of the ATP synthase (58, 59). Results of in vitro assays also showed interaction of Cpn60 with a variety of other newly imported proteins (60–63), although its significance and mechanism largely elusive. Our finding suggests that Cpn60-dependent protein unfolding is needed for proper transport in chloroplasts. The outcomes of the future studies that will identify the components of the 700-kDa complex that incorporate Plsp1 are expected to further our understanding of the functions and specificity of molecular chaperones in chloroplasts. Furthermore, these works have used a heterologous system, i.e. targeting of Arabidopsis Plsp1 to pea chloroplasts/chloroplast membranes. Pea chloroplasts have been used to demonstrate targeting efficiency of various Arabidopsis proteins (64, 65). However, it remains to be determined whether the degree of unassisted mis-sorting in pea chloroplasts is less for the endogenous protein (pea Plsp1) compared with that for the Arabidopsis ortholog.
Similar to the case in LepB, the C-terminal portion of Plsp1 including catalytic residues is predicted to form a hydrophobic surface at the trans-side of the membrane (Fig. 1). This folding is necessary for hydrolysis of peptide bonds near or in the membrane at the trans-side (2), although premature folding would lead to incorrect insertion from the cis-side. In bacteria, this problem is prevented by co-translational transport by the SRP pathway, which initiates translocation of unfolded polypeptide chain, whereas it is still attached to ribosomes (66, 67). This system also exists in chloroplasts, as demonstrated by co-translational insertion of LepB into isolated thylakoids (68). By contrast, Plsp1 is encoded in the nuclear genome, and its targeting occurs post-translationally. Our data suggest that spontaneous insertion of Plsp1 from the cis-side of the membrane is prevented by Cpn60, whose homolog exists in bacteria but does not play a role in co-translational insertion of SPase I. This may represent an adaptive mechanism during organelle evolution, from the ribosome-dependent co-translational insertion to the chaperone-dependent post-translational transport. This is similar to the case with the cpSRP pathway, which has evolved to catalyze post-translational membrane insertion of LHCP during chloroplast evolution with acquisition of a novel chaperone cpSRP43 (69, 70). Together, these findings emphasize the flexibility of protein transport mechanisms.
Author Contributions
J. K. E. and K. I. designed all the experiments except preparation of a plasmid for recombinant cpSecA1. R. S. and D. E. F. established a method to prepare recombinant cpSecA1. J. K. E. conducted all the experiments. J. K. E. and K. I. analyzed the data and wrote the paper. All the authors reviewed and completed the manuscript.
Acknowledgments
We thank Dr. Kenneth Cline (University of Florida) for providing us with antibodies against pea thylakoid translocon components and very helpful suggestions for transport assays, Dr. Gitta Coaker (University of California, Davis) for seeds of Nicotiana benthamiana, Dr. Roger Tsien (University of California, San Diego) for the plasmid containing the Citrine coding sequence, Dr. Anne Knowlton (University of California, Davis) for the HSP60 antibody, and Drs. Steven Theg (University of California, Davis) and Kenneth Keegstra (Michigan State University) for the antibody against Hsp70.
This work was supported in part by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Dept. of Energy through Grant DE-FG02–08ER15963 (to K. I.). Establishment of recombinant cpSecA1 preparation was supported by National Science Foundation Grant MCB-1158173 (to D. E. F.). Purchase of reagents for in vitro transcription and translation was supported by a Henry A. Jastro Research Scholarship (to J. K. E.). The authors declare that they have no conflicts of interest with the contents of this article.
- SPase I
- type I signal peptidase
- BN
- blue native
- Cpn60
- chaperonin 60
- cpSec
- chloroplast Sec
- cpTat
- chloroplast twin-Arg translocation
- cTTS
- cleavable thylakoid-transfer signal
- dp
- degradation product
- IB
- import buffer
- IBM
- IB containing 10 mm MgCl2
- ISP
- Rieske iron/sulfur subunit of cytochrome b6f complexes
- LHCP
- light-harvesting chlorophyll-a/b-binding protein
- Plsp1
- plastidic type I signal peptidase 1
- RUBISCO
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- SE
- stroma extract
- TMD
- transmembrane domain
- HSD
- Tukey honest significant difference
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1.Schatz G., and Dobberstein B. (1996) Common principles of protein translocation across membranes. Science 271, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 2.Paetzel M. (2014) Structure and mechanism of Escherichia coli type I signal peptidase. Biochim. Biophys. Acta 1843, 1497–1508 [DOI] [PubMed] [Google Scholar]
- 3.San Millan J. L., Boyd D., Dalbey R., Wickner W., and Beckwith J. (1989) Use of phoA fusions to study the topology of the Escherichia coli inner membrane protein leader peptidase. J. Bacteriol. 171, 5536–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalbey R. E., Wang P., and van Dijl J. M. (2012) Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol. Mol. Biol. Rev. 76, 311–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paetzel M., Dalbey R. E., and Strynadka N. C. (1998) Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature 396, 186–190 [DOI] [PubMed] [Google Scholar]
- 6.van Klompenburg W., Paetzel M., de Jong J. M., Dalbey R. E., Demel R. A., von Heijne G., and de Kruijff B. (1998) Phosphatidylethanolamine mediates insertion of the catalytic domain of leader peptidase in membranes. FEBS Lett. 431, 75–79 [DOI] [PubMed] [Google Scholar]
- 7.Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., and Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 8.Wolfe P. B., Rice M., and Wickner W. (1985) Effects of two sec genes on protein assembly into the plasma membrane of Escherichia coli. J. Biol. Chem. 260, 1836–1841 [PubMed] [Google Scholar]
- 9.Lee J. I., Kuhn A., and Dalbey R. E. (1992) Distinct domains of an oligotopic membrane protein are Sec-dependent and Sec-independent for membrane insertion. J. Biol. Chem. 267, 938–943 [PubMed] [Google Scholar]
- 10.Paila Y. D., Richardson L. G., and Schnell D. J. (2015) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 427, 1038–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue K., and Glaser E. (2014) Processing and degradation of chloroplast extension peptides. In Plastid Biology (Theg S. M., and Wollman F.-A., eds) pp. 305–323, Springer; New York [Google Scholar]
- 12.Li H. M., and Teng Y. S. (2013) Transit peptide design and plastid import regulation. Trends Plant Sci. 18, 360–366 [DOI] [PubMed] [Google Scholar]
- 13.Dabney-Smith C., and Storm A. (2014) Protein routing processes in the thylakoid. In Plastid Biology (Theg S. M., and Wollman F.-A., eds) pp. 271–289, Springer; New York [Google Scholar]
- 14.Di Cola A., Klostermann E., and Robinson C. (2005) The complexity of pathways for protein import into thylakoids: it's not easy being green. Biochem. Soc. Trans. 33, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 15.Albiniak A. M., Baglieri J., and Robinson C. (2012) Targeting of lumenal proteins across the thylakoid membrane. J. Exp. Bot. 63, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 16.Molik S., Karnauchov I., Weidlich C., Herrmann R. G., and Klösgen R. B. (2001) The Rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts: missing link in the evolution of protein transport pathways in chloroplasts? J. Biol. Chem. 276, 42761–42766 [DOI] [PubMed] [Google Scholar]
- 17.Madueño F., Bradshaw S. A., and Gray J. C. (1994) The thylakoid-targeting domain of the chloroplast Rieske iron-sulfur protein is located in the N-terminal hydrophobic region of the mature protein. J. Biol. Chem. 269, 17458–17463 [PubMed] [Google Scholar]
- 18.Hsu S. C., Endow J. K., Ruppel N. J., Roston R. L., Baldwin A. J., and Inoue K. (2011) Functional diversification of thylakoidal processing peptidases in Arabidopsis thaliana. PLoS One 6, e27258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shipman R. L., and Inoue K. (2009) Suborganellar localization of plastidic type I signal peptidase 1 depends on chloroplast development. FEBS Lett. 583, 938–942 [DOI] [PubMed] [Google Scholar]
- 20.Midorikawa T., Endow J. K., Dufour J., Zhu J., and Inoue K. (2014) Plastidic type I signal peptidase 1 is a redox-dependent thylakoidal processing peptidase. Plant J. 80, 592–603 [DOI] [PubMed] [Google Scholar]
- 21.Inoue K., Baldwin A. J., Shipman R. L., Matsui K., Theg S. M., and Ohme-Takagi M. (2005) Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shipman-Roston R. L., Ruppel N. J., Damoc C., Phinney B. S., and Inoue K. (2010) The significance of protein maturation by plastidic type I signal peptidase 1 for thylakoid development in Arabidopsis chloroplasts. Plant Physiol. 152, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A., and Tsien R. Y. (2001) Reducing the environmental sensitivity of yellow fluorescent protein: mechanism and applications. J. Biol. Chem. 276, 29188–29194 [DOI] [PubMed] [Google Scholar]
- 24.Endow J. K., and Inoue K. (2013) Stable complex formation of thylakoidal processing peptidase and PGRL1. FEBS Lett. 587, 2226–2231 [DOI] [PubMed] [Google Scholar]
- 25.Curtis M. D., and Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouranov A., Wang H., and Schnell D. J. (1999) Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274, 25181–25186 [DOI] [PubMed] [Google Scholar]
- 27.Tripp J., Inoue K., Keegstra K., and Froehlich J. E. (2007) A novel serine/proline-rich domain in combination with a transmembrane domain is required for the insertion of AtTic40 into the inner envelope membrane of chloroplasts. Plant J. 52, 824–838 [DOI] [PubMed] [Google Scholar]
- 28.Cline K., Ettinger W. F., and Theg S. M. (1992) Protein-specific energy requirements for protein transport across or into thylakoid membranes: two lumenal proteins are transported in the absence of ATP. J. Biol. Chem. 267, 2688–2696 [PubMed] [Google Scholar]
- 29.Inoue K., and Keegstra K. (2003) A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 34, 661–669 [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues R. A., Silva-Filho M. C., and Cline K. (2011) FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J. 65, 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnon D. I. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson S. J., Robinson C., and Mant A. (1999) Dual signal peptides mediate the signal recognition particle/Sec-independent insertion of a thylakoid membrane polyprotein, PsbY. J. Biol. Chem. 274, 4059–4066 [DOI] [PubMed] [Google Scholar]
- 33.Emanuelsson O., Nielsen H., Brunak S., and von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 34.Jackson D. T., Froehlich J. E., and Keegstra K. (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273, 16583–16588 [DOI] [PubMed] [Google Scholar]
- 35.Chou M. L., Fitzpatrick L. M., Tu S. L., Budziszewski G., Potter-Lewis S., Akita M., Levin J. Z., Keegstra K., and Li H. M. (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22, 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang C. F., Okou D. T., Griffin T. B., Verret C. R., and Williams M. N. (2001) Green fluorescent protein rendered susceptible to proteolysis: positions for protease-sensitive insertions. Arch. Biochem. Biophys. 394, 229–235 [DOI] [PubMed] [Google Scholar]
- 37.Yuan J., Henry R., McCaffery M., and Cline K. (1994) SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science 266, 796–798 [DOI] [PubMed] [Google Scholar]
- 38.Yuan J., and Cline K. (1994) Plastocyanin and the 33-kDa subunit of the oxygen-evolving complex are transported into thylakoids with similar requirements as predicted from pathway specificity. J. Biol. Chem. 269, 18463–18467 [PubMed] [Google Scholar]
- 39.Cline K., Fulsom D. R., and Viitanen P. V. (1989) An imported thylakoid protein accumulates in the stroma when insertion into thylakoids is inhibited. J. Biol. Chem. 264, 14225–14232 [PubMed] [Google Scholar]
- 40.Woolhead C. A., Thompson S. J., Moore M., Tissier C., Mant A., Rodger A., Henry R., and Robinson C. (2001) Distinct Albino3-dependent and -independent pathways for thylakoid membrane protein insertion. J. Biol. Chem. 276, 40841–40846 [DOI] [PubMed] [Google Scholar]
- 41.Nakai M., Goto A., Nohara T., Sugita D., and Endo T. (1994) Identification of the SecA protein homolog in pea chloroplasts and its possible involvement in thylakoidal protein-transport. J. Biol. Chem. 269, 31338–31341 [PubMed] [Google Scholar]
- 42.Eubel H., Braun H. P., and Millar A. H. (2005) Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellis R. J., and Van Der Vies S. (1988) The Rubisco subunit binding protein. Photosynth. Res. 16, 101–115 [DOI] [PubMed] [Google Scholar]
- 44.Gatenby A. A., Lubben T. H., Ahlquist P., and Keegstra K. (1988) Imported large subunits of ribulose bisphosphate carboxylase/oxygenase, but not imported beta-ATP synthase subunits, are assembled into holoenzyme in isolated chloroplasts. EMBO J. 7, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bloom M. V., Milos P., and Roy H. (1983) Light-dependent assembly of ribulose-1,5-bisphosphate carboxylase. Proc. Natl. Acad. Sci. U.S.A. 80, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy H., Hubbs A., and Cannon S. (1988) Stability and dissociation of the large subunit RuBisCO binding protein complex in vitro and in organello. Plant Physiol. 86, 50–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pace C. N. (1986) Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131, 266–280 [DOI] [PubMed] [Google Scholar]
- 48.Thompson S. J., Kim S. J., and Robinson C. (1998) Sec-independent insertion of thylakoid membrane proteins: analysis of insertion forces and identification of a loop intermediate involving the signal peptide. J. Biol. Chem. 273, 18979–18983 [DOI] [PubMed] [Google Scholar]
- 49.Woolhead C. A., Mant A., Kim S. J., Robinson C., and Rodger A. (2001) Conformation of a purified “spontaneously” inserting thylakoid membrane protein precursor in aqueous solvent and detergent micelles. J. Biol. Chem. 276, 14607–14613 [DOI] [PubMed] [Google Scholar]
- 50.Rocco A. G., Mollica L., Ricchiuto P., Baptista A. M., Gianazza E., and Eberini I. (2008) Characterization of the protein unfolding processes induced by urea and temperature. Biophys. J. 94, 2241–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fincher V., Dabney-Smith C., and Cline K. (2003) Functional assembly of thylakoid deltapH-dependent/Tat protein transport pathway components in vitro. Eur. J. Biochem. 270, 4930–4941 [DOI] [PubMed] [Google Scholar]
- 52.Denks K., Vogt A., Sachelaru I., Petriman N. A., Kudva R., and Koch H. G. (2014) The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84 [DOI] [PubMed] [Google Scholar]
- 53.Xie K., and Dalbey R. E. (2008) Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 6, 234–244 [DOI] [PubMed] [Google Scholar]
- 54.Trösch R., Mühlhaus T., Schroda M., and Willmund F. (2015) ATP-dependent molecular chaperones in plastids: more complex than expected. Biochim. Biophys. Acta 1847, 872–888 [DOI] [PubMed] [Google Scholar]
- 55.Saibil H. R., Fenton W. A., Clare D. K., and Horwich A. L. (2013) Structure and allostery of the chaperonin GroEL. J. Mol. Biol. 425, 1476–1487 [DOI] [PubMed] [Google Scholar]
- 56.Castanié-Cornet M. P., Bruel N., and Genevaux P. (2014) Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta 1843, 1442–1456 [DOI] [PubMed] [Google Scholar]
- 57.Gutteridge S., and Gatenby A. A. (1995) Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell 7, 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen G. G., and Jagendorf A. T. (1994) Chloroplast molecular chaperone-assisted refolding and reconstitution of an active multisubunit coupling factor CF1 core. Proc. Natl. Acad. Sci. U.S.A. 91, 11497–11501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao J., Chi W., Ouyang M., He B., Chen F., and Zhang L. (2015) PAB is an assembly chaperone that functions downstream of chaperonin 60 in the assembly of chloroplast ATP synthase coupling factor 1. Proc. Natl. Acad. Sci. U.S.A. 112, 4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lubben T. H., Donaldson G. K., Viitanen P. V., and Gatenby A. A. (1989) Several proteins imported into chloroplasts form stable complexes with the GroEL-related chloroplast molecular chaperone. Plant Cell 1, 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsugeki R., and Nishimura M. (1993) Interaction of homologues of Hsp70 and Cpn60 with ferredoxin-NADP+ reductase upon its import into chloroplasts. FEBS Lett. 320, 198–202 [DOI] [PubMed] [Google Scholar]
- 62.Bonk M., Hoffmann B., Von Lintig J., Schledz M., Al-Babili S., Hobeika E., Kleinig H., and Beyer P. (1997) Chloroplast import of four carotenoid biosynthetic enzymes in vitro reveals differential fates prior to membrane binding and oligomeric assembly. Eur. J. Biochem. 247, 942–950 [DOI] [PubMed] [Google Scholar]
- 63.Madueno F., Napier J. A., and Gray J. C. (1993) Newly imported Rieske iron-sulfur protein associates with both Cpn60 and Hsp70 in the chloroplast stroma. Plant Cell 5, 1865–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Froehlich J. E., and Keegstra K. (2011) The role of the transmembrane domain in determining the targeting of membrane proteins to either the inner envelope or thylakoid membrane. Plant J. 68, 844–856 [DOI] [PubMed] [Google Scholar]
- 65.Teng Y. S., Chan P. T., and Li H. M. (2012) Differential age-dependent import regulation by signal peptides. PLoS Biol. 10, e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kudva R., Denks K., Kuhn P., Vogt A., Müller M., and Koch H. G. (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534 [DOI] [PubMed] [Google Scholar]
- 67.Dalbey R. E., Wang P., and Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 68.Houben E., de Gier J. W., and van Wijk K. J. (1999) Insertion of leader peptidase into the thylakoid membrane during synthesis in a chloroplast translation system. Plant Cell 11, 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaru-Ampornpan P., Shen K., Lam V. Q., Ali M., Doniach S., Jia T. Z., and Shan S. O. (2010) ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat. Struct. Mol. Biol. 17, 696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dünschede B., Träger C., Schröder C. V., Ziehe D., Walter B., Funke S., Hofmann E., and Schünemann D. (2015) Chloroplast SRP54 was recruited for posttranslational protein transport via complex formation with chloroplast SRP43 during land plant evolution. J. Biol. Chem. 290, 13104–13114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 73.Kim Y. T., Muramatsu T., and Takahashi K. (1995) Identification of Trp300 as an important residue for Escherichia coli leader peptidase activity. Eur. J. Biochem. 234, 358–362 [DOI] [PubMed] [Google Scholar]