Background: Two pancreatic lipases differ in substrate specificities.
Results: Substitution of specific surface loops from one lipase into the other decreases activity against triglycerides and galactolipids.
Conclusion: The β5-loop and lid domain influence lipase substrate specificity.
Significance: Defining the relationship of structure to lipase function is crucial for understanding lipolysis and developing reagents to modulate lipase activity.
Keywords: lipase, lipid-protein interaction, protein chimera, protein structure, site-directed mutagenesis, structure-function, triglyceride, galactolipid
Abstract
Pancreatic triglyceride lipase (PNLIP) is essential for dietary fat digestion in children and adults, whereas a homolog, pancreatic lipase-related protein 2 (PNLIPRP2), is critical in newborns. The two lipases are structurally similar, yet they have different substrate specificities. PNLIP only cleaves neutral fats. PNLIPRP2 cleaves neutral and polar fats. To test the hypothesis that the differences in activity between PNLIP and PNLIPRP2 are governed by surface loops around the active site, we created multiple chimeras of both lipases by exchanging the surface loops singly or in combination. The chimeras were expressed, purified, and tested for activity against various substrates. The structural determinants of PNLIPRP2 galactolipase activity were contained in the N-terminal domain. Of the surface loops tested, the lid domain and the β5-loop influenced activity against triglycerides and galactolipids. Any chimera on PNLIP with the PNLIPRP2 lid domain or β5-loop had decreased triglyceride lipase activity similar to that of PNLIPRP2. The corresponding chimeras of PNLIPRP2 did not increase activity against neutral lipids. Galactolipase activity was abolished by the PNLIP β5-loop and decreased by the PNLIP lid domain. The source of the β9-loop had minimal effect on activity. We conclude that the lid domain and β5-loop contribute to substrate specificity but do not completely account for the differing activities of PNLIP and PNLIPRP2. Other regions in the N-terminal domain must contribute to the galactolipase activity of PNLIPRP2 through direct interactions with the substrate or by altering the conformation of the residues surrounding the hydrophilic cavity in PNLIPRP2.
Introduction
Before the body can utilize dietary fats, the acyl chains must be cleaved from the parent lipid (1, 2). The digestion and absorption of dietary lipids are highly efficient processes involving several integrated steps, including emulsification, hydrolysis by various lipases, dispersion of the released fatty acids into a protein aqueous environment as mixed micelles with bile salts, and uptake by enterocytes (3). The efficient digestion and absorption of dietary fats and fat-soluble vitamins require the concerted action of multiple lipases with different substrate specificities (4, 5). Hydrolysis starts in the stomach where, in humans, gastric lipase cleaves 15–20% of the fatty acids from triglycerides and continues in the duodenum, where pancreatic lipases complete digestion (6, 7).
Pancreatic triglyceride lipase (PNLIP)2 is the major triglyceride lipase in the duodenum as evidenced by the fat malabsorption seen in patients with isolated PNLIP deficiency (8–11). PNLIP is the archetype of a small lipase subfamily within the α/β-hydrolase fold gene family (12). The family includes PNLIP and two related proteins, PNLIPRP1 and PNLIPRP2. The lipases share 70% amino acid identity and have super-imposable α-carbon backbones (Fig. 1A) (13–15). Each lipase has two domains, an N-terminal domain from residues 18 to 353 and a C-terminal domain from residues 354 to 466. The N-terminal domain consists of an α/β-hydrolase fold, which is present in other lipases and esterases (16). This domain also contains the Ser-His-Asp catalytic triad defined by analogy to serine proteases and confirmed by site-directed mutagenesis (15, 17, 18). A surface loop, the lid domain, covers the active site of PNLIP. In the presence of mixed micelles or non-ionic detergents, a 29-Å hinge movement of the PNLIP lid domain and a smaller movement of the β5-loop open and configure the active site (Fig. 1, A and B) (19–23). In contrast, the lid domain of PNLIPRP2 is more mobile and can adopt an open conformation in the absence of amphiphiles (24). The C-terminal domain of each lipase has a β-sandwich structure similar to the C2 domain of other lipid-binding proteins such as 15-lipoxygenase, Clostridium perfringens α-toxin, phospholipase A2, and synaptotagmin I (25).
FIGURE 1.
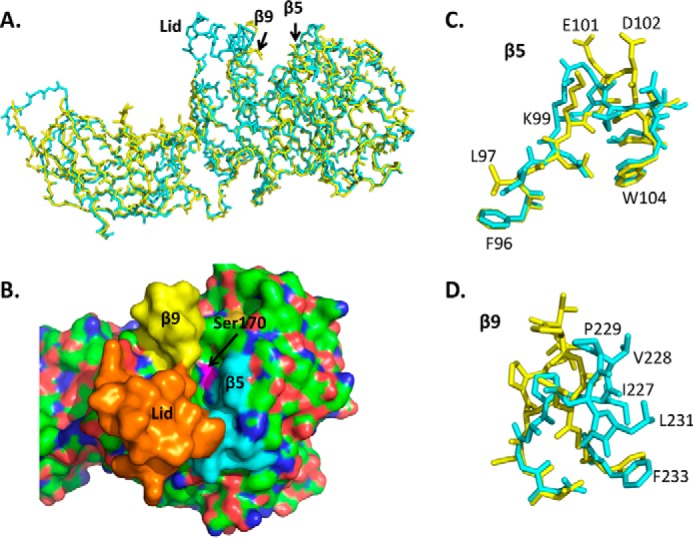
Structure of human PNLIP and PNLIPRP2 and the corresponding lid, β-5, and β-9 loops. A, superimposed α-carbon structure of PNLIP (blue) and PNLIPRP2 (yellow). B, surface structure of PNLIP showing the catalytic site cavity and the location of the lid domain (orange), β5-loop (blue), and β9-loop (yellow). C, superimposed β5-loops of PNLIP (blue) and PNLIPRP2 (yellow). The labeled amino acids are PNLIPRP2 residues. D, superimposed β9-loops of PNLIP (blue) and PNLIPRP2 (yellow). The labeled amino acids are PNLIPRP2 residues.
Despite their marked structural homology, these three homologs differ in their enzymatic properties (12). PNLIP cleaves acyl chains from triglycerides (26, 27). Even though PNLIP can bind phosphatidylcholine in the active site, it has no significant activity against phospholipids or galactolipids (19, 27, 28). In contrast, PNLIPRP2 cleaves acyl chains from triglycerides, galactolipids, and phospholipids (14, 29, 30). PNLIP has an absolute requirement for colipase in the presence of bile salt micelles. PNLIPRP2 does not, although its activity is stimulated by colipase (31–33). PNLIPRP1 has no defined lipase activity (13, 34, 35). The molecular mechanism for the differences in substrate specificity must be determined by specific structural domains in the individual lipases.
Even with tremendous progress in understanding lipase function, the molecular details underlying the binding of a substrate molecule in the active site are limited. Much of our knowledge about pancreatic lipase substrate specificity comes from predictions based on the structures of human PNLIP co-crystallized with substrate analogs. The nucleophilic Ser-160 of PNLIP lies at the bottom of a 916-Å2 hydrophobic canyon, which appears well adapted for the binding of lipid substrates. The β5- and β9-loops and lid domain border the catalytic pocket (Fig. 1). Based on a structure of PNLIP containing a C11 alkyl phosphonate inhibitor, Egloff et al. (36) proposed a model for the binding of lipid substrates. In this model, the canyon contains binding sites for the sn-1 and sn-3 acyl chains of triglycerides. The sn-2 acyl chain, which is not cleaved by PNLIP, is predicted to remain in the lipid emulsion surface. One putative acyl chain-binding site is a hydrophobic channel formed by β9-loop residues Leu-231 and Phe-233 along with Tyr-132, Ala-196, and Pro-198. van Tilbeurgh et al. (19) reported the structure of the PNLIP-colipase complex containing phosphatidylcholine in the active site and noted contacts of the sn-1 acyl chain with these same residues. The other acyl chain-binding site consisted of side chains from residues 269–277 in the lid domain and Ile-96 from the β5-loop. Phe-94 in the β5-loop likely contributes to the oxyanion hole in the active site.
Structures of PNLIPRP2 with substrate or substrate analogs are not available. However, the crystal structure of human PNLIPRP2 with the lid domain in the open conformation was recently described (24). In general, the overall structure of PNLIPRP2 superimposes well with the open conformation of PNLIP. Closer inspection of the structures that include the active site (the β5-loop, β9-loop, and the lid domain) reveals several features pertinent to the differences in substrate specificity (Fig. 1, C and D). First, the PNLIPRP2 lid domain is quite mobile, whereas the PNLIP lid is stable in the closed or open position. Second, several residues in the PNLIPRP2 β5-loop were present at different positions when compared with the analogous region of PNLIP. The position of Leu-97, which corresponds to Leu-96 in PNLIP, could alter the interaction with the acyl chain of bound substrate. Both Glu-91 and Asp-92 occupy different positions in PNLIPRP2 compared with the corresponding residues in PNLIP, Glu-90 and Glu-91. In particular, the position of the Cα carbon atoms of Asp-92 in PNLIPRP2 and Glu-91 in PNLIP differs by 4.7 Å. Consequently, the hydrogen bond observed in PNLIP between Glu-91 and Trp-260 in the open lid does not exist in PNLIPRP2. The conformation of Glu-91 and Asp-92 in PNLIPRP2 could influence the binding of the polar headgroup of phospholipids or galactolipids in the active site of PNLIPRP2. In addition, the conformation of the β9-loop shows differing conformations of several side chains crucial to acyl chain binding. Two hydrophobic residues from the PNLIP β9-loop, Leu-231 and Phe-233, interact with the alkyl chain of a phosphonate inhibitor and likely stabilize the acyl-enzyme intermediate formed during lipolysis (36). These residues are conserved in all pancreatic lipases, but whereas Phe-233 of PNLIP and Phe-234 of PNLIPRP2 superimpose well, the Cα carbons of Leu-232 in PNLIPRP2 and Leu-231 in PNLIP are 3.4 Å apart. These findings suggest the hypothesis that the β5- and β9-loops and the lid domain contribute to the differences in substrate specificity between PNLIP and PNLIPRP2.
In this study, we tested this hypothesis by creating chimeric proteins by exchanging the respective β5- and β9-loops and the lid domain between human PNLIP and PNLIPRP2. We expressed and purified the chimeras and determined their activity against short-, medium-, and long-chain triglycerides and against a galactolipid, digalactosyldiacylglycerol. We found that the lid domain and the β5-loop influence substrate specificity significantly.
Experimental Procedures
DNA Manipulations
The cDNAs encoding for mature human PNLIP and PNLIPRP2 and for PNLIP/PNLIPRP2 and PNLIPRP2/PNLIP chimeras were amplified by regular and overlap PCR, respectively. The amplified cDNAs were subcloned into the yeast protein expression vector pHILSI, in which lipase native secretion signal peptide was replaced by the yeast PHO1 secretion peptide (Invitrogen). All other domain swap mutations were introduced into the parent cDNA in the pHILS1 vector using XL QuikChange site-directed mutagenesis kit (Stratagene). All of the DNA constructs were verified by dideoxynucleotide sequencing.
Recombinant Protein Expression, Production, and Purification
All recombinant lipase proteins were produced in Pichia pastoris yeast strain GS 115 following the manufacturer's manual (Invitrogen). Each plasmid DNA was linearized by BglII and purified by phenol chloroform method. The competent yeast cells were transformed with purified DNAs by electroporation, and the resultant yeast transformants were screened by lipase activity assay and/or immunoblot analysis of culture medium after 24 h of methanol induction as described previously (31, 37). All proteins were robustly secreted indicating that the chimeras were not misfolded.
One highly expressing colony for each of the recombinant lipases was then used to produce a large quantity of recombinant protein (31, 37). After 24–48 h of methanol induction, cell-free culture medium was clarified by filtration and concentrated to ∼50 ml over a Pellicon XL Biomax 10 membrane (Millipore). The concentrated protein sample was then dialyzed at 4 °C overnight against distilled H2O containing 2 mm benzamidine. Each recombinant lipase protein was purified to homogeneity by one-step chromatography using a Mono S FPLC column (GE Healthcare) according to the protocols described previously (31, 37). Fractions of purified lipase were evaluated and analyzed by lipase activity assay and 10% SDS-polyacrylamide gel staining using GelCode Blue Stain Reagent (Pierce). The pooled purified recombinant protein was then concentrated and buffer-exchanged to 25 mm Tris-HCl, pH 8.0. Protein concentration was determined by spectrophotometry at 280 nm. The extinction coefficient of each lipase was calculated using ProtParam program at EXPASY. The final yield ranged from 10 to 40 mg/liter for all of the purified proteins. The homogeneity and integrity of each purified lipase protein was verified by SDS-polyacrylamide gel staining.
Standard Lipase Activity Measurements
The activity of lipases was determined in bulk by measuring the release of fatty acids from mechanically stirred emulsions of tributyrin, trioctanoin, or triolein as described previously (38, 39). Unless otherwise stated, the assay was conducted with or without 5 m excess of purified recombinant human colipase. The lipolytic activities are expressed in international lipase units per mg of enzyme. One unit corresponds to 1 μmol of fatty acid released per min.
Galactolipase activity was also determined using the standard 5-min pH Stat method. However, only 5 mg of digalactosyldiacylglyceride (Larodan Fine Chemicals, Sweden) was emulsified in 15 ml of the standard assay buffer containing 4 mm sodium taurodeoxycholate (NaTDC), and 3 μg of purified recombinant lipase was included. Colipase did not affect the galactolipase activity in the assays; therefore, it was not included.
Colipase Titration
The interactions of lipases with colipase were assayed by adapting the method described previously (40). The assays were performed by measuring lipase activity over a range of colipase concentrations (0–53 nm) with a constant concentration of lipase (2.6 nm) and excess substrate (180 mm trioctanoin) in 4 mm NaTDC assay buffer in a total volume of 15 ml.
Results
Construction of N- and C-terminal Chimeras
Because PNLIP and PNLIPRP2 have distinct N- and C-terminal domains, we constructed chimeras of PNLIP and PNLIPRP2 to determine whether one or both of these domains contributes to substrate specificity. One chimera contained the PNLIP N-terminal domain and the PNLIPRP2 C-terminal domain (PNLIP/PNLIPRP2), and the other chimera included the PNLIPRP2 N-terminal domain and the PNLIP C-terminal domain (PNLIPRP2/PNLIP). Both were expressed in P. pastoris and purified in a single step. We then tested activity against triglycerides with varying acyl chain lengths and against digalactosyldiacylglycerol in various concentrations of NaTDC with and without colipase.
Triglyceride Lipase Activity
First, we tested activity against tributyrin, trioctanoin, and triolein emulsified in 4 mm NaTDC in the presence of a 5-fold molar excess of colipase (Fig. 2). As expected, the activity of PNLIP decreased with increasing acyl chain length, and the activity of PNLIP was significantly higher than the activity of PNLIPRP2 for all substrates. Of note, both of the chimeras had activities that were comparable with the activity of PNLIPRP2 for each of the substrates. These results suggest that the structures of the PNLIPRP2 N- and C-terminal domain influence activity more than the corresponding domains from PNLIP.
FIGURE 2.
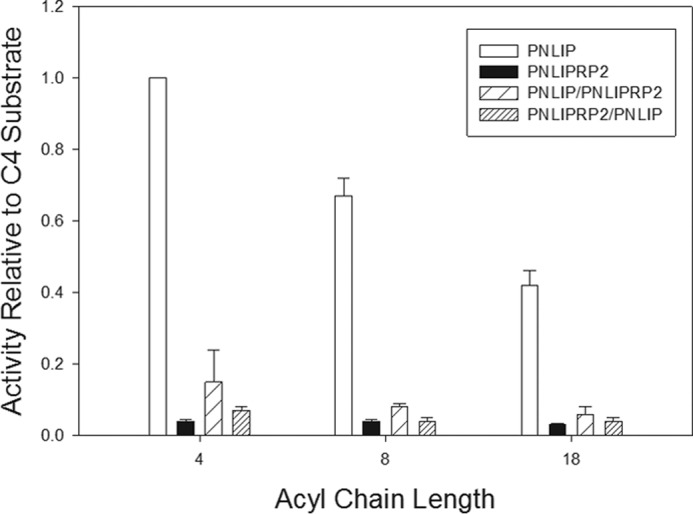
Activity of PNLIP, PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP against various triglyceride substrates. Activity was measured in the pH-stat in the presence of 4 mm NaTDC and a 5 m excess of colipase. Each assay contained 2.6 nm lipase. The activity was determined from the slope of the titration curve. All results are expressed relative to the activity of PNLIP against tributyrin (4-carbon acyl chain) (5063 ± 8.5 units/mg protein). Trioctanoin has an 8-carbon acyl chain; triolein has an 18-carbon acyl chain. The values are the mean ± S.D. of three separate measurements. White bars, PNLIP; black bars, PNLIPRP2; spaced hatched bars, PNLIP/PNLIPRP2; thinly spaced hatched bars, PNLIPRP2/PNLIP. Pairwise comparison of PNLIP activity with the other activities is significantly different for all comparisons (p < 0.001). Pairwise comparisons of PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP were not significantly different.
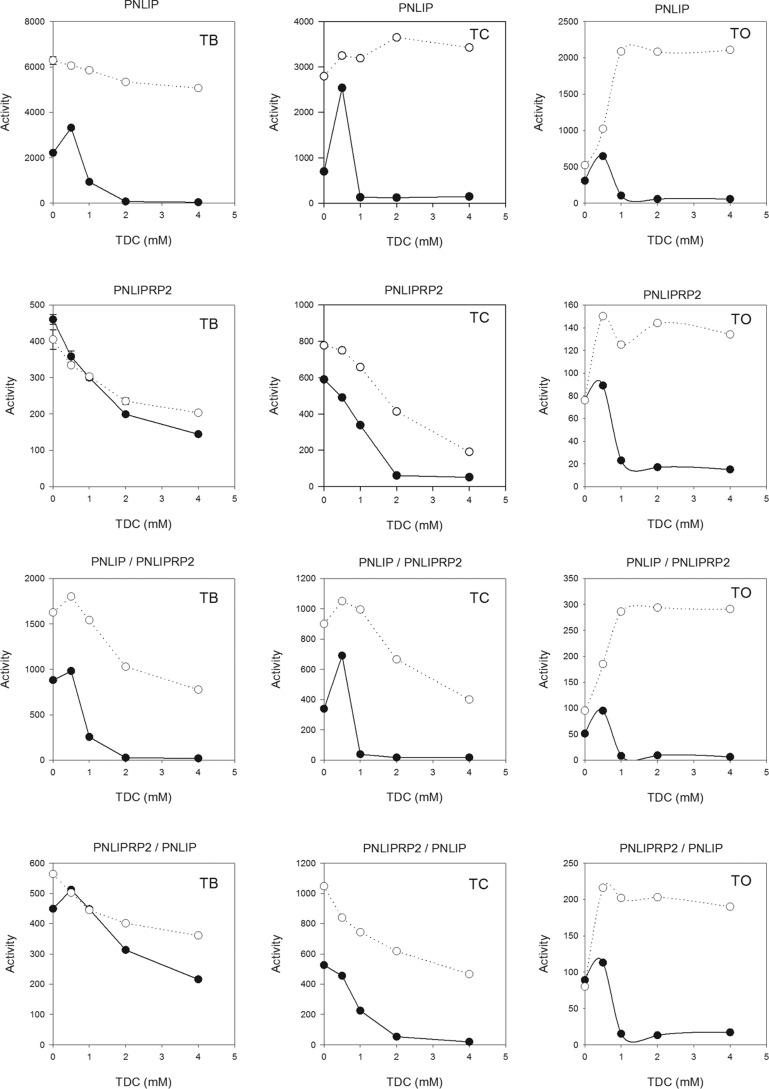
To better understand the effects of the PNLIPRP2 domains on the activity of the chimeric lipases, we determined the activity of all four lipases against tributyrin, trioctanoin, and triolein in various concentrations of NaTDC with and without a 5 m excess of colipase (Fig. 3). As reported before, PNLIPRP2 had lower activity against tributyrin than PNLIP, and the activity of PNLIPRP2 was not stimulated by colipase as was the case with PNLIP (Fig. 3, left-hand panels). The activity of the PNLIP/PNLIPRP2 chimera was intermediate between the activity of PNLIP and PNLIPRP2 (Fig. 3). Like PNLIP, the PNLIP/PNLIPRP2 chimera was completely inhibited at 4 mm NaTDC. Similarly, colipase stimulated the activity of the PNLIP/PNLIPRP2 chimera but did not restore the activity in 4 mm NaTDC to the level of activity in 0.5 mm NaTDC as it did with PNLIP. The PNLIPRP2/PNLIP chimera had an activity curve and colipase response similar to that of PNLIPRP2 (Fig. 3). In the absence of colipase, increasing NaTDC concentrations did not completely inhibit the PNLIPRP2/PNLIP chimera similar to the results with PNLIPRP2.
FIGURE 3.
Effects of NaTDC concentration and colipase on the activity of PNLIP, PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP against tributyrin, trioctanoin, and triolein. The activity of each lipase was measured in the pH-stat in the presence of various concentrations of NaTDC plus and minus a 5 m excess of colipase. Each assay contained 2.6 nm lipase. The activity was determined from the slope of the titration curve. Open circles, with colipase; black circles, no colipase. Each data point is the mean of triplicate assays. TB, tributyrin; TC, trioctanoin; TO, triolein.
With trioctanoin, the PNLIP/PNLIPRP2 chimera had about 2-fold higher activity than PNLIPRP2, but the activity was 9-fold lower than PNLIP (Fig. 3, middle panels). PNLIPRP2 and PNLIPRP2/PNLIP had similar activity (Fig. 3, middle panels). Increasing concentrations of NaTDC inhibited all four lipases, and colipase restored activity. As observed in the tributyrin assays, colipase increased PNLIP activity in 4 mm NaTDC to the level measured in 0.5 mm NaTDC. Colipase-stimulated activity of PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP did not reach the levels measured in 0.5 mm NaTDC.
With triolein, the activity of the two chimeras was similar to the activity of PNLIPRP2 (Fig. 3, right-hand panels). The activity of PNLIP was about 10-fold higher than the activity of the other lipases. For all lipases, micellar concentrations of TDC inhibited activity, and colipase restored the activity to the level measured below the critical micellar concentration for TDC (1.9 mm). As reported previously, PNLIPRP2 required oleic acid to overcome a long lag time (31). Neither of the chimeras had an appreciable lag time, and oleic acid was not required for full activity.
Colipase Activation of the Chimeras
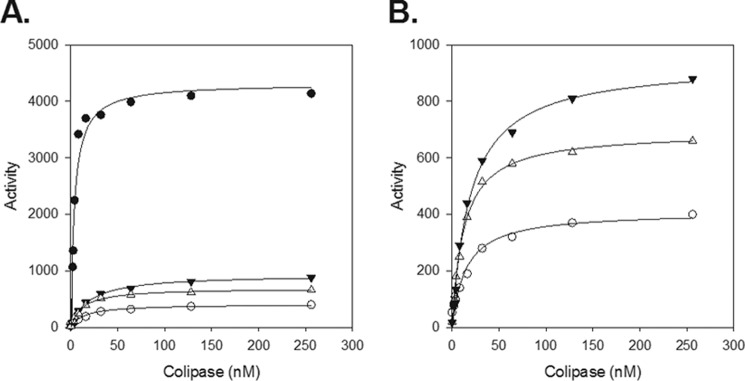
Because colipase appeared to activate the two chimeras to a lesser extent than it activated PNLIP with tributyrin and trioctanoin, we next measured the effect of colipase over a range of colipase concentrations in the presence of a large excess of trioctanoin emulsified in 4.0 mm NaTDC and a constant concentration of each lipase (Fig. 4). The activity of each lipase saturated with increasing concentrations of colipase suggested a specific interaction of colipase with each lipase and the substrate. We then used nonlinear regression to determine the concentration of colipase that restored half-maximal activity to each lipase (apparent Kd). Given the large excess of substrate in the assays, the apparent Kd value likely reflects the interaction between colipase and lipase rather than the interaction of colipase and the substrate emulsion, although the interaction probably occurs at the substrate interface. The apparent Kd value for PNLIPRP2 and the two chimeras was 3–5-fold higher than the apparent Kd value for PNLIP, indicating that the chimeras have a lower affinity for colipase (Table 1).
FIGURE 4.
Colipase dependence of PNLIP, PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP. A, activity was measured in pH-stat in the presence of 4 mm NaTDC. Each assay included 2.6 nm lipase, 180 mm trioctanoin, and the indicated concentrations of colipase. B, only the activities of PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP from A are shown. Each assay was performed in triplicate. Black circles, PNLIP; open circles, PNLIPRP2; black triangles, PNLIP/PNLIPRP2; open triangles, PNLIPRP2/PNLIP. The nonlinear regression lines were fitted by a rectangular hyperbola function and are given as solid lines.
TABLE 1.
Apparent Kd value of the PNLIP and PNLIPRP2 chimeras for colipase with 4 mm NaTDC
Data from three separate experiments were fit by nonlinear regression with a rectangular hyperbolic function. The Kd is reported as nm ± S.D.
| Lipase | Apparent Kd |
|---|---|
| PNLIP | 3.9 ± 0.66 |
| PNLIPRP2 | 14.6 ± 2.79 |
| PNLIP/PNLIPRP2 | 19.0 ± 1.32 |
| PNLIPRP2/PNLIP | 12.4 ± 0.68 |
Galactolipase Activity of the Chimeras
We then determined the activity of the lipases against a galactolipid. We performed the bulk phase assays using a pH-stat method and varying amounts of NaTDC in the presence of colipase. Neither PNLIP nor PNLIP/PNLIPRP2 had detectable galactolipase activity at any NaTDC concentration (Fig. 5). In contrast, PNLIPRP2 and PNLIPRP2/PNLIP had identical activity at all bile salt concentrations tested. The activity of both was stimulated by bile salts below the critical micellar concentration of NaTDC. At 4 mm NaTDC, removing colipase or including 5–10-fold molar excess colipase in the reaction mixture had no effect on the activity of PNLIPRP2 or PNLIPRP2/PNLIP against galactolipids (data not shown). The results clearly show that the structural determinants for galactolipase activity reside in the N-terminal domain of PNLIPRP2.
FIGURE 5.
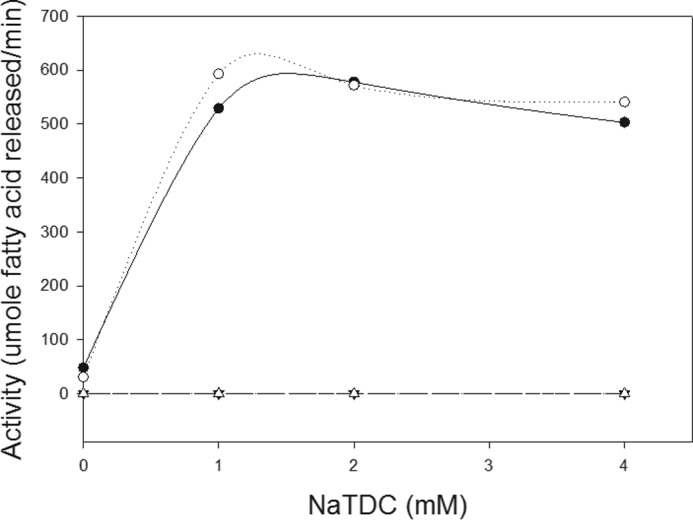
Galactolipase activity of PNLIP, PNLIPRP2, PNLIP/PNLIPRP2, and PNLIPRP2/PNLIP. Activity was measured in the pH-stat in the presence of 4 mm NaTDC. Each assay contained 5 mg of digalactosyldiacylglyceride, 2.6 nm each lipase, and 5-fold molar excess colipase. Black circles, PNLIPRP2; open circles, PNLIPRP2/PNLIP; black triangles, PNLIP; open triangles, PNLIP/PNLIPRP2. Each point represents a single assay.
Exchange of the Lid Domain and the β5- and β9-Loops
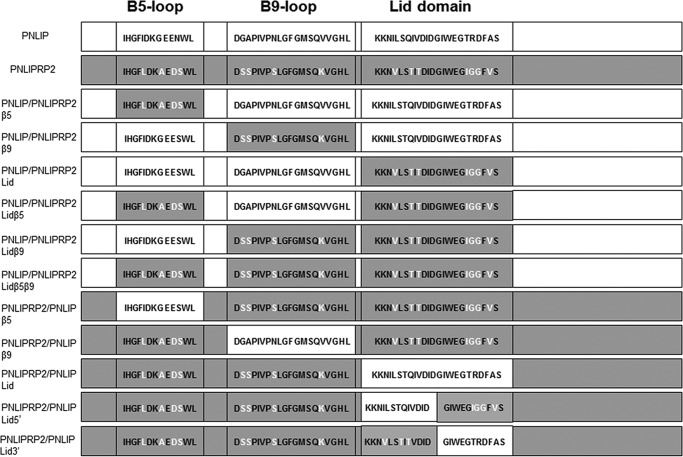
To further define the molecular structures responsible for the galactolipase activity of PNLIPRP2 lipases, we performed switches of smaller domains between PNLIP and PNLIPRP2. We focused on the surface loops (lid domain, the β5-loop, and the β9-loop) surrounding the active site (Fig. 1). First, we substituted the regions in PNLIP with the corresponding sequence from PNLIPRP2 to create a chimera of PNLIP with each of the PNLIPRP2 domains singly or in combination (Fig. 6). Each construct was expressed in P. pastoris and purified as described under “Experimental Procedures.”
FIGURE 6.
Schematic representation of the domain chimeras of PNLIP and PNLIPRP2. Regions from PNLIP are shown in light gray. Regions from PNLIPRP2 are shown in dark gray. The amino acid sequence of the targeted regions, the β5-loop, the β9-loop, and the lid domain, are given in the single-letter code. Differences between the PNLIP and PNLIPRP2 sequences are highlighted as white letters in the PNLIPRP2 sequence.
Triglyceride Lipase Activity of the Lid Domain and the β5- and β9-Chimeras
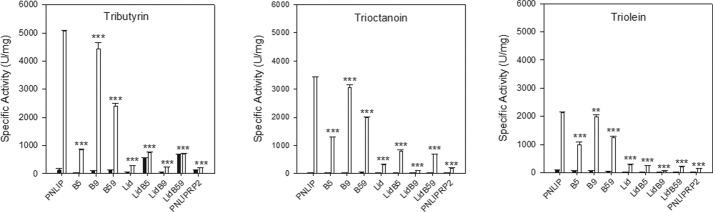
Before testing galactolipase activity, we determined the effect of the domain switches on the activity against triglycerides in the presence of 4 mm NaTDC and excess colipase (Fig. 7). All of the chimeras with substitution of the PNLIPRP2 surface loops into PNLIP significantly decreased activity against all three triglycerides. Substitution of the PNLIPRP2 β5-loop or lid domain into PNLIP had a larger effect on activity than substituting the β-9 loop (p < 0.001). The largest effect on activity occurred when the PNLIPRP2 lid domain was substituted into PNLIP. The presence of the PNLIPRP2 lid domain decreased activity against all three triglycerides to the level of activity measured for PNLIPRP2. Including the β5-loop and β9-loop singly or together with the lid domain made little difference in activity compared with the lid domain substitution alone.
FIGURE 7.
Activity of the N-terminal domain chimeras of PNLIP against various triglyceride substrates. Chimeras of PNLIP containing domains from PNLIPRP2 were expressed and purified. The activity of each lipase was measured in the pH-stat in the presence of 4 mm NaTDC plus and minus a 5 m excess of colipase. Each assay contained 2.6 nm lipase. The activity was determined from the slope of the titration curve. The labels on the x axis state which PNLIP domains were exchanged with the corresponding domain from PNLIPRP2. Black bar, no colipase; white bar, with colipase. The results are the mean ± S.D. of three determinations. Statistical analysis was done by one-way analysis of variance with a Bonferroni t test method for multiple comparisons to PNLIP (α = 0.005). There is a statistically significant difference among the mean values of the different assays in the presence of colipase. Tributyrin (F(8,18) = 1514, p < 0.001); trioctanoin (F(8,18) = 2754, p < 0.001); triolein (F(8,18) = 1123, p < 0.001). **, p < 0.01, and ***, p < 0.001.
Galactolipase Activity of the Lid Domain and the β5- and β9-Chimeras of PNLIP
We then tested the activity of the various chimeras of PNLIP with PNLIPRP2 domains against digalactosyldiacylglycerol. None of the PNLIPRP2 domains, either singly or in combination, conferred detectable galactolipase activity to PNLIP (Table 2).
TABLE 2.
Galactolipase activity of PNLIP chimeras with PNLIPRP2 surface loops
All assays were done by the pH-stat method with digalactosyldiacylglyceride as described under “Experimental Procedures.” The values are ± 1 S.D. ND = none detected.
| Lipase | Activity |
|---|---|
| units/mg | |
| PNLIPRP2 | 110 ± 7.0a |
| PNLIP | ND |
| PNLIPRP2/PNLIP | 150 ± 14a |
| PNLIP/PNLIPRP2 | ND |
| PNLIP/PNLIPRP2β5 | ND |
| PNLIP/PNLIPRP2β9 | ND |
| PNLIP/PNLIPRP2Lid | ND |
| PNLIP/PNLIPRP2Lidβ5 | ND |
| PNLIP/PNLIPRP2Lidβ9 | ND |
| PNLIP/PNLIPRP2Lidβ5β9 | ND |
a p = 0.011 by Student's t test.
Galactolipase Activity of the Lid Domain and the β5- and β9-Chimeras of PNLIPRP2
We then took another approach and determined whether substitution of the PNLIP lid domain, β5-loop, or β9-loop into PNLIPRP2 affected galactolipase and neutral lipase activity compared with the activity of PNLIPRP2 (Table 3). Exchange of the PNLIPRP2 β5-loop with the β5-loop from PNLIP impaired galactolipase activity completely. The chimera also had significantly decreased activity against tributyrin and trioctanoin. Unexpectedly, exchange of the β9-loop from PNLIP increased activity against galactolipids about 1.6-fold but had no effect on activity against the neutral lipids. Substitution of the PNLIPRP2 lid domain with the analogous domain from PNLIP decreased activity against galactolipids about 10-fold. In contrast, the lid domain chimera had preserved activity against tributyrin and higher activity against trioctanoin. Most of the effect resided in the 3′-half of the lid domain because PNLIPRP2/PNLIPLid3′galactolipase activity was decreased about 5-fold, and the activity against tributyrin and trioctanoin was decreased 1.5- and 2-fold, respectively. In contrast, substitution of the 5′-half of the PNLIP lid into PNLIPRP2 (PNLIPRP2/PNLIPLid5′) increased activity against both the galactolipid and neutral lipids.
TABLE 3.
Lipase activity of PNLIPRP2 chimeras with PNLIP surface loops
All assays were done by the pH-stat method with digalactosyldiacylglyceride (DGDG) as described under “Experimental Procedures.” Activity was in units/mg. The values are ± 1 S.D. ND = none detected. Statistical analysis was done by one-way analysis of variance with a Bonferroni t test method for multiple comparisons with PNLIPRP2 (α = 0.010). There is a statistically significant difference among the mean values of the different assays. Digalactosyldiacylglyceride (F(4,10) = 79.8, p < 0.001); tributyrin (F(5,12) = 629, p < 0.001); trioctanoin (F(5,12) = 1718, p < 0.001). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
| Lipase | Substrate |
||
|---|---|---|---|
| DGDG | Tributyrin | Trioctanoin | |
| PNLIPRP2 | 127 ± 24 | 203 ± 9.0 | 191 ± 6.0 |
| PNLIPRP2/PNLIPβ5 | ND | 45 ± 3.0*** | 29 ± 5.0*** |
| PNLIPRP2/PNLIPβ9 | 154 ± 9.0 | 242 ± 5.0*** | 190 ± 8.0 |
| PNLIPRP2/PNLIPLid | 12 ± 5.0*** | 184 ± 2.0* | 315 ± 6.0*** |
| PNLIPRP2/PNLIPLid5′ | 190 ± 21** | 355 ± 13*** | 498 ± 11*** |
| PNLIPRP2/PNLIPLid3′ | 26 ± 7.0*** | 130 ± 5.0*** | 100 ± 3.0*** |
Discussion
In this study, we utilized domain swaps between PNLIP and PNLIPRP2 to provide additional and novel insight into the molecular mechanisms for the differences in substrate specificity between PNLIP and PNLIPRP2 (12). First, the differences in activity against neutral lipids are dominated by the properties of the PNLIPRP2 N- or C-terminal domains. The decreased activity of the N- and C-terminal chimeras is largely explained by impaired interactions with colipase and by structural differences in the lid domain and the β5-loop. Second, the structural determinants of galactolipase activity reside solely in the N-terminal domain of PNLIPRP2. The lid domain and the β5-loop in the N-terminal domain of PNLIPRP2 strongly influence galactolipase activity.
Regardless of whether the N- or C-terminal domain originated from PNLIPRP2, the triglyceride lipase activity of the chimeras more closely resembled that of PNLIPRP2 than that of PNLIP. This finding agrees with the report of similar domain chimeras for the horse lipases (41). The presence of the horse PNLIPRP2 C- or N-terminal domain in a chimera with the opposite PNLIP domain resulted in activity against tributyrin that was similar to PNLIPRP2 and 20-fold lower than horse PNLIP. Similarly, a chimera between the N-terminal domain of guinea pig PNLIPRP2 and the C-terminal domain of human PNLIP had a specific activity close to the activity of the native guinea pig PNLIPRP2 (42).
The decreased activity of the human domain chimeras is partly explained by impaired interactions with colipase. Both PNLIP/PNLIPRP2 and PNLIPRP2/PNLIP have an apparent Kd value for colipase that is similar to the value for PNLIPRP2 and about 3–5-fold higher than the value for PNLIP. Thus, both the N- and C-terminal domains contribute to the interaction of the lipases with colipase. Our results are similar to the data reported in a previous publication (41) on the functional properties of domain chimeras between horse PNLIP and horse PNLIPRP2. Colipase did not stimulate the activity of either chimera suggesting the interaction with colipase was impaired by the domains derived from PNLIPRP2.
Because binding of colipase to PNLIP is mediated through a pincer mechanism by residues in the C-terminal domain and the lid domain, it is not surprising that both domains contribute to binding (25). Earlier studies identified interactions between residues in colipase and the lid domain that are critical for the interaction between colipase and PNLIP (40, 43). Although the critical lid domain residues (Asn-258 and Val-264) are conserved in the PNLIPRP2 lid domain, the increased mobility of the lid domain likely weakens or prevents the interaction of lid domain residues with colipase (24). Consequently, the presence of the PNLIPRP2 N-terminal domain could result in weaker interactions with colipase.
Similarly, differences in the interaction of colipase with the C-terminal domain could decrease affinity of colipase for PNLIPRP2. Among the 12 C-terminal residues in PNLIP that interact with colipase, eight are conserved in all PNLIPRP2 lipases, and four residues are substituted in human PNLIPRP2 (20, 44). Of these four, only Tyr-421 has been tested for its contribution to the lipase-colipase interaction (45). Mutations to the corresponding amino acid in PNLIPRP2, Y421N, decreased the affinity of colipase for Y421N PNLIP about 3-fold. Although the Y421N change in PNLIPRP2 does not fully explain the decreased affinity of the PNLIP/PNLIPRP2 chimera for colipase, the result suggests that the loss of the interactions with some or all four differing residues might decrease colipase affinity for the PNLIP/PNLIPRP2.
To determine whether domains around the active site also contribute to the activity of the two lipases against triglycerides, we investigated the contributions of the β5-loop, β9-loop, and the lid domain to activity against neutral lipids (46). We first replaced these domains in PNLIP with the corresponding domains from PNLIPRP2 to make single, double, and triple mutants. The exchange of the lid domain in PNLIP with the PNLIPRP2 lid domain decreased specific activity for all three triglycerides to a level that was similar to PNLIPRP2. The combination of the PNLIPRP2 β5-loop and β9-loop singly or together with the PNLIPRP2 lid domain on the PNLIP backbone had smaller effects on activity compared with the PNLIPRP2 lid domain alone.
The large effect of the human PNLIPRP2 lid domain on the neutral lipase activity of human PNLIP mirrors that reported for lid domain exchanges between the homologous rat lipases (39). The explanation for the lid domain influence on activity likely rests on the observation that the PNLIPRP2 lid is more mobile in the open position (14, 24). Because the open PNLIP lid domain contributes to one acyl chain-binding site, any change in the confirmation of the lid domain can potentially alter the binding affinity of the mutant lipase for substrate (36, 46).
In contrast, exchange of the β5-loop, β9-loop, and the lid domain in PNLIPRP2 with the corresponding domains from PNLIP had little effect on the activity of the chimeras against tributyrin or trioctanoin. The largest effect was a 4–6-fold decrease in activity when the PNLIPRP2 β5-loop was replaced with the PNLIP loop. Exchanges involving the β9-loop or lid domain had smaller effects on activity.
As opposed to the effects on activity against neutral lipids, activity against galactolipids was totally conferred by the PNLIPRP2 N-terminal domain, and colipase had no effect on activity. Likewise, the phospholipase activity of guinea pig PNLRP2 is determined by structures in the N-terminal domain (42). The molecular structure around the active site of PNLIPRP2 lipases must differ from the structure of PNLIP in a way that allows accommodation of the polar headgroup of galactolipids and phospholipids.
By analyzing chimeras of the β5-loop, β9-loop, and lid domain from one lipase on the backbone of the other lipase, we were able to show that both the 3′-half of the lid domain and β5-loop contribute to the galactolipase activity of PNLIPRP2. Our finding that the β5-loop is crucial for galactolipase activity is similar to a report demonstrating that the β5-loop influences substrate specificity in extracellular phospholipase A1 (47).
Molecular modeling of digalactosyldiglyceride suggests the digalactose polar headgroup can fit into a cavity in the active site of PNLIPRP2 (46). In lipases lacking activity against polar lipids, the cavity does not form. In particular, human PNLIP does not have a cavity for the polar headgroup because the region is occupied by Asp-265 and Arg-274 of the lid domain. The steric hindrance of these residues would prevent productive binding of polar lipids into the active site of human PNLIP.
Rather than hinder binding of polar lipids, the properties of the β5-loop may increase the hydrophilicity of the active site and enhance binding of polar lipids. It was noted that the water-accessible area of the β5-loop in guinea pig PNLIPRP2 is more hydrophilic than the β5-loop in PNLIP, and it was suggested that this property contributes to the hydrophilic cavity where the polar headgroup of digalactosyldiglyceride is predicted to reside (28, 46). To test this hypothesis, we calculated the water-accessible area of the human PNLIPRP2 β5-loop. The hydrophilic/hydrophobic balance of the exposed residues was slightly higher (2.3) than that measured in human PNLIP (1.4) (28). The value is lower than that measured for the β5-loop of guinea pig PNLIPRP2 (5.4) and may not provide an adequate explanation for the effect of replacing the β5-loop in human PNLIPRP2 with the corresponding loop from human PNLIP (28).
Another possibility is suggested by a polar interaction that forms between Glu-101 in the β5-loop and Trp-269 in the open lid domain of human PNLIP (36). The corresponding residue in PNLIPRP2, Glu-102, assumes a different orientation, and the interaction with Trp-269 may not form (24). If Glu-102 assumes the same position in PNLIPRP2/PNLIPβ5 as the position of Glu-101 in PNLIP, the interaction with Trp-269 may form and bring other lid domain residues into the hydrophilic crevice as occurs in the open conformation of PNLIP. The resolution of the different possibilities would be aided by a crystal structure of the PNLIPRP2/PNLIPβ5 chimera.
In summary, our study suggests that the differences in activity against neutral lipids between PNLIP and PNLIPRP2 are influenced by the alterations in the interaction of colipase with each lipase and by the structure of the lid domain and β5-loop. These same structures mediate the ability of PNLIPR2 to hydrolyze galactolipids. However, the lid domain and β5-loop do not completely account for the activity. Other regions in the N-terminal domain must contribute to the galactolipase activity of PNLIPRP2 by direct interactions with the substrate or by altering the conformation of the residues surrounding the hydrophilic cavity in PNLIPRP2.
Author Contributions
X. X. constructed, expressed, purified, and assayed all of the recombinant proteins described in this study. He contributed to the writing of the manuscript. M. E. L. directed all aspects of this work. He conceived the project, designed the chimeras, and helped interpret the data. He was primarily responsible for writing the manuscript.
Acknowledgments
We thank Leah Ross for providing the colipase used in assays and Margaret Haughney for technical assistance in several of the lipase assays.
This work was supported by National Institutes of Health Grant DK080820 (to M. E. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PNLIP
- pancreatic lipase
- NaTDC
- sodium taurodeoxycholate.
References
- 1.Hofmann A. F., and Borgstrom B. (1964) The intraluminal phase of fat digestion in man: the lipid content of the micellar and oil phases of intestinal content obtained during fat digestion and absorption. J. Clin. Invest. 43, 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofmann A. F., and Mekhijan H. S. (1971) in The Bile Acids (Nair P. P., and Kritchevsky D., eds) pp. 103–152, Plenum Publishing Corp, New York [Google Scholar]
- 3.Carey M. C., and Hernell O. (1992) Digestion and absorption of fat. Semin. Gastrointest. Dis. 3, 189–208 [Google Scholar]
- 4.Bernbäck S., Bläckberg L., and Hernell O. (1990) The complete digestion of human milk triacylglycerol in vitro requires gastric lipase, pancreatic colipase-dependent lipase, and bile salt-stimulated lipase. J. Clin. Invest. 85, 1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borgström B., and Erlanson-Albertsson C. (1982) Hydrolysis of milk fat globules by pancreatic lipase. Role of colipase, phospholipase A2, and bile salts. J. Clin. Invest. 70, 30–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carriere F., Barrowman J. A., Verger R., and Laugier R. (1993) Secretion and contribution to lipolysis of gastric and pancreatic lipases during a test meal in humans. Gastroenterology 105, 876–888 [DOI] [PubMed] [Google Scholar]
- 7.Carrière F., Renou C., Lopez V., De Caro J., Ferrato F., Lengsfeld H., De Caro A., Laugier R., and Verger R. (2000) The specific activities of human digestive lipases measured from the in vivo and in vitro lipolysis of test meals. Gastroenterology 119, 949–960 [DOI] [PubMed] [Google Scholar]
- 8.Figarella C., De Caro A., Leupold D., and Poley J. R. (1980) Congenital pancreatic lipase deficiency. J. Pediatr. 96, 412–416 [DOI] [PubMed] [Google Scholar]
- 9.Figarella C., Negri G. A., and Sarles H. (1972) Presence of colipase in a congenital pancreatic lipase deficiency. Biochim. Biophys. Acta 280, 205–211 [DOI] [PubMed] [Google Scholar]
- 10.Ghishan F. K., Moran J. R., Durie P. R., and Greene H. L. (1984) Isolated congenital lipase-colipase deficiency. Gastroenterology. 86, 1580–1582 [PubMed] [Google Scholar]
- 11.Szabó A., Xiao X., Haughney M., Spector A., Sahin-Tóth M., and Lowe M. E. (2015) A novel mutation in PNLIP causes pancreatic triglyceride lipase deficiency through protein misfolding. Biochim. Biophys. Acta 1852, 1372–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe M. E. (2002) The triglyceride lipases of the pancreas. J. Lipid Res. 43, 2007–2016 [DOI] [PubMed] [Google Scholar]
- 13.Roussel A., de Caro J., Bezzine S., Gastinel L., de Caro A., Carrière F., Leydier S., Verger R., and Cambillau C. (1998) Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations. Proteins 32, 523–531 [PubMed] [Google Scholar]
- 14.Roussel A., Yang Y., Ferrato F., Verger R., Cambillau C., and Lowe M. (1998) Structure and activity of rat pancreatic lipase-related protein 2. J. Biol. Chem. 273, 32121–32128 [DOI] [PubMed] [Google Scholar]
- 15.Winkler F. K., D'Arcy A., and Hunziker W. (1990) Structure of human pancreatic lipase. Nature 343, 771–774 [DOI] [PubMed] [Google Scholar]
- 16.Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., and Schrag J. (1992) The α/β hydrolase fold. Protein Eng. 5, 197–211 [DOI] [PubMed] [Google Scholar]
- 17.Lowe M. E. (1992) The catalytic site residues and interfacial binding of human pancreatic lipase. J. Biol. Chem. 267, 17069–17073 [PubMed] [Google Scholar]
- 18.Lowe M. E. (1996) Mutation of the catalytic site Asp177 to Glu177 in human pancreatic lipase produces an active lipase with increased sensitivity to proteases. Biochim. Biophys. Acta 1302, 177–183 [DOI] [PubMed] [Google Scholar]
- 19.van Tilbeurgh H., Egloff M. P., Martinez C., Rugani N., Verger R., and Cambillau C. (1993) Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by x-ray crystallography. Nature 362, 814–820 [DOI] [PubMed] [Google Scholar]
- 20.van Tilbeurgh H., Gargouri Y., Dezan C., Egloff M. P., Nésa M. P., Ruganie N., Sarda L., Verger R., and Cambillau C. (1993) Crystallization of pancreatic procolipase and of its complex with pancreatic lipase. J. Mol. Biol. 229, 552–554 [DOI] [PubMed] [Google Scholar]
- 21.Belle V., Fournel A., Woudstra M., Ranaldi S., Prieri F., Thomé V., Currault J., Verger R., Guigliarelli B., and Carrière F. (2007) Probing the opening of the pancreatic lipase lid using site-directed spin labeling and EPR spectroscopy. Biochemistry 46, 2205–2214 [DOI] [PubMed] [Google Scholar]
- 22.Hermoso J., Pignol D., Kerfelec B., Crenon I., Chapus C., and Fontecilla-Camps J. C. (1996) Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex. J. Biol. Chem. 271, 18007–18016 [DOI] [PubMed] [Google Scholar]
- 23.Hermoso J., Pignol D., Penel S., Roth M., Chapus C., and Fontecilla-Camps J. C. (1997) Neutron crystallographic evidence of lipase-colipase complex activation by a micelle. EMBO J. 16, 5531–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eydoux C., Spinelli S., Davis T. L., Walker J. R., Seitova A., Dhe-Paganon S., De Caro A., Cambillau C., and Carrière F. (2008) Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation. Biochemistry 47, 9553–9564 [DOI] [PubMed] [Google Scholar]
- 25.van Tilbeurgh H., Bezzine S., Cambillau C., Verger R., and Carrière F. (1999) Colipase: structure and interaction with pancreatic lipase. Biochim. Biophys. Acta 1441, 173–184 [DOI] [PubMed] [Google Scholar]
- 26.Verger R. (1984) in Lipases (Borgstrom B., and Brockman H. L., eds) 1st Ed., pp. 84–150, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 27.Andersson L., Carriére F., Lowe M. E., Nilsson A., and Verger R. (1996) Pancreatic lipase-related protein 2 but not classical pancreatic lipase hydrolyzes galactolipids. Biochim. Biophys. Acta 1302, 236–240 [DOI] [PubMed] [Google Scholar]
- 28.Withers-Martinez C., Carrière F., Verger R., Bourgeois D., and Cambillau C. (1996) A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig. Structure 4, 1363–1374 [DOI] [PubMed] [Google Scholar]
- 29.Jayne S., Kerfelec B., Foglizzo E., Chapus C., and Crenon I. (2002) High expression in adult horse of PLRP2 displaying a low phospholipase activity. Biochim. Biophys. Acta 1594, 255–265 [DOI] [PubMed] [Google Scholar]
- 30.Sias B., Ferrato F., Grandval P., Lafont D., Boullanger P., De Caro A., Leboeuf B., Verger R., and Carrière F. (2004) Human pancreatic lipase-related protein 2 is a galactolipase. Biochemistry 43, 10138–10148 [DOI] [PubMed] [Google Scholar]
- 31.Xiao X., Mukherjee A., Ross L. E., and Lowe M. E. (2011) Pancreatic lipase-related protein-2 (PLRP2) can contribute to dietary fat digestion in human newborns. J. Biol. Chem. 286, 26353–26363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao X., Ross L. E., Miller R. A., and Lowe M. E. (2011) Kinetic properties of mouse pancreatic lipase-related protein-2 suggest the mouse may not model human fat digestion. J. Lipid Res. 52, 982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao X., Ross L. E., Sevilla W. A., Wang Y., and Lowe M. E. (2013) Porcine pancreatic lipase related protein 2 has high triglyceride lipase activity in the absence of colipase. Biochim. Biophys. Acta 1831, 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crenon I., Foglizzo E., Kerfelec B., Vérine A., Pignol D., Hermoso J., Bonicel J., and Chapus C. (1998) Pancreatic lipase-related protein type I: a specialized lipase or an inactive enzyme. Protein Eng. 11, 135–142 [DOI] [PubMed] [Google Scholar]
- 35.Crenon I., Jayne S., Kerfelec B., Hermoso J., Pignol D., and Chapus C. (1998) Pancreatic lipase-related protein type 1: a double mutation restores a significant lipase activity. Biochem. Biophys. Res. Commun. 246, 513–517 [DOI] [PubMed] [Google Scholar]
- 36.Egloff M. P., Marguet F., Buono G., Verger R., Cambillau C., and van Tilbeurgh H. (1995) The 2.46 A resolution of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate. Biochemistry 34, 2751–2762 [DOI] [PubMed] [Google Scholar]
- 37.Yang Y., and Lowe M. E. (1998) Human pancreatic triglyceride lipase expressed in yeast cells: purification and characterization. Protein Expr. Purif. 13, 36–40 [DOI] [PubMed] [Google Scholar]
- 38.Cordle R. A., and Lowe M. E. (1998) The hydrophobic surface of colipase influences lipase activity at an oil-water interface. J. Lipid Res. 39, 1759–1767 [PubMed] [Google Scholar]
- 39.Yang Y., and Lowe M. E. (2000) The open lid mediates pancreatic lipase function. J. Lipid Res. 41, 48–57 [PubMed] [Google Scholar]
- 40.Crandall W. V., and Lowe M. E. (2001) Colipase residues Glu64 and Arg65 are essential for normal lipase-mediated fat digestion in the presence of bile salt micelles. J. Biol. Chem. 276, 12505–12512 [DOI] [PubMed] [Google Scholar]
- 41.Berton A., Sebban-Kreuzer C., and Crenon I. (2007) Role of the structural domains in the functional properties of pancreatic lipase-related protein 2. FEBS J. 274, 6011–6023 [DOI] [PubMed] [Google Scholar]
- 42.Carrière F., Thirstrup K., Hjorth S., Ferrato F., Nielsen P. F., Withers-Martinez C., Cambillau C., Boel E., Thim L., and Verger R. (1997) Pancreatic lipase structure-function relationships by domain exchange. Biochemistry 36, 239–248 [DOI] [PubMed] [Google Scholar]
- 43.Lowe M. E. (1997) Colipase stabilizes the lid domain of pancreatic triglyceride lipase. J. Biol. Chem. 272, 9–12 [DOI] [PubMed] [Google Scholar]
- 44.Egloff M. P., Sarda L., Verger R., Cambillau C., and van Tilbeurgh H. (1995) Crystallographic study of the structure of colipase and of the interaction with pancreatic lipase. Protein Sci. 4, 44–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bezzine S., Ferrato F., Ivanova M. G., Lopez V., Verger R., and Carrière F. (1999) Human pancreatic lipase: colipase dependence and interfacial binding of lid domain mutants. Biochemistry 38, 5499–5510 [DOI] [PubMed] [Google Scholar]
- 46.Carrière F., Withers-Martinez C., van Tilbeurgh H., Roussel A., Cambillau C., and Verger R. (1998) Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. Biochim. Biophys. Acta 1376, 417–432 [DOI] [PubMed] [Google Scholar]
- 47.Arima N., Inoue A., Makide K., Nonaka T., and Aoki J. (2012) Surface loops of extracellular phospholipase A(1) determine both substrate specificity and preference for lysophospholipids. J. Lipid Res. 53, 513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]