Background: The lipoprotein NisI confers immunity to Lactococcus lactis against its own lantibiotic nisin.
Results: NisI contains a membrane and a nisin binding domain both with the same fold as SpaI from Bacillus subtilis.
Conclusion: The C-terminal domain of NisI interacts directly and specifically with nisin.
Significance: Our results help to understand the mechanism of the NisI mediated immunity against nisin on a structural level.
Keywords: antibiotic resistance, antibiotics, lipoprotein, nuclear magnetic resonance (NMR), protein structure, lantibiotic, nisin binding, small angle x-ray scattering
Abstract
Many Gram-positive bacteria produce lantibiotics, genetically encoded and posttranslationally modified peptide antibiotics, which inhibit the growth of other Gram-positive bacteria. To protect themselves against their own lantibiotics these bacteria express a variety of immunity proteins including the LanI lipoproteins. The structural and mechanistic basis for LanI-mediated lantibiotic immunity is not yet understood. Lactococcus lactis produces the lantibiotic nisin, which is widely used as a food preservative. Its LanI protein NisI provides immunity against nisin but not against structurally very similar lantibiotics from other species such as subtilin from Bacillus subtilis. To understand the structural basis for LanI-mediated immunity and their specificity we investigated the structure of NisI. We found that NisI is a two-domain protein. Surprisingly, each of the two NisI domains has the same structure as the LanI protein from B. subtilis, SpaI, despite the lack of significant sequence homology. The two NisI domains and SpaI differ strongly in their surface properties and function. Additionally, SpaI-mediated lantibiotic immunity depends on the presence of a basic unstructured N-terminal region that tethers SpaI to the membrane. Such a region is absent from NisI. Instead, the N-terminal domain of NisI interacts with membranes but not with nisin. In contrast, the C-terminal domain specifically binds nisin and modulates the membrane affinity of the N-terminal domain. Thus, our results reveal an unexpected structural relationship between NisI and SpaI and shed light on the structural basis for LanI mediated lantibiotic immunity.
Introduction
To protect their habitat many bacteria and archaea produce peptides with antimicrobial functions to inhibit the growth of putative competitors (1). In an age of widespread (over) use of antibiotics and the rapid ascent of resistant pathogens, these so-called bacteriocins have found increasing attention (2). A very promising group of bacteriocin candidates for clinical applications are the lantibiotics (3). Lantibiotics are genetically encoded and ribosomally translated as precursor peptides that undergo extensive posttranslational modifications (4). They contain several unusual amino acids and the characteristic thioether lanthionine and methyllanthionine bridges (5). Nisin, the most prominent lantibiotic, belongs to the so-called class I lantibiotics that are modified by a dehydratase LanB and a cyclase LanC (6–9). After modification the precursor peptide is exported by an ABC transporter LanT and the leader sequence is cleaved by a LanP protease resulting in an active lantibiotic (10). Lantibiotics are only produced by Gram-positive bacteria but are also active against Gram-positive bacteria. Therefore, it is vitally important for the producer strains to protect themselves against the antimicrobial activity of their own lantibiotics. Thus, lantibiotic producer strains possess a so-called immunity system. The immunity system of many lantibiotic producers consists of an ABC transporter LanFEG and an immunity protein LanI (11–13). Its expression is regulated by the extracellular concentration of the lantibiotic through a two-component system with a sensor histidine kinase LanK and a response regulator LanR (14, 15).
Nisin produced by Lactococcus lactis is one of the best-studied and most widely applied lantibiotics. For more than 40 years nisin is used as a natural food preservative (16). Nisin inhibits cell wall biosynthesis by binding to and sequestering of the essential precursor lipid II (17, 18). In addition, lipid II-nisin complexes form pores in the membrane of Gram-positive bacteria and thereby disrupt the membrane potential. The solution NMR structure of a nisin-lipid II complex showed that nisin specifically binds the pyrophosphate moiety of lipid II (19). To protect themselves against their own lantibiotic, nisin producer strains of L. lactis express the specific immunity proteins NisI, a lipoprotein (Fig. 1A), and NisFEG, an ABC transporter. Knock-out experiments of either one system showed that both confer nisin immunity. However, full immunity is only reached when both systems are co-expressed (11, 12, 20, 21). Because the immunity mechanism against lantibiotics is still unclear, a detailed study of the participating proteins on a structural and functional level is required as a prerequisite for developing novel lantibiotics. Structural information for lantibiotic immunity proteins in general and LanI proteins in particular is so far very limited. Only a solution structure for a LanI protein has been solved (22) so far, the structure of SpaI from Bacillus subtilis conferring immunity against subtilin. In addition, the structure for the unrelated lipoprotein MlbQ conferring immunity to actinomycetes against NAI-107 has been reported (23). The SpaI structure revealed a novel three-dimensional fold with a structured core of six antiparallel twisted β-sheets and an unusually long β-hairpin. The flexible N terminus of SpaI was shown to interact with membranes, an interaction that is functionally important for lantibiotic immunity. Interestingly, the lantibiotics nisin and subtilin have a high level of sequential and structural homology. However, the lantibiotic immunity proteins SpaI and NisI differ significantly in size (16.8 versus 25.8 kDa) and show only very limited sequence homology in the C-terminal part (Fig. 1B). In fact, NisI is only able to confer immunity against nisin and SpaI confers only immunity against subtilin (24). This lack of cross-immunity suggests that the two LanI proteins are highly specific for their cognate lantibiotics. It was also shown by native PAGE that in vitro NisI specifically binds to nisin but not subtilin (24). A nisin binding affinity in the low micromolar range was determined for lipid-free NisI by surface plasmon resonance (25). Furthermore, experiments with C-terminal truncation variants of NisI suggested that the C-terminal 22 amino acids of NisI are involved in nisin immunity (26, 27). For a better understanding of the specificity of NisI and SpaI and the lantibiotic immunity mechanism in general a detailed structural analysis of NisI is required.
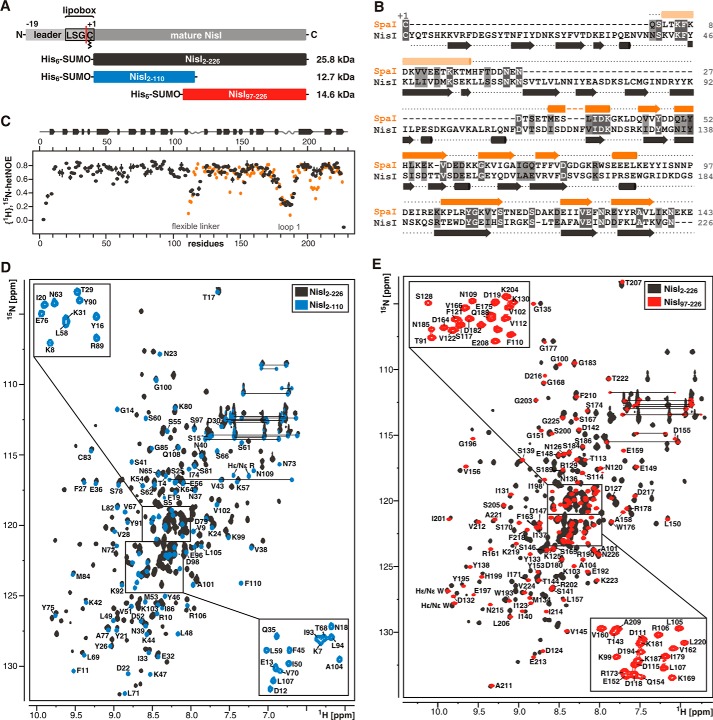
FIGURE 1.
NisI is a two-domain protein with a SpaI-like domain in its C-terminal half. A, schematic representation of the lipoprotein NisI and NMR constructs used in this study. NisI contains an N-terminal leader sequence for export across the cytoplasmic membrane followed by a lipobox motif where a diacylglyceryl moiety is covalently attached to the cysteine indicated. The red line indicates the cleavage site for the leader sequence. The NMR construct for full-length NisI starts at residue +2, where the +1 residue is the cysteine of the lipobox. B, sequence alignment of SpaI and NisI. Identical residues are highlighted with dark gray boxes and residues with similar chemical properties are shown in light gray boxes. The secondary structure based on chemical shift analysis for both proteins is indicated above and below the sequence with arrows representing β-sheets and cylinders α-helices. C, {1H},15N-hetNOE values for backbone amide groups of NisI-(2–226) in gray and SpaI-(18–143) in orange. The secondary structure of NisI is indicated above. D and E show overlays of the 15N-TROSY-HSQC spectra of NisI-(2–226) in gray and the 15N-HSQC spectra of NisI-(2–110) in blue (D) and NisI-(97–226) in red (E), respectively.
Here we describe the solution structure of the lantibiotic immunity protein NisI and characterize its interaction with nisin and membrane vesicles in vitro. NisI consists of two structurally independent domains with very different biophysical and functional properties. Unexpectedly, both domains are structurally very similar to SpaI suggesting a close evolutionary relationship between the two proteins. Importantly, our results suggest that the two domains are functionally specialized. Although the N-terminal domain binds to the membrane the C-terminal domain interacts specifically with nisin.
Experimental Procedures
Expression and Purification
For structure determination of NisI from L. lactis the two domains of NisI were separately expressed in Escherichia coli. Functional analyses of NisI were carried out with the full-length protein lacking the leader sequence and the lipobox (starting at residue +2, the +1 residue is the cysteine of the lipobox). The coding sequences were cloned into a modified pET11a vector containing an N-terminal His6-SUMO tag. For NMR experiments all NisI constructs were expressed and purified as described before (28). Cysteine mutants were constructed by in vitro mutagenesis. Additional cysteines were introduced at the following positions: S114C, S15C, and S41C. For the extended linker mutant a 10-fold glycine serine linker was introduced at amino acid position 114 of NisI-(2–226) (NisI-(2–226)-10xGS). A NisI variant with a shortened linker (NisI-(2–226)-Δ114–117) was also constructed by in vitro mutagenesis.
NMR Spectroscopy
For NMR measurements NisI-(2–226) (400 μm) and the isolated NisI domains were dissolved in 50 mm sodium phosphate buffer, pH 6.5, 100 mm NaCl, 1 mm DTT, and 10% D2O (NMR buffer). For NMR titration experiments a protein concentration of 50–200 μm was used. NMR spectra were acquired at 25 °C on Bruker AVANCE 600, 700, 800, 900, and 950 MHz spectrometers equipped with cryogenic triple resonance probes. The proton chemical shifts were internally referenced to 2.2-dimethyl-2-silapentane-5-sulfonic acid and the heteronuclear 13C and 15N chemical shifts were indirectly referenced with the appropriate conversion factors (29). All spectra were processed using Bruker TopSpinTM 2.1/3.2 and analyzed using the programs CARA (30) and CcpNmr Analysis (31). The standard set of triple resonance experiments was used for the backbone resonance assignments of NisI-(2–226) and the backbone and side chain resonance assignments of NisI-(2–110) and NisI-(97–226) as described in detail elsewhere (28).
To evaluate NMR titration experiments the chemical shifts were determined using the peak picking function of CcpNmr Analysis (31). The chemical shift differences were calculated using the following function (32),
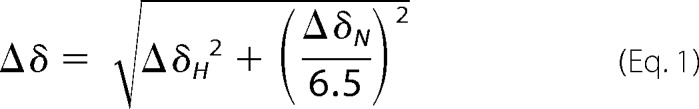 |
{1H},15N-heteronuclear nuclear Overhauser effect (hetNOE) experiments (34) for 2H,15N-labeled NisI-(2–226) (28) as well as for 15N-labeled NisI-(2–110) and NisI-(97–226), respectively, were recorded using Bruker standard pulse sequences. Experiments were run twice in an interleaved fashion with and without proton saturation during the recovery delay. Peak intensity differences were obtained using Bruker TopSpinTM 2.1/3.2 for peak integration and the peak intensity difference was calculated as ΔI = IX/I0.
For paramagnetic relaxation enhancement measurements a site-directed spin label was introduced in NisI-(2–226) cysteine mutants. Wild type NisI-(2–226) contains a natural cysteine in the core of the N-terminal domain, which, however, is only marginally accessible to labeling even at extended reaction times as tested with fluorescein-5-maleimide (Sigma) under the same conditions as for spin labeling. All 15N-labeled cysteine mutants were expressed and purified as described for the wild type protein. The correct folding of the mutants was tested with 15N-HSQCs. Prior to spin labeling the buffer was exchanged via a PD-10 desalting column (GE Healthcare) to a non-reducing environment (25 mm HEPES, 100 mm NaCl, pH 7.0). The protein was incubated for 3 h at room temperature with a 10-fold excess of the spin label 4-maleimido-TEMPO (Sigma). After the reaction, excess of spin label was removed by gel filtration with NMR buffer without DTT. 15N-HSQC4 spectra were recorded with spin-labeled protein at a concentration of ∼100 μm. To record spectra without paramagnetic effects the spin label was reduced with 1 mm ascorbic acid for 1 h.
To measure amide residual dipolar couplings IPAP-[1H,15N]-HSQC spectra with and without alignment media were recorded on a 600 MHz spectrometer for 15N-labeled NisI-(2–110) and NisI-(97–226) (35). For the positively charged NisI-(2–110) a lyotrophic liquid crystalline phase (3.8% PEG-hexanol, (36) and for the negatively charged NisI-(97–226) pf1 phage (15 mg/ml, Hyglos GmbH) were used as alignment media. 1DNH values were extracted where signals could be tracked unambiguously and peak maxima were determined by using the peak picking function in the program CcpNmr Analysis (36). Residual dipolar coupling values for NisI-(2–226) were not determined due to interactions of the protein with all tested alignment media.
Structure Calculation
Distance restraints based on nuclear Overhauser effect (NOE) were obtained from 15N-NOESY-HSQC, 13C-NOESY-HSQC (aliphatic carbons), and 13C-NOESY-HSQC (aromatic carbons) experiments in H2O with mixing times of 120 ms. Torsion angle restraints were calculated using TALOS-N (37) based on backbone H, N, Cα, Cβ, and CO chemical shifts and used for all residues with {1H},15N-hetNOE values >0.5.
Peak picking and NOE assignment was performed with the ATNOS/CANDID module in UNIO (38) in combination with CYANA (39) using the three-dimensional NOESY spectra. Peak lists were reviewed manually and corrected in case of artifacts. Distance restrains were obtained using the automated NOE assignment and structure calculation protocol available in CYANA (39, 38). For all NOESY spectra an assignment of more than 87% of the observable NOESY cross-peaks was achieved. In the final round of structure calculation 72 and 76 residual dipolar coupling restraints for backbone amide groups in NisI-(2–110) and NisI-(97–226), respectively, were included for residues with well resolved signals and a hetNOE value >0.5. The 20 structures with the lowest target functions were used for restrained energy refinement with OPALp (40) and the AMBER94 force field (41). With the CYANA “regularized”-macro a single representative structure was obtained (42). This representative mean structure as well as the 19 conformers with the lowest CYANA target function were used for the structure validation with the Protein Structure Validation Software suite 1.5 (43) restricted to residues with hetNOE values >0.6.
Electrostatic surface potential calculations were conducted with the PDB2PQR web server (44) using the PARSE force field and visualized with the APBS plug-in (45) for PyMOL (46). All figures of structures were prepared with PyMOL.
Small-angle X-ray Scattering Measurements and Data Analysis
SAXS data for solutions of NisI, NisI-(2–226)-10xGS, and NisI-(2–226)-Δ114–117 were recorded on an in-house SAXS instrument (SAXSess mc2, Anton Paar, Graz, Austria) equipped with a Kratky camera, a sealed x-ray tube source and a two-dimensional Princeton Instruments PI·SCX:4300 (Roper Scientific) CCD detector. The scattering patterns were measured with a 90-min exposure time (540 frames, each 10 s) for several solute concentrations in the range from 1 to 9.0 mg/ml. Measurements for NisI were carried out at NaCl concentrations of 25, 100, and 500 mm in NMR buffer. NisI linker mutants were measured in NMR buffer containing 100 mm NaCl. For the nisin-binding experiments SAXS data were recorded for NisI (70, 140, and 210 μm) and NisI-nisin (70 + 270 μm) in an optimized nisin titration buffer (200 mm NaAc, pH 4.5, 25 mm NaCl, 1 mm DTT). Radiation damage was excluded based on a comparison of individual frames of the 90/180-min exposures, where no changes were detected. A range of momentum transfer of 0.012 < s < 0.63 Å-1 was covered (s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ = 1.5 Å is the x-ray wavelength). All SAXS data were analyzed with the package ATSAS (version 2.5). The data were processed with the SAXSQuant software (version 3.9), and desmeared using the programs GNOM (47) and GIFT (48). The forward scattering, I(0), the radius of gyration, Rg, the maximum dimension, Dmax, and the inter-atomic distance distribution functions, (P(R)), were computed with the program GNOM. The masses of the solutes were evaluated by comparison of the forward scattering intensity with that of a human serum albumin reference solution (molecular mass 69 kDa) and using the Porod volume. Ensemble optimization method calculations were carried out using the ensemble optimization method program (49) with default settings. A random pool of 100,000 independent structures was generated using the primary sequence and the averaged and energy-minimized NMR structures of the individual domains as input. The disordered regions in the termini and connecting linkers were randomized (residues 1–9 and 111–118). Using the built-in genetic algorithm and using default settings a subset of a few independent structures was selected that describes the experimental SAXS best, and used to prepare the figures showing Rg/Dmax distributions.
Liposome Preparation
Multilamellar liposomes for NMR titration experiments were prepared using 1,2-dioleoyl-sn-glycero-3-phospho-(1-rac-glycerol) and cardiolipin from Avanti Polar Lipids (supplied in chloroform) in a 1:3 ratio mimicking the phospholipid composition of L. lactis membranes (50). Lipids were dried in a vacuum concentrator and hydrated in NMR buffer. Following hydration liposomes were prepared by 5 to 6 freeze/thaw cycles. For the floating assay small unilamellar vesicle liposomes were prepared by using a Mini-Extruder (Avanti Polar Lipids) equipped with membranes of a pore size of 0.4 μm.
Floating Assay
50 μg of protein and 4 mg of liposomes were mixed with a sucrose stock solution (70% (w/v) in NMR buffer) to generate 0.75 ml of a 50% (w/v) sucrose liposome protein solution, which was overlaid with 3.5 ml of 40% (w/v) sucrose and 0.75 ml of NMR buffer. After centrifugation at 10,000 × g for 17 h at 4 °C, 1-ml fractions were collected from top to bottom. Each fraction was analyzed by SDS-PAGE followed by Coomassie Blue staining.
Nisin Titration
Nisin was purified from commercial available nisin powder (Sigma, 2.5% nisin, N5764). 40 mg/ml of the lyophilized powder were dissolved in 50 mm lactic acid, pH 3.0, and filtered through a 0.45-μm membrane filter. 500 μl were applied to a Phenomenex Gemini-NX 250 × 10-mm HPLC column. Nisin was eluted by increasing acetonitrile concentrations from 5 to 100% with 0.1% TFA. The elution fractions were dried in a vacuum concentrator and washed twice with H2O. Nisin was then dissolved in an optimized NMR buffer with 200 mm NaAc, pH 4.5, 25 mm NaCl, 1 mm DTT (nisin titration buffer) at a concentration of 2 mm. For nisin titrations the NisI constructs were dissolved in the same buffer as nisin. A 60 μm NisI-(2–226) and 60 and 100 μm NisI-(97–226) protein samples were used for titration with an up to 10-fold excess of nisin. 15N-TROSY-HSQC spectra for NisI-(2–226) and 15N-HSQC spectra for NisI-(2–110), and NisI-(97–226) were recorded and peak intensities were extracted using Topspin 3.2. For KD determination the binding curves were fitted to the following equation using Origin Pro 2015,
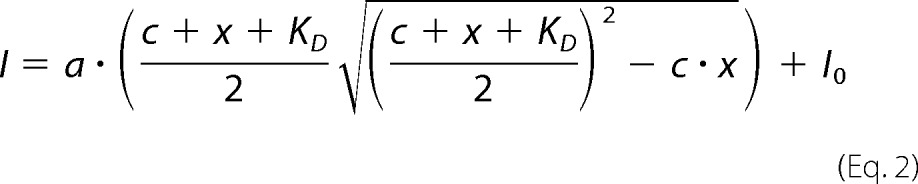 |
I peak is intensity (normalized), I0, initial peak intensity, KD, dissociation constant, a normalization factor, c, protein concentration, x, concentration of nisin.
Determination of NisI-mediated Immunity in Vivo
For the determination of LanI-mediated immunity, B. subtilis cells were grown in TY medium supplemented with 0.3 m NaCl and 1% xylose for the induction of lanI expression. Spectinomycin (100 μg/ml) and erythromycin (1 μg/ml) were used for the selection of B. subtilis transformants. B. subtilis TMB299 strains expressing different NisI variants were used to determine immunity against nisin. Overnight cultures of the corresponding strains were diluted to a final A600 of 0.1 in TY medium with added 0.3 m NaCl, and 1% xylose. Cells were incubated until an A600 of 1.0 was reached and were split into 2-ml aliquots. Different amounts of a stock solution containing 0.08 μg/ml of purified nisin were added and A600 was measured after 30 and 210 min. Strains expressing the wt-nisI gene or no lanI gene were used as positive and negative controls. Protein expression and localization was verified by Western blot analysis.
Results
NisI Is a Two-domain Protein
A NisI variant lacking the N-terminal leader sequence and the lipobox motif (NisI-(2–226), Fig. 1A) (28) was used for initial NMR studies. A nearly complete backbone resonance assignment was obtained for this 25.8-kDa protein (BMRB accession number 25193). The secondary structure of NisI-(2–226) was determined using HN, N, Cα, Cβ, and CO chemical shifts and indicated that NisI is mainly a β-sheet protein. A comparison of the secondary structure derived for full-length NisI with that of SpaI showed that the C-terminal half of NisI has an arrangement of secondary structure elements very similar to SpaI (Fig. 1B). In addition, the limited sequence homology of NisI and SpaI is restricted to the C-terminal half of NisI (Fig. 1B). Furthermore, hetNOE measurements revealed that the C-terminal half of NisI contains a large flexible loop that is also present at a similar position in SpaI (Fig. 1C). Taken together, these data suggested that the C-terminal half of NisI contains a domain with a SpaI-like three-dimensional structure. In addition, hetNOE measurements showed that the N-terminal part of NisI and the C-terminal SpaI-like part are connected via a flexible linker (amino acids 112–119) indicating that NisI consists of two separate domains (Fig. 1C). Expression of the isolated C-terminal SpaI-like part of NisI (NisI-(97–226)) resulted in a stable, well folded protein with the same secondary structure as seen in the full-length protein. Furthermore, the isolated N-terminal part (NisI-(2–110)) could be expressed as a well folded and stable protein. Thus, NisI is composed of two structurally independent domains. Comparison of the 15N-HSQC spectra of full-length NisI with the 15N-HSQC spectra for the isolated domains (Fig. 1, D and E) showed overlap for most signals in agreement with similar structures of the domains in isolation and as part of the full-length protein. A fraction of the signals in the 15N-HSQC spectra show small chemical shift differences suggesting a weak interaction between the two domains (see below). Consequently, we used the isolated 12.7-kDa N-terminal (NisI2–110) and the isolated 14.6-kDa C-terminal domain (NisI-(97–226)) of NisI, respectively, for structure determination.
Structure of the Individual NisI Domains
The NMR-derived solution structures for the two domains were based on 2336 and 2277 NOE-derived proton-proton distance restrains for NisI-(2–110) and NisI-(97–226), respectively. In addition, 156 and 162 chemical shift derived dihedral angle restrains were used for NisI-(2–110) and NisI-(97–226), respectively. In the final round of structure calculations, residual dipolar coupling restraints were included for both domains. For the C-terminal domain (NisI-(97–226)) the first 22 amino acids were excluded from structure calculations, because residues 97–118 are flexible in solution as indicated by low hetNOE values and the absence of long-range NOEs cross-peaks for those residues. Additionally, a comparison of 15N-HSQC spectra of NisI-(97–226) and a shortened construct starting at amino acid 119 (NisI-(119–226)) did not show any chemical shift differences (data not shown). The ensembles of the 19 lowest energy structures calculated with CYANA (39) and an energy minimized representative mean structure (39) for both domains are shown in Fig. 2A. A schematic depiction of the domain structures is shown in Fig. 2B.
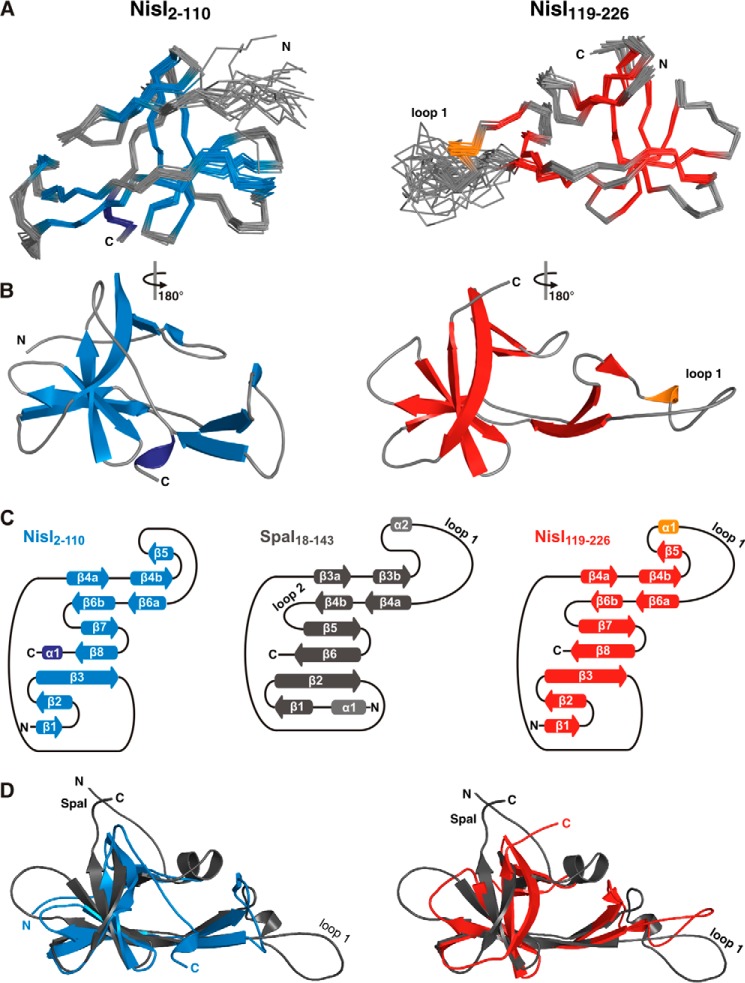
FIGURE 2.
Both domains of NisI have a SpaI-like three-dimensional structure. A, backbone traces of the 19 lowest energy conformers and the regularized mean structure for NisI-(2–110) (left) and NisI-(119–226) (right). β-Sheets are shown in blue for NisI-(2–110) and red for NisI-(119–226). α-Helices are shown in dark blue for NisI-(2–110) and orange for NisI-(119–226). B, schematic representations of the energy minimized mean structures of Nis-(2–110) (left) and NisI-(119–226) (right) with the same color-coding as in A. C, schematic representation of the secondary structure of NisI-(2–110) (left), SpaI (middle), and NisI-(119–226) (right) with arrows representing β-sheets and cylinders representing α-helices. D, superpositions of the mean structure of SpaI colored gray with the mean structures of NisI-(2–110) (blue, left) and NisI-(119–226) (red, right).
Both domain ensembles have a low backbone r.m.s. deviation of ∼0.3 Å for the rigid parts of the proteins. The NMR statistics for the NisI domain structures are listed in Table 1 according to the recommendations of the NMR-VTF (51). Both structures have been deposited in the Protein Data Bank under ID codes 2n32 (NisI-(2–110)) and 2n2e (NisI-(119–226)).
TABLE 1.
Structural statistics for the NMR solution structures of NisI-(2–110) and NisI-(119–226)
| NisI-(2–110) | NisI-(119–226) | |
|---|---|---|
| Conformational restricting restraints | ||
| Total NOE distance restraints | 2336 | 2277 |
| Intraresidue |i = j| | 526 | 428 |
| Sequential |i - j| = 1 | 616 | 634 |
| Medium range 1 <|i - j| <5 | 290 | 316 |
| Long-range |i - j| ≥5 | 904 | 899 |
| Dihedral angle restraints (Talos+) | 156 | 162 |
| Residual dipolar couplings (DNH) | 72 | 76 |
| No. of restraint per residue | 24.1 | 23.6 |
| No. of long-range restraints per residue | 8.4 | 8.3 |
| Residual restraint violationsa | ||
| Average no. of distance violations per structure | ||
| 0.1–0.2 Å | 4.65 | 5.4 |
| 0.2–0.5 Å | 1 | 1.1 |
| >0.5 Å | 1 | 3 |
| Average No. of dihedral angle violations per structure | ||
| 1–10° | 50.05 | 49.45 |
| >10° | 1 | 1 |
| Model quality (ordered residues)a | ||
| R.m.s. deviation backbone atoms (Å) | 0.3 | 0.3 |
| R.m.s. deviation heavy atoms (Å) | 0.7 | 0.7 |
| R.m.s. deviation bond lengths (Å) | 0.011 | 0.011 |
| R.m.s. deviation bond angles (°) | 2.2 | 2.2 |
| MolProbity Ramachandran statisticsa | ||
| Most favored regions | 94.7% | 94% |
| Allowed regions | 4.4% | 5.7% |
| Disallowed regions | 0.9% | 0.4% |
| Global quality scores (raw/Z score)a | ||
| Verify3D | 0.39/−1.12 | 0.43/−0.48 |
| Prosall | 0.76/ 0.45 | 0.67/ 0.08 |
| PROCHECK (ϕ-ψ) | −0.58/−1.97 | −0.59/−2.01 |
| PROCHECK (all) | 0.67/−3.96 | −0.64/−3.78 |
| MolProity clash score | 6.08/ 0.48 | 4.82/ 0.7 |
| Model contents | ||
| Ordered residue ranges (hetNOE >0.6) | 9–110 | 121–178,191–224 |
| Total No. of residues | 109 | 108 |
| BMRB accession number | 25175 | 25194 |
| PDB code | 2n32 | 2n2e |
a Calculated using PSVS 1.5 for using ordered residues (HetNOE > 0.6) (43). Average distance violations were calculated using the sum over r−6.
As already expected based on the NMR-derived secondary structure NisI-(119–226) shows the same three-dimensional fold as SpaI. Similar to SpaI, NisI-(119–226) is mainly a β-sheet protein with an elongated shape. The core of the domain is formed by a seven-stranded antiparallel twisted β-sheet with the strand order β1-β2-β3-β8-β7-β6b-β4a. In comparison to the six-stranded β-sheet of SpaI there is an additional N-terminal β-strand β1. An extended β-hairpin is formed by strands β4b and β6a and is stabilized by hydrophobic packing interactions with residues from β1 and β2. In addition, the β-hairpin is flanked by a short 310 helix. The characteristic large flexible loop of SpaI at the apex of the long β-hairpin is also seen in NisI-(119–226). Low hetNOE values specify this loop as highly flexible in solution (Fig. 1C, loop 1). In contrast to the structure of SpaI, the second flexible internal loop between β4b and β5 is not found to be flexible in NisI-(119–226) according to the hetNOE data (Fig. 1C). In summary, the overall structure of NisI-(119–226) closely resembles the three-dimensional-fold found in SpaI. This is reflected in a DALI Z-score of 7.4 (52) and a Cα-r.m.s. deviation to SpaI-(18–143) of 3.9 Å.
Surprisingly, the N-terminal domain of NisI also adopts a fold centered around a seven-stranded antiparallel twisted β-sheet, which is similar to SpaI (Cα-r.m.s. deviation to SpaI-(18–143): 3.5; DALI Z-score 6.3) and to the C-terminal domain of NisI (Cα-r.m.s. deviation to NisI-(119–226): 2.6; DALI Z-score: 9.7). However, the individual β-strands are shorter in comparison to SpaI and NisI-(119–226) (Fig. 2D). The long β-hairpin comprised of strands β4b and β6a is flanked by a short additional β-strand (β5) similar to NisI-(119–226). The large flexible loop 1 is not found in NisI-(2–110) but a shorter loop with a rigid structure indicated by hetNOE values >0.6 is present at the tip of the β-hairpin. The secondary structure pattern (Fig. 2C) and the three-dimensional fold of NisI-(2–110) is very similar to both SpaI (Fig. 2D) and the NisI C-terminal domain but the hetNOE data (Fig. 1C) reveal an overall rigid and globular structure of NisI-(2–110). However, whereas the three-dimensional structures of the two NisI domains both strongly resemble the structure of the folded core of SpaI, NisI lacks the long unstructured basic N terminus of SpaI, which interacts with the membrane. Furthermore, the two NisI domains display strong differences in their surface properties. The surface of the N-terminal domain is highly positively charged, whereas the C-terminal domain has a highly negatively charged surface with hydrophobic areas (see below and Fig. 3D). In this respect, the C-terminal domain of NisI also resembles the folded core domain of SpaI. However, the exact location of the negatively charged surface patches of the NisI C-terminal domain differs from SpaI.
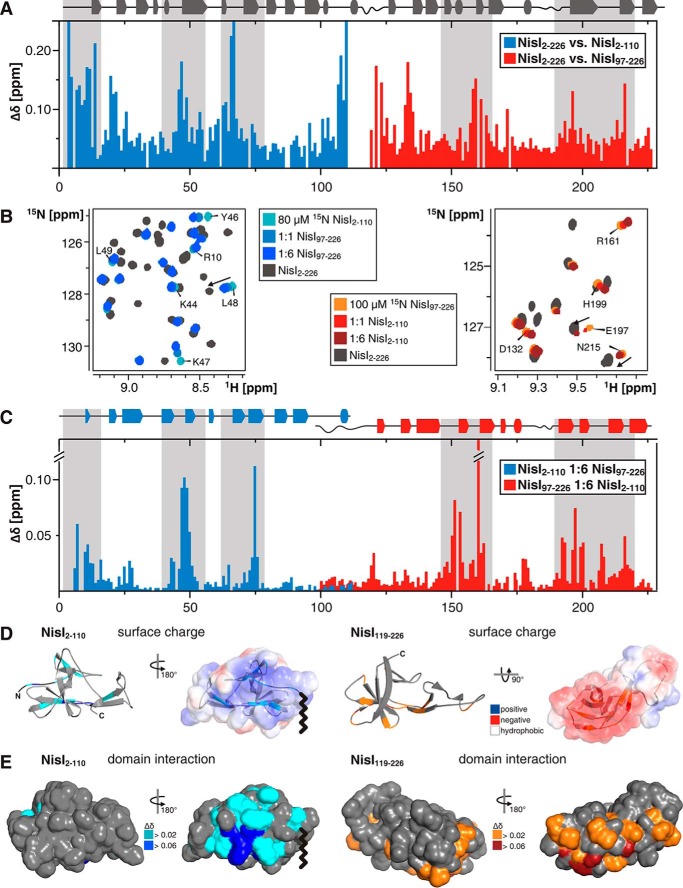
FIGURE 3.
Interdomain interactions in NisI. A, histogram of the measured amide backbone 1H,15N chemical shift differences between full-length NisI (NisI-(2–226)) and the isolated N-terminal (NisI-(2–110)) and C-terminal (NisI-(97–226)) domains. The secondary structure for full-length NisI is indicated by arrows and tubes atop of the histogram. B, overlay of a part of the 15N-HSQC spectra of 80 μm 15N-labeled NisI-(2–110) (left) and 100 μm 15N-labeled NisI-(97–226) (right) in the absence (light blue and light red) and presence of increasing amounts of the other domain in its unlabeled form. The molar ratios of the two domains are 1:1 (blue and red) and 1:6 (dark blue and dark red). The 15N-TROSY-HSQC of full-length NisI is shown in gray for comparison. Signals of residues of the individual domains, which move toward the chemical shift of the same residue in the full-length protein are indicated with arrows. C, histogram of the chemical shift differences of NisI-(2–110) (blue) and NisI-(97–226) (red) upon addition of a 6-fold molar excess of the other domain in its unlabeled form. D and E, show the chemical shift changes of NisI-(2–110) and NisI-(97–226) upon addition of the other unlabeled domain in 6-fold excess mapped either onto the schematic representation (D) or a solvent accessible surface representation (E) of the structure of NisI-(2–110) (left) and NisI-(119–226) (right) in two different orientations. For comparison, D also shows the electrostatic surface potentials of each domain mapped on the solvent accessible surfaces of each domain with negatively charged surface areas colored in red, positively charged areas colored in blue and white areas corresponding to hydrophobic surfaces. A wavy line indicates the approximate position of the diacylglycerol anchor connected to the cysteine of the lipobox in membrane bound NisI. The orientation of the electrostatic surface maps in D correspond to the orientation of the chemical shift maps shown on the right-hand side in each panel in E.
Domain Interaction
The comparison of the 15N-TROSY-HSQC spectra of NisI-(2–226) and the 15N-HSQC spectra of the individual domains showed chemical shift differences for some residues not located in or near the interdomain linker. This indicates an interaction of the two domains in the context of the full-length protein. Significant chemical shift differences between the domains and the full-length protein cluster to two structurally equivalent areas of each domain, β-strands β4 and β6, which are part of the β-hairpin (Fig. 3A). Additionally, the N-terminal residues of NisI-(2–110) show significant chemical shift differences between the free domain and the full-length protein (Fig. 3A).
To further investigate the interdomain interaction the 15N-labeled N-terminal domain of NisI was titrated with the unlabeled C-terminal domain and vice versa. In both cases, the 15N-HSQC spectra of the labeled domain show chemical shift changes upon the addition of increasing amounts of the unlabeled domain (Fig. 3B). Thus, the two isolated domains form an intermolecular complex that is in fast exchange on the NMR time scale in agreement with a weak interaction. To identify the surface areas involved in the interaction between the two separated domains the observed chemical shift changes were mapped onto the sequence (Fig. 3C) and the structures of the two domains (Fig. 3, D and E). The chemical shift changes observed in the titration experiments cluster to the same surface areas as the chemical shift differences observed between the full-length protein and the isolated domains. Thus, both data sets identify similar interaction surfaces for the two domains. In line with this observation, in the NMR titration experiments with the two isolated NisI domains a number of signals are shifting toward their corresponding chemical shift in the full-length protein (Fig. 3B).
The two NisI domains clearly show opposite electrostatic surface properties (Fig. 3D). Although the N-terminal domain is overall positively charged with 17 basic and only 13 acidic residues, the C-terminal domain has an excess of 11 negatively charged residues. Mapping the electrostatic properties onto the surface of the two domains reveals that the positive charges of NisI-(2–110) and the negative charges of NisI-(119–226) cluster to defined surface regions on each domain (Fig. 3D). Interestingly, the interdomain interaction surface of NisI-(2–110) is dominated by positive charges, whereas the interdomain interaction site of NisI-(97–226) is predominantly negatively charged (Fig. 3, D and E). Additionally some hydrophobic residues flank the interaction surface in both domains. These residues are part of β-strands β4 and β6 in both domains and are solvent-exposed in the structures of the individual domains. Thus, the interaction between the two domains of NisI is dominated by complementary electrostatic surface features and supplemented by matching hydrophobic patches. The importance of electrostatic complementarity for the interdomain interaction is borne out by the effects of increasing salt concentrations on the NMR spectra of the full-length protein. An increase of the NaCl concentration to 500 mm leads to chemical shift changes of the full-length protein for many residues that are located in the interacting domain surfaces (Fig. 4A). Importantly, the 15N-TROSY-HSQC spectrum of NisI-(2–226) at 500 mm NaCl is more similar to the 15N-HSQC spectra of the isolated domains as the one recorded with the standard buffer containing 100 mm NaCl. This indicates the weakening or the loss of the interdomain interaction at higher salt concentrations.
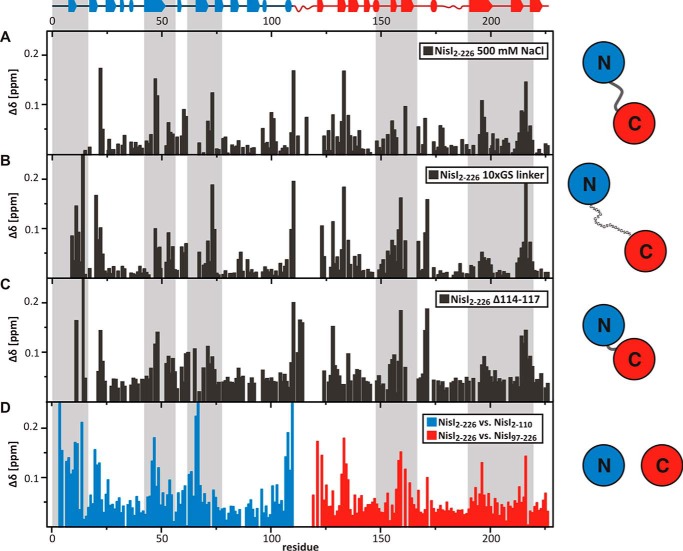
FIGURE 4.
Chemical shift differences between full-length NisI (WT) in NMR buffer (100 mm NaCl) and full-length NisI (WT) in buffer containing 500 mm NaCl (A) or NisI constructs with either an extended 10xGS linker insertion (B) or with a truncated linker (C). For comparison, D shows the chemical shift differences between full-length NisI and the two isolated domains as shown in Fig. 3A and serves as a reference. Gray boxes highlight residues involved in interdomain interactions.
To define the orientation of the two domains with respect to each other, we analyzed 15N- and 13C-NOESY-HSQC spectra of the full-length protein. However, there were no observable NOE interactions between the two domains. Furthermore, many residues located in the interdomain interface showed broadened NMR signals. This is in agreement with a weak and dynamically disordered interface. Alternatively, we employed chemical shift driven docking using HADDOCK (53) to generate models for the domain orientation in the full-length protein. However, multiple structural families with different relative domain orientations were obtained in these calculations. Residual dipolar couplings can in principle deliver the information required to orient domains in a multidomain protein with respect to each other (54). We successfully used residual dipolar couplings to refine the structures of the isolated NisI domains using media matched to the physical properties of the two domains to prevent an interaction of the aligning medium with the protein. However, for full-length NisI all tested alignment media including uncharged media (36) induced chemical shift changes also involving residues in the interdomain interfaces suggesting that the alignment media changed the interdomain orientation and therefore could not be used to derive the interdomain orientation in NisI by residual dipolar couplings. Finally, paramagnetic relaxation enhancements measured in spin-labeled proteins could provide long-range distance constraints that might be helpful in defining interdomain orientations (55, 33). To this end cysteine mutants of the protein are required. We tested a number of cysteine point mutations with mutation sites chosen based on the structures of the isolated domains and not located in the putative interdomain interaction surface as identified by chemical shift changes. However, many of these mutations lead to chemical shift changes not restricted to the vicinity of the mutation sites but often including residues in the interdomain interface. We were able to only measure the paramagnetic relaxation enhancement for three cysteine mutants, all located in the N-terminal domain. Nonetheless, structure calculations with these paramagnetic relaxation enhancements did not result in converged structures with a defined interdomain orientation.
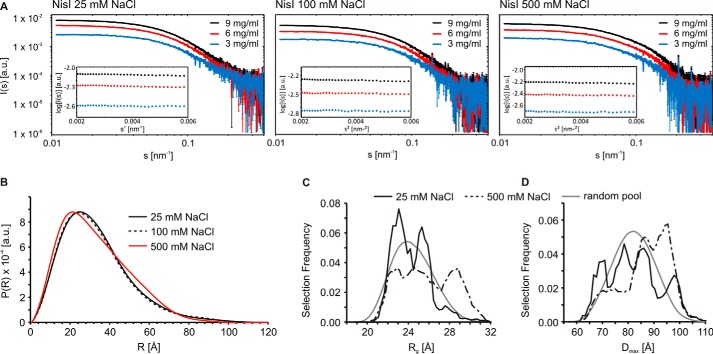
To further address the conformation of NisI in solution at different NaCl concentrations, we performed SAXS experiments. Scattering curves recorded at several solute concentrations (Fig. 5A) shows that NisI adopts a compact conformation at low salt concentrations (25 and 100 mm) and a more extended conformation at high salt concentrations (500 mm NaCl) as we observed an increase in radial density at higher distances (Fig. 5B). We analyzed the conformational space sampled by NisI using the ensemble optimization method (49). Here, a pool of independent structures based on the amino acid sequence and available structural information for the individual domains is first generated. A genetic algorithm is then used to select an ensemble of structures that best describes the experimental SAXS data (for more details see “Experimental Procedures”). We identified the best ensembles concerning the agreement of experimental and back-calculated data and compared the Rg and Dmax distributions of the random pool of structures to the selected ensembles (Fig. 5, C and D). At a low NaCl concentration, the selected NisI ensemble clearly shows a preference for conformations with a higher degree of compaction compared with the random pool (Fig. 5, C and D). This suggests that there must be inter-domain interactions within NisI that lead to a compaction of the molecule. At higher NaCl concentrations, NisI partly loses this compaction, as regions of the conformational ensemble with higher Rg and Dmax become more populated (Fig. 5).
FIGURE 5.
A dynamic global conformation of full-length NisI in solution as seen by SAXS. A, raw SAXS scattering data for NisI-(2–226) at three different salt concentrations and measured for three different protein concentrations under each condition as indicated. The insets show the corresponding Guinier plots, which demonstrate the absence of protein aggregation. B, SAXS curves show an extension of NisI at higher salt concentrations. SAXS data showing a comparison of the experimental radial density distributions of NisI with increasing salt concentrations as indicated. C and D, salt-dependent changes in the conformational ensemble of NisI. Rg (C) and Dmax (D) distributions of the ensemble optimization method models of full-length NisI at 25 and 500 mm NaCl in the initial pool of structures with randomized interdomain linkers (gray lines) and in the ensembles selected to represent the experimental data (solid and dashed black lines).
Taken together, SAXS data, salt sensitivity of the interdomain interaction, the absence of interdomain NOEs, the line broadening for residues in the interdomain interface, and the long-range chemical shift changes observed upon cysteine mutations suggest that the interaction of the N- and C-terminal domains of NisI in the context of the full-length protein is weak and dynamic. Consequently, we refrain here from presenting a structure of full-length NisI with a defined interdomain orientation.
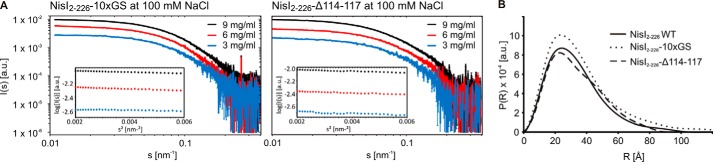
To explore a putative biological function of the interdomain orientation in NisI we constructed linker mutants of NisI-(2–226). The flexible linker connecting the two domains was either shortened by 4 residues (NisI-(2–226)-Δ114–117) or extended with a flexible GS-linker ((NisI-(2–226)-10xGS). For both linker mutants we observed 15N-TROSY-HSQC spectra with significant chemical shift differences to WT-NisI in NMR buffer (100 mm NaCl) for residues located in the interdomain interfaces (Fig. 4B, 4C). Furthermore, for both mutants the 15N-TROSY-HSQC spectra are more similar to the spectra of the free domains than to WT-NisI in NMR buffer (100 mm NaCl) consistent with a weakened interdomain interaction in the mutants. The SAXS data recorded for both mutants are in good agreement with the NMR results. Both the extension and deletion of the linker residues resulted in more open conformations (Fig. 6). Extension of the interdomain linker in the NisI-(2–226)-10xGS mutant leads to an increase in the radial density distribution at higher distances and in the Dmax. This would be in agreement with weakened interactions between the N- and C-terminal domains. Deletion of linker residues in the NisI-(2–226)-Δ114–117 mutant induces structural rearrangement of the N- and C-terminal domains and an increase in the radial density distribution at larger distances. Despite these structural rearrangements, in growth tests both linker mutations were able to confer immunity to B. subtilis to a level similar to the wild type protein (data not shown). Thus, a well defined relative orientation of the two domains of NisI with respect to each other is apparently not important for the biological function of NisI.
FIGURE 6.
Changes in the global conformation in NisI linker mutants NisI-(2–226)-10xGS and NisI-(2–226)-Δ114–117. A, raw SAXS scattering data for both NisI linker mutants at three different protein concentrations recorded at a NaCl concentration of 100 mm. The insets show the corresponding Guinier plots. B, comparison of the experimental radial density distributions of the two NisI linker mutants with WT-NisI.
Membrane Interaction
SpaI and NisI are both lipoproteins, which are attached to the outer membrane via a diacylglycerol anchor. For SpaI it was shown that the unstructured basic N terminus is able to bind to membranes even in the absence of a lipid anchor (22). NisI does not contain an unstructured N terminus like SpaI but an N-terminal domain with a highly positively charged surface. To test if NisI is able to interact with membranes even in the absence of a covalent diacylglycerol anchor the interaction of NisI with liposomes composed of the two major phospholipids found in L. lactis 1,2-dioleoyl-sn-glycero-3-phospho-(1-rac-glycerol) and cardiolipin in an 1:3 molar ratio (22) was investigated. Both phospholipids harbor a negatively charged head group, which might interact with residues of the positively charged surface of the NisI N-terminal domain.
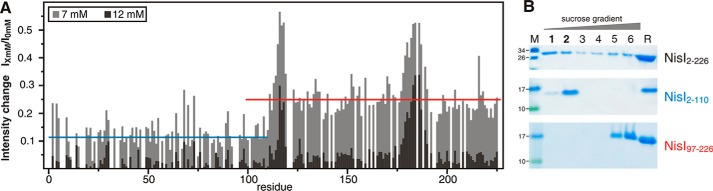
In NMR titration experiments with these liposomes all 15N-TROSY-HSQC signals of full-length NisI undergo intensity changes. Such a decrease in signal intensities is due to signal broadening caused by binding to the large liposomes and the resulting increase of the rotational correlation time of the complex. However, a quantitative analysis of the changes in the signal intensities for full-length NisI showed that the signal intensities for the residues of the N-terminal domain decrease more strongly than those for residues of the C-terminal domain (Fig. 7A). Thus, the positively charged N-terminal domain of NisI is the major determinant for liposome binding. NMR-based liposome titration experiments with the isolated domains confirmed these results because they showed a strong decrease in signal intensities for the N-terminal domain and no signal intensity or chemical shift changes for the isolated C-terminal domain (data not shown).
FIGURE 7.
The interaction of NisI with membranes. A, NMR signal intensity changes in 15N-TROSY-HSQC spectra of full-length NisI after addition of liposomes in increasing concentrations mapped onto its sequence. Missing data points correspond to prolines or overlapping signals. Blue and red lines correspond to the average intensity changes observed for the N-terminal (blue) and the C-terminal (red) domain, respectively. B, SDS-PAGE analysis of the liposome floating assay for either full-length NisI (top) or the isolated N-terminal (middle) and C-terminal (bottom) domains. Lanes 1–6 correspond to fractions from the top (fraction 1) to the bottom (fraction 6) of the gradient after ultracentrifugation. Fractions 1 and 2 contain floated liposomes. Lane R corresponds to the protein-liposome mixture before ultracentrifugation as a reference.
In addition, we tested the membrane interaction of full-length NisI (NisI-(2–226)) and its isolated N- and C-terminal domains in floating assays with unilamellar liposomes. The different membrane binding capabilities of full-length NisI and the isolated N- and C-terminal domains of NisI are obvious (Fig. 7B). After ultracentrifugation NisI-(2–110) is completely located in the floated liposome fraction. In contrast, the negatively charged NisI-(97–226) does not float together with the liposomes and therefore does not bind to the liposomes. The full-length NisI is found in both the liposome containing and the liposome-free fractions of the gradient. In agreement with the NMR results presented above the floating assay shows that only the N-terminal domain binds to liposomes. Interestingly, full-length NisI binds weaker to liposomes than the isolated N-terminal domain. This suggests that the C-terminal domain partially shields the membrane binding surface of the N-terminal domain, which is therefore not fully accessible for membrane interactions in the context of the full-length protein.
Nisin Binding
Previously it was shown by native PAGE that NisI directly binds to nisin (24) although the interaction is rather weak with a KD for the interaction in the micromolar range (25). Stein et al. (24) also showed that NisI does not bind the structurally closely related subtilin. In vivo studies with truncated NisI variants (26, 27) implied the C terminus of NisI and in particular the last 22 amino acids in nisin binding. To further elucidate the NisI/nisin interaction we titrated NisI with purified nisin in 15N-HSQC experiments. According to solubility studies by us and others nisin has a poor solubility at higher pH and higher salt concentrations. For our NMR binding studies we therefore chose buffer conditions where both the protein and the lantibiotic were soluble and stable. Both were dissolved in 200 mm NaAc buffer, pH 4.5, with 25 mm NaCl and 1 mm DTT. The assignments for NisI were transferred to this new buffer condition by comparison of 15N-HSQC spectra.
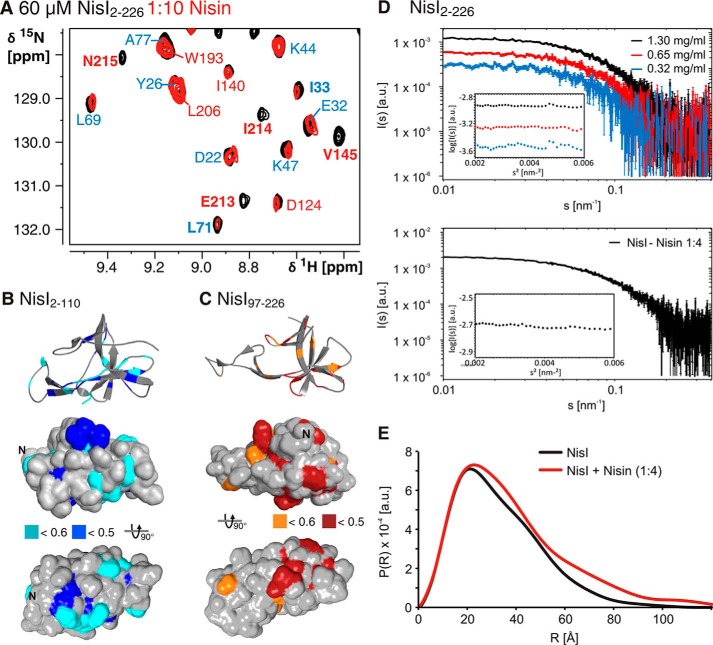
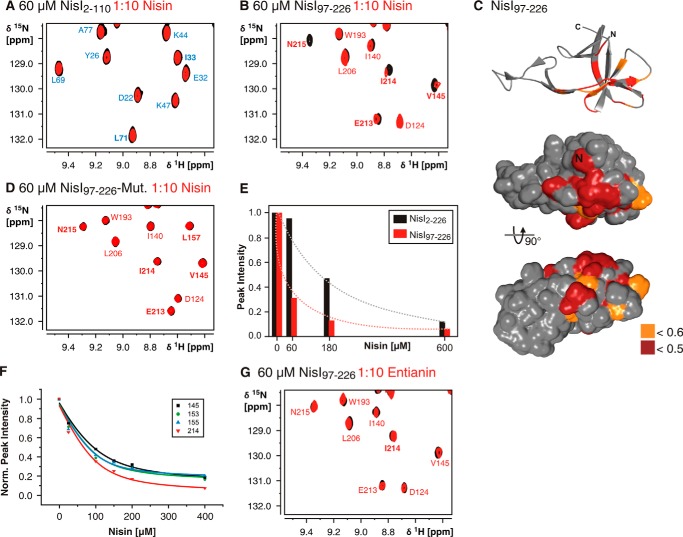
In TROSY-HSQC-based nisin titration experiments with full-length NisI we observed chemical shift changes or significant reductions in signal intensities for a number of residues (Fig. 8A). This confirms the earlier observations of a direct nisin/NisI interaction. The reduction in signal intensities observed for a number of NisI residues suggests that the complex is in intermediate exchange with the free components on the NMR time scale. Interestingly, signals for residues of both domains are affected by the binding of nisin (Fig. 8, B and C). Notably, some of the residues affected by nisin binding are also part of the interaction surface of both domains. This could be explained by two different models. Either nisin is sandwiched between residues of both domains and both domains form a composite binding interface or nisin binds to only one of the two domains and disrupts the interdomain interaction leading to subsequent chemical shift changes in the other domain. To distinguish between these two models we first recorded SAXS data for NisI alone and NisI in the presence of a 4-fold excess of nisin (Fig. 8D). Analysis of the SAXS data showed that the binding of nisin changes the interdomain orientation leading to more open and elongated NisI conformations (Fig. 8E). In addition, we titrated the isolated NisI domains with nisin. The spectra of the N-terminal domain NisI-(2–110) do not show any differences upon nisin titration demonstrating that the isolated N-terminal domain is not able to interact with nisin (Fig. 9A). In contrast, the C-terminal domain of NisI (NisI-(97–226)) showed chemical shift changes and significant changes in signal intensities for a number of signals (Fig. 9B). Thus, the isolated C-terminal domain of NisI retains the ability to bind to nisin. The nisin interacting residues are located in β-strands β4a, β6, and β7 and overlap with the interdomain interaction site (Fig. 9C). Surprisingly, only three of 28 residues affected by nisin binding are located in the C-terminal 22 amino acids. This is in contrast to previous proposals that these C-terminal residues are very important for nisin binding and specificity (26, 27) (see below). To confirm that the residues of the C-terminal domain, which show chemical shift changes upon nisin addition are functionally important for nisin binding we constructed a double mutant including two of these residues located in β-strand β4a in the center of the putative binding site. This double mutant Y153A,D155K is stably folded because it yielded a 15N-HSQC spectrum with well dispersed signals but was no longer able to bind nisin (Fig. 9D).
FIGURE 8.
Nisin binding of full-length NisI. A, overlay of a part of the 15N-TROSY-HSQC spectra of NisI-(2–226) in the absence (black) or presence of a 10-fold molar excess of nisin (red). Nisin binding causes a reduction in signal intensities in agreement with complex formation in intermediate exchange on the NMR time scale. Assignments for residues in the N-terminal domain are indicated in blue and assignments for residues of the C-terminal domain are indicated in red. The residues showing intensity changes are mapped onto the structure of the N-terminal domain (B) or the C-terminal domain (C). Large intensity changes with a relative intensity in the bound state of <0.5 are highlighted in darker colors, unaffected residues are colored gray. D, raw SAX scattering data for NisI-(2–226) in nisin titration buffer at three different concentrations and NisI-(2–226) (70 μm) upon addition of a 4-fold excess of nisin. The corresponding Guinier plots are shown as insets. E, a comparison of the experimental radial density distributions of NisI in the absence and presence of nisin.
FIGURE 9.
Nisin binds to the isolated C-terminal domain of NisI (NisI-(97–226)). A, the isolated N-terminal domain of NisI (NisI-(2–110)) is not interacting with nisin. An overlay of 15N-HSQC spectra of the domain in the absence (black) and presence of a 10-fold molar excess of nisin shows no differences. B, an overlay of 15N-HSQC spectra of the isolated C-terminal domain in the absence (black) and presence of a 10-fold molar excess of nisin shows clear differences in a number of peak intensities indicative of nisin binding. C, mapping of the NMR signal intensity changes in 15N-HSQC spectra upon nisin addition to the isolated NisI C-terminal domain onto the solvent accessible surface map that is shown in the same orientations as in Fig. 8C. D, the NisI C-terminal domain double mutant Y153A,D155K has lost the ability to bind nisin. No changes in the 15N-HSQC spectra are observable upon addition of nisin. E, comparison of the NMR signal intensity changes for residue Tyr-153 as a function of nisin concentration in full-length NisI (black bars) and the isolated C-terminal domain (red bars). F, determination of the KD for the interaction of the NisI C-terminal domain for nisin by plotting the NMR signal intensities for exemplary residues in the nisin binding site as indicated as a function of nisin concentration. G, the C-terminal domain of NisI does not bind to the subtilin variant entianin as seen from an overlay of the 15N-HSQC spectra of the C-terminal domain in the absence (black) and presence (red) of a 10-fold molar excess of entianin, which shows no chemical shift differences.
Because the same protein concentrations and the same nisin stock solution were used in the nisin titration experiments with full-length NisI and the NisI C-terminal domain the change in NMR signal intensity as a function of nisin concentration can be analyzed qualitatively to compare the affinity of full-length NisI and its isolated C-terminal domain for nisin (Fig. 9E). Here, it is obvious that for the same residues the decrease in signal intensity is much more pronounced for the isolated C-terminal domain in comparison to the full-length protein. Thus, the isolated C-terminal domain binds nisin with significantly higher affinity than the full-length protein. Although full-length NisI in the presence of nisin was too unstable to carry out the nisin titration experiments in a quantitative manner we were able to obtain a full NMR titration curve for the NisI C-terminal domain. An analysis of peak intensities as a function of nisin concentration for a number of NMR signals in the nisin binding site yielded a KD for nisin of 22 ± 10 μm (Fig. 9F).
Finally, we tested the specificity of the C-terminal domain for nisin in NMR titration experiments. NisI does not confer immunity against subtilin and its close analog entianin (56), whereas SpaI confers immunity against both of these lantibiotics but not against nisin. We therefore titrated NisI-(97–226) with entianin, which in contrast to subtilin can be purified and dissolved in the amounts required for NMR titration experiments. In contrast to nisin entianin does not induce chemical shift changes or changes in signal intensity even at high molar excess (Fig. 9G).
C-terminal Truncated NisI
In 2006 Takala and Saria (26) proposed that the C terminus of NisI is important for nisin binding and therefore plays a major role in immunity. In growth tests, a deletion of 5 amino acids at the C terminus of NisI already results in a decrease of conferred immunity of ∼78% compared with native NisI in L. lactis. A deletion of 21 amino acids and even further truncated constructs resulted in a remaining immunity of only 14%. Later Al-Khatib et al. (27) showed that a deletion of the last 22 amino acids at the C terminus of NisI resulted in a decrease of the IC50 value for nisin by two-thirds. Our structural studies on NisI reveal that the 22 C-terminal amino acids are part of the structured core of the C-terminal domain of NisI. These amino acids form two β-strands central to the 7-stranded antiparallel twisted β-sheet. A deletion of these amino acids could therefore destabilize the structured core of the domain.
We used NMR spectroscopy to investigate the folding state of a C terminally truncated version of NisI. 15N-TROSY-HSQC spectra of C-terminal truncated NisI-(2–204) show a partly unfolded protein. All signals from the N-terminal domain are well dispersed and overlap perfectly with signals from the individual N-terminal domain NisI-(2–110). All remaining signals from the C-terminal domain are in the center of the spectrum and therefore indicate an unfolded domain (Fig. 10). The 15N-HSQC spectrum of C terminally truncated NisI-(97–204) alone overlaps perfectly with the unfolded peaks in the spectrum of NisI-(2–204). Thus, deletion of the 22 C-terminal amino acids of NisI leads to unfolding of the entire C-terminal domain of NisI. The decreased immunity of the truncated protein is most likely caused by the unfolding of the C-terminal domain.
FIGURE 10.
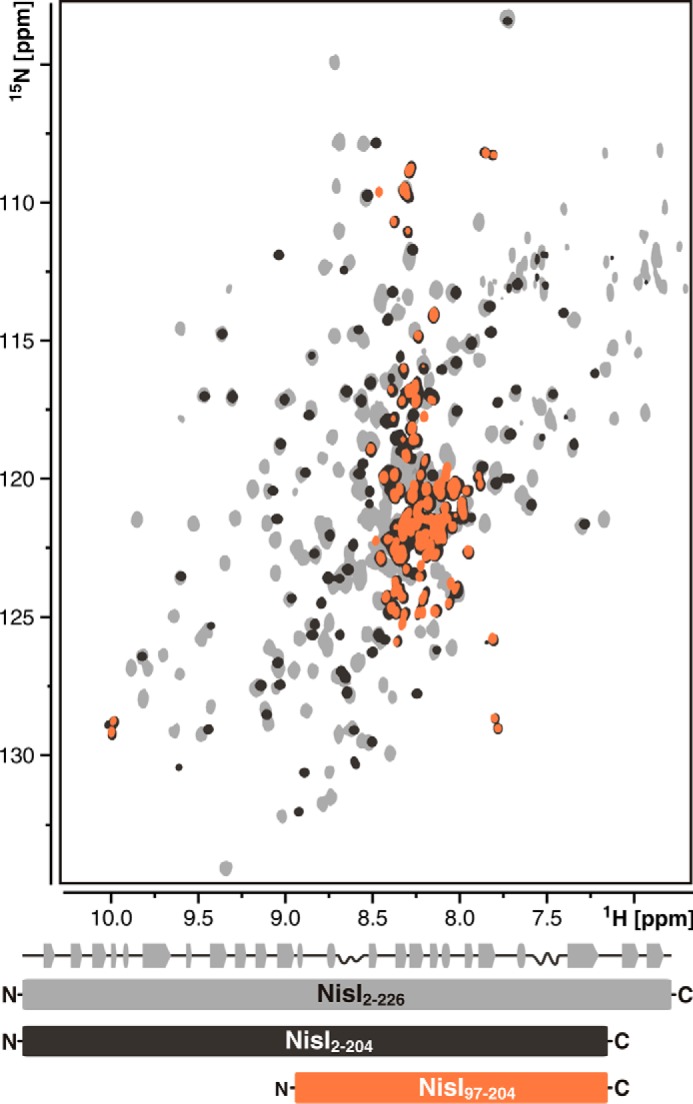
Deletion of the 22 C-terminal amino acid residues leads to unfolding of the C-terminal domain of NisI. Overlay of 15N-TROSY-HSQC spectra of full-length NisI (NisI-(2–226)) (light gray), the C-terminal deletion mutant of NisI (NisI-(2–204)) (black), and the C-terminal deletion mutant of the isolated C-terminal domain (NisI-(97–204)) (orange). A schematic representation of the secondary structure of full-length NisI is shown atop a schematic representation of the NMR constructs below the spectral overlay with the same color-coding as used for the spectra.
Discussion
Here we present a structural characterization of the lantibiotic immunity (LanI) protein NisI from L. lactis, which confers immunity against the lantibiotic nisin. Structural insights into the organization of LanI proteins are so far limited to the solution structure of SpaI (22), which protects B. subtilis against the lantibiotic subtilin. Although nisin and subtilin are structurally very similar the two LanI proteins differ significantly in size and only very limited sequence homology was observed. Furthermore, NisI only confers immunity against nisin, whereas SpaI is only active against subtilin and both proteins are not able to confer cross-immunity against the other lantibiotic (24). The structure of SpaI revealed a long, disordered, positively charged N terminus able to interact with membranes and a structured core domain with a central six-stranded β-sheet establishing a novel three-dimensional fold class (22). Our solution NMR studies of NisI demonstrate that NisI is a two-domain protein. Both domains are structurally independent from each other. Surprisingly, both fold into three-dimensional structures very similar to the SpaI core domain and to each other. Together, the structures of SpaI and the two domains of NisI establish a three-dimensional fold class typical for LanI proteins.
However, despite their similar three-dimensional structure both domains of NisI differ significantly with regard to their surface properties. The N-terminal domain of NisI has a basic pI and a strongly positively charged surface patch. In contrast, the C-terminal domain of NisI is highly negatively charged as is the core domain of SpaI. Our membrane interaction experiments (Fig. 7) show that the basic N-terminal domain of NisI is functionally equivalent to the basic disordered N-terminal tail of SpaI because it is also able to bind to membranes. On the other hand, the C-terminal domain of NisI represents the major determinant for nisin binding (Fig. 9). The negatively charged surface of the C-terminal domain might also play a role in keeping it away from the membrane and thereby increase its accessibility for nisin. Taken together, these observations suggest a possible evolutionary relationship between NisI-like and SpaI-like proteins. NisI-like proteins could have arisen from SpaI-like predecessors by domain duplication and subsequent functional specialization of the two domains. On the other hand, the disordered SpaI N terminus might represent a remnant of a basic N-terminal NisI-like domain as indicated by a short homologous stretch of sequence between the SpaI N terminus and β-sheet β4 of the NisI N-terminal domain (Fig. 1B). However, a SpaI-like domain organization appears to be prevalent among other functionally characterized LanI proteins from many firmicutes conferring immunity to nisin-like peptides.
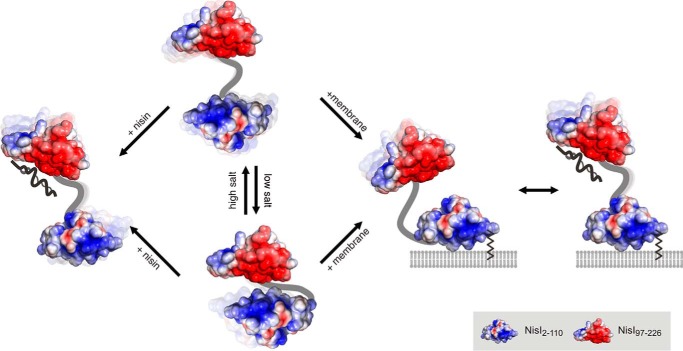
The two domains of NisI, which are separated from each other by a flexible linker, nevertheless, interact through an interface dominated by electrostatic complementarity and hydrophobic contacts. However, this domain interface apparently corresponds to a rather weak interaction with considerable dynamics with regard to the relative interdomain interactions. Consequently, no single preferred orientation of the two domains with respect to each other could be defined by the NMR and SAXS data. This is often observed for dynamic multidomain proteins, which sample a diverse continuum of conformations in solution involving encounter-like interdomain contacts (57–59). Unexpectedly, the amino acid residues in the C-terminal domain involved in interactions with the N-terminal domain are similar to those amino acids involved in nisin binding. In addition, based on qualitative NMR titration experiments the isolated C-terminal domain binds nisin with a higher affinity compared with the full-length protein. Similarly, the isolated N-terminal domain binds with higher affinity to membrane vesicles than the full-length protein. This suggests that the lipid-free NisI-protein predominantly exists as a family of conformers where the two domains interact with each other and thereby partially mask the membrane binding surface of the N-terminal domain as well as the nisin binding surface of the C-terminal domain. Membrane binding to the N-terminal domain or nisin binding to the C-terminal domain then interferes with the interdomain interactions leading to a family of (more) open conformations (Fig. 11). In vivo NisI most likely exhibits a predominantly open conformation because it is attached to the membrane via its N-terminal diacylglyceryl anchor thereby bringing the highly positively charged surface patch of the NisI N-terminal domain in close contact with the membrane surface. The sequestration of the N-terminal domain by the membrane would also lead to an enhanced affinity of the C-terminal domain for the lantibiotic. For the non-lipidated, secreted subpopulation of NisI (25) on the other hand nisin binding might promote the accessibility of the membrane-binding surface of the N-terminal domain.
FIGURE 11.
Model representation of the solution conformational ensemble of NisI in different functional states. Free NisI consists of a conformational ensemble with prevalent interdomain interactions at low salt concentrations. The membrane interaction and nisin binding surfaces of the N- and C-terminal domains are partially shielded in this state. At higher salt concentrations, more open conformations without interdomain interactions dominate the conformational ensemble. Nisin binding or membrane interactions compete with interdomain interactions and might thereby lead to conformational ensembles with reduced interdomain interactions.
Our NMR- and SAXS-based titration experiments with nisin yielded the first structural insights about the interaction of any lantibiotic with its corresponding immunity proteins. Previous biochemical experiments employing mainly truncated variants of NisI had pointed to a major role of the C-terminal 22 amino acids (NisI residues 205–226) for nisin recognition. However, in our NMR titration experiments only amino acids 214, 215, and 216 of this part of the protein were affected by nisin binding suggesting the C terminus of NisI is not the major determinant for nisin binding. Instead, we could show by NMR spectroscopy that in a 22-amino acid C-terminal truncation of NisI the entire C-terminal domain is unfolded explaining the previously reported loss of NisI function in these mutants. The complex of NisI and nisin or the NisI C-terminal domain and nisin is in intermediate exchange between free and complexed states on the NMR time scale causing signal loss and thereby preventing a further structural characterization of the nisin-protein complex. Nevertheless, our SAXS data show that NisI elongates in the presence of a stoichiometric excess of nisin. On the other hand, these experiments suggest that nisin binds to NisI with a KD in the micromolar range in agreement with previous biochemical experiments (25). This rather low affinity of NisI for nisin is unexpected because nisin is affecting cell growth already at nanomolar concentrations (MIC50 = 50 nm for L. lactis subsp. cremoris (60). Thus, one would expect that an immunity protein able to prevent nisin toxicity through simple binding and sequestering of nisin should have a higher affinity for its ligand. This suggests that NisI might confer immunity against nisin by a more complex mechanism than just simple binding to the lantibiotic. Additional components of the cell membrane such as lipid II might be required for an effective immunity mechanism mediated by NisI. However, the C-terminal domain of NisI apparently contributes significantly to the specificity of the immunity mechanism because it interacts exclusively with nisin but not with the close structural analog entianin (a subtilin variant) in NMR titration experiments (Fig. 9).
In conclusion, with our structural studies of NisI and SpaI we establish a unique three-dimensional fold class as prevalent in LanI proteins. We show that NisI contains two structurally highly similar but functionally specialized domains, lay the foundations to understand the interaction of lantibiotics with the associated LanI immunity proteins in molecular detail, and rationalize previous biochemical data regarding the determinants of lantibiotic binding and specificity.
Author Contributions
C. H. and N. A. C. conducted most of the experiments, analyzed the results, and wrote most of the paper together with J. W. E. D.-F. supervised NMR experiments and NMR data analysis. S. K. conducted growth assays with different NisI variants in B. subtilis. S. D. purified nisin. L. B. provided expression vectors for NisI. U. A. H. helped in design, setup, and data analysis for experiments analyzing the membrane interaction of NisI. C. G. and T. B. designed and conducted SAXS measurements together with C. H. and analyzed SAXS data. P. K., K. D. E., and J. W. conceived the idea for the project, designed experiments, and analyzed data. All authors read and commented on the manuscript.
Acknowledgments
We are grateful to the Center of Biomolecular Magnetic Resonance (BMRZ), Goethe-University Frankfurt for providing the NMR infrastructure and Dr. Christian Richter for spectrometer maintenance and helpful discussions. We acknowledge the Leibniz Supercomputing Center for providing computing time on the Linux Cluster.
This work was supported in part by Grants Wo 901/4-1, En 134/11-1, and MA 5703/1-1 (Emmy-Noether program) from the Deutsche Forschungsgemeinschaft (DFG) (to J. W., K. D. E., and T. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- HSQC
- heteronuclear single quantum coherence
- hetNOE
- heteronuclear nuclear Overhauser effect
- SAXS
- small-angle x-ray scattering
- r.m.s.
- root mean square.
References
- 1.Klaenhammer T. R. (1988) Bacteriocins of lactic acid bacteria. Biochimie 70, 337–349 [DOI] [PubMed] [Google Scholar]
- 2.Cotter P. D., Ross R. P., and Hill C. (2013) Bacteriocins: a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105 [DOI] [PubMed] [Google Scholar]
- 3.Dischinger J., Basi Chipalu S., and Bierbaum G. (2014) Lantibiotics: promising candidates for future applications in health care. Int. J. Med. Microbiol. 304, 51–62 [DOI] [PubMed] [Google Scholar]
- 4.Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., Dorrestein P. C., Entian K.-D., Fischbach M. A., Garavelli J. S., Göransson U., Gruber C. W., Haft D. H., Hemscheidt T. K., Hertweck C., Hill C., Horswill A. R., Jaspars M., Kelly W. L., Klinman J. P., Kuipers O. P., Link A. J., Liu W., Marahiel M. A., Mitchell D. A., Moll G. N., Moore B. S., Müller R., Nair S. K., Nes I. F., Norris G. E., Olivera B. M., Onaka H., Patchett M. L., Piel J., Reaney M. J., Rebuffat S., Ross R. P., Sahl H.-G., Schmidt E. W., Selsted M. E., Severinov K., Shen B., Sivonen K., Smith L., Stein T., Süssmuth R. D., Tagg J. R., Tang G.-L., Truman A. W., Vederas J. C., Walsh C. T., Walton J. D., Wenzel S. C., Willey J. M., and van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnell N., Entian K. D., Schneider U., Götz F., Zähner H., Kellner R., and Jung G. (1988) Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333, 276–278 [DOI] [PubMed] [Google Scholar]
- 6.Knerr P. J., and van der Donk W. A. (2012) Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81, 479–505 [DOI] [PubMed] [Google Scholar]
- 7.Klein C., Kaletta C., Schnell N., and Entian K. D. (1992) Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl. Environ. Microbiol. 58, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., Yu J. P., Brunzelle J. S., Moll G. N., van der Donk W. A., and Nair S. K. (2006) Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311, 1464–1467 [DOI] [PubMed] [Google Scholar]
- 9.Ortega M. A., Hao Y., Zhang Q., Walker M. C., van der Donk W. A., and Nair S. K. (2015) Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee C., Paul M., Xie L., and van der Donk W. A. (2005) Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684 [DOI] [PubMed] [Google Scholar]
- 11.Kuipers O. P., Beerthuyzen M. M., Siezen R. J., and De Vos W. M. (1993) Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216, 281–291 [DOI] [PubMed] [Google Scholar]
- 12.Siegers K., and Entian K. D. (1995) Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alkhatib Z., Abts A., Mavaro A., Schmitt L., and Smits S. H. (2012) Lantibiotics: how do producers become self-protected? J. Biotechnol. 159, 145–154 [DOI] [PubMed] [Google Scholar]
- 14.Engelke G., Gutowski-Eckel Z., Kiesau P., Siegers K., Hammelmann M., and Entian K. D. (1994) Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl. Environ. Microbiol. 60, 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuipers O. P., Beerthuyzen M. M., de Ruyter P. G., Luesink E. J., and de Vos W. M. (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304 [DOI] [PubMed] [Google Scholar]
- 16.Cotter P. D., Hill C., and Ross R. P. (2005) Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788 [DOI] [PubMed] [Google Scholar]
- 17.Brötz H., Josten M., Wiedemann I., Schneider U., Götz F., Bierbaum G., and Sahl H. G. (1998) Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30, 317–327 [DOI] [PubMed] [Google Scholar]
- 18.Hasper H. E., Kramer N. E., Smith J. L., Hillman J. D., Zachariah C., Kuipers O. P., de Kruijff B., and Breukink E. (2006) An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–1637 [DOI] [PubMed] [Google Scholar]
- 19.Hsu S.-T., Breukink E., Tischenko E., Lutters M. A., de Kruijff B., Kaptein R., Bonvin A. M., and van Nuland N. A. (2004) The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967 [DOI] [PubMed] [Google Scholar]
- 20.Ra R., Beerthuyzen M. M., de Vos W. M., Saris P. E., and Kuipers O. P. (1999) Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145, 1227–1233 [DOI] [PubMed] [Google Scholar]
- 21.Stein T., Borchert S., Kiesau P., Heinzmann S., Klöss S., Klein C., Helfrich M., and Entian K.-D. (2002) Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44, 403–416 [DOI] [PubMed] [Google Scholar]
- 22.Christ N. A., Bochmann S., Gottstein D., Duchardt-Ferner E., Hellmich U. A., Düsterhus S., Kötter P., Güntert P., Entian K.-D., and Wöhnert J. (2012) The first structure of a lantibiotic immunity protein, SpaI from Bacillus subtilis, reveals a novel fold. J. Biol. Chem. 287, 35286–35298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pozzi R., Coles M., Linke D., Kulik A., Nega M., Wohlleben W., and Stegmann E. (2015) Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024. Environ. Microbiol. 10.1111/1462-2920.12892 [DOI] [PubMed] [Google Scholar]
- 24.Stein T., Heinzmann S., Solovieva I., and Entian K.-D. (2003) Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278, 89–94 [DOI] [PubMed] [Google Scholar]
- 25.Takala T. M., Qiao M., and Saris P. E. (2004) Lipid-free NisI: interaction with nisin and contribution to nisin immunity via secretion. FEMS Microbiol. Lett. 237, 171–177 [DOI] [PubMed] [Google Scholar]
- 26.Takala T. M., and Saris P. E.(2006) C terminus of NisI provides specificity to nisin. Microbiology 152, 3543–3549 [DOI] [PubMed] [Google Scholar]
- 27.AlKhatib Z., Lagedroste M., Fey I., Kleinschrodt D., Abts A., and Smits S. H. (2014) Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS ONE 9, e102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hacker C., Christ N. A., Duchardt-Ferner E., Korn S., Berninger L., Kötter P., Entian K.-D., and Wöhnert J. (2015) NMR resonance assignments of the lantibiotic immunity protein NisI from Lactococcus lactis. Biomol. NMR Assign. 9, 293–297 [DOI] [PubMed] [Google Scholar]
- 29.Markley J. L., Bax A., Arata Y., Hilbers C. W., Kaptein R., Sykes B. D., Wright P. E., and Wüthrich K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J. Biomol. NMR 12, 1–23 [DOI] [PubMed] [Google Scholar]
- 30.Keller R. (2004) The Computer Aided Resonance Tutorial, CANTINA Verlag, Goldau [Google Scholar]
- 31.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 32.Mulder F. A., Schipper D., Bott R., and Boelens R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
- 33.Kosen P. A., Scheek R. M., Naderi H., Basus V. J., Manogaran S., Schmidt P. G., Oppenheimer N. J., and Kuntz I. D. (1986) Two-dimensional 1H NMR of three spin-labeled derivatives of bovine pancreatic trypsin inhibitor. Biochemistry 25, 2356–2364 [DOI] [PubMed] [Google Scholar]
- 34.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., and Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 35.Cordier F., Dingley A. J., and Grzesiek S. (1999) A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J. Biomol. NMR 13, 175–180 [DOI] [PubMed] [Google Scholar]
- 36.Rückert M., and Otting G. (2000) Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122, 7793–7797 [Google Scholar]
- 37.Shen Y., and Bax A. (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrmann T., Güntert P., and Wüthrich K. (2002b) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 39.Güntert P. (2009) Automated structure determination from NMR spectra. Eur. Biophys. J. 38, 129–143 [DOI] [PubMed] [Google Scholar]
- 40.Koradi R., Billeter M., and Güntert P. (2000) Point-centered domain decomposition for parallel molecular dynamics simulation. Comp. Phys. Commun. 124, 139–147 [Google Scholar]
- 41.Ponder J. W., and Case D. A. (2003) Force fields for protein simulations. Adv. Protein Chem. 66, 27–85 [DOI] [PubMed] [Google Scholar]
- 42.Gottstein D., Kirchner D. K., and Güntert P. (2012) Simultaneous single-structure and bundle representation of protein NMR structures in torsion angle space. J. Biomol. NMR 52, 351–364 [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharya A., Tejero R., and Montelione G. T. (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]
- 44.Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delano W. (2010) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 47.Svergun D. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 48.Bergmann A., Fritz G., and Glatter O. (2000) Solving the generalized indirect Fourier transformation (GIFT) by Boltzmann simplex simulated annealing (BSSA). J. Appl. Crystallogr. 33, 1212–1216 [Google Scholar]
- 49.Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., and Svergun D. I. (2007) Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 50.O'Leary W. M., Ratledge C., and Wilkinson S. G. (1988) Microbial Lipids, Academic Press, London [Google Scholar]
- 51.Montelione G. T., Nilges M., Bax A., Güntert P., Herrmann T., Richardson J. S., Schwieters C. D., Vranken W. F., Vuister G. W., Wishart D. S., Berman H. M., Kleywegt G. J., and Markley J. L. (2013) Recommendations of the wwPDB NMR Validation Task Force. Structure 21, 1563–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominguez C., Boelens R., and Bonvin A. M. (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 54.Göbl C., Madl T., Simon B., and Sattler M. (2014) NMR approaches for structural analysis of multidomain proteins and complexes in solution. Prog. Nucl. Magn. Reson. Spectrosc. 80, 26–63 [DOI] [PubMed] [Google Scholar]
- 55.Schmidt P. G., and Kuntz I. D. (1984) Distance measurements in spin-labeled lysozyme. Biochemistry 23, 4261–4266 [DOI] [PubMed] [Google Scholar]
- 56.Fuchs S. W., Jaskolla T. W., Bochmann S., Kötter P., Wichelhaus T., Karas M., Stein T., and Entian K.-D. (2011) Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl. Environ. Microbiol. 77, 1698–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J. R., Warner L. R., Sanchez C., Gabel F., Madl T., Mackereth C. D., Sattler M., and Blackledge M. (2014) Transient electrostatic interactions dominate the conformational equilibrium sampled by multidomain splicing factor U2AF65: a combined NMR and SAXS study. J. Am. Chem. Soc. 136, 7068–7076 [DOI] [PubMed] [Google Scholar]
- 58.Mackereth C. D., Madl T., Bonnal S., Simon B., Zanier K., Gasch A., Rybin V., Valcárcel J., and Sattler M. (2011) Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475, 408–411 [DOI] [PubMed] [Google Scholar]
- 59.Volkov A. N., Worrall J. A., Holtzmann E., and Ubbink M. (2006) Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc. Natl. Acad. Sci. U.S.A. 103, 18945–18950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan S. M., O'connor P. M., Cotter P. D., Ross R. P., and Hill C. (2005) Sequential actions of the two component peptides of the lantibiotic lacticin 3147 explain its antimicrobial activity at nanomolar concentrations. Antimicrob. Agents Chemother. 49, 2606–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]