Background: Atg7 is an essential autophagic enzyme. Disrupted autophagy has been implicated in retinal degeneration.
Results: Mice with RPE-specific Atg7 deletion exhibit normal retinoid recycling, histology, and A2E accumulation, but display hypertrophy and cytosolic debris.
Conclusion: Atg7 deficiency does not severely affect the health of RPE cells in mice.
Significance: A2E accumulation and retinoid recycling are independent of Atg7-mediated autophagy in RPE cells.
Keywords: aging, autophagy, autophagy-related protein 7 (ATG7), retina, retinal degeneration, A2E, ABCA4, Stargardt disease
Abstract
Autophagy is an evolutionarily conserved catabolic mechanism that relieves cellular stress by removing/recycling damaged organelles and debris through the action of lysosomes. Compromised autophagy has been implicated in many neurodegenerative diseases, including retinal degeneration. Here we examined retinal phenotypes resulting from RPE-specific deletion of the autophagy regulatory gene Atg7 by generating Atg7flox/flox;VMD2-rtTA-cre+ mice to determine whether autophagy is essential for RPE functions including retinoid recycling. Atg7-deficient RPE displayed abnormal morphology with increased RPE thickness, cellular debris and vacuole formation indicating that autophagy is important in maintaining RPE homeostasis. In contrast, 11-cis-retinal content, ERGs and retinal histology were normal in mice with Atg7-deficient RPE in both fasted and fed states. Because A2E accumulation in the RPE is associated with pathogenesis of both Stargardt disease and age-related macular degeneration (AMD) in humans, deletion of Abca4 was introduced into Atg7flox/flox;VMD2-rtTA-cre+ mice to investigate the role of autophagy during A2E accumulation. Comparable A2E concentrations were detected in the eyes of 6-month-old mice with and without Atg7 from both Abca4−/− and Abca4+/+ backgrounds. To identify other autophagy-related molecules involved in A2E accumulation, we performed gene expression array analysis on A2E-treated human RPE cells and found up-regulation of four autophagy related genes; DRAM1, NPC1, CASP3, and EIF2AK3/PERK. These observations indicate that Atg7-mediated autophagy is dispensable for retinoid recycling and A2E deposition; however, autophagy plays a role in coping with stress caused by A2E accumulation.
Introduction
The retinal pigmented epithelium (RPE)2 is a single layer of post mitotic cells located in the back of the eye directly adjacent to the photoreceptor layer of the retina. This layer is responsible for maintaining many functions needed to preserve retinal health including retinoid recycling, nutrient exchange, ion balance, blood-retinal-barrier integrity, and phagocytosis of daily shed photoreceptor outer segments (POS) (1, 2).
Daily shedding of POS and subsequent phagocytosis by the RPE occurs in a cyclic manner, and is required for the maintenance of retinal health since photoreceptor cells continually renew their outer segments (3, 4). Defects in RPE phagocytosis cause the accumulation of undigested POS which can ultimately cause progressive death of photoreceptor cells and vision loss (5).
Autophagy is the major mechanism for renewing organelles and cytoplasmic constituents in post-mitotic cells. Mammalian cells possess three types of autophagy: microautophagy, chaperone-mediated autophagy, and macroautophagy, each with distinct machinery and targeted substrates (6). Macroautophagy, herein known as autophagy, is an evolutionarily conserved pathway involving lysosome-dependent degradation of cytoplasmic material. This process begins with the sequestration and enclosure of cytoplasmic materials by double-membrane vacuoles termed autophagosomes. Autophagosomes then fuse with lysosomes and the contents are degraded by lysosomal enzymes, which may be recycled within the cell. Functional autophagy is thought to play an important role in maintaining homeostasis of various tissues, such as liver, brain, heart, and skeletal muscle (7–11). Dysfunctional autophagy pathways have been implicated in many degenerative diseases such as Alzheimer, Huntington, and Parkinson disease (12–15).
The autophagy process is controlled by a core group of over 20 highly conserved autophagy-related (Atg) genes first identified in budding yeast (16). Atg7 is required for activation of the ubiquitin-related proteins Atg8 and Atg12 and is essential for autophagosome formation (17).
Autophagic proteins are highly expressed in both retinal and RPE tissue (2, 18, 19). In addition, cellular stressors in the eye, such as A2E accumulation, reactive oxygen species production and mitochondrial dysfunction can increase autophagy (20, 21). Autophagic activity has also been observed to follow a cyclic pattern similar to the daily rhythm of POS phagocytosis detected in RPE cells; this suggests a role for autophagy in retinoid recycling and daily maintenance of photoreceptors (2, 22).
Continual phagocytosis of indigestible material by the RPE results in the accumulation of autofluorescent granules called lipofuscin. The number and size of lipofuscin granules increase with age, and elevated levels are found in some retinal diseases such as Stargardt disease and age-related macular degeneration (AMD) (23). A2E is the major fluorophore identified in lipofuscin, and A2E oxidation products have been implicated in complement activation and inflammation in retinal disease (24, 25).
The ABCA4 gene encodes a photoreceptor-specific protein responsible for the transport of all-trans-retinal from the disc lumen to the cytoplasmic space (26). Moreover, mutations in ABCA4 are associated with Stargardt disease. Stargardt disease patients exhibit accumulation of lipofuscin and A2E as well as early-onset macular degeneration (27). Abca4−/− mice display similar features, including A2E accumulation in the RPE, and therefore provide an appropriate model for studying retinal dystrophy.
In this study, we used a RPE-specific Atg7-deficient mouse Atg7flox/flox;VMD2-rtTA-cre+ (abbreviated to Atg7flox/flox;cre+)to address whether autophagy-deficiency results in RPE dysfunction including impaired POS phagocytosis and abnormal A2E accumulation during aging, which could lead to retinal degeneration (28–31).
Experimental Procedures
Animals
Mice were housed in the animal facility at the School of Medicine, Case Western Reserve University, where they were maintained either under complete darkness or in a 12 h light (∼5 lux)/12 h dark cyclic environment. Manipulations were done in the dark under dim red light transmitted through a Kodak No. 1 safe light filter (transmittance >560 nm). All animal procedures and experiments were approved by the Case Western Reserve University Animal Care Committees and conformed to recommendations of both the American Veterinary Medical Association Panel on Euthanasia and the Association of Research for Vision and Ophthalmology. Atg7flox/flox mice were a gift from Dr. Kiji Tanaka at the Laboratory of Frontier Science, Tokyo Metropolitan Institute of Medical Science, Tokyo (7). VMD2-rtTA-cre+ mice (32) and Abca4−/− mice (33) were generated as previously described. Atg7 gene deletion was induced by oral gavage of 0.4 mg/g doxycycline for two consecutive days in mice at 4 weeks of age. Littermate Atg7flox/flox;VMD2-rtTA-cre- animals with doxycycline treatment were used as controls. Abca4−/−Atg7flox/flox;VMD2-rtTA-cre+ mice were generated by crossing Abca4−/− with Atg7flox/flox;VMD2-rtTA-cre+ mice.
Quantitative RT-PCR
All procedures for quantitative RT-PCR (qRT-PCR) were described previously (34). qRT-PCR was performed with the following primers: mouse Gapdh, forward (5′-GTGTTCCTACCCCCAATGTG-3′), reverse (5′- AGGAGACAACCTGGTCCTCA-3′); mouse Atg7 forward (5′-ACCATGCAGGGAGCTAGAGA-3′) and reverse (5′-CCACTGAGGTTCACCATCCT-3′); mouse CFH, forward (5′-TGGACTTCCTTGTGGACCTC-3′), reverse (5′-TGGGTCAGACCACTTTCCTC-3′); human RPE65 forward (5′-AAAAATGCCAGAAAGGCTCC-3′) and reverse (5′-AGTTGTATTGGGGAGCGTGA-3′); human MERTK forward (5′-GAAATTACAGTCCGCAGCC-3′) and reverse (5′-TCTTCCCTTGCCTCAGTGAT-3′); human BEST1 forward (5′-GCTGCTATATGGCGAGTTCTT-3′) and reverse (5′-CAGCTGTTGTTCTTCCGTGA-3′) and human MITF forward (5′-CAAATACGTTGCCTGTCTCGGG-3′) and reverse (5′-GGGTGGACAGGAGTTGCTGA-3′). Relative expression of genes was normalized to the housekeeping gene Gapdh, and sample group data were normalized to data collected from wild-type (WT) age-matched controls.
Retinoid Analyses
Retinoid extraction was performed on eye samples from dark-adapted and light-exposed mice as previously described (35). Briefly, eyes were homogenized in 1.2 ml of retinoid analysis buffer (50 mm Mops, 10 mm NH2OH, and 50% (v/v) ethanol in 50% (v/v) H2O, pH 7.0). Retinoids were extracted twice with 4 ml of hexane. The organic layer, containing non-polar retinoids, was dried down in a SpeedVac. Retinoids then were suspended in 0.3 ml of hexane and separated by normal-phase HPLC (Ultrasphere-Si, 4.6 μm 3 × 250 mm; Agilent) with 10% ethyl acetate and 90% hexane at a flow rate of 1.4 ml/min. Specific retinoid peaks were identified by their elution times and peak wave length absorbance.
A2E Analysis
A2E was extracted twice from two eyes with 1 ml of acetonitrile in a glass/glass homogenizer. After evaporation of solvent, extracts were dissolved in 150 μl acetonitrile with 0.1% TFA and then passed through a Teflon syringe filter (National Scientific Company, Rockwood, TX). Samples (100 μl) were loaded onto a C18 column (Phenomenex, Torrance, CA) and analyzed by normal-phase HPLC with mobile-phase gradient of acetonitrile-H2O, 100:0, to acetonitrile-H2O, 80:20 with 0.1% TFA for 30 min. Quantification of A2E by HPLC was achieved by comparison with known concentrations of pure synthetic A2E (35).
ERG Recordings
All ERG experimental procedures were performed under dim red light transmitted through a Kodak No. 1 safelight filter (transmittance >560 nm) as previously described (35). Briefly, after dark-adaptation overnight prior to recording; mice were anesthetized by an intraperitoneal injection of 20 μl/g body weight of 6 mg/ml ketamine and 0.44 mg/ml xylazine diluted with 10 mm sodium phosphate, pH 7.2, containing 100 mm NaCl. Pupils were dilated with a mixture of 0.5% tropicamide and 0.5% phenylephrine hydrochloride. A contact lens electrode was placed on the eye, and a reference electrode and ground electrode were positioned on the ear and tail, respectively. ERGs were recorded by the universal testing and electrophysiological system with UTAS E-300 (LKC Technologies Gaithersburg, MD).
Recovery of dark adaptation was investigated as previously published (35). Briefly, dark-adapted mice were bleached with the background light of a Ganzfeld chamber (500 cd·m−2) for 3 min. After bleach, a single-flash ERG at −0.2 cd·s·m−2 was used to monitor recovery of a-wave amplitude every 5 min for 60 min in the dark-condition. The recovery ratio was calculated by normalizing single flash a-wave amplitude responses at various times following bleaching to the dark-adapted a-wave response at the identical flash intensity of −0.2 cd·s·m−2.
Histology and Immunohistochemistry
Histological and immunohistochemical procedures employed were previously reported (35). Briefly, eyecups for electron microscopy imaging were fixed in 2% glutaraldehyde, 4% paraformaldehyde, and processed for embedding in Epon. Sections for routine histology were cut at 1 μm and stained with toluidine blue. For immunocytochemistry, eyes were immersion-fixed for 2 h with freshly prepared 4% paraformaldehyde in 0.1 mm phosphate buffer, pH 7.4, and processed for optimal cutting temperature (Miles) embedding. Sections were cut at 12 μm and viewed through a Zeiss LSM 510 inverted laser scan confocal microscope. Fourteen bit images were analyzed with Metamorph Imaging Software (Molecular Devices, Downington, PA). For RPE surface area experiments, images were flattened, and then subjected to thresholding with areas inside the polygon shaped borders of the RPE cells being targeted. Resulting areas then were filtered based on both size and shape and areas of intact RPE cells were recorded.
Immunoblotting
Cell/tissue extracts were lysed in ice-cold lysis buffer (150 mm NaCl, 1 mm EDTA, 0.2% Nonidet P-40 and 20 mm Tris-HCl, pH7.5) containing protease inhibitor mixture (Sigma-Aldrich). Protein from each sample was transferred onto Immobilon-P membranes (Millipore, Bedford, MA) after SDS-PAGE gel electrophoresis. Membranes were incubated in 1% BSA solution containing a 1:1000 dilution of either anti-Atg7 rabbit polyclonal antibody (Cell Signaling Technology, Boston, MA), anti-p62 polyclonal antibody (Novus, Littleton, CO), anti-ZO-1 rabbit polyclonal antibody (Invitrogen, Camarillo, CA) or anti-β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Targeted molecules were visualized with alkaline phosphatase (Promega, Madison, WI) at a dilution of 1: 5000.
Primary RPE Isolation and Cell Culture
RPE cells were isolated from eyes of 10–12-day-old mice as described previously (36). Briefly, between 9 AM and 10 AM, enucleated eyes were harvested from animals and incubated in 2% dispase (Invitrogen) solution for 45 min in a 37 °C water bath with gentle mixing every 15 min. After removal of the neural retina, sheets of RPE were peeled away from the choroid and pipetted into a tube containing DMEM plus streptomycin/penicillin and 10% fetal bovine serum (FBS). Primary isolated RPE cells were either plated into a 96-well dish and grown in DMEM/10% FBS for 5 days or pelleted and frozen for further analysis. Human RPE cells were differentiated from pluripotent stem cells and cultured according to procedures reported previously (37, 38).
Microarray Analysis
The Human Autophagy RT2 Profiler PCR Array (SABiosciences, Frederick, MD) was used to study autophagy-specific gene expression profiles in accordance with the manufacturer's recommendations. Briefly 60 ng of RNA was isolated from cells/tissue with a RNeasy mini Kit (Qiagen, Valencia, CA) and was reverse transcribed by using a RT2 First Stand Kit (Qiagen, Valencia, CA). The PCR array was run with the recommended RT2 SYBR Green ROX qPCR Mastermix (Qiagen) according to manufacturer's instructions. Data were analyzed with Qiagen software. In all qPCR experiments, amplification data (fold changes in Ct values of all the genes) were analyzed by the ΔΔCt method.
Cell Culture Treatments
A2E, prepared as previously described (39), was conjugated to BSA (bovine serum albumin, Sigma-Aldrich) by adding 20 μm A2E to 0.2% BSA solution. The A2E-BSA mixture was incubated at 4 °C for 2 days and then centrifuged to remove insoluble debris.
Lactate Dehydrogenase (LDH) Colorimetric Assay
LDH measurements were carried out with the LDH-Cytotoxicity Colorimetric Assay Kit II (BioVision, Milpitas, CA) on cultured human RPE cells in 96-well plates following manufacturer's instructions. Optical density values from the 96-well plate at 450 nm were measured with a microplate reader (Multiscan FC Microplate Reader, Fisher Scientific).
Statistical Analyses
Data representing the means ± S.D. for the results of at least five independent experiments were compared by one-way ANOVA with p < 0.05 considered statistically significant.
Results
Loss of Atg7 in Atg7flox/flox;cre+ RPE
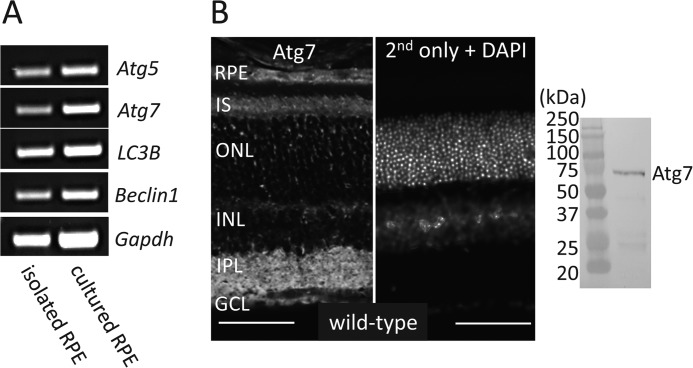
Autophagy is a ubiquitous pathway in many tissues. To confirm its presence in the RPE we performed RT-PCR for essential autophagy genes, Atg5, Atg7, LC3B, and Beclin1. These genes were found to be expressed in both freshly isolated RPE from mouse eye cups and from cultured mouse RPE cells (Fig. 1A). Atg7 is a required component of the autophagosome maturation pathway and is ubiquitously expressed in the retina, specifically in the RPE, inner segment of photoreceptors (IS), inner plexiform layer (IPL), and the ganglion cell layer (GCL) (Fig. 1B, left). Immunoblot with lysates of primary mouse RPE cells demonstrated no major cross-reactivity of Atg7 antibody (Fig. 1B, right). To test the effects of autophagy loss in the RPE we used the Atg7flox/flox;VMD2-rtTA-cre+ mouse in which the Atg7 gene is flanked with loxP sites and cre expression is driven by a VMDR2-rtTA promoter. Atg7flox/flox;VMD2-rtTA-cre- (Atg7flox/flox;cre-) mice, that do not express cre and therefore retain Atg7 expression in the RPE, were used as littermate controls for all experiments unless otherwise noted. To validate the loss of Atg7 in the RPE, Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice were euthanized at 5 weeks of age and their RPE flat-mounts were dissected. Atg7flox/flox;cre+ RPE displayed a decrease in Atg7 protein compared with Atg7flox/flox;cre- RPE measured by immunoblot (Fig. 2A). A 58% decrease in Atg7 mRNA transcript was detected in freshly isolated RPE from Atg7flox/flox;cre+ compared with Atg7flox/flox;cre- animals (Fig. 2B). To assess the loss of autophagy in our model we performed immunohistochemistry for p62, a cytoplasmic protein which aggregates upon autophagy disruption (40). Loss of Atg7 in the RPE resulted in increased detection of p62 punctate in the RPE (Fig. 2C).
FIGURE 1.
Expression of autophagic molecules including Atg7 in the RPE of wild-type mice. A, essential autophagy genes, Atg5, Atg7, LC3B, and Beclin1, were analyzed by RT-PCR from freshly isolated mouse RPE or primary cultured mouse RPE maintained in DMEM/10% FBS for 5 days. B, representative immunohistochemistry image showing Atg7 localization in the retina is presented with a control stained with secondary only and DAPI nuclear staining (left panel). Immunoblot with lysates of primary mouse RPE demonstrated specificity of Atg7 antibody (right panel). IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bar 50 μm.
FIGURE 2.
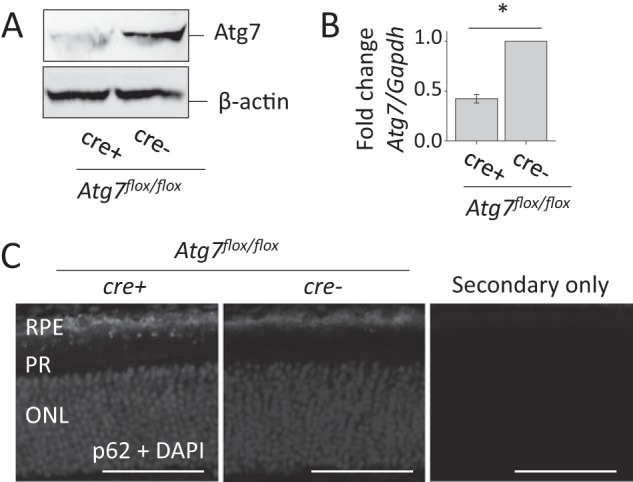
Loss of Atg7 increases p62 accumulation in RPE of Atg7flox/flox;cre+ mice. A, representative immunoblot of Atg7 protein expression was examined in 5-week-old Atg7flox/flox;cre+ mice. β-actin was used as a loading control, n = 5 animals per group. B, qRT-PCR analysis was performed on freshly isolated RPE from 5-week-old Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice. Atg7 mRNA expression was normalized to Gapdh, *, p < 0.001, n = 3 per group. Error bar indicates the mean ± S.D. C, immunohistochemistry against p62, a marker of increased autophagy, was performed on retinal sections of Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice at 5 weeks of age and a secondary only control. Nuclei were stained with DAPI. PR, photoreceptor; ONL, outer nuclear layer. Scale bar 50 μm.
RPE-Specific Atg7 Loss Does Not Affect the Visual Cycle of Mice in a Fed or Fasted State
As Atg7-mediated autophagy plays an important role in the starvation response in quiescent cells (7), we tested the effect of Atg7 loss in the RPE during a 24 h fast in mice. Freshly isolated RPE tissue from WT mice was shown to have increased p62 accumulation following a 24 h fast (Fig. 3A). Atg7flox/flox;cre+ and Atg7flox/flox;cre- mouse eyes were analyzed for differences in 11-cis-retinal concentrations to determine whether autophagy contributes to retinoid recycling. Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice, at 5 weeks of age, were dark-adapted for 24 h and then were either given unlimited access to chow, or starved for 24 h to induce increased autophagic processing. Retinoid analysis showed no remarkable differences in 11-cis-retinal concentrations in Atg7flox/flox;cre+ and Atg7flox/flox;cre- eyes in the fed or fasted conditions (Fig. 3B). Similar kinetics for 11-cis-retinal and all-trans-retinal metabolism after light illumination were also observed in fed 5-week-old Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice (Fig. 3C). Retinas from 5-week-old Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice displayed normal histology and photoreceptor morphology in both fed and fasted states (Fig. 3D).
FIGURE 3.
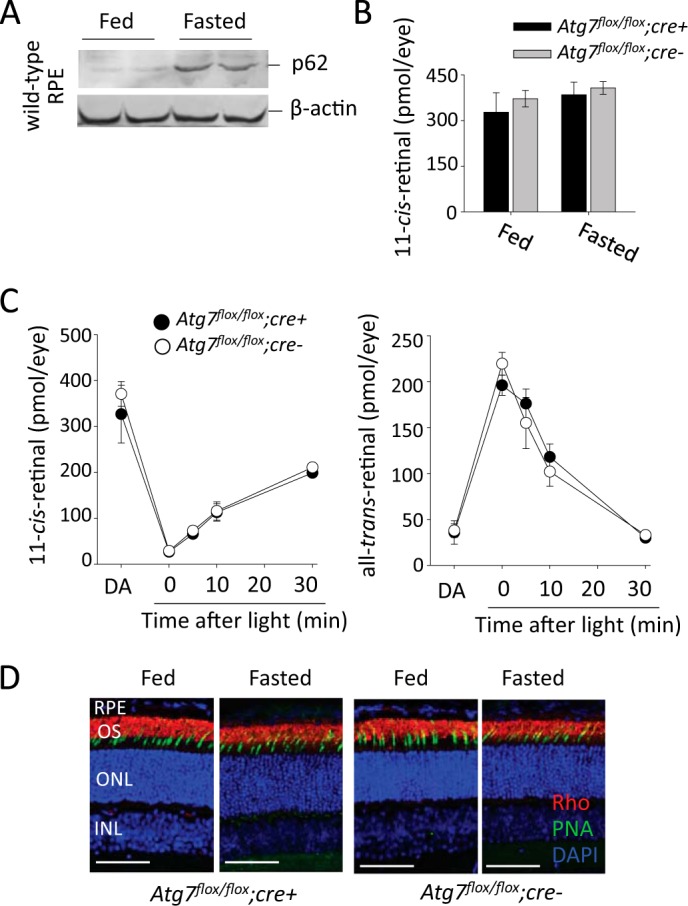
RPE specific loss of Atg7 does not affect retinoid cycling in fed or fasted mice A, representative immunoblots demonstrating the accumulation of p62, a marker of increased autophagy, in freshly isolated RPE from 24 h fasted WT mice, n = 2. B, 11-cis-retinal concentrations (pmol/per eye) of dark-adapted Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice (5 weeks of age) in fed and 24-h fasted states measured by HPLC, n ≥ 3 animals per group. Error bars indicate the mean ± S.D. C, kinetics of 11-cis-retinal and all-trans-retinal metabolism minutes after dark adaptation post light illumination at 10,000 lux for 3 min were examined in 5-week-old fed Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice. n ≥ 3 animals per group. Error bars indicate the mean ± S.D. D, representative images of retinal histology showing immunofluorescent staining of rhodopsin (red), PNA (green), and DAPI (blue) in sections from Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice (5 weeks of age) in fed or 24-h fasted states, n = 5. OS, outer segment; ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar 50 μm.
Electroretinogram (ERG) analysis was performed on Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice at 5 weeks of age to determine if loss of Atg7 in the RPE disrupts normal visual function in mice. Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice were dark-adapted for 24 h prior to ERGs and scotopic b-waves and a-waves were recorded (Fig. 4, A and B). Likewise we tested if lack of Atg7 in the RPE together with up-regulation of autophagy seen in the starvation response resulted in any changes of visual function in mice. Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice at 5 weeks of age were dark adapted and kept without food for 24 h prior to b-wave and a-wave recordings (Fig. 4, C and D). No significant difference was observed in a-waves or b-waves between Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice in the fed state. A trend of decreased amplitudes was recorded in Atg7flox/flox;cre+ mice as compared with control Atg7flox/flox;cre- mice after mice were fasted for 24 h.
FIGURE 4.
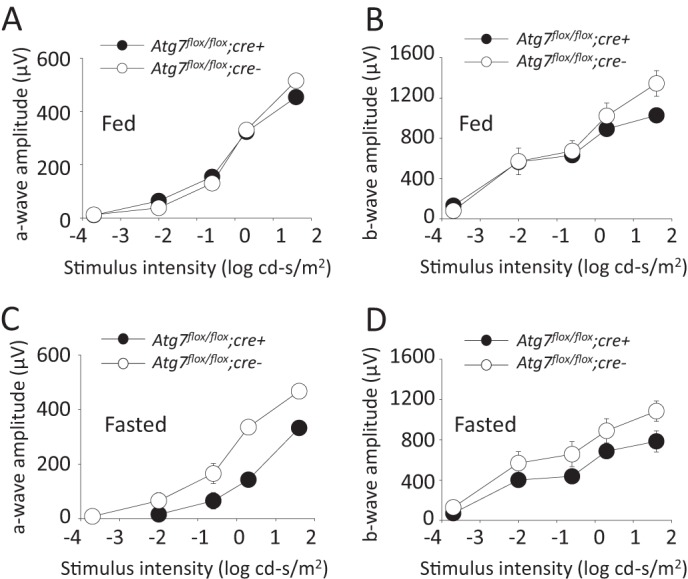
Intact photoreceptor function is present in mice lacking Atg7 in the RPE in both fed and fasted states A, ERG measurements of a-wave amplitudes from dark adapted Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice in the fed state, n = 5 animals per group. Error bars indicate the mean ± S.D. B, ERG measurements of b-wave amplitude from dark adapted Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice in the fed state, n = 5 animals per group. Error bars indicate the mean ± S.D. C, ERG measurements of a-wave amplitudes from dark-adapted Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice in a 24 h fasted state, n = 3 animals per group. Error bars indicate the mean ± S.D. D, ERG measurements of b-wave amplitudes from dark adapted Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice in a 24 h fasted state, n = 3 animals per group. Error bars indicate the mean ± S.D.
Loss of Atg7-mediated Autophagy in the RPE Does Not Lead to Age-related Retinal Degeneration
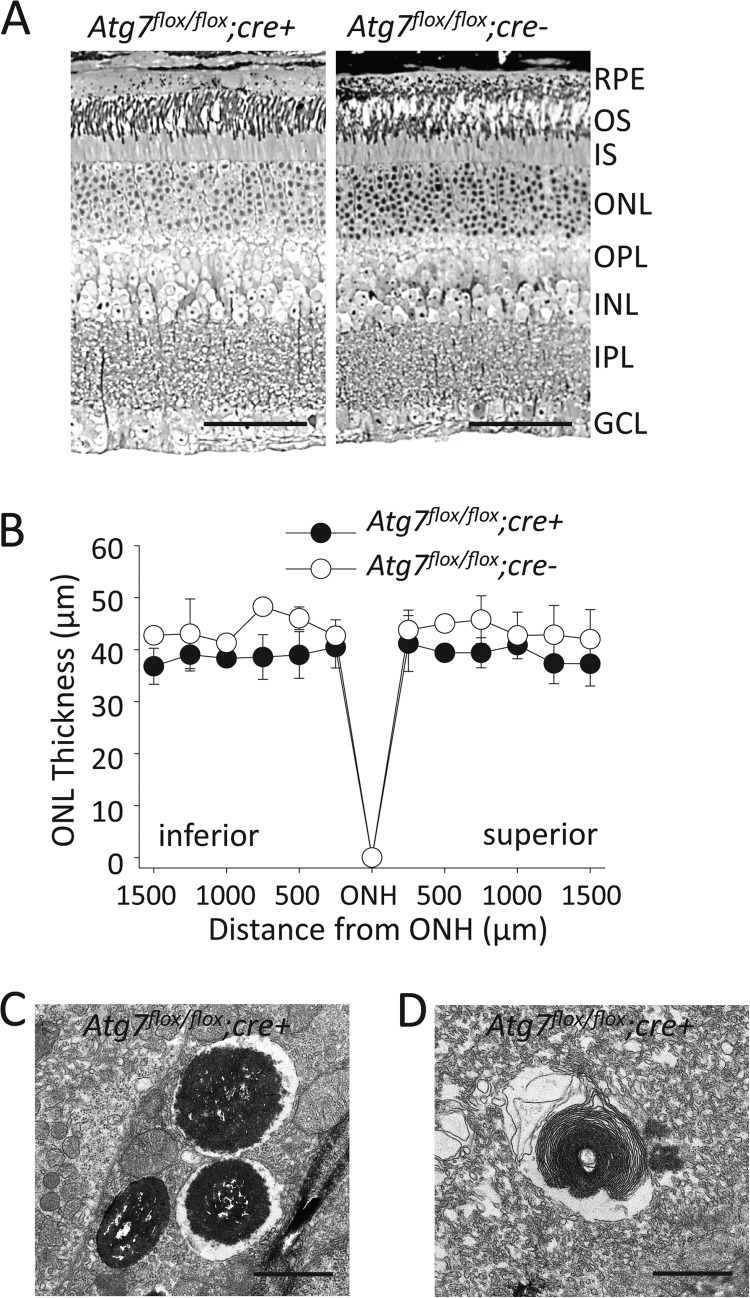
Normal retinal histology (Fig. 5A) and outer nuclear layer (ONL) thickness (Fig. 5B) was maintained in 6- and 12-month-old Atg7flox/flox;cre+ mice which were maintained under normal lighting conditions (∼5 lux 12/12 day/night cycle). Abnormal autophagic/lysosomal processing, including vacuoles containing melanosomes (Fig. 5C) and partially digested POS (Fig. 5D) were observed in Atg7flox/flox;cre+ RPE.
FIGURE 5.
Loss of Atg7 in the RPE does not lead to age-associated retinal disease A, toluene blue staining of retinal sections from 12-month-old Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice. OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bar 50 μm. B, outer nuclear layer (ONL) thickness was measured on DAPI stained retinal sections from 12-month-old Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice, n = 4 animals per group. Error bars indicate the mean ± S.D. ONH, optic nerve head. C and D, electron microscopic images displaying dysfunctional autophagy in the RPE of Atg7flox/flox;cre+ mice, including partial digestion of melanosomes (C) and rod outer segments (D). Scale bar 1 μm.
Hypertrophy and Vacuole-like Structures in Atg7-deficient RPE
Though normal retinal histology was observed in Atg7flox/flox;cre+ mice at the age of 12 months, increased RPE thickness was noted in both 6- and 12-month-old Atg7flox/flox;cre+ mice as revealed by electron microscopy (Fig. 6A). This finding was documented in other tissue-specific deficits of Atg7 including liver (7) and myocytes (41). Electron microscopy data revealed large vacuole-like structures (asterisk) commonly associated with cell-to-cell boarders (black arrows) within Atg7flox/flox;cre+ RPE (Fig. 6B). Abnormal vacuole structures were not found in Atg7flox/flox;cre- mice, but were observed at an average density of 2.6 vacuoles per 4.0 μm2 when measured in EM images of 12 month old Atg7flox/flox;cre+ mice (n = 22). Similar aberrant structures were noted in Atg7-deficient neuronal axons (42) and myocytes (41). Abnormal autofluorescent signals were also observed in Atg7flox/flox;cre+ RPE (Fig. 6C). To examine whether Atg7 deficiency interferes with normal RPE cellular morphology, we prepared flat mounts stained for the tight junction protein, zonula occludens protein 1 (ZO-1), and measured the surface area of individual RPE cells. No difference in RPE cell surface area or ZO-1 staining was noted between 6-month-old Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice (Fig. 6, D and E) and no obvious difference was seen in ZO-1 protein expression between Atg7flox/flox;cre- and Atg7flox/flox;cre+ RPE (Fig. 6F).
FIGURE 6.
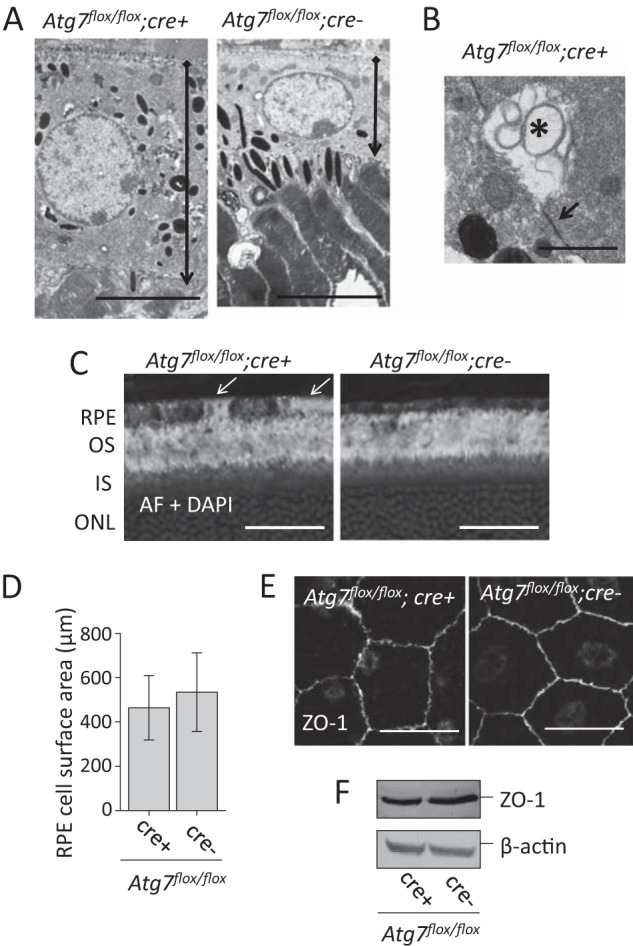
Loss of Atg7 in the RPE leads to cell hypertrophy and vacuolization. A, electron microcopic imaging demonstrates RPE cell hypertrophy in Atg7flox/flox;cre+ mice. Black and diamond arrowheads indicate apical and basal sides of the RPE, respectively, Scale bar 5 μm. B, electron microcopy revealed the presence of large vacuole-like structures containing concentric contents (asterisk) commonly associated with cell-to-cell boundaries (black arrow) in Atg7flox/flox;cre+ RPE. Scale bar 1 μm. (C). Autofluorescent (AF) imaging showed higher AF signals (white arrows) in some RPE cells of Atg7flox/flox;cre+ mice. D, RPE surface area was measured on whole RPE flat mounts after ZO-1 staining; no significant difference in size was observed between Atg7flox/flox;cre+ and Atg7flox/flox;cre- mice, n = 5 animals per group. Error bars indicate the mean ± S.D. E, representative ZO-1 staining performed on whole RPE flat mounts from Atg7flox/flox;cre- and Atg7flox/flox;cre+ mice. Scale bar 20 μm. F, representative immunoblots of ZO-1 protein expression in Atg7flox/flox;cre+ and Atg7flox/flox;cre- RPE cell lysates.
A2E Accumulation in the RPE Is Not Affected by the Absence of Atg7
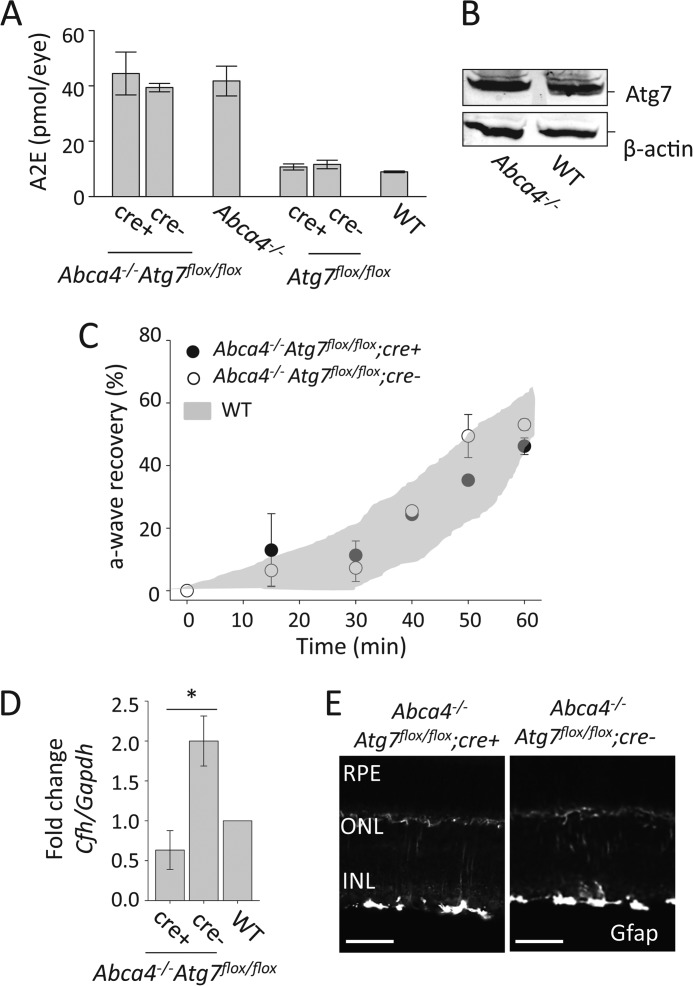
Because accumulation and noxious effects of A2E in the RPE are associated with lysosomal dysfunction and impaired degradation processes (28, 29, 43, 44), we hypothesize that autophagy deficiency, due to loss of Atg7, affects the phenotype caused by A2E accumulation in mice. To determine whether loss of autophagy in the RPE affects A2E accumulation, we measured the amounts of A2E present in the eyes of Atg7flox/flox;cre+ mice by HPLC. To increase the rate of A2E buildup in the RPE, we crossed our Atg7flox/flox;cre+ mice with the Abca4−/− mouse line (which accumulate high levels of A2E with age) to generate Abca4−/−Atg7flox/flox;cre+ mice. A2E levels then were found to be equal between Atg7flox/flox;cre+, Atg7flox/flox;cre- and WT mice, as well as between Abca4−/−Atg7flox/flox;cre+ and Abca4−/−Atg7flox/flox;cre- and Abca4−/− mice at 6 months of age (Fig. 7A). Likewise comparable protein levels for Atg7 were observed between Abca4−/− and WT RPE at 6 months of age, suggesting A2E accumulation does not affect Atg7 protein expression (Fig. 7B). Normal retinal morphology was observed in Abca4−/−Atg7flox/flox;cre+ and Abca4−/−Atg7flox/flox;cre- mice. The rate of dark adaptation was not altered in Abca4−/−Atg7flox/flox;cre+ and Abca4−/−Atg7flox/flox;cre- mice as compared with WT mice, suggesting that Atg7 deficiency in the RPE and A2E accumulation do not affect dark adaptation in mice (Fig. 7C). These data suggest that Atg7-mediated autophagy is not involved in age-related A2E accumulation in the RPE.
FIGURE 7.
A2E accumulation and retinal inflammation is not affected by the loss of Atg7. A, A2E accumulation was measured by HPLC in 6-month-old Abca4−/−Atg7flox/flox;cre+, Abca4−/−Atg7flox/flox;cre-, Abca4−/−, Atg7flox/flox;cre+, Atg7flox/flox;cre-, and WT mouse eyes, n ≥ 6 animals per group. Error bars indicate the mean ± S.D. B, representative immunoblots demonstrating that the presence of A2E in the RPE did not alter the expression of Atg7 in 6-month-old Abca4−/− as compared with WT mice. C, rate of dark adaptation was monitored by measuring a-wave recovery by the ERG. Abca4−/−Atg7flox/flox;cre+, Abca4−/−Atg7flox/flox;cre-, Abca4−/−, Atg7flox/flox;cre+, Atg7flox/flox;cre-, and WT mice at 3 months of age were used. n = 3 per group. Error bars indicate the mean ± S.D. D, qRT-PCR analysis was performed to measure the expression of Cfh normalized to Gapdh in 6-month-old Abca4−/−Atg7flox/flox;cre+, Abca4−/−Atg7flox/flox;cre-, and WT whole eye lysates. Expression in Abca4−/−Atg7flox/flox;cre+ and Abca4−/−Atg7flox/flox;cre- samples was normalized to the WT value, *, p < 0.001, n = 3 animals per group. Error bars indicate the mean ± S.D. E, immunohistochemistry of Gfap in retinal sections from 6-month-old Abca4−/−Atg7flox/flox;cre+ and 6-month-old Abca4−/−Atg7flox/flox;cre- mice. Scale bar = 50 μm.
Loss of Autophagy in the RPE Modulates CFH but Fails to Induce Inflammation in a Mouse Model of Stargardt Disease
A2E accumulation is thought to increase inflammatory responses in RPE cells. Moreover dysregulation of complement factors have been observed in mouse models of Stargardt disease, and certain alleles for complement factor H (CFH) have been identified as risk factors for developing AMD (45, 46). Furthermore studies on retinal microglia demonstrated that A2E accumulation increases microglial activation but decreases their migration and neuroprotective functions in the retina (25). We sought to address whether autophagy in the RPE contributes to retinal inflammation by measuring the expression of both CFH, a negative regulator of the alternative complement cascade, and glial fibrillary acidic protein (GFAP), a marker of inflammation and gliosis in the retina. Previous work revealed that A2E accumulation triggers complement activation in RPE cells resulting in the prediction that Abca4−/−Atg7flox/flox;cre- mice would have a higher expression of Cfh compared with WT mice (24). A significant down-regulation (>3 fold) of Cfh was noted in the eyes of 6-month-old Abca4−/−Atg7flox/flox;cre+ animals compared with Abca4−/−Atg7flox/flox;cre- mice suggesting that autophagy regulates CFH expression in the RPE (Fig. 7D). Gfap was not found to be highly expressed in the retina of 6-month-old Abca4−/−Atg7flox/flox;cre+ and Abca4−/−Atg7flox/flox;cre- mice suggesting the absence of inflammation in the retina of these animals (Fig. 7E).
Up-regulation of Autophagy-related Stress Genes in Human RPE Incubated with A2E
We asked if autophagy-related genes other than Atg7 were involved in A2E accumulation. To answer this question, we used human RPE derived from pluripotent stem cells as previously described (37, 38). These cells express the RPE-specific markers, RPE65, microphthalmia-associated transcription factor (MITF), c-mer proto-oncogene tyrosine kinase (MERTK) and Bestrophin 1 (BEST1) (Fig. 8, A and B). Human RPE were incubated with either BSA (0.1%) or 10 μm A2E-BSA conjugate for 16 h in complete growth medium. A2E-BSA conjugate incubation was found not to have a significant effect on cell toxicity measured by the LDH assay (Fig. 8C). RNA from human RPE cells was used to perform the gene array analysis. Of the 84 genes included in this array, four genes were increased 3-fold or higher in A2E-BSA conjugate-treated samples relative to BSA-treated samples (shaded circles)(Fig. 8D). These included a 4.58-fold up-regulation of DNA-damage regulated autophagy modulator 1 (DRAM1), a 4.46-fold up-regulation of Niemann-Pick disease, type C1 (NPC1), a 4.28-fold up- regulation of apoptosis-related cysteine peptidase (CASP3), and a 3.29-fold up-regulation of eukaryotic translation initiation factor 2-α kinase 3 (EIF2AK3/PERK).
FIGURE 8.
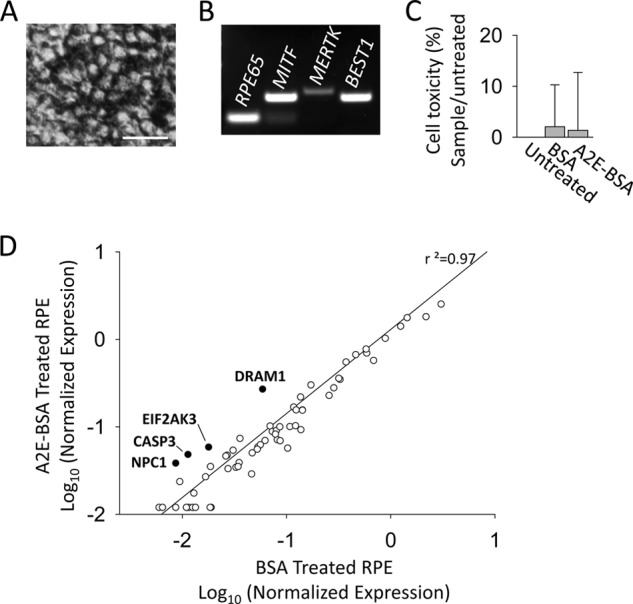
Up-regulation of autophagy-related genes in A2E treated human-derived RPE cells. A, phase contrast image of human derived RPE cells in culture. Scale bar = 10 μm. B, RT-PCR analysis of RPE-specific gene expression markers, RPE65, MITF, MERTK, and BEST1, in cultured human-derived RPE. C, LDH assay comparing cell viability in BSA (0.1%) and 10 μm A2E-BSA conjugate-treated human-derived RPE cells, n = 4 replicates per group. Error bars indicate the mean ± S.D. D, scatter blot of differential gene expression in response to 10 μm A2E-BSA conjugate treatment versus BSA (0.1%) treatment analyzed by the RT2 Human Autophagy™ PCR Expression Array. Genes highlighted in black were up-regulated >3 fold. No genes were significantly down-regulated.
Discussion
Atg7 Maintains RPE Cellular Health
Autophagy, a dynamic process by which contents can be degraded and recycled within a cell, is most active during conditions of cellular stress such as starvation. The RPE is an essential post mitotic tissue in the eye required to maintain the health of the retina. Many post-mitotic tissues are highly reliant upon autophagy to relieve stress and remove damaged organelles through lysosomal degradation mechanisms. Yet loss of Atg7-mediated autophagy in the RPE did not show impaired POS phagocytosis and abnormal accumulation of A2E which could cause retinal degeneration (28–31).
RPE lacking Atg7 did develop signs of increased cellular stress such as hypertrophy and increased undigested cellular debris. Cell hypertrophy resulting from cellular stress was observed in Atg7-deficient RPE as early as 6 weeks of age. Similar findings were reported in a study of mice with RPE-selective postnatal loss of mtDNA transcription and replication that displayed increased RPE size, reduced tight junction integrity, vacuole formation and mTOR activation (47). Accumulation of cellular debris in Atg7flox/flox;cre+ RPE suggests a role for Atg7 in the elimination of cellular content via autophagy. Additionally Atg7-deficient RPE were observed to have numerous vacuole-like structures adjacent to cell membranes indicative of decreased membrane integrity associated with the loss of autophagy. The loss of RPE integrity in models with inadequate autophagy, including our study, suggests that autophagy could be important for maintaining basal RPE health and promoting cell survival during stress.
The Visual Cycle Operates Independently of Atg7-mediated Autophagy
The RPE is responsible for retinoid recycling in the eye, and therefore is required for phototransduction and visual perception. Autophagy has been observed to play a role in the retinoid cycle based on ERG and retinoid measurements made in mice lacking autophagy-related protein 5 (Atg5) in the RPE (2). RPE-specific deficiency of Atg5 was shown to reduce levels of 11-cis-retinal, the visual chromophore required for visual function, and deficient animals demonstrated attenuated ERG signals, which were rescued by 9-cis-retinal administration (2). In contrast to Atg5 deficiency, the normal content of retinoids in Atg7flox/flox;cre+ mice suggest that Atg7 is not required for retinoid recycling, implying different roles for Atg5 and Atg7 in the RPE. Normal ERG a-wave responses, indicative of photoreceptor function also were found in Atg7flox/flox;cre+ mice whereas attenuated amplitudes were measured in Atg5 deficient mice. Of particular note, the study of Atg5 deficiency by Kim et al. reported ERGs in both 12- and 16-week-old mice, whereas we used 5-week-old animals for our ERG analysis. Because younger mice could have minimal abnormal cellular accumulation related to disturbed autophagic processing, our 5-week-old animals might have failed to develop ERG changes observed in Atg5-deficient mice. Additionally the different mouse strains used between our group and Kim et al. (RPE65 variant Leu-450 and Met-450, respectively) could account for the differences in retinoid kinetics because Leu-450 variants regenerate rhodopsin at a faster rate compared with Met-450 mice (48). Moreover, Kim et al. found that Atg5 promotes the visual cycle through non-canonical autophagy instead of degradation via autophagosomes in which both Atg5 and Atg7 are involved. Their observation and the normal visual cycle in mice with Atg7-deficiency in the RPE could suggest another Atg5-dependent and Atg7-independent non-canonical mechanism in the RPE.
Additionally, autophagic proteins not only influence autophagy pathways but also apoptosis, more specifically caspase dependent cell death pathways. In this way Atg7 and Atg5 can act synergistically in autophagy but have opposing effects on apoptosis (49). Atg5 and Atg7 are targets of regulation and themselves regulate different caspase proteins, caspase 6/8 and caspase 9 respectively, in the cell death pathway. Therefore it is feasible that our studies in Atg7 knock-out RPE tissue lead to different conclusions when compared to other studies in which different autophagy genes were deleted.
Notably, we detected a difference in measured b-wave ERG amplitude between the fasted and fed states in mice. Our data indicate that fasting reduces b-wave amplitudes, but that loss of Atg7 does not significantly compound this phenomenon. We have shown that RPE cells respond to the fasted state by increasing autophagic flux, as measured by p62 accumulation. Therefore this increased autophagy induced by starvation could interfere with normal retinoid processing in the RPE.
There is considerable discrepancy in the literature regarding the phenotypes of autophagy-deficient models in various tissues. For example, various studies involving deletion of autophagy-related molecules in the lens have not produced consistent data. In lens development, deficiency of Atg5 or phosphatidylinositol 3-kinase catalytic subunit type (Pik3c3) failed to abolish organelle-free zone (ONF) formation required for lens transparency (50). But more recent data indicate that the mammalian target of rapamycin complex 1 (mTOR-1) signaling is required for proper development of the OFZ in lens (51). These contrasting observations could be explained by differences in gene targeting approaches (choice of exon deletion), cell type (context) dependence, influence of genetic background, and timing of examinations. Because of the nature of autophagy in coping with stress, background differences can significantly modulate phenotypes and hence should be considered when interpreting experimental findings.
Autophagy pathways also appear to have redundant modulators and many pathways share the same enzymes, making it difficult to study a single individual pathway. Pharmacological inhibition of autophagy and genetic deletion of Atg genes have been shown to regulate complementary pathways. In fibroblast cells decreased macroautophagy, due to loss of Atg5, stimulates chaperone-mediated autophagy during basal and stressed conditions (52). Likewise, selective inhibition of chaperone-mediated autophagy has been shown to increase overall macroautophagy suggesting cross-talk and possibly the existence of compensatory mechanisms between these two pathways (53). Of particular interest are the more recently discovered Atg5/Atg7 independent macroautophagy pathways regulated by Unc-51-like kinase 1 (Ulk1) and Beclin1 (54). These pathways do not involve LC3B and therefore examining lipidated LC3 (a common indicator of macroautophagy) may not suffice to determine the lack of autophagy.
A2E Accumulation Does Not Require Atg7-mediated Autophagy but Activates Other Autophagy-related Pathways
RPE cells are phagocytic cells required for the digestion of shed photoreceptor outer segments and thus may accumulate non-degradable visual byproducts such as A2E, with increasing age and retinal disease. Increased A2E accumulation is observed in many such diseases, including Stargardt disease (55), and AMD (56). A2E is considered hazardous to the RPE and the mechanism by which RPE cells accumulate A2E is still not well understood. Numerous studies have examined the association between lysosomes and A2E accumulation in the RPE (29, 43, 57). As autophagic pathways converge upon lysosomal processes, it can be speculated that autophagy is involved in A2E accumulation. In human RPE incubated with A2E, we observed an up-regulation of NCP1 involved in intracellular lipoprotein trafficking (namely cholesterol) to late endosomal/lysosomal compartments. A2E accumulation has previously been shown to affect cholesterol trafficking and phospholipid degradation in the RPE (57–59). Therefore the up-regulation of NCP1 observed in our study could be a cellular mechanism by which the RPE attempts to eliminate lipids by lysosomes to retain retinal function. In agreement with our findings, a previous study with BALB/c mice, which carry the mutant Npc1nih allele, found that these animals display retinal degeneration and accumulation of lipofuscin deposits with age (60).
Autophagy has been noted to influence cell toxicity when cultured RPE cells are laden with A2E. A study by Saadat et al. observed that blocking autophagy greatly increased A2E- induced cell death in ARPE-19 cells (20). Interestingly, genes involved in both autophagy and apoptosis such as CASP3, DRAM1 and PERK were up-regulated after A2E incubation in our gene array. Impaired autophagy has also been linked to retinal disease in humans. A study by Mitter et al. analyzed donor human eyes from AMD patients and found that autophagy proteins LC3, ATG7, and ATG9 were down-regulated in AMD samples. They concluded that autophagy plays an important role in protecting RPE cells from oxidative stress and lipofuscin accumulation (61). Although Atg7-deficient mice in our study failed to demonstrate a direct link between A2E accumulation and autophagy, modified expression of autophagy-related molecules in A2E-challenged human RPE cells suggests an important relationship between A2E and autophagy in human retinal diseases.
In summary, our results indicate that RPE-specific loss of Atg7 does not affect normal retinoid recycling or visual function. Furthermore, A2E levels were unaffected by Atg7 deficiency in Abca4−/− mice suggesting that accumulation of A2E occurs independent of Atg7-mediated autophagy. Loss of autophagy in the RPE did result in debris accumulation and vacuole formation, suggesting a role for autophagy as a cellular protective mechanism in aging RPE cells.
Author Contributions
L. P. and A. M. designed experiments. L. P., B. S., T. P, H. M., E. A., and Y. C. conducted experiments and acquired data. L. P., B. S., T. P, H. M., E. A., Y. Z. L. C. S., Y. C., K. P, and A. M. analyzed and interpreted data. L. P. and A. M. wrote the manuscript. All authors gave final approval of the version to be published.
Acknowledgments
We thank Dr. Leslie T. Webster, Jr, Vipulkumar Parmar (Case Western Reserve University) for helpful comments on the manuscript, the Visual Sciences Research Center Core Facility (supported by P30 EY11373), in particular Scott Howell and Sarah Lindstrom for their help with microscopy, and the flow cytometry facility in the Department of Dermatology's Skin Diseases Research Center (SDRC) for assistance with confocal microscopy. The Department of Dermatology's Skin Diseases Research Center (SDRC) is supported in part by the National Institutes of Health Grant (5P30AR039750) and the Ohio Department of Development - Center for Innovative Immunosuppressive Therapeutics (TECH 09-023).
This work was supported by funding from the National Institutes of Health (Grants EY022658, EY021126, and EY11373), Research to Prevent Blindness Foundation, Foundation Fighting Blindness, and Ohio Lions Eye Research Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
- RPE
- retinal pigmented epithelium
- ABCA4
- ATP-binding cassette transporter 4
- Atg7
- autophagy-related gene 7
- AMD
- age-related macular degeneration
- A2E
- di-retinoid-pyridinium-ethanolamine
- ERG
- electroretinogram
- POS
- photoreceptor outer segments
- ZO-1
- zonula occludens protein 1.
References
- 1.Strauss O. (2005) The retinal pigment epithelium in visual function. Physiol. Rev. 85, 845–881 [DOI] [PubMed] [Google Scholar]
- 2.Kim J. Y., Zhao H., Martinez J., Doggett T. A., Kolesnikov A. V., Tang P. H., Ablonczy Z., Chan C. C., Zhou Z., Green D. R., and Ferguson T. A. (2013) Noncanonical autophagy promotes the visual cycle. Cell 154, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young R. W. (1967) The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaVail M. M. (1976) Rod outer segment disk shedding in rat retina: relationship to cyclic lighting. Science 194, 1071–1074 [DOI] [PubMed] [Google Scholar]
- 5.Duncan J. L., LaVail M. M., Yasumura D., Matthes M. T., Yang H., Trautmann N., Chappelow A. V., Feng W., Earp H. S., Matsushima G. K., and Vollrath D. (2003) An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest. Ophthalmol. Vis. Sci. 44, 826–838 [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N., and Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., and Chiba T. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., and Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 9.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 10.Taneike M., Yamaguchi O., Nakai A., Hikoso S., Takeda T., Mizote I., Oka T., Tamai T., Oyabu J., Murakawa T., Nishida K., Shimizu T., Hori M., Komuro I., Takuji Shirasawa T. S., Mizushima N., and Otsu K. (2010) Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 6, 600–606 [DOI] [PubMed] [Google Scholar]
- 11.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., and Plotz P. (2008) Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 17, 3897–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., and Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., and Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubinsztein D. C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 15.Jo C., Gundemir S., Pritchard S., Jin Y. N., Rahman I., and Johnson G. V. (2014) Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat. Commun. 5, 3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klionsky D. J., Cregg J. M., Dunn W. A. Jr., Emr S. D., Sakai Y., Sandoval I. V., Sibirny A., Subramani S., Thumm M., Veenhuis M., and Ohsumi Y. (2003) A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545 [DOI] [PubMed] [Google Scholar]
- 17.Noda N. N., Satoo K., Fujioka Y., Kumeta H., Ogura K., Nakatogawa H., Ohsumi Y., and Inagaki F. (2011) Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475 [DOI] [PubMed] [Google Scholar]
- 18.Kunchithapautham K., and Rohrer B. (2007) Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy 3, 433–441 [DOI] [PubMed] [Google Scholar]
- 19.Besirli C. G., Chinskey N. D., Zheng Q. D., and Zacks D. N. (2011) Autophagy activation in the injured photoreceptor inhibits fas-mediated apoptosis. Invest. Ophthalmol. Vis. Sci. 52, 4193–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saadat K. A., Murakami Y., Tan X., Nomura Y., Yasukawa T., Okada E., Ikeda Y., and Yanagi Y. (2014) Inhibition of autophagy induces retinal pigment epithelial cell damage by the lipofuscin fluorophore A2E. FEBS 4, 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S. Y., Oh J. S., Rho J. H., Jeong N. Y., Kwon Y. H., Jeong W. J., Ryu W. Y., Ahn H. B., Park W. C., Rho S. H., Yoon Y. G., Jeong S. Y., Choi Y. H., Kim H. Y., and Yoo Y. H. (2014) Retinal pigment epithelial cells undergoing mitotic catastrophe are vulnerable to autophagy inhibition. Cell Death Dis. 5, e1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao J., Jia L., Shelby S. J., Ganios A. M., Feathers K., Thompson D. A., and Zacks D. N. (2014) Circadian and noncircadian modulation of autophagy in photoreceptors and retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 55, 3237–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birnbach C. D., Järveläinen M., Possin D. E., and Milam A. H. (1994) Histopathology and immunocytochemistry of the neurosensory retina in fundus flavimaculatus. Ophthalmology 101, 1211–1219 [DOI] [PubMed] [Google Scholar]
- 24.Zhou J., Jang Y. P., Kim S. R., and Sparrow J. R. (2006) Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A. 103, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma W., Coon S., Zhao L., Fariss R. N., and Wong W. T. (2013) A2E accumulation influences retinal microglial activation and complement regulation. Neurobiol. Aging 34, 943–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illing M., Molday L. L., and Molday R. S. (1997) The 220-kDa Rim protein of retinal rod outer segments is a member of the ABC transporter superfamily. J. Biol. Chem. 272, 10303–10310 [DOI] [PubMed] [Google Scholar]
- 27.Molday R. S. (2007) ATP-binding cassette transporter ABCA4: molecular properties and role in vision and macular degeneration. J. Bioenerg. Biomembr. 39, 507–517 [DOI] [PubMed] [Google Scholar]
- 28.Eldred G. E., and Lasky M. R. (1993) Retinal age pigments generated by self-assembling lysosomotropic detergents. Nature 361, 724–726 [DOI] [PubMed] [Google Scholar]
- 29.Sparrow J. R., Parish C. A., Hashimoto M., and Nakanishi K. (1999) A2E, a lipofuscin fluorophore, in human retinal pigmented epithelial cells in culture. Invest. Ophthalmol. Vis. Sci. 40, 2988–2995 [PubMed] [Google Scholar]
- 30.Holz F. G., Schütt F., Kopitz J., Eldred G. E., Kruse F. E., Völcker H. E., and Cantz M. (1999) Inhibition of lysosomal degradative functions in RPE cells by a retinoid component of lipofuscin. Invest. Ophthalmol. Vis. Sci. 40, 737–743 [PubMed] [Google Scholar]
- 31.Schütt F., Davies S., Kopitz J., Holz F. G., and Boulton M. E. (2000) Photodamage to human RPE cells by A2-E, a retinoid component of lipofuscin. Invest. Ophthalmol. Vis. Sci. 41, 2303–2308 [PubMed] [Google Scholar]
- 32.Le Y. Z., Zheng W., Rao P. C., Zheng L., Anderson R. E., Esumi N., Zack D. J., and Zhu M. (2008) Inducible expression of cre recombinase in the retinal pigmented epithelium. Invest. Ophthalmol. Vis. Sci. 49, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda A., Maeda T., Golczak M., and Palczewski K. (2008) Retinopathy in mice induced by disrupted all-trans-retinal clearance. J. Biol. Chem. 283, 26684–26693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohno H., Chen Y., Kevany B. M., Pearlman E., Miyagi M., Maeda T., Palczewski K., and Maeda A. (2013) Photoreceptor proteins initiate microglial activation via Toll-like receptor 4 in retinal degeneration mediated by all-trans-retinal. J. Biol. Chem. 288, 15326–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda A., Maeda T., Imanishi Y., Kuksa V., Alekseev A., Bronson J. D., Zhang H., Zhu L., Sun W., Saperstein D. A., Rieke F., Baehr W., and Palczewski K. (2005) Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J. Biol. Chem. 280, 18822–18832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diemer T., Gibbs D., and Williams D. S. (2008) Analysis of the rate of disk membrane digestion by cultured RPE cells. Adv. Exp. Med. Biol. 613, 321–326 [DOI] [PubMed] [Google Scholar]
- 37.Maeda T., Lee M. J., Palczewska G., Marsili S., Tesar P. J., Palczewski K., Takahashi M., and Maeda A. (2013) Retinal pigmented epithelial cells obtained from human induced pluripotent stem cells possess functional visual cycle enzymes in vitro and in vivo. J. Biol. Chem. 288, 34484–34493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osakada F., Ikeda H., Sasai Y., and Takahashi M. (2009) Stepwise differentiation of pluripotent stem cells into retinal cells. Nat. Protoc. 4, 811–824 [DOI] [PubMed] [Google Scholar]
- 39.Iriyama A., Fujiki R., Inoue Y., Takahashi H., Tamaki Y., Takezawa S., Takeyama K., Jang W. D., Kato S., and Yanagi Y. (2008) A2E, a pigment of the lipofuscin of retinal pigment epithelial cells, is an endogenous ligand for retinoic acid receptor. J. Biol. Chem. 283, 11947–11953 [DOI] [PubMed] [Google Scholar]
- 40.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., and Sandri M. (2009) Autophagy Is Required to Maintain Muscle Mass. Cell Metab. 10, 507–515 [DOI] [PubMed] [Google Scholar]
- 42.Komatsu M., Wang Q. J., Holstein G. R., Friedrich V. L. Jr., Iwata J. I., Kominami E., Chait B. T., Tanaka K., and Yue Z. Y. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guha S., Liu J., Baltazar G., Laties A. M., and Mitchell C. H. (2014) Rescue of compromised lysosomes enhances degradation of photoreceptor outer segments and reduces lipofuscin-like autofluorescence in retinal pigmented epithelial cells. Adv. Exp. Med. Biol. 801, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holz F. G., Schutt F., Kopitz J., and Volcker H. E. (1999) [Introduction of the lipofuscin-fluorophor A2E into the lysosomal compartment of human retinal pigment epithelial cells by coupling to LDL particles. An in vitro model of retinal pigment epithelium cell aging]. Ophthalmologe. 96, 781–785 [DOI] [PubMed] [Google Scholar]
- 45.Radu R. A., Hu J., Yuan Q., Welch D. L., Makshanoff J., Lloyd M., McMullen S., Travis G. H., and Bok D. (2011) Complement system dysregulation and inflammation in the retinal pigment epithelium of a mouse model for Stargardt macular degeneration. J. Biol. Chem. 286, 18593–18601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haines J. L., Hauser M. A., Schmidt S., Scott W. K., Olson L. M., Gallins P., Spencer K. L., Kwan S. Y., Noureddine M., Gilbert J. R., Schnetz-Boutaud N., Agarwal A., Postel E. A., and Pericak-Vance M. A. (2005) Complement factor H variant increases the risk of age-related macular degeneration. Science 308, 419–421 [DOI] [PubMed] [Google Scholar]
- 47.Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., Ahern K., Snyder M., Bok D., Dunaief J. L., LaVail M. M., and Vollrath D. (2011) mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 121, 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wenzel A., Reme C. E., Williams T. P., Hafezi F., and Grimm C. (2001) The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J. Neurosci. 21, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H., Che X., Zheng Q., Wu A., Pan K., Shao A., Wu Q., Zhang J., and Hong Y. (2014) Caspases: a molecular switch node in the crosstalk between autophagy and apoptosis. Int. J. Biol. Sci. 10, 1072–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morishita H., Eguchi S., Kimura H., Sasaki J., Sakamaki Y., Robinson M. L., Sasaki T., and Mizushima N. (2013) Deletion of autophagy-related 5 (Atg5) and Pik3c3 genes in the lens causes cataract independent of programmed organelle degradation. J. Biol. Chem. 288, 11436–11447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basu S., Rajakaruna S., Reyes B., Van Bockstaele E., and Menko A. S. (2014) Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells. Autophagy 10, 1193–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaushik S., Massey A. C., Mizushima N., and Cuervo A. M. (2008) Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell 19, 2179–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massey A. C., Kaushik S., Sovak G., Kiffin R., and Cuervo A. M. (2006) Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U.S.A. 103, 5805–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., and Shimizu S. (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 [DOI] [PubMed] [Google Scholar]
- 55.Mata N. L., Weng J., and Travis G. H. (2000) Biosynthesis of a major lipofuscin fluorophore in mice and humans with ABCR-mediated retinal and macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 97, 7154–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yannuzzi L. A., Ober M. D., Slakter J. S., Spaide R. F., Fisher Y. L., Flower R. W., and Rosen R. (2004) Ophthalmic fundus imaging: today and beyond. Am. J. Ophthalmol. 137, 511–524 [DOI] [PubMed] [Google Scholar]
- 57.Finnemann S. C., Leung L. W., and Rodriguez-Boulan E. (2002) The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A. 99, 3842–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toops K. A., Tan L. X., Jiang Z., Radu R. A., and Lakkaraju A. (2015) Cholesterol-mediated activation of acid sphingomyelinase disrupts autophagy in the retinal pigment epithelium. Mol. Biol. Cell 26, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lakkaraju A., Finnemann S. C., and Rodriguez-Boulan E. (2007) The lipofuscin fluorophore A2E perturbs cholesterol metabolism in retinal pigment epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 104, 11026–11031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Claudepierre T., Paques M., Simonutti M., Buard I., Sahel J., Maue R. A., Picaud S., and Pfrieger F. W. (2010) Lack of Niemann-Pick type C1 induces age-related degeneration in the mouse retina. Mol. Cell Neurosci. 43, 164–176 [DOI] [PubMed] [Google Scholar]
- 61.Mitter S. K., Song C. J., Qi X. P., Mao H. Y., Rao H., Akin D., Lewin A., Grant M., Dunn W., Ding J. D., Bowes Rickman C. B., and Boulton M. (2014) Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 10, 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]