Abstract
Norovirus (NoV) is now recognized as a leading cause of nonbacterial acute gastroenteritis; however, the NoV GII.17 genotype has rarely been reported as the predominant genotype in clinical diarrhea cases. During the winter of 2014–2015, the GII.17 genotype, together with the NoV GII.4 genotype, dominated in sporadic adult patients with gastroenteritis in Shanghai. Phylogenetic analysis based on full-length VP1 amino acid sequences showed that the GII.17 strains that emerged in Shanghai have close evolutionary relationships with strains recently collected in the Hong Kong area, Guangdong province of China, and Japan during the same period. This cluster in the phylogenetic tree may represent a novel NoV GII.17 lineage recently circulating in East Asia. Pairwise distances between clusters also revealed the evolution of the NoV GII.17 genotype in previous decades. Our study emphasizes the importance of combined surveillance of NoV-associated infections.
Keywords: gastroenteritis, genotypes, norovirus
Introduction
Diarrhea disease morbidity and mortality have been in decline globally, but approximately 1.7–5 billion cases of diarrhea and nearly 1.7 million diarrhea-associated deaths still occur each year,1 the great majority of which are among young children in developing countries.2 Human norovirus (NoV) is an important cause of acute gastroenteritis and is responsible for sporadic and epidemic infection worldwide. Recent studies have revealed that it has the capacity to cause more severe complications than previously expected.1 NoV is not routinely tested for in clinical settings due to requirements for highly specific molecular methods. The characterization of NoV epidemiology has been primarily performed through the analysis of outbreak data. However, there is an overrepresentation of studies merely in children and a lack of sporadic data on NoV infections.3
NoV can be divided into at least six genogroups, designated G1–GVI, three of which infect humans (GI, GII, and GIV).4 The GII.4 genotype has been associated with the majority of viral gastroenteritis outbreaks worldwide since the mid-1990s,5 and GII.4_Sydney_2012 has replaced GII.4_New_Orleans_2009 as the predominant subtype in circulation since 2012.6
The NoV GII.17 genotype was previously found in individual cases or in the waters of Central and South America, Korea, Thailand, Kenya, and China7,8,9,10,11,12,13 but was rarely reported as the predominant genotype in clinical diarrhea cases. Recently, two provincial CDCs (Guangdong CDC and Jiangsu CDC) of China reported outbreaks associated with NoV GII.17 genotype infections during the winter of 2014–2015.14,15 A similar prevalence was also observed in Hong Kong, Taiwan, and Japan during the same winter season.16,17,18 Although the NoV GII.17 genotype was found in recent outbreaks, the distribution of NoV GII.17 among sporadic cases remains unknown, and the clinical and epidemiologic features of this emerging NoV have not been fully characterized.
Materials and methods
Case definition and specimen processing methods
Surveillance subjects were defined as those who visited the enteric disease clinics of sentinel hospitals and presented with three or more loose or watery stools per day (the definition of diarrhea by the World Health Organization).19 NoV-affected patients were defined as those whose stool samples were NoV-positive, including patients with single infections and coinfections. All surveillance subjects were interviewed by doctors. Epidemiological and medical information was obtained and recorded. According to “The diagnosis and treatment standard of NoV-leaded gastroenteritis and determination of suspected NoV-associated outbreak in China” (http://www.chinacdc.cn/jkzt/crb/nrbdjxwcy/jszl_2273/200701/t20070111_24754.htm), cases suspected of outbreak settings were excluded via inquiry.
Stool samples were collected from surveillance subjects at designated intervals by trained medical staff of the sentinel hospitals distributed in different regions, including Shanghai Public Health Clinical Centre (SPHCC), Shanghai Tongren Hospital and Children's Hospital of Fudan University.
One stool specimen was collected from each patient before treatment. A 10% stool suspension (e.g., 100 mg of feces sample in 900 µL of phosphate-buffered saline solution) was prepared. Sera, urines, and feces suspensions were frozen at −80 °C until use.
Extraction of total RNA and one-step RT-PCR for GI and GII genotyping
Template RNA was extracted from patient fecal suspensions using a QIAamp Viral RNA Mini Kit (Qiagen, Germany) following the manufacturer's instruction.
Two sets of primers G1-SKF/G1-SKR19 and CoG2F/G2-SKR20,21 were used for amplifying the capsid gene at the C region (Supplementary Table S1). The products were sequenced and typed using the NoV Genotyping Tool (http://www.rivm.nl/mpf/norovirus/typingtool)22 followed by phylogenetic analysis.
A one-step PrimeScript RT-PCR Kit (Takara, Japan) was used with a 25-μL total volume containing 12.5 μL of 2× one-step RT-PCR Buffer III, 2.5 U TaKaRa Ex Taq HS, 2.5 U PrimeScript RT Enzyme Mix II, 0.32 µM forward/reverse primers, and 2.5 μL of extracted RNA. Reverse transcription polymerase chain reaction (RT-PCR) was carried out at 42 °C for 20 min, followed by 95 °C for 1 min, and 35 cycles of PCR at 94 °C for 30 s, 54 °C for 30 s (G1-SKF/G1-SKR, CoG2F/G2-SKR), 72 °C for 40 s (G1-SKF/G1-SKR, CoG2F/G2-SKR), and a final incubation at 72 °C for 10 min.17 The RT-PCR amplification was performed using the ABI StepOne Plus System (Applied Biosystems), and the products were separated by electrophoresis on 1.5% agarose gels and visualized under UV light.
RT-PCR of VP1 and the full-length genome of the NoV GII.17 genotype
The primers II.17-VP1F/II.17-VP1R (Supplementary Table S1) were designed and used for full-length VP1 amplification of NoV GII.17 strains. RT-PCR amplification was carried out at 42 °C for 20 min, followed by 95 °C for 1 min, and 35 cycles of PCR at 94 °C for 30 s, 57 °C for 30 s, 72 °C for 110 s, and a final incubation at 72 °C for 10 min.
The representative NoV GII.17 strain selected from the Shanghai area was subjected to complete genome amplification using a one-step RT-PCR kit (TaKaRa, Dalian, China), using the overlapping primers (Supplementary Table S2) designed based primarily on the sequence of NoV GII/Hu/JP/2002/GII.P16_GII.17/Saitama/T87 (GenBank NO KJ196286).
Sequencing
The products were directly sequenced on an ABI 3730XL automatic DNA analyzer using an ABI Prism BigDye Terminator cycle sequencing kit 3.1 (Applied Biosystems). Each amplicon was sequenced in both the forward and reverse directions. The sequences were assembled using BioEdit software and deposited into GenBank (the accession numbers are listed in Supplementary Table S3).
Phylogenetic and pairwise distance analysis
The viral amino acid and reference sequences obtained from the GenBank database were subjected to phylogenetic analysis using the MEGA 5.1 program (http://www.megasoftware.net/). A phylogenetic tree was constructed using the neighbor-joining method with a bootstrap analysis of 1000 replicates in the MEGA6.0 program. The evolutionary distances were computed using the Poisson model as implemented. Numbers next to nodes indicate bootstrap value percentages (>70%).
Pairwise distances between full-length VP1 amino acid sequences were calculated in MEGA 6.0 program using the Poisson model. All ambiguous positions were removed for each sequence pair. If divergence of an amino acid exceeded 5%, the strain was defined as a GII.17 novel variant relative to the earlier strains.
Ethics
The study protocol was reviewed and approved by the Human Research Ethics Committee of the Shanghai Public Health Clinical Center.
Statistical analysis
The nonparametric Mann–Whitney U test was used for comparing the distributions of two unmatched groups and Fisher's exact test was used for categorical variables. Statistical analyses were performed using Prism 6.0 (GraphPad). Two-tailed P < 0.05 were considered statistically significant.
Results
Distribution of NoV genotypes
Overall, 529 outpatients (375 adults aged >15 years and 154 children aged <15 years) diagnosed as having acute gastroenteritis in three sentinel hospitals were enrolled in our study from January 2014 to January 2015. NoV was detected in 90 (17.0%) out of 529 patients. Genotyping was successful for 78 (86.67%) cases. Among them, 50 (13.3%) out of 375 adults and 40 (26.0%) out of 154 children were infected with NoV. The GII genogroup accounted for most of the NoV-infected cases (90%, 81/90), followed by the GI strain (10%, 9/90). Six genotypes of the GII genogroup were found, including NoV GII.17, GII.4, GII.3, GII.2, GII.6, and GII.13 (Figure 1).
Figure 1.
Genotype distribution of NoV genogroupII (GII) in sporadic patients with NoV infection from 2014–2015 in Shanghai, China. (A) Genogroup II of GII.2, GII.3, GII.4 Sydney_2012, GII.6, GII.13, and GII.17 in all patients. (B) Genogroup II of GII.2, GII.4 Sydney_2012, GII.6, GII.13, and GII.17 in adult patients. (C) Genogroup II of GII.3, GII.4, and GII.6 in pediatric patients.
However, the distribution of genotypes was different between adults and children. Five genotypes, specifically GII.17 (51.2%, 22/43), GII.4 (41.9%, 18/43), GII.2 (2.3%, 1/43), GII.6 (2.3%, 1/43), and GII.13 (2.3%, 1/43), were detected in adults, while only three genotypes, GII.4 (21/35, 60%), GII.3 (13/35, 37.1%), and GII.6 (1/35, 2.9%), were found in children. GII.4 was detected in both adults and children, while NoV GII.17 was detected only in adults and the GII.3 genotype was detected only in children, which may be due to the limited number of cases in our study.
NoV GII.17 was first detected in November 2014. It should be noted that this time point coincided with the first report of NoV GII.17 that caused outbreaks in Guangdong province14 and Japan.17 A similar observation was made in Jiangsu province, except that the first cases were identified in October 2014.15 In addition, starting in November 2014, the monthly detection rate of NoV GII.17 outnumbered GII.4 (Figure 2) in our adult patients and rapidly became the predominant genotype (NoV GII.17 vs. GII.4: 15.38% vs. 7.69% in November 2014, 20% vs. 15% in December 2014, and 28.57% vs. 16.67% in January 2015).
Figure 2.
Monthly detection rate of NoV in adult outpatients (≥15 years of age) with acute gastroenteritis from 2014 to 2015 in Shanghai, China.
General characterization of cases
We also compared the demographic and clinical characteristics of adult patients infected with either GII.4 or GII.17 genotype. As shown in Table 1, there were no significant differences in patient gender and median age between the two groups. Moreover, we did not find a statistically significant difference (P = 0.10) in the maximum number of stools per day between GII.4 (6.5, IQR = 5.375–10.50) and GII.17 (10, IQR = 7.25–13).
Table 1. Comparison of demographic and clinical information between the NoV GII.4 and GII.17 genogroups in adult patients.
| Characteristics | GII.4 group (n = 18) | GII.17 group (n = 22) | P value |
|---|---|---|---|
| Gender (Male) | 9 | 14 | Pa = 0.39 |
| Median age (IQR) | 57.5 (30.75–62.50) | 35.5 (25.75–59.50) | Pb = 0.34 |
| Median of maximum number of stools per day (IQR) | 6.5 (5.375–10.50) | 10 (7.25–13) | Pb = 0.10 |
| Case number with vomiting | 6 | 3 | Pa = 0.14 |
Fisher's exact test.
Mann–Whitney U test.
Phylogenetic analysis of the VP1 gene
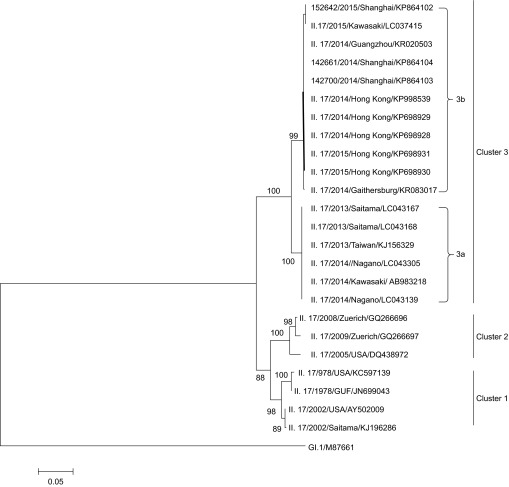
Three NoV GII.17 strains, including 142700, 142661, and 152642, were selected as the monthly representative strains from November 2014 to January 2015. Phylogenetic analysis based on full-length amino acid sequences of the VP1 gene of NoV GII.17 available from GenBank database showed that all the sequences could be divided into three clusters (Figure 3), and cluster 3 further led to two sub-clusters (cluster 3a and cluster 3b). The Shanghai representative strains clustered together with four Hong Kong strains (NS-463, NS-491, NS-494, and NS-511) collected from 2014 to 2015, a Guangzhou strain (Hu/GII.17/41621/Guangzhou/2014/CHN) and a Japan strain (Kawasaki308) collected in 2015 to constitute cluster 3b. Another five Japan strains collected from 2013 to 2014 and a Taiwan strain collected in 2013 formed cluster 3a, while earlier strains collected in 1978, 2002, 2005, 2008 and 2009 formed clusters 1 and 2. Although recent GII.17 strains have emerged within the past two years, two lineages could be distinguished easily on the tree, which hinted at their different evolutionary histories. A similar observation was obtained by research groups from Japan and Jiangsu CDC, China.15,17 Additionally, comparison of the complete genome sequence of the Shanghai representative strain 142700 to the reference strains from Hong Kong (GII/Hu/HKG/2014/GII.17/CUHK-NS-463) and Japan (Hu/GII/JP/2015/GII.P17GII.17/Kawasaki308) indicated high sequence identities of 99.7% and 99.6%, respectively, which showed their close relationship and confirmed the cluster classification based on VP1 gene.
Figure 3.
Phylogenetic analysis of full-length VP1 amino acid sequences of the NoV GII.17 genotype. The full-length VP1 sequences of three representative NoV GII.17 strains from November and December 2014 and January 2015 were sequenced (primers listed in Supplementary Table S1; GenBank accession NOs KP864102–KP864104). The GenBank accession numbers of the NoV GII.17 reference strains are displayed in the phylogenetic tree, followed by the genotypes, years, and countries.
To clarify the relationships between the NoV GII.17 strains from sporadic patients in Shanghai and the outbreak strains in Guangdong during the winter 2014–2015, sequence alignments were carried out with all 22 Shanghai strains (Supplementary Table S3) and the representative strain from Guangdong (Hu/15F98/ZQ/GD/CHN/2015) based on a 282-bp fragment of the VP1 gene. The high similarity of the nucleotide sequences was observed with identities of 98.9%–100%, which may support a link between the outbreak events in Guangdong and the prevalence in the Shanghai area.
Pairwise distance between three clusters
The pairwise distance of the VP1 amino acid sequence was calculated between all of the strains in three clusters, and a 5% amino acid difference cut-off was used to define GII.17 new variants (Supplementary Table S4). The distances between the strains from 1978 to 2002 ranged from 2.1% to 2.6%, indicating the stability of the VP1 gene over a long period of time. However, the distances between cluster 2 (the strains obtained from 2005 to 2009) and cluster 1 were 7%–8%. The distances between recent strains (cluster 3a and 3b, 2013–2015) to earlier strains (cluster 1 and 2) increased by 10%–15%, suggesting that the recently emerged GII.17 virus is a new variant, and the distance among recent strains (cluster 3a vs. cluster 3b) has been close to 5% only within the past two years, suggesting that the new GII.17 variants have different evolutionary histories but likely share a more distant parent, which was supported by a time-scaled phylogenetic tree constructed by a Japanese group.17
Amino acid substitutions and indels within the VP1 protein
Differences in the major capsid protein (VP), including 1-219 amino acids in the shell (S) domain and 220-544 amino acids in the protruding (P) domain, were analyzed among NoV GII.17 strains. One or two representative strain from each cluster, specifically II.17/1978/USA/KC597139, II.17/2002/USA/AY502009, II.17/2005/USA/DQ438972, II.17/2013/Saitama/LC043167, and II.17/2014/Shanghai/KP864103, was selected for amino acid sequence alignment of the VP1 sequence.
Sequence analysis of the VP1 sequence in the strains revealed a number of amino acid substitutions that were unique to each cluster (92/543). Three deletions (residues 295, 296, and 384) and three insertions (344, 380, and 396) were present in the strains from cluster 3a and cluster 3b. Comprehensive alignments have been provided by other researchers,15,17,18,23 which indicated three deletions and at least one insertion compared with previous GII.17 strains. Moreover, cluster 3b differed by 22 amino acid site substitutions (sites N169S, E293Q, T294I, D297N, H298Q, K301R, V336M, A337V, H353Q, E354Q, V356W, I357V, F373L, S375I, -380D, I394V, G398D, −400G, D410N, L414E, V452I, Y517H) compared with cluster 3a, and cluster 3b had 14 unique amino acid sites, including sites 169, 298, 336, 337, 353, 354, 356, 373, 375, 380, 394, 398, 414, and 517 (Figure 4). Most of these substitutions and indels in the VP1 protein were located in the P2 domain, which contains the antigenic epitopes and host receptor binding domain.24
Figure 4.
Amino acid variations in the VP1 protein of NoV GII.17 strains. Amino acid sequence alignment of the VP1 protein was performed for representative strains from each of the GII.17 clusters described in Figure 3. Amino acid substitutions and indel mutations are indicated with different colors. The shell (S) domain, protruding (P) domain, and five speculated epitopes (A–E) are also labeled in the figure. The epitopes are putatively identified based on analogy to putative GII.4 epitopes.
Discussion
From October 2012 to December 2012, we reported that a variant of the GII.e/GII.4 strain (GII.4_Sydney_2012) led to an outbreak of NoV-associated infections in Shanghai.25 This genotype has dominated in adult patients with acute gastroenteritis since then. However, the uncommon NoV GII.17 genotype emerged in November 2014 and became the predominant genotype in adult outpatients. Interestingly, NoV GII.17 was only detected in adults and the reason for this must be further confirmed.
Our study showed that the NoV GII.17 genotype emerged first in November of 2014 and rapidly became the major genotype in the Shanghai area. Similar findings were reported in Guangdong and Jiangsu provinces, in which the NoV GII.17 genotype first emerged in November 2014 and October 2014, respectively.14,15 Moreover, NoV GII.17-related outbreaks occurred in 10 cities within only two months in Guangdong.14 Combining these observations of patients, outbreak events, and viral sequence analyses, it could be speculated that NoV GII.17 exhibited high epidemic activity during the winter of 2014–2015 in several areas of China. The clinical data for the NoV GII.17 genotype compared with the GII.4_Sydney_2012 genotype showed no significant differences between the two groups in our study. The full scale of GII.17 prevalence in China remains unclear.
In addition, phylogenetic and pairwise distance analysis based on VP1 sequences showed that NoV GII.17 strains in Shanghai were composed of a novel NoV GII.17 lineage and had close evolutionary relationships not only with Guangdong strains but also with very recent strains from Japan and Hong Kong obtained from 2014 to 2015. This suggested that the novel GII.17 variants might have emerged and circulated in East Asia in recent years. As the majority of the NoV sequences have been reported from coastal regions,26 it was not surprising that the NoV GII.17 lineage appeared in Shanghai city, Guangdong province, Hong Kong, and Japan. However, the origin and transmission chain of this new lineage remain unknown.
From the limited data available on the full-length VP1 gene in GenBank, the GII.17 genotype remained stable for several decades prior to 2002, but at least two replacement events in the predominant strains may have occurred during the period from 2002 to 2005 and the period from 2009 to 2013, although there were no outbreak reports in those years. NoV GII.17 strains in Shanghai had close evolutionary relationships not only with Guangdong strains but also with very recent strains from Japan and Hong Kong obtained during the period from 2014 to 2015, which comprised a novel NoV GII.17 lineage. A Japanese group also reported that the novel GII.17 variants might have emerged and circulated in East Asia in recent years and the emerging GII.17 variants have different evolutionary histories than GII.17 strains identified earlier based on a time-scaled phylogenetic tree.17
We employed a 5% amino acid difference cut-off value to define GII.4 variants and delineate clusters 1–3 based on GII.17 sequences. The pairwise distances of VP1 amino acid sequences among the recent GII.17 strains were 4.2%–4.4%, which were close to but did not reach the cut-off value. Thus, cluster 3 should be divided into two sub-clusters (cluster 3a and 3b). Cluster 3b mainly comprised the predominant genotype in Asia during the winter of 2014–2015, which had 14 unique amino acid site mutants compared with earlier GII.17 strains (Figure 4). Most (12/14, 85.7%) of these substitutions and indels in the VP1 protein were located in the P2 domain, which suggests that the recent prevalence of NoV GII.17 in the population induced protective herd immunity, consequently driving molecular evolution.2
The rapid evolution of the novel GII.17 may lead to escaping from the host immune response, driving changes in HBGA affinities and altering population susceptibility patterns, but this speculation could not be easily confirmed due to the lack of a cell culture system for isolating antibody escape mutants.3 A crystallography study and a binding blockade assay with a GII.4 monoclonal antibody suggested that the P domain of the VP1 protein contained antigenic epitopes and a receptor-binding domain.24 The features of the VP1 protein were not extensively studied in NoV GII.17, but our study indicated that the amino acid changes over time are primarily located in the P2 subdomain, which is similar to observations in GII.4 and other genotypes of the GII genogroup.3,18 Five blockade epitopes on VP1 in GII.17 NoV have been hypothesized based on the known epitopes on the VP1 protein of GII.4.23 These are Epitope A (residues 294–301, 374, and 378), Epitope B (336 and 395), Epitope C (343 and 382), Epitope D (residues 400–402), and Epitope E (residues 410, 414–416) (Figure 4). Matsushima et al.17 predicted B-cell epitopes associated with humoral immunity at the amino acid positions 217–225 (I), 291–298 (II), 359–363 (III), 371–379 (IV), and 390–396 (V) in the VP1 protein of the Kawasaki323 strain. Although the predicted methods were different, the predicted epitopes were relatively close in the P2 domain. Most of the substitutions and indel mutations we observed in the VP1 sequences of recent GII.17 strains mapped in or near these epitopes, which may lead to herd immune escape and/or alter human susceptibility to the GII.17 genotype, thus partially explaining the recent prevalence of this novel variant. Further investigation should be carried out to reveal greater detail.
In conclusion, we report the emergence of a novel NoV GII.17 lineage during the winter of 2014–2015. As Shanghai is one of the most dynamic and cosmopolitan cities in the eastern part of Asia, our study here emphasizes the importance of continual and combined surveillance of NoV-associated infections among the large coastal cities in Eastern Asia. This collaborative surveillance will monitor the emergence of novel NoV strains and help control new outbreaks in the coming epidemic season.
Acknowledgments
This work was supported by the Shanghai Health Bureau Project (20124467 and 20134151), the Shanghai City Science and Technology Commission (134119a9000), the National Natural Science Funds of China (81341004, 31200108 and 81470829), and the National Megaprojects of China for Infectious Disease (2014ZX10004002-003-004 and 2012ZX10004-211).
Footnotes
Supplementary information of this article can be found on the Emerging Microbes and Infections's website: http://www.nature.com/emi.
Supplementary Information
References
- 1Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol 2009; 44: 1–8. [DOI] [PubMed] [Google Scholar]
- 2Lindesmith LC, Beltramello M, Donaldson EF et al. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog 2012; 8: e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Debbink K, Lindesmith LC, Donaldson EF, Baric RS. Norovirus immunity and the great escape. PLOS Pathog 2012; 8: e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Green KY. Caliciviridae: the noroviruses. In: Knipe DM, Howley PM, Cohen JI, Griffin DI, Lamb RA, Martin MA, Racaniello VR, Roizman B (eds.) Fields virology. 6th ed. Philadelphia, PA: Lippincott Williams & Wilkins, 2013: 582–608. [Google Scholar]
- 5Karst SM, Baric RS. What is the reservoir of emergent human norovirus strains? J Virol 2015; 89: 5756–5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6van Beek J, Ambert-Balay K, Botteldoorn N et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill 2013; 18: 8–9. [PubMed] [Google Scholar]
- 7Ferreira MS, Xavier Mda P, Tinga AC et al. Assessment of gastroenteric viruses frequency in a children's day care center in Rio De Janeiro, Brazil: a fifteen year study (1994–2008). PLoS One 2012; 7: e33754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Park SH, Kim EJ, Oh SA et al. Viral agents associated with acute gastroenteritis in Seoul, Korea. Clin Lab 2011; 57: 59–65. [PubMed] [Google Scholar]
- 9Kittigul L, Pombubpa K, Taweekate Y et al. Norovirus GII-4 2006b variant circulating in patients with acute gastroenteritis in Thailand during a 2006–2007 study. J Med Virol 2010; 82: 854–860. [DOI] [PubMed] [Google Scholar]
- 10Mans J, Murray TY, Kiulia NM et al. Human caliciviruses detected in HIV-sero-positive children in Kenya. J Med Virol 2014; 86: 75–81. [DOI] [PubMed] [Google Scholar]
- 11Wang YH, Zhou DJ, Zhou X et al. Molecular epidemiology of noroviruses in children and adults with acute gastroenteritis in Wuhan, China, 2007–2010. Arch Virol 2012; 157: 2417–2424. [DOI] [PubMed] [Google Scholar]
- 12Kiulia NM, Mans J, Mwenda JM, Taylor MB. Norovirus GII.17 predominates in selected surface water sources in Kenya. Food Environ Virol 2014; 6: 221–231. [DOI] [PubMed] [Google Scholar]
- 13Lee BR, Lee SG, Park JH et al. Norovirus contamination levels in ground water treatment systems used for food-catering facilities in South Korea. Viruses 2013; 5: 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Lu J, Sun L, Fang L et al. Gastroenteritis outbreaks caused by norovirus GII.17, Guangdong Province, China, 2014–2015. Emerg Infect Dis 2015; 21: 1240–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Fu J, Ai J, Jin M et al. Emergence of a new GII.17 norovirus variant in patients with acute gastroenteritis in Jiangsu, China, September 2014 to March 2015. Euro Surveill 2015; 20: 21157. [DOI] [PubMed] [Google Scholar]
- 16Taipei Times. CDC probing strain of Taichung norovirus. Taipei: The Taipei Times, 2015. Available at http://www.taipeitimes.com/News/taiwan/archives/2015/02/25/2003612208. [Google Scholar]
- 17Matsushima Y, Ishikawa M, Shimizu T et al. Genetic analyses of GII.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan reveal a novel polymerase sequence and amino acid substitutions in the capsid region. Euro Surveill 2015; 20: 21173. [DOI] [PubMed] [Google Scholar]
- 18de Graaf M, van Beek J, Vennema H et al. Emergence of a novel GII.17 norovirus – End of the GII.4 era? Euro Surveill 2015; 20: 21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Hansman GS, Biertumpfel C, Georgiev I et al. Crystal structures of G II.10 and G II.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J Virol 2011; 85: 6687–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Kojima S, Kageyama T, Fukushi S et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods 2002; 100: 107–114. [DOI] [PubMed] [Google Scholar]
- 21Shinohara M, Kageyama T. Rapid and efficient detection method of Norwalk virus. Nihon Rinsho 2002; 60: 1181–1187. [PubMed] [Google Scholar]
- 22Kroneman A, Vennema H, Deforche K et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol 2011; 51: 121–125. [DOI] [PubMed] [Google Scholar]
- 23Parra GI, Green KY. Genome of emerging norovirus G II.17, United States, 2014. Emerg Infect Dis 2015; 21: 1477–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol 2010; 8: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Shen Z, Qian F, Li Y et al. Novel Norovirus GII.4 Variant, Shanghai, China, 2012. Emerg Infect Dis 2013; 19: 1337–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Yu Y, Yan S, Li B, Pan Y, Wang Y. Genetic diversity and distribution of human norovirus in China (1999–2011). Biomed Res Int 2014; 2014: 196169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.