Dear Editor,
Among the clinically significant carbapenemases, the New Delhi metallo-β-lactamase (NDM) is one of the most formidable. NDM efficiently hydrolyses β-lactams and last-resort carbapenems. Hence, therapeutic options for infections by NDM-producers are restricted to a handful of antibiotics, such as colistin, tigecycline, and fosfomycin.1 NDM is predominantly associated with Enterobacteriaceae. This carbapenemase has also been described in Acinetobacter spp. but has been much less frequently detected in Pseudomonas aeruginosa.1
The first report of NDM-1 production in P. aeruginosa came from Serbia in 2011.2 It is now acknowledged that the NDM-1 gene is endemic to the Balkan states.3,4 NDM-producing P. aeruginosa has since been isolated from other European countries,4,5,6,7 as well as in India8 and Egypt.9 Local hospital laboratory surveillance suggests that approximately 12% of P. aeruginosa isolates are not susceptible to carbapenems.10 Previously, only producers of the metallo-β-lactamases (MBLs) VIM and IMP had been sporadically detected.11 Here, we describe the first observed case of NDM-1-producing P. aeruginosa in Southeast Asia.
P. aeruginosa was cultured from the endotracheal aspirate of a 90-year-old female patient with colon cancer in March 2015. The isolate exhibited multidrug resistance to carbapenems (meropenem, imipenem, minimum inhibitory concentrations (MICs) > 32 mg/L), cephalosporin (ceftazidime, cefepime, MICs > 256 mg/L), aminoglycosides (gentamicin, amikacin, MICs > 256 mg/L), and fluoroquinolones (ciprofloxacin, levofloxacin, MICs > 32 mg/L). The isolate was partially resistant to aztreonam (MIC 16 mg/L). Colistin susceptibility was observed at an MIC of 1 mg/L. Phenotypic testing for carbapenemases using the KPC/MBL Confirm Kit (Rosco Diagnostica A/S, Taastrup, Denmark) indicated the presence of an MBL.
Comprehensive polymerase chain reaction (PCR) screening for β-lactamase genes was performed.12 The isolate was positive for blaNDM and determined to be blaNDM-1 by full-length gene sequencing. The isolate was negative for other MBLs (IMP, VIM, SPM, DIM, AIM) and for genes encoding class A carbapenemases (KPC, GES). TEM and CTX-M extended spectrum β-lactamases (ESBLs) were found to be present.
Plasmid analysis using S1 nuclease pulsed-field gel electrophoresis13 and spin column plasmid extractions (QIAprep Spin Miniprep Kit, QIAGEN, Valencia, CA, USA) did not the reveal the presence of plasmids. Southern blot analysis with a blaNDM-1 probe of S1 nuclease-treated DNA agarose plugs indicated that the probe hybridized to high molecular weight chromosomal DNA, suggesting that blaNDM-1 was situated on the chromosome (data not shown). Furthermore, solid media conjugation assays were performed to assess the transferability of blaNDM-1 from the clinical isolate to the azide-resistant recipient Escherichia coli J53. No transconjugants were obtained, suggesting the non-transmissibility of blaNDM-1 (at least to E. coli), again suggesting a chromosomal position. Because the isolate was highly resistant to most antibiotics, including rifampicin, this excluded the use of the rifampicin-resistant laboratory strain of P. aeruginosa for conjugation. Hence, we were unable to assess the intra-species transmissibility of blaNDM-1. P. aeruginosa blaNDM-1 may be present chromosomally2,5,6 or on a plasmid; in the latter context, blaNDM-1 is transmissible.8
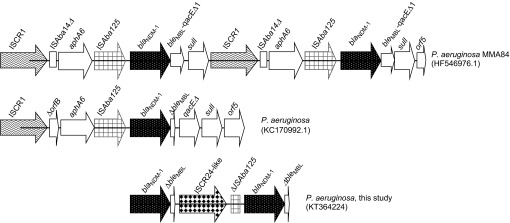
The nucleotide sequences immediately flanking blaNDM-1 were determined by an inverse PCR and primer walking approach. Two copies of blaNDM-1 were detected and separated by an Insertion Sequence Common Region (ISCR) element (Figure 1). Sequencing of this ISCR element revealed a 97% nucleotide homology to ISCR24. ISCR24 has been identified in the genetic environment of a novel blaPME-1 (Pseudomonas aeruginosa ESBL 1) and implicated in the acquisition of the ESBL by P. aeruginosa.15 This is not surprising because ISCR elements mediate the mobilization of almost every class of antibiotic resistance genes, including those encoding ESBLs and carbapenemases.16 ISCR elements, such as ISCR1, have been found to be associated with NDM-1 from P. aeruginosa.3,5,7 Jovcic et al.3 reported two copies of blaNDM-1 in P. aeruginosa (Figure 1), where it is presumed that the ISCR1 element, as part of its rolling-circling mechanism of transposition, duplicates adjacent genetic segments.16 Because ISCR elements are known to construct extended clusters of antibiotic resistance genes on plasmids as well as on chromosomes,16 it would be interesting to investigate the presence of other resistance determinants surrounding blaNDM-1 and the ISCR24-like region that may contribute to its multidrug resistance.
Figure 1.
Comparative schematic diagrams of the diverse genetic organizations of blaNDM-1 in P. aeruginosa. The selected blaNDM-1 sequences are chromosomally located and associated with ISCR elements and class I integrons.3,5 Typical common genetic features surrounding blaNDM-1 are the insertion sequence ISAba125 and the bleomycin resistance gene (bleMBL).14
After this isolate was identified, two other NDM-1-producing P. aeruginosa isolates with antibiograms identical to the initial isolate were cultured from the sputum samples of two other patients in April and May 2015. One of the isolates was from a 58-year-old male with intracranial bleeding from a ruptured aneurysm, and the other was from an 84-year-old female with a femur fracture, complicated by pancreatitis and small intestine perforation. The genetic relatedness of the three NDM-1 P. aeruginosa isolates was investigated by DiversiLab rep-PCR fingerprinting (bioMérieux, Marcy l'Etoile, France), which revealed that the rep-PCR profiles were indistinguishable, suggesting that the three isolates were clonal in nature. PCR mapping and sequencing of the two latter isolates reveal a blaNDM genetic context identical to that of the first isolate. Because all three patients stayed in the same surgical intensive care unit, the detection of indistinguishable NDM-1-positive P. aeruginosa suggested a transmission event.
In summary, this is the first report of the emergence of NDM-1 in P. aeruginosa in Southeast Asia in an unusual genetic context. The apparent intra-ward transmission of this extremely drug-resistant isolate highlights the gravity of this escalating public health issue.
Nucleotide sequence accession number
The blaNDM-1 sequence from the initial P. aeruginosa isolate has been deposited into Genbank under the accession number KT364224.
Acknowledgments
This work was supported by a National University Health Engineering Medicine Seed Grant (#N171-000-099-001) provided to Jeanette WP Teo.
References
- 1Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014; 2014: 249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Jovcić B, Lepsanovic Z, Suljagic V et al. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother 2011; 55: 3929–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Jovcić B, Lepsanović Z, Begović J et al. The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob Agents Chemother 2013; 57: 3405–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Kulkova N, Babalova M, Sokolova J, Krcmery V. First report of New Delhi metallo-β-lactamase-1-producing strains in Slovakia. Microb Drug Resist 2015; 21: 117–120. [DOI] [PubMed] [Google Scholar]
- 5Janvier F, Jeannot K, Tessé S et al. Molecular characterization of blaNDM-1 in a sequence type 235 Pseudomonas aeruginosa isolate from France. Antimicrob Agents Chemother 2013; 57: 3408–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Carattoli A, Fortini D, Galetti R et al. Isolation of NDM-1-producing Pseudomonas aeruginosa sequence type ST235 from a stem cell transplant patient in Italy, May 2013. Euro Surveill 2013; 14: 20633. [DOI] [PubMed] [Google Scholar]
- 7Wang M, Borris L, Aarestrup FM, Hasman H. Identification of a Pseudomonas aeruginosa co-producing NDM-1, VIM-5 and VIM-6 metallo-β-lactamases in Denmark using whole-genome sequencing. Int J Antimicrob Agents 2015; 45: 324–325. [DOI] [PubMed] [Google Scholar]
- 8Khajuria A, Praharaj AK, Kumar M, Grover N. Emergence of NDM - 1 in the clinical isolates of Pseudomonas aeruginosa in India. J Clin Diagn Res 2013; 7: 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Zafer MM, Amin M, El Mahallawy H, Ashour MS, Al Agamy M. First report of NDM-1-producing Pseudomonas aeruginosa in Egypt. Int J Infect Dis 2014; 29: 80–81. [DOI] [PubMed] [Google Scholar]
- 10Tan TY, Hsu LY, Koh TH et al. Antibiotic resistance in gram-negative bacilli: a Singapore perspective. Ann Acad Med Singapore 2008; 37: 819–825. [PubMed] [Google Scholar]
- 11Koh TH, Khoo CT, Tan TT et al. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-beta-lactamase genes, including the novel bla(IMP-26) gene. J Clin Microbiol 2010; 48: 2563–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J 2012; 3: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem 1995; 10: 235–240. [DOI] [PubMed] [Google Scholar]
- 14Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetics features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 2011; 55: 5403–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Tian GB, Adams-Haduch JM, Bogdanovich T, Wang HN, Doi Y. PME-1, an extended-spectrum β-lactamase identified in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2011; 55: 2710–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 2006; 70: 296–316. [DOI] [PMC free article] [PubMed] [Google Scholar]