Abstract
Recognition and correct interpretation of facial emotion is essential for social interaction and communication. Previous studies have shown that impairments in this cognitive domain are common features of several psychiatric disorders. Recent association studies identified CACNA1C as one of the most promising genetic risk factors for psychiatric disorders and previous evidence suggests that the most replicated risk variant in CACNA1C (rs1006737) is affecting emotion recognition and processing. However, studies investigating the influence of rs1006737 on this intermediate phenotype in healthy subjects at the behavioral level are largely missing to date. Here, we applied the “Reading the Mind in the Eyes” test, a facial emotion recognition paradigm in a cohort of 92 healthy individuals to address this question. Whereas accuracy was not affected by genotype, CACNA1C rs1006737 risk-allele carries (AA/AG) showed significantly slower mean response times compared to individuals homozygous for the G-allele, indicating that healthy risk-allele carriers require more information to correctly identify a facial emotion. Our study is the first to provide evidence for an impairing behavioral effect of the CACNA1C risk variant rs1006737 on facial emotion recognition in healthy individuals and adds to the growing number of studies pointing towards CACNA1C as affecting intermediate phenotypes of psychiatric disorders.
The CACNA1C gene is one of the most promising candidate genes for psychiatric disorders. CACNA1C encodes the alpha 1C subunit of the L-type voltage-gated calcium channel and is primarily expressed in the cardiovascular system but also the brain1. L-type voltage-gated calcium channels play an important role in the nervous system as they assimilate the electrical activity of neurons and muscle cells in ordered to regulate physiologically relevant processes from coupling excitation to contraction in muscle cells to modulating synaptic transmission and regulating gene expression in neurons. Single nucleotide polymorphisms (SNPs) in CACNA1C have repeatedly been identified as risk variants for bipolar disorder, schizophrenia, and major depressive disorder in genome-wide association studies2,3,4,5,6,7. Furthermore, missense mutations in CACNA1C are causative for the Timothy syndrome, a multiorgan dysfunction accompanied by behavioral abnormalities including autistic traits8.
Impairments in emotion recognition and processing are common features of several psychiatric disorders e.g. autism spectrum disorders, bipolar disorder, schizophrenia, and major depression9,10,11,12,13,14,15,16. Several studies demonstrated an association between the most replicated CACNA1C risk variant rs1006737 and emotion recognition and processing, suggesting an influence of CACNA1C not only on the risk for psychiatric disorders themselves, but also on intermediate phenotypes underlying them. Previous findings include modified activation of several brain regions involved in emotion processing and memory formation (hippocampus, inferior occipital gyrus, fusiform gyrus, right ventrolateral prefrontal cortex, and amygdala)17,18,19,20, and decreased brain connectivity21. On a behavioral level, the results are more inconsistent. Two studies found delayed responses during facial emotion recognition and negative face matching in bipolar and schizophrenia patients compared to healthy control individuals18,19 but did not discover an effect of rs1006737 genotype18,19. A third study reported contradictory results. Here, an influence of CACNA1C rs1006737 genotype on facial emotion recognition was discovered in bipolar patients. In healthy individuals, no such effect was found22. Schizophrenia patients were not included in the study and the limited sample size (39 patients and 40 control individuals) might have been too small to allow the detection of effects in healthy subjects.
These previous behavioral results confirm impairments in emotion recognition and processing as an intermediate phenotype of bipolar disorder and schizophrenia, but given the limited number of studies conducted thus far, the contribution of CACNA1C rs1006737 to emotion recognition and processing still needs to be determined.
Therefore, we designed the present study to clarify whether an effect of CACNA1C on emotion recognition can also be detected at the behavioral level in healthy individuals. We hypothesized that carrier of the CACNA1C rs1006737 risk variant will show an impaired performance in the revised version of the “reading the mind in the eyes” task (RMET)23. We have chosen the RMET as it is a sensitive test which is able to reveal subtle difficulties in emotional recognition and is therefore suitable to study healthy individuals.
Results
Genotypes
Genotype frequency was as follows: 52 individuals where risk-allele carrier (5 AA and, 47 AG) and 40 individuals were homozygous for the G allele. Both genotype groups (AA/AG vs. GG individuals) did not differ significantly with respect to age (AA/AG: 25.808 ± 7.257, GG: 26.525 ± 5.487, p = 0.604), gender ratio (AA/AG: 51.923% male, GG: 42.5% male; p = 0.375), and education (100% secondary school education).
Reading the mind in the eyes task (RMET)
CACNA1C genotype was significantly associated with performance in the RMET
[F(2,89) = 4.224, p = 0.018, partial η2 = 0.087]. Subsequent pairwise comparisons revealed significant longer mean reaction times in risk-allele carriers compared to non-risk allele homozygous individuals [F(1,90) = 8.129, p = 0.005, partial η2 = 0.083] (Fig. 1). To investigate whether the reaction time depends upon the risk-allele dose, we additionally examined the three genotype groups independently. Due to the small group size for risk-allele homozygous individuals (n = 5), the data are presented descriptively. A gene dose effect could not be observed (GG homozygous individuals [n = 40]: 6263.912 ms ± 225.884; GA heterozygous individuals [n = 47]: 7457.046 ms ± 319.317; AA homozygous individuals [n = 5]: 6965.178 ms ± 1172.396), most likely due to the small number of AA-homozygote individuals. The CACNA1C genotype had no effect on the number of errors [F(1,90) = 0.023, p = 0.880] (Fig. 2).
Figure 1.
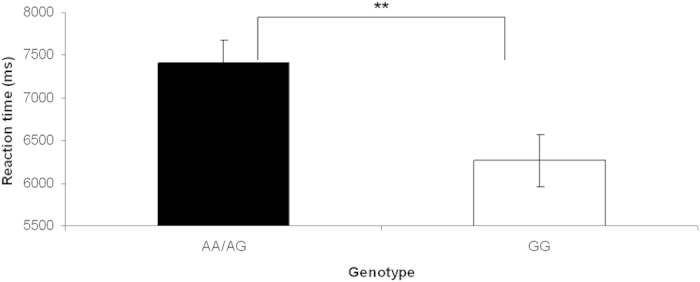
Effects of CACNA1C genotype on reaction time. Mean reaction time (in ms) is shown for the two genotype groups. Risk-allele carriers (AA/AG) have significantly longer mean reaction times compared to non-risk allele homozygous individuals.
Figure 2. Effects of CACNA1C genotype on the number of errors.
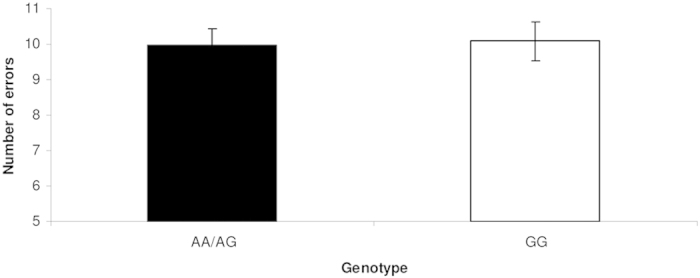
The number of errors is shown for the two genotype groups. CACNA1C genotype had no effect on the number of errors. **p ≤ 0.01, error bars represent standard error of the mean.
Discussion
We investigated the influence of CACNA1C, a strong candidate gene for psychiatric disorders, on facial emotion recognition in healthy individuals to test its effect at the level of this intermediate phenotype. We found that carriers of the CACNA1C rs1006737 risk genotype (AA/AG) show a prolonged reaction time in the RMET compared to individuals homozygous for the non-risk genotype (GG), whereas the accuracy was not significantly different in both groups. Our study thus provides new evidence for an involvement of the psychiatric risk gene CACNA1C in cognitive processes underlying facial emotion recognition in healthy individuals.
We have chosen a cohort of healthy individuals as the investigation of genetic effects on intermediate phenotypes in non-affected individuals is a useful approach since it is not limited by medication and other confounding factors e.g. duration and severity of the illness. Facial emotion recognition can be viewed as a valid intermediate phenotype especially for bipolar disorder as previous studies showed that deficits in facial emotion recognition are present independent from mood states and are also present in healthy first degree relatives24, suggesting that these deficits are heritable. Our data contribute to that notion, as we were able to demonstrate, that healthy carriers of a genetic variant associated with psychiatric disorders are also impaired in their facial emotion recognition abilities.
The RMET was applied as a sensitive test suitable to investigate inter-individual differences of facial emotion recognition in healthy subjects. Deficits in this task have been observed in bipolar disorder, schizophrenia, major depression, and autism23,25,26,27. Furthermore, impaired performance in the RMET predicts higher scores on measures of autistic traits in clinical samples as well as healthy control individuals23,28.
In this study, we only investigated the effects of the most replicated and best described variant in CACNA1C (rs1006737) without considering the contribution of other polymorphisms in CACNA1C or additional genes potentially interacting with CACNA1C. Rs1006737 is located in an intron and is therefore not causing an amino acid substitution in the encoded protein. Nevertheless, rs1006737 potentially has a functional impact, as a proxy variant for rs1006737 (rs2159100) is associated with increased gene expression in the dorsolateral prefrontal cortex17. We cannot establish definitively whether the effect we observed in our study can be attributed to rs1006737, the proxy variant rs2159100, or any other genetic variant in linkage disequilibrium to rs1006737.
A further limitation of our study is that we cannot distinguish whether the observed prolonged reaction time in the RMET is based on an impairment specific for emotional recognition or rather reflects a more general underlying deficit in complex signal detection and processing as we did not check for slower reaction times of risk-allele carriers in other neuropsychological tasks. Healthy carriers of the risk genotype were already found to be impaired in alerting and orienting29, in verbal fluency30, and in working memory31. However, other studies contradict these findings22,32 or did not find impairments of healthy CACNA1C risk genotype carriers in other aspects of cognition for instance in attention32, verbal learning and memory32,33,34,35,36 or logical memory37.
We have decided to take the mean reaction time for all trials including errors, to give a valid estimate of the processing time a subject requires to identify and react to a facial emotion. Delays in this cognitive domain can severely impair social interactions and competence and are associated with core symptomatology especially in schizophrenia38. For instance, it has been shown that schizophrenia patients require more visual information, and therefore more time to correctly identify emotional expression in faces, compared to controls39,40. Therefore, differences in emotion recognition between the risk-allele carrier and the other subjects may be more prominent in speed of performance than in accuracy of the RMET41. Since we did not detect differences in accuracy, our data most likely also reflect differences in the amount of information needed to correctly identify a facial emotion. In healthy individuals, this characteristic is not associated with obvious impairments. However, in complex and challenging situations, this subtle impairment might significantly exert negative effects on social interactions, and in psychiatric patients it might add to further deficits and contribute to psychosocial dysfunction.
The effect of CANA1C on facial emotion recognition is most likely not specific, as deficits in a variety of other intermediate phenotypes have been reported in CANA1C risk-allele carriers (reviewed in42,43). However, our data provide clear support that CACNA1C has a subtle but functionally relevant influence on facial emotion recognition as an intermediate phenotype involved in psychiatric disorders.
In conclusion, the present study is the first to provide evidence for an impairing effect of the CACNA1C risk genotype rs1006737 AA/AG on facial emotion recognition in healthy individuals. Our finding that risk-allele carriers are subtly challenged in facial emotion recognition adds to the growing body of knowledge on the role of CACNA1C in cognitive functions affected in various psychiatric disorders. However, future studies are needed to replicate our finding in independent cohorts as well as to investigate the specificity of the effect.
Material and Methods
Subjects
Ninety-two healthy volunteers (44 males, 48 females; mean age = 26.12, SD = 6.523) participated in this study and gave written informed consent to the experimental procedure approved by the University of Tübingen local ethics committee. All experiments were carried out in accordance with the approved guidelines. None of the subjects had a history of physical, mental, or neurological illness and none of the subjects had performed the “Reading the mind in the eyes” (REM) task before.
Reading the mind in the eyes task (RMET)
The RMET was used to assess emotional recognition. The task was conducted as previously described23. In brief, 36 black-and-white photographs (15 cm × 6 cm) of the area of the face including and immediately surrounding the eyes were presented together with a word correctly characterizing the emotional state of the person in the photograph as well as three distracter words of the same emotional valence. Participants were asked to choose the most suitable adjective and to quickly click it on the screen. The next picture appears immediately after the response. The number of incorrect choices was measured as number of errors, the time interval between presentation of the photograph and the click on the word was recorded as reaction time.
Genotyping
Genomic DNA was extracted from ethylenediaminetetraacetic acid (EDTA) anti-coagulated venous blood according to standard protocols. CACNA1C rs1006737 was genotyped on a StepOne system (life technologies; Darmstadt; Germany) using TaqMan®SNP Genotyping Assay C___2584015_10 (life technologies; Darmstadt; Germany) and the standard protocol for allelic discrimination. Accuracy was assessed by duplicating 15% of the original sample, and reproducibility was 100%. The genotype frequencies did not deviate from Hardy–Weinberg equilibrium (HWE; p = 0.06).
Statistical analysis
Number of errors and mean reaction time were treated as outcome variables. The data were analysed with SPSS (IBM SPSS Statistics 22.0; Ehningen; Germany). The effect of rs1006737 on the outcome variables was evaluated by applying a multivariate analysis of variance (MANOVA) with the between factor genotypeA allele carrier vs. G homozygout. Results were considered significant when p ≤ 0.05. For significant interaction F-values, post hoc pairwise comparisons were performed.
Additional Information
How to cite this article: Nieratschker, V. et al. CACNA1C risk variant affects facial emotion recognition in healthy individuals. Sci. Rep. 5, 17349; doi: 10.1038/srep17349 (2015).
Acknowledgments
We thank Danuta Altpaß for excellent technical assistance and Dr. Daniel Bucher for critical reading of the manuscript. C.P. is supported by the German Federal Ministry of Education and Research (research consortia ESPRIT / FKZ 01EE1407H and GCBS / FKZ 01EE1403D). The funding source had no role in study design, the collection, analysis and interpretation of data, the writing of the manuscript and the decision to submit the article for publication. The authors declare no conflict of interest.
Footnotes
Author Contributions V.N. and C.P. designed the study. C. B. performed the laboratory analysis. V.N. analyzed the data and wrote the first draft of the manuscript. All authors participated in the preparation, modification and revision of the manuscript.
References
- Striessnig J. et al. Role of voltage-gated L-type Ca2+ channel isoforms for brain function. Biochemical Society transactions 34, 903–909, doi: 10.1042/BST0340903 (2006). [DOI] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study, C. Genome-wide association study identifies five new schizophrenia loci. Nature genetics 43, 969–976, doi: 10.1038/ng.940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric G. C. B. D. W. G. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics 43, 977–983, doi: 10.1038/ng.943 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M. A. et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nature genetics 40, 1056–1058, doi: 10.1038/ng.209 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379, doi: 10.1016/S0140-6736(12)62129-1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S. et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature genetics 45, 1150–1159, doi: 10.1038/ng.2742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P. et al. Whole-genome association study of bipolar disorder. Molecular psychiatry 13, 558–569, doi: 10.1038/sj.mp.4002151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I. et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 119, 19–31, doi: 10.1016/j.cell.2004.09.011 (2004). [DOI] [PubMed] [Google Scholar]
- Ashwin C., Baron-Cohen S., Wheelwright S., O’Riordan M. & Bullmore E. T. Differential activation of the amygdala and the ‘social brain’ during fearful face-processing in Asperger Syndrome. Neuropsychologia 45, 2–14, doi: 10.1016/j.neuropsychologia.2006.04.014 (2007). [DOI] [PubMed] [Google Scholar]
- Deeley Q. et al. An event related functional magnetic resonance imaging study of facial emotion processing in Asperger syndrome. Biological psychiatry 62, 207–217, doi: 10.1016/j.biopsych.2006.09.037 (2007). [DOI] [PubMed] [Google Scholar]
- Wegbreit E. et al. Developmental meta-analyses of the functional neural correlates of bipolar disorder. JAMA psychiatry 71, 926–935, doi: 10.1001/jamapsychiatry.2014.660 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miu A. C., Pana S. E. & Avram J. Emotional face processing in neurotypicals with autistic traits: implications for the broad autism phenotype. Psychiatry research 198, 489–494, doi: 10.1016/j.psychres.2012.01.024 (2012). [DOI] [PubMed] [Google Scholar]
- Wessa M. & Linke J. Emotional processing in bipolar disorder: behavioural and neuroimaging findings. International review of psychiatry 21, 357–367, doi: 10.1080/09540260902962156 (2009). [DOI] [PubMed] [Google Scholar]
- Morris R. W., Weickert C. S. & Loughland C. M. Emotional face processing in schizophrenia. Current opinion in psychiatry 22, 140–146, doi: 10.1097/YCO.0b013e328324f895 (2009). [DOI] [PubMed] [Google Scholar]
- Kohler C. G., Walker J. B., Martin E. A., Healey K. M. & Moberg P. J. Facial emotion perception in schizophrenia: a meta-analytic review. Schizophrenia bulletin 36, 1009–1019, doi: 10.1093/schbul/sbn192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke C., Douglas K. & Porter R. Processing of facial emotion expression in major depression: a review. The Australian and New Zealand journal of psychiatry 44, 681–696, doi: 10.3109/00048674.2010.496359 (2010). [DOI] [PubMed] [Google Scholar]
- Bigos K. L. et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Archives of general psychiatry 67, 939–945, doi: 10.1001/archgenpsychiatry.2010.96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima D. et al. Independent modulation of engagement and connectivity of the facial network during affect processing by CACNA1C and ANK3 risk genes for bipolar disorder. JAMA psychiatry 70, 1303–1311, doi: 10.1001/jamapsychiatry.2013.2099 (2013). [DOI] [PubMed] [Google Scholar]
- Tesli M. et al. CACNA1C risk variant and amygdala activity in bipolar disorder, schizophrenia and healthy controls. PloS one 8, e56970, doi: 10.1371/journal.pone.0056970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogia J. et al. The impact of the CACNA1C gene polymorphism on frontolimbic function in bipolar disorder. Molecular psychiatry 16, 1070–1071, doi: 10.1038/mp.2011.49 (2011). [DOI] [PubMed] [Google Scholar]
- Radua J. et al. The impact of CACNA1C allelic variation on effective connectivity during emotional processing in bipolar disorder. Molecular psychiatry 18, 526–527, doi: 10.1038/mp.2012.61 (2013). [DOI] [PubMed] [Google Scholar]
- Soeiro-de-Souza M. G. et al. The impact of the CACNA1C risk allele on limbic structures and facial emotions recognition in bipolar disorder subjects and healthy controls. Journal of affective disorders 141, 94–101, doi: 10.1016/j.jad.2012.03.014 (2012). [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S., Wheelwright S., Hill J., Raste Y. & Plumb I. The “Reading the Mind in the Eyes” Test revised version: a study with normal adults, and adults with Asperger syndrome or high-functioning autism. Journal of child psychology and psychiatry, and allied disciplines 42, 241–251 (2001). [PubMed] [Google Scholar]
- Seidel E. M. et al. Risk or resilience? Empathic abilities in patients with bipolar disorders and their first-degree relatives. Journal of psychiatric research 46, 382–388, doi: 10.1016/j.jpsychires.2011.11.006 (2012). [DOI] [PubMed] [Google Scholar]
- Bora E. et al. Evidence for theory of mind deficits in euthymic patients with bipolar disorder. Acta psychiatrica Scandinavica 112, 110–116, doi: 10.1111/j.1600-0447.2005.00570.x (2005). [DOI] [PubMed] [Google Scholar]
- Lee L., Harkness K. L., Sabbagh M. A. & Jacobson J. A. Mental state decoding abilities in clinical depression. Journal of affective disorders 86, 247–258, doi: 10.1016/j.jad.2005.02.007 (2005). [DOI] [PubMed] [Google Scholar]
- Craig J. S., Hatton C., Craig F. B. & Bentall R. P. Persecutory beliefs, attributions and theory of mind: comparison of patients with paranoid delusions, Asperger’s syndrome and healthy controls. Schizophrenia research 69, 29–33, doi: 10.1016/S0920-9964(03)00154-3 (2004). [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S., Wheelwright S., Skinner R., Martin J. & Clubley E. The autism-spectrum quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. Journal of autism and developmental disorders 31, 5–17 (2001). [DOI] [PubMed] [Google Scholar]
- Thimm M. et al. Effects of a CACNA1C genotype on attention networks in healthy individuals. Psychological medicine 41, 1551–1561, doi: 10.1017/S0033291710002217 (2011). [DOI] [PubMed] [Google Scholar]
- Krug A. et al. Effect of CACNA1C rs1006737 on neural correlates of verbal fluency in healthy individuals. NeuroImage 49, 1831–1836, doi: 10.1016/j.neuroimage.2009.09.028 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. The effects of CACNA1C gene polymorphism on spatial working memory in both healthy controls and patients with schizophrenia or bipolar disorder. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology 37, 677–684, doi: 10.1038/npp.2011.242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts B., Simons C. J. & Os J. Evidence for the impact of the CACNA1C risk allele rs1006737 on 2-year cognitive functioning in bipolar disorder. Psychiatric genetics 23, 41–42, doi: 10.1097/YPG.0b013e328358641c (2013). [DOI] [PubMed] [Google Scholar]
- Roussos P., Giakoumaki S. G., Georgakopoulos A., Robakis N. K. & Bitsios P. The CACNA1C and ANK3 risk alleles impact on affective personality traits and startle reactivity but not on cognition or gating in healthy males. Bipolar disorders 13, 250–259, doi: 10.1111/j.1399-5618.2011.00924.x (2011). [DOI] [PubMed] [Google Scholar]
- Erk S. et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Archives of general psychiatry 67, 803–811, doi: 10.1001/archgenpsychiatry.2010.94 (2010). [DOI] [PubMed] [Google Scholar]
- Dietsche B. et al. The impact of a CACNA1C gene polymorphism on learning and hippocampal formation in healthy individuals: a diffusion tensor imaging study. NeuroImage 89, 256–261, doi: 10.1016/j.neuroimage.2013.11.030 (2014). [DOI] [PubMed] [Google Scholar]
- Erk S. et al. Hippocampal and frontolimbic function as intermediate phenotype for psychosis: evidence from healthy relatives and a common risk variant in CACNA1C. Biological psychiatry 76, 466–475, doi: 10.1016/j.biopsych.2013.11.025 (2014). [DOI] [PubMed] [Google Scholar]
- Hori H. et al. Effects of the CACNA1C risk allele on neurocognition in patients with schizophrenia and healthy individuals. Scientific reports 2, 634, doi: 10.1038/srep00634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C. G., Bilker W., Hagendoorn M., Gur R. E. & Gur R. C. Emotion recognition deficit in schizophrenia: association with symptomatology and cognition. Biological psychiatry 48, 127–136 (2000). [DOI] [PubMed] [Google Scholar]
- Clark C. M., Gosselin F. & Goghari V. M. Aberrant patterns of visual facial information usage in schizophrenia. Journal of abnormal psychology 122, 513–519, doi: 10.1037/a0031944 (2013). [DOI] [PubMed] [Google Scholar]
- Lee J., Gosselin F., Wynn J. K. & Green M. F. How do schizophrenia patients use visual information to decode facial emotion? Schizophrenia bulletin 37, 1001–1008, doi: 10.1093/schbul/sbq006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhof E., de Sonneville L. M., Meijer C. J. & de Haan L. Processing of facial and nonsocial information is differentially associated with severity of symptoms in patients with multiepisode schizophrenia. The Journal of nervous and mental disease 203, 112–119, doi: 10.1097/NMD.0000000000000246 (2015). [DOI] [PubMed] [Google Scholar]
- Bhat S. et al. CACNA1C (Cav1.2) in the pathophysiology of psychiatric disease. Progress in neurobiology 99, 1–14, doi: 10.1016/j.pneurobio.2012.06.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X. et al. CACNA1C rs1006737 genotype and bipolar disorder: Focus on intermediate phenotypes and cardiovascular comorbidity. Neuroscience and biobehavioral reviews 55, 198–210, doi: 10.1016/j.neubiorev.2015.04.022 (2015). [DOI] [PubMed] [Google Scholar]


