Abstract
Molecular epidemiological research suggests that interleukin-10 (IL-10) polymorphisms may be associated with an increased risk of head and neck cancer (HNC), but results remain controversial. To derive a more precise evaluation, we performed a meta-analysis focused on genetic polymorphisms of IL-10. PubMed, Embase, CNKI and Wanfang databases were searched for studies that examined the relationship between IL-10 polymorphisms or haplotypes and HNC risk. The odds ratio (OR) and 95% confidence interval (CI) were applied to assess the relationship strength. Publication bias, sensitivity and cumulative analyses were conducted to measure the robustness of our findings. Overall, nine related studies involving 2,258 patients and 2,887 control samples were analyzed. Significant associations between the IL-10-1082A > G polymorphism and HNC risk were observed (G vs. A: OR = 1.56, 95% CI = 1.27–1.92, P < 0.01, I2 = 69.4%; AG vs. AA: OR = 1.64, 95% CI = 1.32–2.05, P < 0.01, I2 = 55.6%; GG vs. AA: OR = 2.24, 95% CI = 1.69–2.97, P < 0.01, I2 = 38.5%; AG + GG vs. AA: OR = 1.70, 95% CI = 1.36−2.14, P = 0.02, I2 = 61.8%; GG vs. AA + AG: OR = 1.89, 95% CI = 1.23−2.90, P = 0.01, I2 = 46.3%) in the total population, as well as in subgroup analysis. Moreover, increased HNC risks were also associated with the IL-10 −819T > C polymorphism and the GCC haplotype. In conclusion, our meta-analyses suggest that IL-10 polymorphisms, specifically the −1082A > G polymorphism, may be associated with increased risk of HNC development.
Head and neck cancer (HNC) is one of the most common malignancies and derives from the malignant transformation of the epithelial cells that line the upper respiratory tract and digestive system1,2. HNC accounts for a large proportion of cancer-related deaths and constitutes approximately 3.31% of all cancers in the United States, with 55,070 new cases and 12,000 deaths in 20143. Significant improvement in the 5-year disease-specific survival (DSS) was achieved due to recent advances in treatment, from 55% in 1992–1996 to 66% in 2002–20064. The development of prophylactic strategies remains critical for managing HNC. For example, many studies have demonstrated that HNC is associated with complex environmental factors, such as tobacco use, alcohol consumption, and intake of vitamins and minerals. In addition, infection with human papillomavirus (HPV) and dental trauma are also thought to be potential HNC risk factors5,6,7,8,9,10.
Interestingly, these factors do not explain the variation in susceptibility observed within different populations. Over the past decade, many studies have suggested that leukocytes and their relevant cytokines may play a central role in inflammatory infiltration and malignant transformation11,12,13. Interleukin-10 (IL-10) is a cytokine produced by monocytes, macrophages, lymphocytes and other human Thl cells14, that plays a functional role in inflammatory and immunoregulatory activities15. During carcinogenesis, IL-10 functions in both immune suppression (cancer promotion) and anti-angiogenesis (cancer inhibition)16.
IL-10 is an important inflammatory cytokine with anti-inflammatory properties. The IL-10 gene is located on chromosome 1 at q31-32, contains five exons and four introns that span a length of 4.8 kb and encode a 178-amino-acid protein17,18,19. Mutations in IL-10 have been detected at several loci, and the association between IL-10 SNPs and disease risk has been heavily studied. The three most common IL-10 SNPs are located in the promoter region (−1082A > G (rs1800870), −819T > C (rs1800871) and −592A > C (rs1800872)) and have been reported to regulate IL-10 transcription and expression20,21,22. These mutations influence IL-10 gene transcription and translation, resulting in abnormal cell proliferation and cancer development23,24. Numerous molecular epidemiological studies have investigated the association betweenIL-10 gene polymorphisms and cancer risk, such as breast cancers25, lung cancer26, cervical cancer27, and digestive cancer28. In 2006, Pratesi et al.29 published the first study describing the negative association between three IL-10 promoter polymorphisms and the risk of nasopharyngeal cancer in Italian populations. However, the results were not consistent with subsequent studies. To date, no published genome-wide association studies (GWAS) have been performed to explore the precise association between IL-10 polymorphisms and HNC risk. Given the important role of IL-10 in the development of HNC cancer, we conducted a meta-analysis to assess the relationship between IL-10 polymorphisms (−1082A > G, −819T > C and −592A > C) and HNC risk.
Methods
This meta-analysis was designed according to the guidelines described in the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA Compliant) statement30.
Search strategy
The Pubmed, Embase CNKI and Wanfang databases were searched to identify studies that examined the association between HNC and IL-10 polymorphisms, using the following search terms: “head and neck cancer”, “oral cancer”, “nasopharyngeal cancer”, “pharynx cancer”, “larynx cancer”, “Interleukin-10”, “IL-10”, “polymorphism”, and “variant”.
Eligible criteria
All selected studies met the following criteria: 1) the research design was a case-control study; 2) the study focused on the association between the IL-10 polymorphisms and HNC susceptibility; 3) the study included sufficient genotype distribution data to calculate odds ratios (ORs) and 95% confidence intervals (CIs); 4) the study was published in either Chinese or English. The largest sample size or most recent publication was preferentially selected in cases of overlapping data or duplicate publications.
Data extraction
The following information was extracted from all qualified studies by two independent researchers (Niu and Cai): first author, publication year, country, racial descent (categorized as either Asian or Caucasian), source of controls, number of cases and controls with different genotypes, genotyping method, Hardy-Weinberg equilibrium (HWE) for controls, cancer location and study quality assessment.
Quality assessment
Two independent authors (Niu and Du) assessed the quality of the included studies using quality scoring criteria modified from previous meta-analyses (Supplemental Table S1 online, which demonstrated the scale for quality assessment.)31,32. Modified criteria were based on traditional quality scoring used for observational studies in genetic epidemiological issues and ranged from 0 points (worst) to 9 points (best). Studies with a score of 6 or higher were classified as high quality, whereas studies with a score of 6 or less were classified as low quality.
Statistical analysis
ORs with 95% CIs were used to assess the strength of the association between the IL-10-1082A > G, −819T > C and −592A > C polymorphisms and HNC risk. For the IL-10–1082A > G polymorphism, pooled ORs were obtained for allele contrast (G vs. A), co-dominant model (AG vs. AA; GG vs. AA), dominant model (GA + GG vs. AA), and recessive model (GG vs. AA + GA). These similar genetic models were also used to assess the IL-10 −819T > C and −592A > C polymorphisms. Subgroup analyses were performed based on cancer location, HWE status of controls, ethnicity, and study design. ORs were calculated using the random-effects model (DerSimonian and Laird method) when the P-value was less than 0.10 or I2 was greater than 40%33. Otherwise, a fixed-effect model (the Mantel-Haenszel method) was adopted34. Cumulative meta-analyses and sensitivity analysis were conducted to evaluate the overall robustness of the study’s results. Publication bias was analyzed using Egger’s linear regression and Begg’s funnel plots35. In addition, heterogeneity was assessed using Cochran’s Q statistic and the I2 method, and meta-regression was conducted to analyze heterogeneity36. Statistical analysis was performed using STATA version 11.0 (Stata Corporation, College Station, TX, USA) with a two-sided P-value, P < 0.05 was considered significant.
Results
Study characteristics
A systematic review of the literature identified 111 relevant studies. Figure 1 and PRISMA flow diagram (Supplemental Table S2 and S3 online) show a flow chart of the studies selection procedure. One hundred and two studies were excluded. Ultimately, nine studies29,37,38,39,40,41,42,43,44 satisfied the outlined inclusion criteria and characteristics (Table 1 and Supplemental Table S4 online), including the quality assessment score that was generated based on the Newcastle-Ottawa Scale of studies. Each of the nine studies focused on the association of the −1082A > G polymorphism with HNC risk29,37,38,39,40,41,42,43,44, and six studies investigated the association of the −819T > C and −592A > C variants with HNC risk29,37,39,42,43,44. Three studies focused on Caucasian populations29,38,40, and six studies focused on Asian populations37,39,41,42,43,44. Within the distribution of genotypes in the control groups, only three studies exhibited HWE in the −1082A > G polymorphism29,41,44, and only one study deviated from HWE in the −819T > C and −592A > Cvariants42.
Figure 1. Flow diagram of the study selection process.
Table 1. Characteristics of case-control studies on IL-10 −1082A > G and −819T > C and −592A > C polymorphisms and cancer risk included in the meta-analysis.
| Firstauthor | Year | Country | Racialdescent | Source of controls | Case | Control | Genotype distribution |
PforHWEa | Genotypingmethod | CancerLocation | Qualityscore | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case |
Control |
|||||||||||||||
| A/A | A/G | G/G | A/A | A/G | G/G | |||||||||||
| Pratesi | 2006 | Italy | Caucasian | Population-control | 89 | 130 | 29 | 41 | 19 | 46 | 58 | 26 | 0.33 | Direct sequencing | Nasopharyngeal | 7 |
| Wei | 2007 | China | Asian | Population-control | 198 | 210 | 123 | 61 | 14 | 167 | 38 | 5 | 0.01 | PCR-RFLP | Nasopharyngeal | 6 |
| Farhat | 2008 | Qatar | Caucasian | Population-control | 160 | 156 | 58 | 80 | 22 | 70 | 60 | 26 | 0.04 | AS-PCR | Nasopharyngeal | 6 |
| Yao | 2008 | China | Asian | Population-control | 280 | 300 | 184 | 75 | 21 | 234 | 56 | 10 | 0.01 | PCR–RFLP | Oral | 6 |
| Vairaktaris | 2008 | Greek/Germany | Caucasian | Population-control | 144 | 141 | 46 | 96 | 2 | 81 | 60 | 0 | <0.01 | PCR–RFLP | Oral | 6 |
| Jeong | 2010 | Korean | Asian | Hospital-control | 278 | 350 | 238 | 38 | 2 | 304 | 45 | 1 | 0.62 | TaqMan | HN | 6 |
| Tsai1 | 2013 | China | Asian | Hospital-control | 176 | 522 | 117 | 49 | 10 | 419 | 92 | 11 | 0.03 | PCR–RFLP | Nasopharyngeal | 6 |
| Tsai2 | 2014 | China | Asian | Hospital-control | 788 | 956 | 522 | 217 | 49 | 766 | 168 | 22 | <0.01 | PCR–RFLP | Oral | 6 |
| Hsu | 2015 | China | Asian | Population-control | 145 | 112 | 130 | 14 | 1 | 96 | 16 | 0 | 0.42 | PCR-SSP | Oral | 8 |
| T/T | T/C | C/C | T/T | T/C | C/C | |||||||||||
| Pratesi | 2006 | Italy | Caucasian | Population-control | 89 | 130 | 5 | 36 | 48 | 6 | 54 | 70 | 0.27 | Direct sequencing | Nasopharyngeal | 7 |
| Wei | 2007 | China | Asian | Population-control | 198 | 210 | 82 | 81 | 35 | 94 | 92 | 24 | 0.84 | PCR-RFLP | Nasopharyngeal | 7 |
| Yao | 2008 | China | Asian | Population-control | 280 | 300 | 113 | 120 | 47 | 129 | 134 | 37 | 0.66 | PCR–RFLP | Oral | 7 |
| Tsai1 | 2013 | China | Asian | Hospital-control | 176 | 522 | 88 | 69 | 19 | 285 | 185 | 52 | 0.01 | PCR–RFLP | Nasopharyngeal | 6 |
| Tsai2 | 2014 | China | Asian | Hospital-control | 788 | 956 | 418 | 288 | 82 | 528 | 335 | 93 | <0.01 | PCR–RFLP | Oral | 6 |
| Hsu | 2015 | China | Asian | Population-control | 145 | 112 | 33 | 101 | 11 | 53 | 51 | 8 | 0.36 | PCR-SSP | Oral | 8 |
| A/A | A/C | C/C | A/A | A/C | C/C | |||||||||||
| Pratesi | 2006 | Italy | Caucasian | Population-control | 89 | 130 | 5 | 36 | 48 | 6 | 54 | 70 | 0.27 | Direct sequencing | Nasopharyngeal | 7 |
| Wei | 2007 | China | Asian | Population-control | 198 | 210 | 82 | 81 | 35 | 94 | 92 | 24 | 0.84 | PCR-RFLP | Nasopharyngeal | 7 |
| Yao | 2008 | China | Asian | Population-control | 280 | 300 | 113 | 120 | 47 | 129 | 134 | 37 | 0.66 | PCR–RFLP | Oral | 7 |
| Tsai1 | 2013 | China | Asian | Hospital-control | 176 | 522 | 93 | 66 | 17 | 261 | 205 | 56 | 0.10 | PCR–RFLP | Nasopharyngeal | 7 |
| Tsai2 | 2014 | China | Asian | Hospital-control | 788 | 956 | 408 | 301 | 79 | 484 | 374 | 98 | 0.04 | PCR–RFLP | Oral | 6 |
| Hsu | 2015 | China | Asian | Population-control | 145 | 112 | 33 | 101 | 11 | 53 | 51 | 8 | 0.36 | PCR-SSP | Oral | 8 |
MAF: Minor allele frequency in control group.
HN: All cancer locates in head and neck region, no specific description in original article.
aHWE in control.
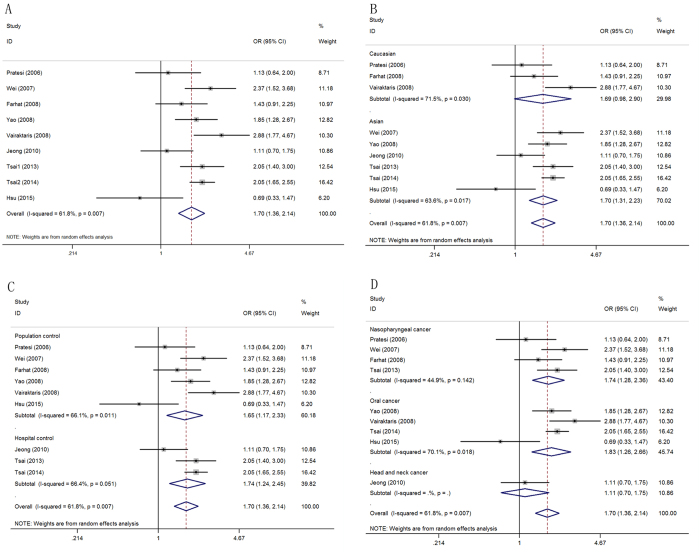
Association between the IL-10 −1082A > G polymorphism and HNC risk A total of 9 relevant studies, consisting of 2,258 patients and 2,887 controls, were examined for the association between the IL-10 −1082A > G polymorphism and HNC risk. The combined analyses revealed a significantly increased risk of HNC risk for this mutation in all five genetic models (G vs. A: OR = 1.56, 95% CI = 1.27–1.92, P < 0.01, I2 = 69.4%; AG vs. AA: OR = 1.64, 95% CI = 1.32–2.05, P < 0.01, I2 = 55.6%; GG vs. AA: OR = 2.24, 95% CI = 1.69–2.97, P < 0.01, I2 = 38.5%; AG + GG vs. AA: OR = 1.70, 95% CI = 1.36–2.14, P = 0.02, I2 = 61.8%, Fig. 2; GG vs. AA + AG: OR = 1.89, 95% CI = 1.23–2.90, P = 0.01, I2 = 46.3%; Table 2). Subsequent analyses accounting for ethnicity revealed similar results in Asian populations, using all five genotype models. Enhanced HNC risk was also observed in Caucasians for the AG vs. AA model and the dominant model. Significant correlations with increased HNC risk were also observed with all five genetic models in the hospital control group and four genetic models (except for the recessive model) in population-based control groups. Moreover, elevated risks of oral cancer (G vs. A: OR = 1.76, 95% CI = 1.36–2.27, P < 0.01, I2 = 55.2%; AG vs. AA: OR = 1.71, 95% CI = 1.15–2.54, P = 0.01, I2 = 71.1%; GG vs. AA: OR = 3.13, 95% CI = 2.06–4.77, P < 0.01, I2 = 0%; AG + GG vs. AA: OR = 1.83, 95% CI = 1.26–2.66, P < 0.01, I2 = 70.1%; GG vs. AA + AG: OR = 2.69, 95% CI = 1.77–4.09, P < 0.01, I2 = 0%) and nasopharyngeal cancer (G vs. A: OR = 1.53, 95% CI = 1.06–2.20, P = 0.02, I2 = 76.5%; AG vs. AA: OR = 1.75, 95% CI = 1.38–2.21, P < 0.01, I2 = 5.2%; AG + GG vs. AA: OR = 1.74, 95% CI = 1.28–2.36, P = 0.02, I2 = 44.9%; Table 2) were detected.
Figure 2. Calculated OR and 95% CIs for the associations between IL-10 −1082A > G polymorphism and HNC risk in the AG + GG vs. AA model ((A) for overall populations; (B) for ethnicity subgroup; (C) for control sources subgroup; (D) for cancer location subgroup).
Table 2. Summary ORs and 95% CI of IL-10 −1082A > G and −819T > C and −592A > C polymorphisms and head and neck cancer risk.
| −1082 A>G | N* | G vs. A |
AG vs. AA |
GG vs. AA |
AG + GG vs. AA |
GG vs. AA + AG |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | |||
| Total | 9 | 1.56 | 1.27–1.92 | <0.01 | 69.4 | 1.64 | 1.32–2.05 | <0.01 | 55.6 | 2.24 | 1.69–2.97 | <0.01 | 38.5 | 1.70 | 1.36–2.14 | <0.01 | 61.8 | 1.89 | 1.23–2.90 | 0.01 | 46.3 | |
| HWE-yes | 3 | 1.05 | 0.81–1.37 | 0.71 | 0 | 0.99 | 0.71–1.38 | 0.95 | 0 | 1.29 | 0.64–2.58 | 0.49 | 0 | 1.02 | 0.74–1.41 | 0.88 | 0 | 1.18 | 0.63–2.22 | 0.60 | 0 | |
| HWE-no | 6 | 1.82 | 1.51–2.19 | <0.01 | 54.4 | 1.94 | 1.67–2.25 | <0.01 | 0 | 2.54 | 1.57–4.12 | <0.01 | 48.0 | 2.04 | 1.77–2.35 | <0.01 | 0.4 | 2.12 | 1.24–3.60 | 0.01 | 58.5 | |
| Ethnicity | ||||||||||||||||||||||
| Caucasian | 3 | 1.34 | 0.93–1.93 | 0.12 | 67.9 | 1.76 | 1.05–2.94 | 0.03 | 65.3 | 1.17 | 0.72–1.90 | 0.53 | 0 | 1.69 | 0.98–2.90 | 0.06 | 71.5 | 0.95 | 0.61–1.49 | 0.84 | 0 | |
| Asian | 6 | 1.72 | 1.37–2.15 | <0.01 | 60.7 | 1.59 | 1.22–2.07 | <0.01 | 58.7 | 3.15 | 2.21–4.51 | <0.01 | 0 | 1.70 | 1.31–2.23 | <0.01 | 63.6 | 2.73 | 1.92–3.89 | <0.01 | 0 | |
| Design | ||||||||||||||||||||||
| PC | 6 | 1.47 | 1.10–1.96 | 0.01 | 70.5 | 1.62 | 1.15–2.28 | 0.01 | 62.2 | 1.72 | 1.18–2.49 | <0.01 | 37.1 | 1.65 | 1.17–2.33 | <0.01 | 66.1 | 1.51 | 0.89–2.57 | 0.13 | 41.4 | |
| HC | 3 | 1.73 | 1.28–2.34 | <0.01 | 65.4 | 1.64 | 1.19–2.26 | <0.01 | 58.4 | 3.24 | 2.09–5.02 | <0.01 | 0 | 1.74 | 1.24–2.45 | <0.01 | 66.4 | 2.80 | 1.81–4.33 | <0.01 | 0 | |
| Cancer location | ||||||||||||||||||||||
| Oral | 4 | 1.76 | 1.36–2.27 | <0.01 | 55.2 | 1.71 | 1.15–2.54 | 0.01 | 71.1 | 3.13 | 2.06–4.77 | <0.01 | 0 | 1.83 | 1.26–2.66 | <0.01 | 70.1 | 2.69 | 1.77–4.09 | <0.01 | 0 | |
| Nasopharyngeal | 4 | 1.53 | 1.06–2.20 | 0.02 | 76.5 | 1.75 | 1.38–2.21 | <0.01 | 5.2 | 1.81 | 0.94–3.48 | 0.07 | 60.3 | 1.74 | 1.28–2.36 | <0.01 | 44.9 | 1.52 | 0.79–2.90 | 0.21 | 64.2 | |
| −819 T>C | C vs. T | TC vs. TT | CC vs. TT | TC + CC vs.TT | CC vs. TT + TC | |||||||||||||||||
| Total | 6 | 1.15 | 1.04–1.21 | 0.01 | 20.1 | 1.24 | 0.93–1.65 | 0.14 | 65.8 | 1.28 | 1.03–1.59 | 0.03 | 0 | 1.27 | 0.99–1.64 | 0.06 | 62.0 | 1.19 | 0.98–1.45 | 0.08 | 0 | |
| HWE-yes | 4 | 1.24 | 1.07–1.45 | 0.01 | 33.3 | 1.33 | 0.75–2.37 | 0.33 | 78.3 | 1.53 | 1.08–2.15 | 0.02 | 0 | 1.41 | 0.86–2.32 | 0.18 | 73.8 | 1.31 | 0.99–1.75 | 0.06 | 0 | |
| HWE-no | 2 | 1.09 | 0.96–1.24 | 0.20 | 0 | 1.11 | 0.93–1.33 | 0.24 | 0 | 1.13 | 0.85–1.50 | 0.40 | 0 | 1.12 | 0.95–1.32 | 0.19 | 0 | 1.08 | 0.82–1.42 | 0.57 | 0 | |
| Ethnicity | ||||||||||||||||||||||
| Asian | 5 | 1.16 | 1.05–1.28 | <0.01 | 29.7 | 1.27 | 0.94–1.71 | 0.12 | 72.0 | 1.30 | 1.04–1.61 | 0.02 | 0 | 1.30 | 0.99–1.70 | 0.06 | 68.7 | 1.22 | 0.99–1.51 | 0.07 | 0 | |
| Design | ||||||||||||||||||||||
| PC | 4 | 1.24 | 1.07–1.45 | 0.01 | 33.3 | 1.33 | 0.75–2.37 | 0.33 | 78.3 | 1.53 | 1.08–2.15 | 0.02 | 0 | 1.41 | 0.86–2.32 | 0.18 | 73.8 | 1.31 | 0.99–1.75 | 0.06 | 0 | |
| HC | 2 | 1.09 | 0.96–1.24 | 0.20 | 0 | 1.11 | 0.93–1.33 | 0.24 | 0 | 1.13 | 0.85–1.50 | 0.40 | 0 | 1.12 | 0.95–1.32 | 0.19 | 0 | 1.08 | 0.82–1.42 | 0.57 | 0 | |
| Cancer location | ||||||||||||||||||||||
| Oral | 3 | 1.23 | 0.98–1.54 | 0.08 | 63.6 | 1.44 | 085–2.44 | 0.18 | 85.5 | 1.6 | 0.97–1.63 | 0.09 | 2.4 | 1.45 | 0.90–2.35 | 0.13 | 84.3 | 1.17 | 0.91–1.50 | 0.22 | 0 | |
| Nasopharyngeal | 3 | 1.15 | 0.96–1.37 | 0.13 | 0 | 1.10 | 0.84–1.44 | 0.49 | 0 | 1.33 | 0.90–1.96 | 0.15 | 0 | 1.16 | 0.90–1.49 | 0.25 | 0 | 1.22 | 0.89–1.67 | 0.22 | 0 | |
| –592 A>C | C vs. A | AC vs. AA | CC vs. AA | AC+CC vs. AA | CC vs. AA+AC | |||||||||||||||||
| Total | 6 | 1.11 | 0.94–1.31 | 0.22 | 55.6 | 1.14 | 0.84–1.55 | 0.41 | 71.2 | 1.14 | 0.92–1.41 | 0.24 | 22.3 | 1.17 | 0.88–1.57 | 0.29 | 71.3 | 1.11 | 0.92–1.36 | 0.28 | 0 | |
| HWE-yes | 5 | 1.16 | 0.95–1.42 | 0.14 | 52.3 | 1.21 | 0.79–1.87 | 0.38 | 74.9 | 1.31 | 0.98–1.76 | 0.07 | 8.9 | 1.26 | 0.85–1.88 | 0.26 | 73.4 | 1.22 | 0.94–1.56 | 0.13 | 0 | |
| Ethnicity | ||||||||||||||||||||||
| Asian | 5 | 1.13 | 0.94–1.36 | 0.22 | 64.0 | 1.16 | 0.84–1.62 | 0.36 | 76.7 | 1.15 | 0.92–1.43 | 0.21 | 35.1 | 1.20 | 0.88–1.63 | 0.25 | 76.8 | 1.13 | 0.92–1.40 | 0.25 | 8.5 | |
| Design | ||||||||||||||||||||||
| PC | 4 | 1.24 | 1.07–1.45 | 0.01 | 33.3 | 1.33 | 0.75–2.37 | 0.33 | 78.3 | 1.53 | 1.08–2.15 | 0.02 | 0 | 1.41 | 0.86–2.32 | 0.18 | 73.8 | 1.31 | 0.99–1.75 | 0.06 | 0 | |
| HC | 2 | 0.95 | 0.84–1.08 | 0.47 | 0 | 0.94 | 0.79–1.12 | 0.51 | 0 | 0.93 | 0.70–1.24 | 0.62 | 0 | 0.94 | 0.80–1.11 | 0.46 | 0 | 0.95 | 0.73–1.26 | 0.74 | 0 | |
| Cancer location | ||||||||||||||||||||||
| Oral | 3 | 1.20 | 0.90–1.60 | 0.22 | 77.2 | 1.09 | 0.92–1.28 | 0.32 | 87.8 | 1.24 | 0.82–1.88 | 0.30 | 45.5 | 1.40 | 0.82–2.40 | 0.22 | 87.5 | 1.10 | 0.86–1.41 | 0.46 | 0 | |
| Nasopharyngeal | 3 | 1.03 | 0.87–1.24 | 0.71 | 16.3 | 0.94 | 0.72–1.23 | 0.65 | 0 | 1.14 | 0.77–1.69 | 0.50 | 27.8 | 0.99 | 0.77–1.27 | 0.91 | 0 | 1.14 | 0.83–1.57 | 0.42 | 25.5 | |
PC: Population control HC: Hospital control.
*Numbers of comparisons.
Heterogeneity was observed in the following four models: G vs. A, AG vs. AA, AG + GG vs. AA and GG vs. AA + AG. Meta-regression analyses highlighted ethnicity as a major driver of heterogeneity in the G vs. A model (τ2 = 100%, P = 9 × 10−3) and HWE deviation responsible for other models (AG vs. AA: τ2 = 100%, P = 6 × 10−3; AG + GG vs. AA: τ2 = 100%, P = 9 × 10−3; and GG vs. AA + AG: τ2 = 70.12%, P = 0.02). Importantly, heterogeneity was relieved in the subgroup analysis.
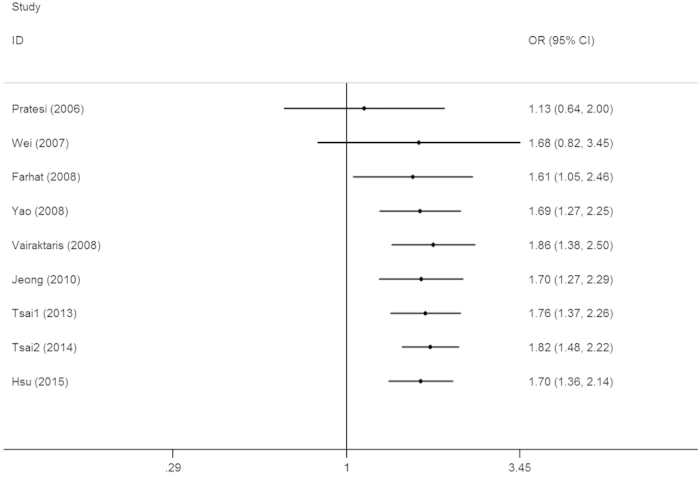
Sensitivity analysis revealed that no single study qualitatively changed the pooled ORs, indicating that the results of this meta-analysis were stable (Fig. 3 for AG + GG vs. AA model). Cumulative analysis by publication date demonstrated that cancer risk increased gradually and became positive following the study conducted by Farhat et al. in 2008 (Fig. 4 for AG + GG vs. AA model).
Figure 3. Sensitivity analysis via deletion of each individual study reflects the relative influence of each individual dataset on the pooled ORs in the AG + GG vs. AA model ofIL-10 −1082A > G polymorphism.
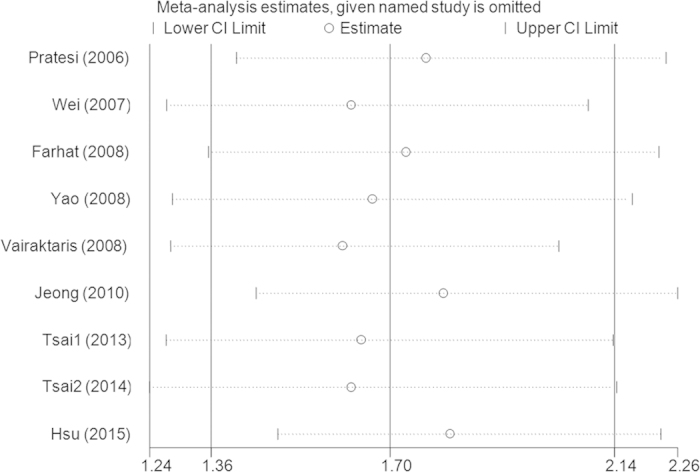
Figure 4. Cumulative meta-analyses according to publication year in the AG + GG vs. AA model of IL-10 −1082A > G polymorphism.
Funnel plots were conducted to assess the publication bias, and no evidence of asymmetry was observed (Fig. 5 for AG + GG vs. AA model). This result was further supported by the analysis using Egger’s test (G vs. A: P = 0.09; AG vs. AA: P = 0.17; GG vs. AA: P = 0.69; AG + GG vs. AA: P = 0.12; GG vs. AA + AG: P = 0.57).
Figure 5. Funnel plot analysis to detect publication bias for AG + GG vs. AA model of IL-10 −1082A > G polymorphism.
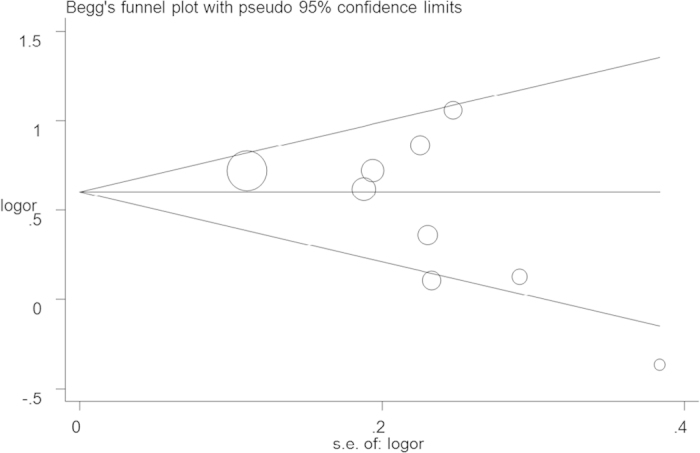
Circles represent the weight of the studies.
Association between the IL-10 −819T > C polymorphism and HNC risk Six studies consisting of 1,676 cases and 2,230 controls were included in the analysis to determine whether the IL-10 −819T > C polymorphism was associated with HNC risk. A significant increase in HNC risk was observed in the overall population (C vs. T, OR = 1.15, 95% CI = 1.04–1.21, P = 0.01, I2 = 20.1%; CC vs. TT, OR = 1.28, 95% CI = 1.03–1.59, P = 0.03, I2 = 0%), as well as among Asian populations (C vs. T, OR = 1.16, 95% CI = 1.05–1.28, P < 0.01, I2 = 29.7%; CC vs. TT, OR = 1.30, 95% CI = 1.04–1.61, P = 0.02, I2 = 0%; Table 2). Furthermore, analysis restricted to HWE studies revealed elevated cancer risk in homozygous comparison (OR = 1.24, 95% CI = 1.07–1.45, P = 0.01, I2 = 33.3%) and recessive model (OR = 1.53, 95% CI = 1.08–2.15, P = 0.02, I2 = 0%). Moreover, these results were consistent with subgroup analysis of the population control group (Table 2). Sensitivity analysis and cumulative analysis were conducted, and no conspicuous change of the pooled ORs was detected. No publication bias was observed, indicating that the results are statistically robust (C vs. T: P = 0.32; TC vs. TT: P = 0.64; CC vs. TT: P = 0.82; TC + CC vs.TT: P = 0.82; CC vs. TT + TC: P = 0.71).
Association between the IL-10 −592A > C polymorphism and HNC risk Six studies consisting of 1,676 cases and 2,230 controls were included in the analysis to determine whether the IL-10–592A > C polymorphism was associated with HNC risk. Overall, no significant association was observed in all five models (Table 2). Only two genetic models (for CC vs. AA, OR = 1.24, 95% CI = 1.07–1.45, P = 0.01, I2 = 33.3%; for CC vs. AA + AC, OR = 1.53, 95% CI = 1.08–2.15, P = 0.02, I2 = 0%) revealed increased risk of HNC in the population control group. Further subgroup analysis of ethnicity and cancer type was conducted, and no significant association was identified. The pooled ORs did not exhibit any change with sensitivity and cumulative analysis, and no publication bias was observed (C vs. A: P = 0.26; AC vs. AA: P = 0.45; CC vs. AA: P = 0.43; AC + CC vs. AA: P = 0.85; CC vs. AA + AC: P = 0.52).
Association between the IL-10 haplotype and HNC risk Of the selected studies, only four described the association between the IL-10 −1082A > G, −819T > C and −592A > C haplotype and HNC risk (Table 3). Quantitative synthesis indicated that only the GCC haplotype was associated with a significant increase in HNC risk in the overall population (GCC vs. ATA, OR = 1.44, 95% CI = 1.04–2.00, P = 0.03, I2 = 52.6%; Table 4).
Table 3. Characteristics of case-control studies on IL-10 −1082A > G and −819T > C and −592A > C haplotype and head and neck cancer risk included in the meta-analysis.
| Firstauthor | Year | Case | Control | Haplotype distribution |
Cancer Location | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case |
Control |
|||||||||||
| ATA | GCC | ACC | GTA | ATA | GCC | ACC | GTA | |||||
| Pratesi | 2006 | 89 | 130 | 46 | 79 | 53 | 0 | 66 | 110 | 84 | 0 | Nasopharyngeal |
| Wei | 2007 | 198 | 210 | 235 | 84 | 69 | 8 | 278 | 51 | 86 | 5 | Nasopharyngeal |
| Yao | 2008 | 280 | 300 | 319 | 106 | 104 | 31 | 377 | 73 | 125 | 25 | Oral |
| Hsu | 2015 | 145 | 112 | 167 | 16 | 107 | 0 | 157 | 16 | 51 | 0 | Oral |
Table 4. Meta-analysis of the IL-10 −1082A > G, −819T > C and −592A > C haplotype and head and neck cancer risk.
| Contrast | OR | 95% CI | P | I2 | PEgger’s test | |
|---|---|---|---|---|---|---|
| GCC vs. ATA | Total | 1.44 | 1.04–2.00 | 0.03 | 52.6 | 0.21 |
| Caucasian | 1.37 | 0.83–2.25 | 0.22 | 66.3 | ||
| Asian | 1.45 | 0.72–2.92 | 0.30 | 66.7 | ||
| Oral | 1.39 | 0.79–2.44 | 0.25 | 54.1 | ||
| Nasopharyngeal | 1.44 | 0.77–2.68 | 0.25 | 75.9 | ||
| ACC vs. ATA | Total | 1.14 | 0.80–1.61 | 0.47 | 69.9 | 0.83 |
| Caucasian | 0.96 | 0.74–1.25 | 0.77 | 0 | ||
| Asian | 1.36 | 0.66–2.79 | 0.40 | 85.9 | ||
| Oral | 1.37 | 0.70–2.72 | 0.36 | 86.6 | ||
| Nasopharyngeal | 0.93 | 0.70–1.25 | 0.65 | 0 | ||
| GTA vs. ATA | Total | 1.54 | 0.94–2.52 | 0.09 | 0 | NA |
NA: not available.
Discussion
Genetic factors have been shown to influence the susceptibility of patients to various diseases and have attracted increasing attention45,46. Inflammation and immune cytokines play an important role during the malignant progression of normal epithelium to cancer by driving angiogenesis, cell metastasis and immune-suppression47,48,49. Genetic mutations influence the transcription and translation of genes, resulting in abnormal expression of corresponding mRNAs and proteins and deregulation of various cellular processes, such as apoptosis and proliferation.
To date, numerous molecular epidemiological studies have been conducted to evaluate the association between polymorphisms of IL-10 and the risk of HNC development, but results have remained conflicting. Regarding the IL-10 −1082A > G polymorphism, Wei et al. reported that the AG and GG genotypes were associated with a significant increase in HNC risk compared with the AA genotype in a Chinese population (95% CI = 1.37–3.48 and 1.33–10.84)37. A similar increase in HNC risk was also observed in other studies that focused on different ethnicities39,40,42,43, whereas other studies identified no significant association between the IL-10 −1082A > G polymorphism and HNC risk29,38,41.
In 2014, a meta-analysis that included four studies focused on nasopharyngeal cancer was published and reported an increased risk with the IL-10 −1082A > G polymorphism50. To our knowledge, this is the first and most comprehensive meta-analysis to date that has explored the association between the IL-10 polymorphisms and HNC risk, more relative studies of HNC and more polymorphisms of IL-10 are collected to make a precise conclusion. Our meta-analysis includes nine studies, consisting of 2,258 patients and 2,887controls, and revealed that the IL-10-1082A > G polymorphism is associated with a significant increased risk of HNC. In subgroup analyses by ethnicity, we found that individuals with the G allele and mutated genotypes had a significant HNC risk compared with healthy individuals in Asian populations, suggesting that the increased HNC risk may be ethno-specific. Six case-control studies of IL-10 −819T > C and−592A > C polymorphisms were examined, including 1,676 cases and 2,230 controls. Positive correlations were observed in the −819T > C polymorphism analyses but not for the −592 A > C polymorphism.
In the current analysis, we found that the frequency of GCC haplotype was higher in the HNC patients than in controls and was associated with a significant increased HNC risk. The GCC haplotype of the IL-10 promoter may result in increased transcription and IL-10 expression. These effects down-regulate the expression of Th1 cytokines, thus allowing tumor cells to escape immune surveillance37. However, due to the small sample size and the limited number of studies examined, it remains unclear whether the GCC haplotype is indeed a marker of HNC patients. Further research is warranted to investigate this relationship.
Stratified analysis was successfully used to relieve moderate heterogeneity bias in the IL-10–1082A > G polymorphism analysis within the Asian population and the oral cancer group, suggesting that ethnicity and cancer location may influence heterogeneity. Moreover, we did not observe any publication bias in the three polymorphisms, demonstrating that the results of this meta-analysis are stable.
It is important to note the limitations of our meta-analysis. First, all of our results may be influenced by casualness due to the small number of studies included and the limited sample size of each study. Larger sample sizes are necessary to accurately clarify the association between the IL-10 polymorphisms and HNC risk. Second, only articles published in English or Chinese were selected, potentially causing a language bias. Third, HNC is a multi-factorial malignant tumor that likely arises from complex interactions between genetic mutations, environmental changes, lifestyle, diet, age and gender. Meta-analysis is a retrospective approach51, and the fundamental underlying mechanisms cannot be explained clearly due to unadjusted databases.
Conclusion
Despite these limitations, our results suggest that the IL-10 −1082A > G polymorphism is a risk factor for HNC, especially in Asian populations. Our findings also indicate that the IL-10 −819T > C polymorphism also plays an important role in HNC development. No significant association was detected between the IL-10 −592A > C polymorphism and HNC risk. Moreover, the GCC haplotype was associated with an increased risk of HNC. In the future, additional studies with larger sample sizes are needed to identify the precise pathogenesis of IL-10 polymorphisms in HNC.
Additional Information
How to cite this article: Niu, Y.-M. et al. Increased risks between Interleukin-10 gene polymorphisms and haplotype and head and neck cancer: a meta-analysis. Sci. Rep. 5, 17149; doi: 10.1038/srep17149 (2015).
Supplementary Material
Acknowledgments
This study was partly supported by the Foundation of Ministry of Education of Hubei Province (D20142102). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions N.Y.M., D.X.Y. and C.H.X. performed the literature search, data extraction, and statistical analysis and wrote the manuscript. Z.C. and Y.R.X. supervised the literature search, data extraction, analysis, Z.X.T. and L.J. reviews the manuscript.
References
- Argiris A., Karamouzis M. V., Raben D. & Ferris R. L. Head and neck cancer. Lancet 371, 1695–1709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiris A. & Eng C. Epidemiology, staging, and screening of head and neck cancer. Cancer Treat Res 114, 15–60 (2003). [DOI] [PubMed] [Google Scholar]
- Siegel R., Ma J., Zou Z. & Jemal A. Cancer statistics, 2014. CA Cancer J Clin 64, 9–29 (2014). [DOI] [PubMed] [Google Scholar]
- Pulte D. & Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncologist 15, 994–1001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye C. et al. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol 15, 1319–1331 (2014). [DOI] [PubMed] [Google Scholar]
- Hashibe M. et al. Evidence for an important role of alcohol- and aldehyde-metabolizing genes in cancers of the upper aerodigestive tract. Cancer Epidemiol Biomarkers Prev 15, 696–703 (2006). [DOI] [PubMed] [Google Scholar]
- Vineis P. et al. Tobacco and cancer: recent epidemiological evidence. J Natl Cancer Inst 96, 99–106 (2004). [DOI] [PubMed] [Google Scholar]
- Ramqvist T., Grun N. & Dalianis T. Human Papillomavirus and Tonsillar and Base of Tongue Cancer. Viruses 7, 1332–1343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry B. J. et al. Sites of origin of oral cavity cancer in nonsmokers vs smokers: possible evidence of dental trauma carcinogenesis and its importance compared with human papillomavirus. JAMA Otolaryngol Head Neck Surg 141, 5–11 (2015). [DOI] [PubMed] [Google Scholar]
- Li Q. et al. Vitamin or mineral supplement intake and the risk of head and neck cancer: pooled analysis in the INHANCE consortium. Int J Cancer 131, 1686–1699 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L. M. & Werb Z. Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F., Allavena P., Sica A., Garlanda C. & Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- Niu Y. et al. Association between cyclooxygenase-2 gene polymorphisms and head and neck squamous cell carcinoma risk. J Craniofac Surg 25, 333–337 (2014). [DOI] [PubMed] [Google Scholar]
- Moore K. W., de Waal Malefyt R., Coffman R. L. & O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19, 683–765 (2001). [DOI] [PubMed] [Google Scholar]
- De Vries J. E. Immunosuppressive and anti-inflammatory properties of interleukin 10. Ann Med 27, 537–541 (1995). [DOI] [PubMed] [Google Scholar]
- Howell W. M. & Rose-Zerilli M. J. Cytokine gene polymorphisms, cancer susceptibility, and prognosis. J Nutr 137, 194S–199S (2007). [DOI] [PubMed] [Google Scholar]
- Fiorentino D. F., Bond M. W. & Mosmann T. R. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med 170, 2081–2095 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. W. et al. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science 248, 1230–1234 (1990). [DOI] [PubMed] [Google Scholar]
- Eskdale J., Kube D., Tesch H. & Gallagher G. Mapping of the human IL10 gene and further characterization of the 5’ flanking sequence. Immunogenetics 46, 120–128 (1997). [DOI] [PubMed] [Google Scholar]
- Kingo K. et al. Influence of genetic polymorphisms on interleukin-10 mRNA expression and psoriasis susceptibility. J Dermatol Sci 37, 111–113 (2005). [DOI] [PubMed] [Google Scholar]
- Turner D. M. et al. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 24, 1–8 (1997). [DOI] [PubMed] [Google Scholar]
- Saxena M., Agrawal C. C., Bid H. K. & Banerjee M. An interleukin-10 gene promoter polymorphism (-592A/C) associated with type 2 diabetes: a North Indian study. Biochem Genet 50, 549–559 (2012). [DOI] [PubMed] [Google Scholar]
- Eskdale J. et al. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA 95, 9465–9470 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson A. W. et al. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol 166, 3915–3922 (2001). [DOI] [PubMed] [Google Scholar]
- Pooja S. et al. Polymorphic variations in IL-1beta, IL-6 and IL-10 genes, their circulating serum levels and breast cancer risk in Indian women. Cytokine 60, 122–128 (2012). [DOI] [PubMed] [Google Scholar]
- Hsia T. C. et al. Interleukin-10 (IL-10) promoter genotypes are associated with lung cancer risk in Taiwan males and smokers. Anticancer Res 34, 7039–7044 (2014). [PubMed] [Google Scholar]
- Singhal P., Kumar A., Bharadwaj S., Hussain S. & Bharadwaj M. Association of IL-10 GTC haplotype with serum level and HPV infection in the development of cervical carcinoma. Tumour Biol 36, 2287–2298 (2015). [DOI] [PubMed] [Google Scholar]
- Kuo W. H. et al. Effects of interleukin-10 polymorphisms and smoking on the risk of gastric cancer in Taiwan. In Vivo 28, 967–971 (2014). [PubMed] [Google Scholar]
- Pratesi C. et al. Interleukin-10 and interleukin-18 promoter polymorphisms in an Italian cohort of patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Immunol Immunother 55, 23–30 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J. & Altman D. G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339, b2535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H. et al. Association between heme oxygenase 1 gene promoter polymorphisms and susceptibility to coronary artery disease: a HuGE review and meta-analysis. Am J Epidemiol 179, 1039–1048 (2014). [DOI] [PubMed] [Google Scholar]
- Thakkinstian A. et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol 162, 201–211 (2005). [DOI] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- Song F., Khan K. S., Dinnes J. & Sutton A. J. Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Int J Epidemiol 31, 88–95 (2002). [DOI] [PubMed] [Google Scholar]
- Huedo-Medina T. B., Sanchez-Meca J., Marin-Martinez F. & Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 11, 193–206 (2006). [DOI] [PubMed] [Google Scholar]
- Wei Y. S. et al. Interleukin-10 gene promoter polymorphisms and the risk of nasopharyngeal carcinoma. Tissue Antigens 70, 12–17 (2007). [DOI] [PubMed] [Google Scholar]
- Farhat K., Hassen E., Gabbouj S., Bouaouina N. & Chouchane L. Interleukin-10 and interferon-gamma gene polymorphisms in patients with nasopharyngeal carcinoma. Int J Immunogenet 35, 197–205 (2008). [DOI] [PubMed] [Google Scholar]
- Yao J. G., Gao L. B., Liu Y. G., Li J. & Pang G. F. Genetic variation in interleukin-10 gene and risk of oral cancer. Clin Chim Acta 388, 84–88 (2008). [DOI] [PubMed] [Google Scholar]
- Vairaktaris E. et al. The interleukin-10 (-1082A/G) polymorphism is strongly associated with increased risk for oral squamous cell carcinoma. Anticancer Res 28, 309–314 (2008). [PubMed] [Google Scholar]
- Jeong S. W. et al. Cox-2 and IL-10 polymorphisms and association with squamous cell carcinoma of the head and neck in a Korean sample. J Korean Med Sci 25, 1024–1028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. W. et al. Association of interleukin-10 (IL10) promoter genotypes with nasopharyngeal carcinoma risk in Taiwan. Anticancer Res 33, 3391–3396 (2013). [PubMed] [Google Scholar]
- Tsai C. W. et al. Significant association of Interleukin-10 genotypes and oral cancer susceptibility in Taiwan. Anticancer Res 34, 3731–3737 (2014). [PubMed] [Google Scholar]
- Hsu H. J. et al. TGF-beta1 and IL-10 single nucleotide polymorphisms as risk factors for oral cancer in Taiwanese. Kaohsiung J Med Sci 31, 123–129 (2015). [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K. W. & Vogelstein B. Genetic instabilities in human cancers. Nature 396, 643–649 (1998). [DOI] [PubMed] [Google Scholar]
- Marshall H. Genetic and epigenetic factors in development of lung cancer. Lancet Oncol 13, 1188 (2012). [DOI] [PubMed] [Google Scholar]
- Germano G., Allavena P. & Mantovani A. Cytokines as a key component of cancer-related inflammation. Cytokine 43, 374–379 (2008). [DOI] [PubMed] [Google Scholar]
- Landskron G., De la Fuente M., Thuwajit P., Thuwajit C. & Hermoso M. A. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res 2014, 149185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R. & Karin M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. et al. A pooled analysis of the IL-10-1082 A/G polymorphism and the nasopharyngeal carcinoma susceptibility. Eur Arch Otorhinolaryngol, 10.1007/s00405-014-3465-9 (2014). [DOI] [PubMed] [Google Scholar]
- Zeng X. et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med 8, 2–10 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.