Abstract
Background
Even though increasing evidences on miRNA involvement in human pathological responses, the distinct roles and related mechanisms of miRNAs in the pathology of osteoarthritis (OA) are not yet fully understood.
Method
RNA levels or protein levels of Apoptotic genes, HDACs, MMP-13, and miRNAs in human chondrocytes isolated from normal biopsy sample and OA cartilages were analyzed by real-time PCR or western blotting. Exogenous modulation of miR-222 level was performed using delivery of its specific precursor or specific inhibitor and target validation assay was applied to identify its potent target. In vivo study using DMM mice model was performed and assessed the degree of cartilage degradation.
Results
According to miRNA profiling, miR-222 was significantly down-regulated in OA chondrocytes. Over-expression of miR-222 significantly suppressed apoptotic death by down-regulating HDAC-4 and MMP-13 level. Moreover, 3′-UTR reporter assays showed that HDAC-4 is a direct target of miR-222. The treatment of chondrocytes with the HDAC inhibitor, trichostatin A (TSA), suppressed MMP-13 protein level and apoptosis, whereas the over-expression of HDAC-4 displayed opposite effects. The introduction of miR-222 into the cartilage of medial meniscus destabilized mice significantly reduced cartilage destruction and MMP-13 level.
Conclusion
Taken together, our data suggest that miR-222 may be involved in cartilage destruction by targeting HDAC-4 and regulating MMP-13 level.
Keywords: miR-222, HDAC-4, MMP-13, Apoptosis, Human chondrocytes, OA chondrocytes
Highlights
-
•
MiR-222 controls OA pathogenesis by targeting HDAC-4.
-
•
HADC-4 regulated by miR-222 modulates MMP-13 expression during cartilage destruction.
-
•
Our study indicates the possibility that miR-222 could act as a protective factor against OA.
1. Introduction
Osteoarthritis (OA), which is a chronic degenerative joint disorder that causes joint pain and dysfunction, is characterized by progressive structural changes in articular cartilage, persistent loss of tissue cellularity, and degeneration of the extracellular matrix (ECM) [1]. Chondrocytes are the only resident cells found in cartilage and are responsible for both synthesis and turnover of the abundant ECM [2]. Therefore, the maintenance of healthy chondrocytes appears to be important for maintaining cartilage integrity. Another significant feature of OA is excessive production of inflammatory mediators [3]. Among them, the pro-inflammatory cytokine, interleukin-1β (IL-1β), plays a crucial role by inducing a cascade of inflammatory and catabolic events. In particular, IL-1β alters chondrocyte anabolism by enhancing matrix metalloproteinase (MMP) production which suppresses cartilage matrix formation. Among the MMPs, the expression of MMP-13 is significantly higher in chondrocytes from late-stage OA cartilage compared with early OA or normal knee cartilage [4]. In addition, transgenic mice over-expressing MMP-13 in their articular chondrocytes develop a joint degradation similar to that seen in human OA [5]. Little and colleagues [6] showed that MMP-13-deficient mice are resistance to osteoarthritic cartilage erosion, which caused by proteolysis of the principal cartilage matrix such as aggrecan and cartilage collagens suggesting cartilage structural damage in OA is highly dependent on MMP-13 activity. Moreover, they have demonstrated that a significant inhibition of cartilage structural damage in DMM-induced MMP-13–knockout is associated with a change in chondrocyte hypertrophy, apoptosis, or osteophyte development not reduction in aggrecanolysis. However, the mechanisms underlying the pathological characteristics of OA are still not fully understood.
Two families of histone deacetylases (HDACs) have been identified: the classical HDAC family and the NAD + -dependent, the so-called SIR2 family (sometimes called class III HDACs). The classical HDACs can be grouped into three classes (I, II, and IV) based on phylogeny [7]. The class I HDACs (HDAC-1, -2, -3, and -8) are related to yeast RPD3, and the class II HDACs (HDAC-4, -5, -6, -7, -9, and -10) are closely related to yeast HDA1 [8]. Trichostatin A (TSA) is an HDAC inhibitor [9] that shows broad-spectrum activity against class I and class II HDACs. Recently, HDACs have emerged as therapeutic targets in cancer and inflammatory diseases, including rheumatoid arthritis (RA) and OA [10], [11], [12], [13]. However, it is still unclear exactly how HDACs are involved in cartilage degradation.
MicroRNAs (miRNAs, miRs) have emerged as fine-tuning regulators for diverse biological processes [14], [15]. During their biogenesis, the miRNA genes are transcribed into primary miRNAs (pri-miR), which are processed by Drosha and Dicer to generate miRNA duplexes consisting of a mature and a passenger miR strand [16]. Recent studies have shown that miRNAs play crucial roles in immune cell development and immune system function [17], [18] and regulate various aspect of cell physiology, including developmental timing, cell differentiation, apoptosis, and anti-viral defense [18], [19], [20]. Given the crucial role of miRNAs in human physiology, the abnormal expression of specific miRNAs may lead to the development of diverse diseases, such as cancer, cardiovascular disorders, mental disorders, musculoskeletal disorders, and lung diseases [21], [22], [23], [24], [25]. Recently, intensive research has focused on delineating the roles of various miRNAs in the development of inflammatory and immune-mediated diseases. Here, we report that miR-222 controls cartilage degradation via HDAC-mediated regulation of MMPs during OA pathogenesis.
2. Materials and methods
2.1. Cartilage procurement and processing
Cartilage was obtained from 6 normal donors (age range 25–49 years) and 20 OA donors (age range 55–75 years). Tissue collection was approved by the Human Subjects Committee of Wonkwang University. Osteoarthritis cartilage was obtained from patients undergoing knee replacement surgery. Cartilage thickness ranged from 1.5 to 2.8 mm. Cartilage surfaces were rinsed with saline, and parallel sections 5 mm apart were cut vertically from the cartilage surface onto subchondral bone with a scalpel.
2.2. Primary cultures of human chondrocytes
Small slices of cartilage were sequentially digested with 0.06% bacterial collagenase (Sigma) then seeded at a density of 1.5 × 104 cells/cm2 in culture medium consisting of DMEM (Gibco-Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco-Invitrogen). Cells were treated with 5 ng/ml for IL-1β (R&D systems, Minneapolis, MN), 3 ng/ml for TGF-βs (R&D systems, Minneapolis, MN), and 300 nM for TSA (Sigma, St Louis, MO, USA) in this study.
2.3. Western blot analysis
Total proteins (30 μg) were electrophoresed and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, Germany). The membranes were individually probed with antibodies specific for Type I, II collagen, PRTG (Calbiochem, La Jolla, CA), (p)AKT, (p)GSK, (p)JNK, GAPDH (Santa Cruz Biotechnology Inc.), Caspase-3, PARP, HADC-1, -2, -3, or -4 (Cell Signaling Technology Inc., Danvers, MA, USA). The blots were developed using a peroxidase-conjugated secondary antibody, and the immunoreactive proteins were visualized with an ECL system (Amersham, UK).
2.4. RNA isolation and quantitative PCR analysis
Total RNA was extracted using the RNeasy Mini Kit (Qiagen). Total RNA was extracted from cells using TRIzol reagent. Isolated RNA was treated with DNase followed by reverse transcription of 1 μg RNA into cDNA using reverse transcriptase, 0.5 μg/μl oligodT primers, and 12.5 mM dNTPs (Invitrogen). Quantitative real-time PCR was performed using StepOne plus system (Applied Biosystems, Foster City, CA). PCR protocol was as follows: 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All PCRs were performed using SYBR green master mix (#436759, Applied biosystems) and 5 μM of forward and reverse primer in a total reaction volume of 25 μl. For mRNA, transcripts were quantified by real-time quantitative polymerase chain reaction (RT-PCR) and normalized to the amount of GAPDH mRNA expressed The oligonucleotides used as primers were as follows: human MMP-2, 5′-acaccaagaacttcgtctg-3′ and 5′-tgcagatc tcaggagtgaca-3′; human MMP-9, 5′-atttctgccaggacc gcttctact-3′ and 5′-atgtcataggt cacgtagcccact-3′; human MMP-12, 5′-gaaccaacgcttgccaaatcctga-3′ and 5′-ttcccacggtagt gacagcatcaa-3′; human MMP-13, 5′-ttgcagagcgctacctgagatcat-3′ and 5′-tttgccagtcacctcta agccgaa-3′.
For analyzing the RNA levels of HDACs, the following primers were used for real-time PCR. HDAC1 (histone deacetylase 1), 5′-gac acgccaagtgtgtggaa-3′ and 5′-cctcccagcatcagca tagg-3′; HDAC10 (histone deacetylase 10), 5′-caacgccggat atcacattg-3′ and 5′-gctgacgcttcct gttgga-3′; HDAC11 (histone deacetylase 11), 5′-acaa cccagctgtaccagcat-3′ and 5′-cgcggcgagta cacgatt-3′; HDAC2 (histone deacetylase 2), 5′-acatgagcaatgcggagaaat-3′ and 5′-tctgccatcttgtgg tacagtga-3′; HDAC3 (histone deacetylase 3), 5′-ccttttccagccg gttatca-3′ and 5′-acaatgcacgt gggttggt-3′; HDAC4 (histone deacetylase 4), 5′-tcagat cgccaacacattcg-3′ and 5′-acgggagcggtt ctgttaga-3′; HDAC5 (histone deacetylase 5), 5′-ccattggagacgtggagtacct-3′ and 5′-gcggagactag gaccacatca-3′; HDAC6 (histone deacetylase 6), 5′-tcgctgcgtgtcctttcag-3′ and 5′-gctgtgaaccaaca tcagctctt-3′; HDAC7 (histone deacetylase 7), 5′-caagag caagcgaagtgctgta-3′ and 5′-ttcagaatca cctccgctagct-3′; HDAC8 (histone deacetylase 8), 5′-tgggaggaggaggctat aacc-3′ and 5′-ccggt caagtatgtccagcat-3′; HDAC9 (histone deacetylase 9), 5′-cacacacgtg cgctctctgt-3′ and 5′-cgagac ggtgtttctctaatcca-3′;
For analyzing the RNA levels of genes involved in apoptotic cell death, the following primers were used for real-time PCR. ABL1 (c-abl oncogene 1, non-receptor tyrosine kinase), 5′-gaagcccaaaccaaaaatgg-3′ and 5′-gactgttga ctggcgtgatgtag-3′; APAF1 (apoptotic peptidase activating factor 1), 5′-tgcgctgctctgccttct-3′ and 5′-gcggagcacaca aatgaaga-3′; ATP6V1G2 (ATPase, H + transporting, lysosomal 13 kDa, V1 subunit G2), 5′-ggaaaacatcctgacttcagtgtct-3′ and 5′-ccagcaagtgacagggtcaa-3′; BAX (BCL2-associated X protein), 5′-ccaaggtgccggaactga-3′ and 5′-cccggagg aagtccaatgt-3′; BCL2L11 (BCL2-like 11), 5′-gctttcccatggtcacaggat-3′ and 5′-ctgca gctggactctgctgta-3′; BIRC2 (baculoviral IAP repeat containing 2), 5′-cctgtggtggg aagctcagt-3′ and 5′-cctccggtgttctgacatagc-3′; CASP1 (caspase 1–apoptosis-related cysteine peptidase) ; 5′-ataccaagaactgcccaagtttg-3′ and 5′-ggcaggcctggatgatga-3′, CASP3 (caspase 3–apoptosis-related cysteine peptidase); 5′-gcctacagcccatttctccat-3′ and 5′-gcgccctggcagcat-3′; CASP6 (caspase 6–apoptosis-related cysteine peptidase), 5′-ggc gtggttactcacacctgta-3′ and 5′-gatccgcccaccttgga-3′; CASP7 (caspase 7–apoptosis-related cysteine peptidase), 5′-ccgccgt gggaacgat-3′ and 5′-cctcaaccccctgctcttc-3′; CASP9 (caspase 9–apoptosis-related cysteine peptidase), 5′-agcagtgggctcactctgaag-3′ and 5′-aacagcattagcgaccctaagc-3′; CD40 (CD40 molecule, TNF receptor superfamily member 5), 5′-tggtgagtgactgcacagagttc-3′ and 5′-cgcttt caccgcaagga-3′, CD40LG (CD40 ligand), 5′-ccaggtgcttcggt gtttgt-3′ and 5′-ccagtgccatggctcactt-3′; CFLAR (CASP8 and FADD-like apoptosis regulator), 5′-gctggcagctgattagatggt-3′ and 5′-ttt gagtcagtggactgggaaa-3′ ; CYLD (cylindromatosis-turban tumor syndrome), 5′-tgtggaggg cttgcaatgt-3′ and 5′-agc tgagatgtccggatcgt-3′; DFFA (DNA fragmentation factor, 45 kDa, alpha polypeptide), 5′-ttgacgctccctgctcaga-3′ and 5′-tggacggtggcacaactct-3′; FAS (Fas-TNF receptor superfamily, member 6), 5′-acccgctcagtacggagttg-3′ and 5′-ccagcatgg ttgttgagcaa-3′; FASLG (Fas ligand-TNF superfamily, member 6), 5′-tgcctcctctt gagcagtca-3′ and 5′-tcc tgtagaggctgaggtgtca-3′; GADD45A (growth arrest and DNA-damage-inducible, alpha), 5′-gatgtggctctgcagatcca-3′ and 5′-atgtcgttct cgcagcaaaa-3′; NOL3 (nucleolar protein 3-apoptosis repressor with CARD domain), 5′-gccca ccacgagcatca-3′ and 5′-cctggactcctaa gggcagat-3′; SPATA2 (spermatogenesis associated 2), 5′-cccctcccattgcaaagc-3′ and 5′-gtggatgtttggt gacctgaag-3′; SYCP2 (synaptonemal complex protein 2), 5′-ttagttgatcttctgct ggtcataca-3′ and 5′-gcgaggtacgaaactttccactac-3′; TNFRSF10A (tumor necrosis factor receptor superfamily, member 10a), 5′-gaacacagcatgtcagtgcaaa-3′ and 5′-ccggcacatctca gcagaat-3′; TNFRSF1A (tumor necrosis factor receptor superfamily, member 1A), 5′-agcctggagtgcacgaagtt-3′ and 5′-tgagtcctca gtgcccttaaca-3′, TNF (tumor necrosis factor), 5′-gcaggtctactttgggatcattg-3′ and 5′-gcgtttggg aaggttgga-3′; TP53 (tumor protein p53), 5′-tgcaataggtgtgcgtcagaa-3′ and 5′-ccccgggacaaagcaaa-3′; human GAPDH, 5′-gatcatcagcaatg cctcct-3′ and 5′-tgtggtcatgagtccttcca-3′.
2.5. MiRNA and mRNA real-time quantitative RT-PCR
MiRNA and mRNA expression were independently quantified using the TaqMan MicroRNA and TaqMan gene expression assays (Applied Biosystems), respectively, according to the manufacturer's protocols. MiRNA expression was normalized to RNU43 small nuclear RNA endogenous controls.
2.6. Reporter vectors and DNA constructs
The 3′-UTR of human HDAC-4 was PCR amplified using the following primers: 5′-TGGGAGCTCCTGGCTCTATT-3′ (bp no. 1616 ~ 1635), 5′-GCTGAGGCTGACTTTGCACT-3′ (bp no. 3088 ~ 3107). It was then cloned downstream of the CMV-driven firefly luciferase cassette in the pMIR-REPORT vector (Ambion). For miRNA target validation, chondroblasts were electroporated with 200 ng of a firefly luciferase reporter construct and 50 pmol of pre-miR-222 or pre-miR-negative (Ambion). The Renilla luciferase vector was used to normalize electroporation efficiency. At 24 h after electroporation, both firefly and Renilla luciferase activity were assayed (Promega). Normalized relative light units represent firefly luciferase activity or Renillar luciferase activity.
2.7. Production of lentiviral particles
The hsa-miR-222 and negative control lentiviruses were transfected with 3rd generation packaging mix from Applied Biological Materials Inc. (ABM) into human 293FT cells using Lentifectin (ABM) in Opti-MEM I medium (Invitrogen, CA) and cultured overnight. The supernatant was collected and lentiviral particles were concentrated using Lenti-X Concentrator (Clontech, CA).
2.8. Arthritic cartilage, experimental OA, and histology of OA cartilage
Human OA cartilage was sourced from individuals undergoing arthroplasty for OA of the knee joint. The Wonkwang University Hospital Institutional Review Board approved the use of these materials, and all individuals provided written informed consent before the operative procedure. Human OA cartilage samples were frozen, sectioned at a thickness of 10 μm, fixed in paraformaldehyde, and stained with safranin O.
Experimental OA was induced by destabilization of the medial meniscus (DMM) surgery 8-week-old male mice. The care and use of experimental animals were approved by our Institutional Animal Care and Use Committee. Sham-operated animals injected with empty lentiviruses (mock transduction) were used as controls for DMM. Mice were killed 8 weeks after DMM surgery or 2 weeks after intra-articular injection (1 × 109 plaque-forming units (PFU)) of miR-222-expressing lentiviruses for histologic and biochemical analyses. Cartilage destruction in mice was examined using safranin O staining. Briefly, knee joints were fixed in 4% paraformaldehyde, decalcified in 0.5 M EDTA (pH 7.4) for 14 days at 4 °C, and embedded in paraffin. The paraffin blocks were sectioned at 6 μm thickness. The sections were deparaffinized in xylene, hydrated with graded ethanol, and stained with safranin O. For MMP-13 staining, sections were incubated with anti-MMP-13 antibody (1:200 dilutions) overnight at 4 °C, followed by incubation with TRITC-conjugated secondary antibody at room temperature for 1 hour. For propidium iodide staining, sections were incubated with 50 μg/ml propidium iodide and 50 μg/ml RNase A in PBS.
2.9. Cell apoptosis assay
MuseTM apoptosis kit (Millipore, Billerica, MA) were used to detect apoptotic cell death.
2.10. RNA Extraction from FFPE (formalin-fixed paraffin-embedded) tissue of mouse cartilages
RNA was extracted from the paraffin samples using the MasterPure kit (Epicentre Biotechnologies, Madison, WI). Briefly, FFPE tissue was resuspended in MasterPure tissue and cell lysis solution with a final concentration of 0.15 mg/ml proteinase K and incubated at 65 °C for 30 min. MasterPure MPC™ protein precipitation reagent was added and nucleic acids were precipitated by adding isopropanol and pelleted by centrifugation at 10,000 ×g for 10 min at 4 °C. The remaining pellet was washed twice with 75% ethanol and allowed to dry before adding 30 μl TE buffer with 40 units of RNase inhibitor.
2.11. Statistical analysis
Statistically significant differences between 2 groups were determined with t tests. Results are presented as mean ± standard deviation (SD). P values of less than 0.05 were considered statistically significant.
3. Results
3.1. Apoptosis and HDAC expression are elevated in OA cartilage
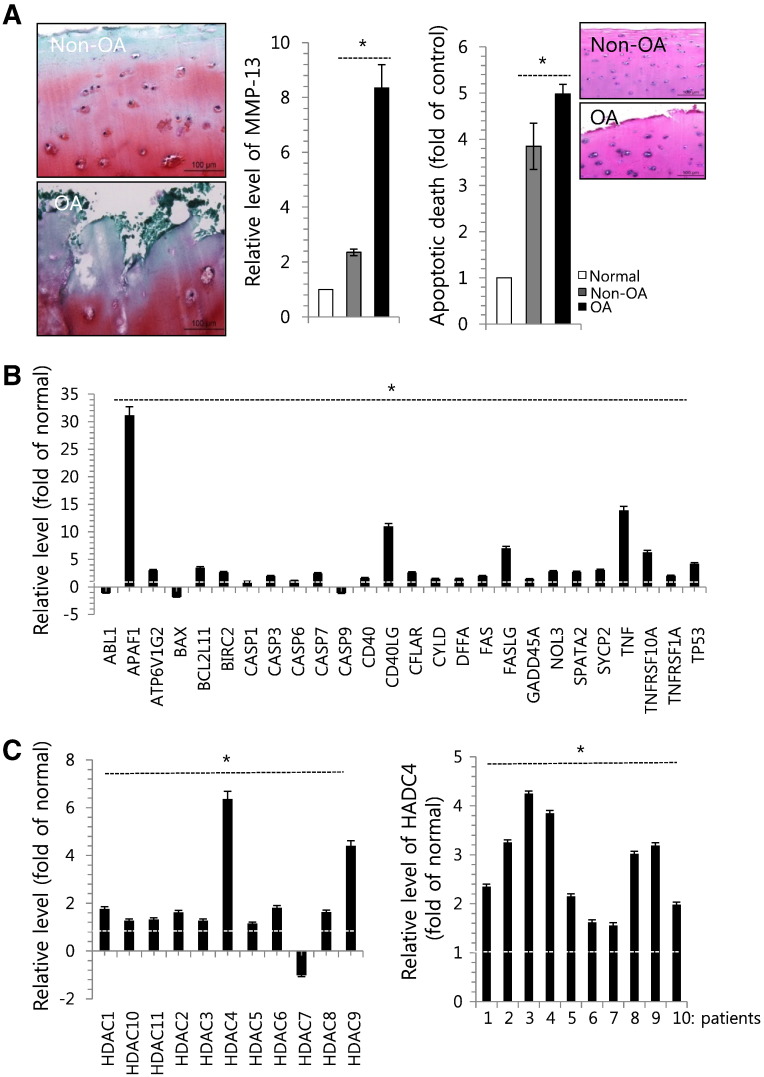
Osteoarthritic (OA) cartilage samples (n = 10) were obtained from patients who underwent joint surgery (mean age, 64.6 years). The cartilage was divided into non-OA and OA areas and stained with safranin O (Fig. 1A, right panel). Proteolytic degradation of cartilage is a hallmark of osteoarthritis (OA), and activated chondrocytes are known to produce matrix-degrading enzymes, such as MMP-13 (also known as collagenase 3) in OA joints [26], [27]. Consistent with these previous reports, the expression of MMP-13 (Fig. 1A middle panel) was increased in the OA cartilage compared to the non-OA cartilage. Apoptotic cell death was also significantly increased in the OA areas (Fig. 1A, left panel). Moreover, we found that various apoptotic genes, including ABL1, ATP6V1GNOL3, CASP-1, -3 and -7, CD40, CYLD, and FAS, were highly induced in OA chondrocytes (Fig. 1B).
Fig. 1.
Apoptosis and histon deacetylation were involved in the pathogenesis of OA. (A) OA cartilage that was divided into 2 classes depending on the progression of OA pathology (Non-OA: healthy zone; OA: severe OA zone) and stained with safranin O (left panel). Chondrocytes were isolated from biopsy sample of normal cartilage (normal) and OA cartilage (Non-OA and OA) and the RNA level of MMP-13 (middle panel) and apoptotic cell death (right panel) were analyzed using MuseTM apoptosis kit. H&E staining was inserted. (B) Changes in the RNA level of genes involved in apoptosis were analyzed by RT-PCR. (C) Changes in the RNA level of HDAC-1 to -11 genes were analyzed by RT-PCR (left panel) and changes in the RNA level of HDAC-4 using OA chondrocytes isolated from 10 different OA patients (right panel). GAPDH was used as control. The mean is plotted, and the error bars represent 95% CI (lower/upper limit). *Statistically different from control cells (p < 0.05).
HDACs balance the activities of histone acetyltransferases (HATs) and epigenetically regulate gene transcription, thereby controlling the acetylation status of histone proteins and non-histone substrates. Recent studies have demonstrated that HDAC inhibitors have therapeutic effects in cancer and inflammatory diseases [10], [11], [12], [13], [28], [29], [30], [31], [32]. To further characterize the molecules involved in OA pathogenesis, we examined the RNA levels of the genes encoding HDAC-1 through HDAC-11 in OA chondrocytes isolated from cartilage of OA patients compared to normal chondrocytes isolated from biopsy sample of normal patients. Our results revealed that the RNA levels of all of these genes were highly up-regulated in OA chondrocytes, particularly those encoding HDAC-4 and -9 (Fig. 1C, left panel). In addition, we observed the significant up-regulation of HDAC-4 expression in 10 different OA chondrocytes compared to normal chondrocytes (Fig. 1C, right panel).
3.2. MiR-222 contributes to OA pathogenesis by modulating the induction of MMPs
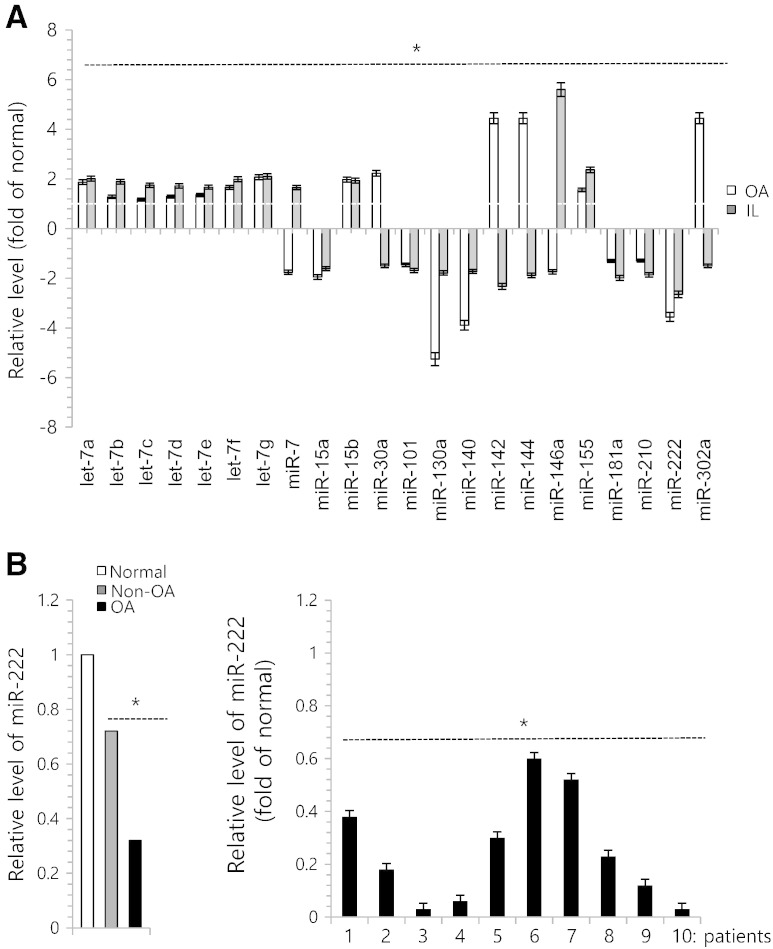
Recent studies have uncovered the important and crucial roles for a family of small regulatory RNA molecules known as microRNAs (miR; miRNAs) in regulating diverse aspect of diseases by acting as a major gene switch [33]. To identify miRNAs that could be involved in modulating HDAC gene expression in OA chondrocytes, we first examined their expression levels in normal, non-OA, and OA cartilage to verify the OA-related miRNAs selected from previous publications. As shown in Fig. 2A, miR-9, -22, -130a, -140-3p, -143, -122, and -222 were down-regulated in OA chondrocytes and degenerated chondrocytes by IL-1β compared to normal chondrocytes, whereas miR-124 was up-regulated in OA chondrocytes and degenerated chondrocytes by IL-1β. Among them, miR-222 was observed to be suppressed by 4-fold in OA chondrocytes and 3-fold in IL-1β-treated chondrocytes (Fig. 2A). We analyzed the expression level of miR-222 in non-OA chondrocyte and OA chondrocytes compared to normal chondrocytes. Suppressed level of miR-222 was observed in both non-OA chondrocytes and OA chondrocytes and its level was gradually decreased as pathogenesis progressed (Fig. 2B, left panel). Moreover, we observed the significant down-regulation of miR-222 expression in 10 different OA chondrocytes (Fig. 2B, right panel).
Fig. 2.
miR-222 is involved in degeneration of human articular chondrocytes. (A) Chondrocytes were isolated from biopsy sample of normal cartilage (normal) and OA cartilage (OA), cultured with or without 5 ng/ml IL-1β. OA. Expression levels of miRs were analyzed. (B) Chondrocytes were isolated from biopsy sample of normal cartilage (normal) and OA cartilage (Non-OA and OA) and expression level of miR-222 (left panel) was analyzed by real-time PCR. Expression level of miR-222 was analyzed using OA chondrocytes isolated from 10 different OA patients (right panel). The mean is plotted and the error bars represent 95% CI (lower/upper limit). *Statistically different from control cells (p < 0.05).
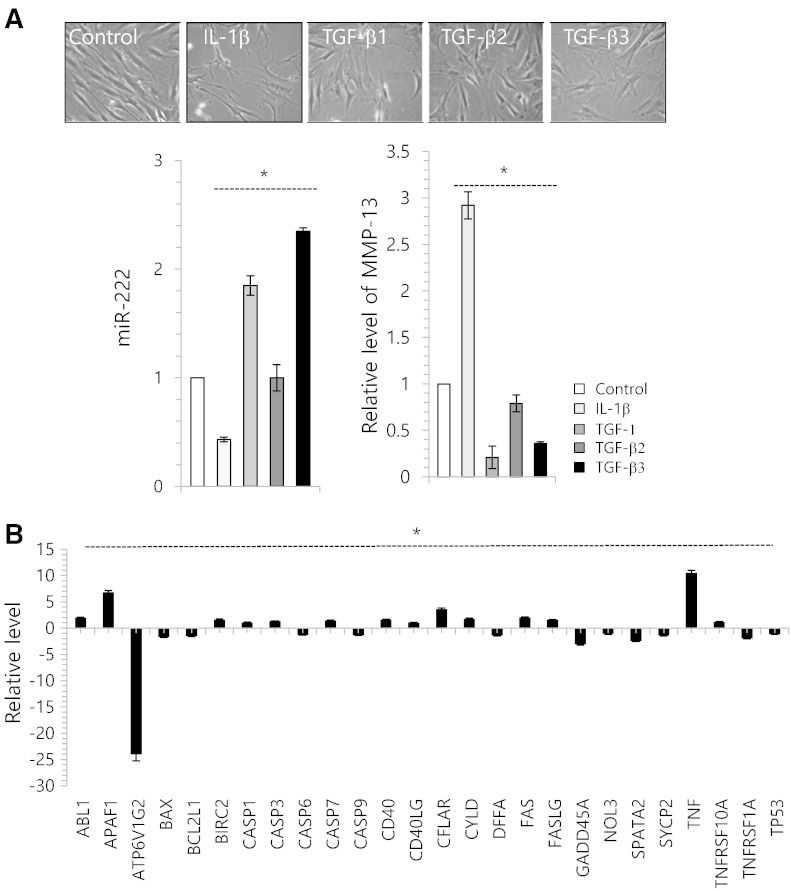
To verify the involvement of miR-222 in the degeneration of human articular chondrocytes, biopsy samples of normal cartilage from seven adult donors without history of joint disease (mean age 40.4 years) were used for the isolation of primary articular chondrocytes and treated with IL-1β or TGF-β1, -2, or -3. With exposure of IL-1β to cells, we observed an increase in cells having a degenerative morphology (Fig. 3A, upper panel). The expression level of miR-222 was significantly decreased by IL-1β and significantly increased by TGF-β1, 2, 3 (Fig. 3A, lower left panel). Moreover, the RNA level of MMP-13 was significantly increased by IL-1β, whereas the MMP-13 was significantly suppressed by TGF-β1, -2, or -3 (Fig. 3A, lower right panel). The RNA levels of the apoptotic genes including ABL-1, CASPs, CD40, and FAS were significantly up-regulated with IL-1β-treatment (Fig. 3B).
Fig. 3.
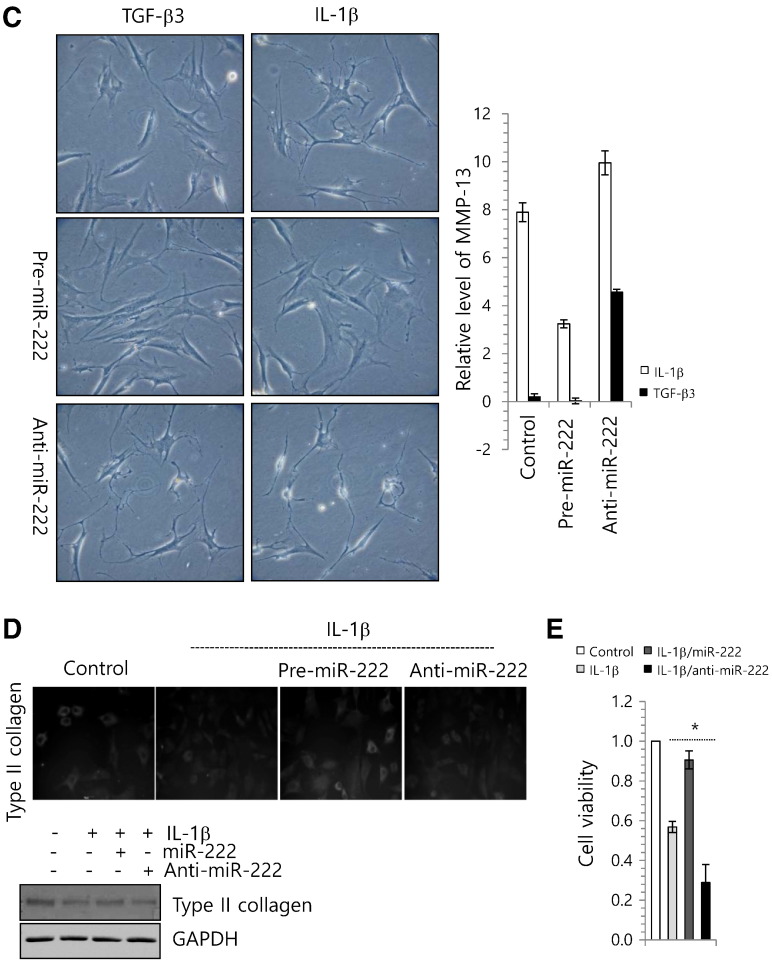
miR-222 is involved in degeneration of human articular chondrocytes. (A) Chondrocytes were isolated from biopsy sample of normal cartilage (control) and cultured with 5 ng/ml IL-1β, 3 ng/ml TGF-β1, -2, or -3. Cell images were captured (upper panel) and changes in the expression levels of miR-222 and MMP-13 were analyzed. (B) Chondrocytes were cultured with 5 ng/ml of IL-1β, and changes in the RNA level of genes involved in apoptosis were analyzed by real-time PCR. (C) Chondrocytes were cultured with 5 ng/ml of IL-1β or 3 ng/ml of TGF-β3 in the presence of miR-222 precursor (pre-miR-222) or miR-222 inhibitor (anti-miR-222). Cell images of each culture were taken (right panel), changes in the RNA levels of MMP-13 was examined by real-time PCR (left panel). (D) Human normal articular chondrocytes were isolated from biopsy cartilage and cultured with 5 ng/ml of IL-1β in the presence of pre-miR-222 or anti-miR-222. The protein level of type II collagen was examined by immunohistochemisty (upper panel) and immunoblotting (lower panel). GAPDH was used as loading control. (e) Cell viability was analyzed. The mean is plotted and the error bars represent 95% CI (lower/upper limit). ***Statistically different from control cells (p < 0.005).
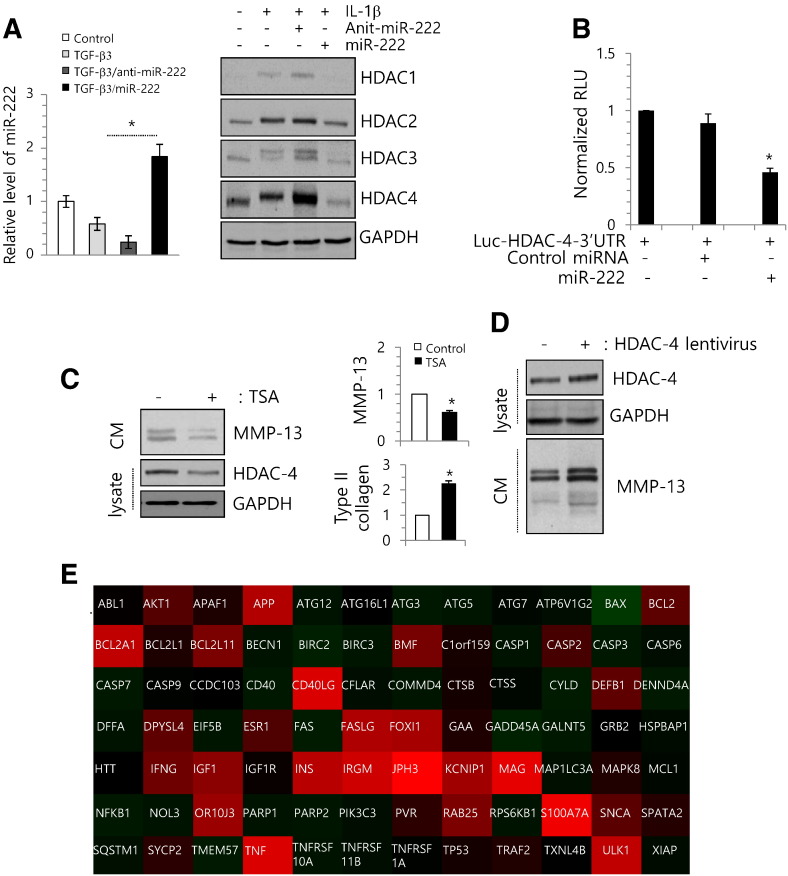
To further investigation to verify the involvement of miR-222 in the degeneration of human articular chondrocyte, normal chondrocytes were treated with miR-222 precursor (miR-222) or miR-222 inhibitor (anti-miR-222) in the presence of IL-1β or TGF-β3, and the RNA level of MMP-13 was analyzed. The introduction of anti-miR-222 dramatically increased the RNA level of MMP-13 in IL-1β-treated and TGF-β3-treated chondrocytes whereas the introduction of miR-222 was dramatically inhibited IL-1β-induced degenerative phenotype and RNA level of MMP-13 (Fig. 3C). In addition, the introduction of miR-222 recovered protein level of type II collagen (Fig. 3D) and rescued cell viability (Fig. 3E), which were suppressed by IL-1β treatment.
3.3. MiR-222 targets HDAC-4 that involved in regulation of MMP-13 and chondrocytes apoptosis
To validate the involvement of miR-222 in up-regulating HDACs in OA chondrocytes, normal chondrocytes were electroporated with miR-222 or anti-miR-222 in the presence of TGF-β3 or IL-1β. The efficiency of miR-222 or anti-miR-222 was confirmed with RT-PCR (Fig. 4A, left panel). The administration of miR-222 significantly suppressed the protein level of HDAC-4 in IL-1β-treated chondrocytes, whereas anti-miR-222 increased HDAC-4 expression (Fig. 4A, right panel). To validate the interaction between miR-222 and HDAC-4, we cloned the entire 3′-UTR of HDAC-4 into a luciferase reporter vector, electroporated the vector into normal chondrocytes along with the precursor of miR-222 or a cognate non-targeting negative control, and assayed cell lysates for luciferase expression. We found that cells transfected with the HDAC-4 3′-UTR-driven vector plus miR-222 exhibited significantly less luciferase activity compared to cells that received the reporter plus the non-targeting negative control (Fig. 4B).
Fig. 4.
miR-222 targets HDAC-4 and regulates MMP-13 activation. (A) Chondrocytes were isolated from biopsy sample of normal cartilage (control) and cultured with 3 ng/ml of TGF-β3 in the presence of miR-222 precursor (pre-miR-222) or miR-222 inhibitor (anti-miR-222). Expression level of miR-222 was analyzed (left panel) and changes in the protein level of HDACs were analyzed by immunoblotting (right panel). GAPDH was used as loading control. (B) Luciferase reporter gene assays of cells expressing the construct containing the human HDAC-4 3′-UTR in the absence or presence of miR-222. (C) OA chondrocytes isolated form OA cartilage were treated with 300 nM TSA, changes in protein level of active MMP-13 and HDAC-4 were analyzed by immunoblotting (left panel), and changes in RNA levels of MMP-13 were analyzed by RT-PCR (right panel). GAPDH was used as loading control. (D) OA chondrocytes were infected with lentiviruses containing HDAC-4. Changes in HDAC-4 and MMP-13 were analyzed by immunoblotting. GAPDH was used as loading control. (E) OA chondrocytes were treated with 300 nM TSA changes in the RNA level of genes involved in apoptosis were analyzed by RT-PCR and results were represented as heat-map using Qiagen program. “Red” indicates high relative expression, and “green” indicates low relative expression. The mean is plotted and the error bars represent 95% CI (lower/upper limit). *Statistically different from control cells (p < 0.005).
To examine the role of histone acetylation in OA pathogenesis, we treated OA chondrocytes with Trichostatin A (TSA), an HDAC inhibitor or infected OA chondrocytes with lentiviruses containing HDAC-4 siRNA (siHDAC-4 lentiviruses). MMP-13 and HDAC protein level were blocked by TSA treatment (Fig. 4C). Furthermore, the introduction of HDAC-4 lentiviruses was increased MMP-13 activation (Fig. 4D). In addition, the RNA levels of the apoptotic genes, such as ABL-1, CASPs, CD40, and FAS that were up-regulated in degenerated chondrocytes in Fig. 3B, were down-regulated by TSA treatment (Fig. 4E).
3.4. Modulation of miR-222 regulates OA pathogenesis in vivo
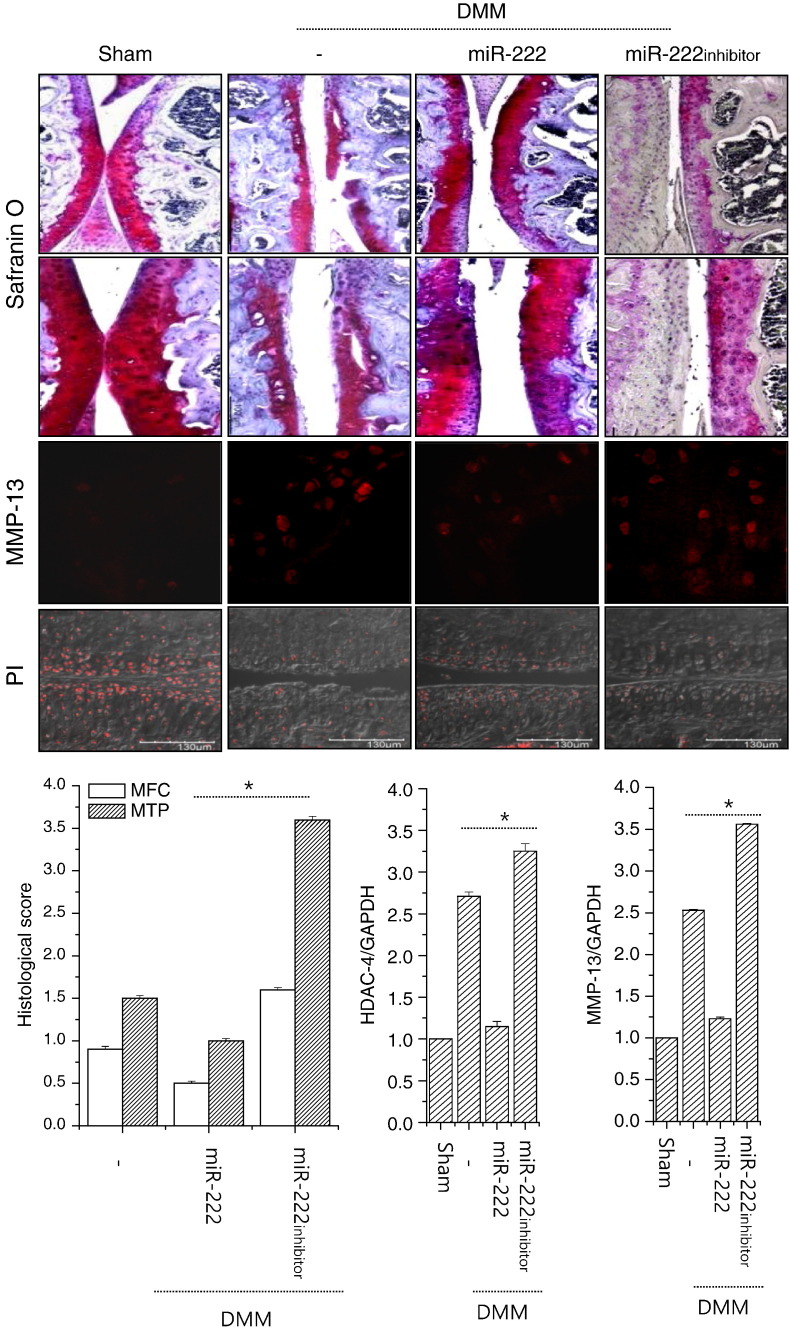
To examine the role of miR-222 in OA and to determine whether its modulation could control the pathogenesis of OA in vivo, we overexpressed or knocked down miR-222 in cartilage tissues by injecting lentiviruses containing miR-222 or miR-222 siRNA into DMM mouse knee joints. As shown in Fig. 5, the cartilage destruction caused by DMM surgery was significantly reduced by the over-expression of miR-222, whereas it was intensified by suppression of miR-222. Consistent with this, enhanced RNA and protein levels of MMP-13 were decreased by the over-expression of miR-222 and increased by suppression of miR-222 in DMM-induced mice. The RNA level of HDAC-4 was also decreased by the over-expression of miR-222 in DMM-induced mice. According to nuclei staining, the introduction of miR-222 recovered cell number which was significantly decreased by DMM surgery. Semi-quantitative scoring of the cartilage destruction seen in safranin-O-stained photomicrographs of the medial femoral condyle (MFC) and medial tibial plateau (MTP) indicated that DMM surgery scored 1.2 for the MFC view and 1.5 for the MTP view. More severe cartilage destruction was observed in photomicrographs of DMM mice that had been infected with lentiviruses encoding the miR-222-specific siRNA (miR-222 inhibitor, MFC score of 3.2, MTP score of 3.7).
Fig. 5.
miR-222 involved in the pathogenesis of OA in DMM mice. Mouse cartilages with OA induced by destabilization of the medial meniscus (DMM) were stained with safranin O and MMP-13 (upper panel). Cartilage destruction was scored according to the guidelines of the OARSI histopathology initiative, the average scores of the MFC and MTP were plotted, and the RNA levels of HDAC-4 and MMP-13 were analyzed using RNAs isolated from paraffin section of mice cartilages (n = 4) (lower panel). Sham-operated (Sham) cartilage was used as control. The mean is plotted, and the error bars represent 95% CI (lower/upper limit). *Statistically different from control cells (p < 0.005).
4. Discussion
The primary pathogenic events in OA include loss and abnormal remodeling of the cartilage ECM. The accumulation of degraded fragments increases MMP-13 synthesis via a positive feedback loop that involves cell-surface integrins, resulting in the destruction of knee joints [34]. Consistent with previous findings [35], we herein confirmed that the RNA and protein level of MMP-13 were increased in OA cartilage. Furthermore, increased protein level of MMP-13 in OA chondrocytes was suppressed by TSA treatment, indicating that TSA may reduce cartilage degradation by suppressing MMP-13. Several reports have shown that HDAC inhibitors can repress the expression of MMPs in human chondrocytes. For example, Young et al. [36] showed that TSA repressed MMP-3 expression in human chondrocytes. Nasu et al. [31] noted that it repressed MMP-13 in the same cells; and multiple reports have shown that TSA decreases MMP-2 expression in different cell types [37], [38]. These results and our present findings indicate that HDAC inhibitors can suppress the expression of MMPs depending on cell types, environmental conditions, and other factors.
The HDACs participate in chromatin remodeling by deacetylating lysine residues and play pivotal roles in the epigenetic regulation of gene expression [39]. Aberrant HDAC activity has been documented in several types of disease, and HDACs have emerged as an attractive therapeutic target. However, the histone modification of chondrocytes has not been extensively studied in OA. Most of the existing studies have examined the ability of HDAC to block cartilage degradation by repressing the induction of cytokines and MMPs [36], [40]. Intra-articular injection of the HDAC inhibitor, TSA, was shown to protect against cartilage degradation during the development of OA in an experimental model [41], suggesting that HDAC activity is crucial for the catabolic activity of chondrocytes. HDAC inhibition is also known to modulate the RNA levels of anabolic cartilage genes, such as COL2A1, COL9A1, COMP, and ACAN [42], [43]. In OA, HDAC-7 is expressed in OA cartilage, where it controls MMP-13 expression [44]. Here, we further found that HDAC-4 is highly up-regulated in OA chondrocytes compared to normal chondrocytes.
Recent studies have shown that miRNAs may exert their activity by interfering with the epigenetic machinery, such as by modulating the expression of enzymes that regulate DNA methylation or histone modification [45], [46]. For example, miRNA-449a regulates histone acetylation status in prostate cancer cells by targeting HDAC-1 [47], and miRNA-9 and -206 contribute to regulating histone acetylation and HDAC-4 activity in Waldenström macroglobulinemia cells [48]. However, we do not yet know which miR genes are altered by HDACs during OA pathogenesis, and only a few miRNAs are known to be involved in the modulation of HDACs in OA. The present study provides crucial information on the regulation of HDAC-4 during the pathogenesis of OA. We focused on upstream regulators of enhanced acetylation and found several miRNAs that were deregulated in OA chondrocytes versus normal chondrocytes.
Previous studies on miRNAs in cartilage have been largely restricted to a few specific miRNAs. Iliopoulos and colleagues [49] showed miRNA profiling in OA cartilage compared to normal cartilage and found 16 osteoarthritic miRNAs including miR-22, miR-140, and miR-483. Unlike to Iliopoulos experiments, OA chondrocytes were isolated from OA cartilages of OA patients who had undergone TKR surgery and cultured prior to osteoarthritic miRNA profiling. Our miRNA profiling shared several miRNAs including miR-140 with Iliopoulos and colleagues. A cartilage-restricted miRNA, miR-140, was shown to target HDAC-4 [50], [51], a repressor of Runx249 and Smad3 [52], [53] in mice, and ADAMTS5 [54], [55] and IGFBP5 [56] in humans. Null mice of miR-140 showed an early-onset OA-like disease [54]. MiR-365 and miR-27 have also been shown to be involved in OA pathogenesis by targeting HDAC-4 and MMP-13, respectively [57].
There are major issues with the therapeutic use of HDAC inhibitors, such as their lack of specificity, their lack of isoform selectivity, and their lack of known targets [58]. Thus, this study has important therapeutic implications and provides a “fine tuning” means of specifically regulating HDAC-4 for the control of OA. In summary, we herein report that miR-222 is one of the miRNAs responsible for cartilage degradation in human OA chondrocytes, where it targets HDAC-4 and thereby alters MMP-13 activity. This suggests that miR-222 may serve as a potential HDAC-4 inhibitor for the therapeutic control of OA.
Author contribution
All authors were involved in drafting the article critically for important intellectual content. Dr. E-J Jin had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: C-H. Chun and E-J. Jin
Acquisition of data: J. Song and D. Kim,
Analysis and interpretation of data: C-H. Chun and E-J. Jin
Conflict of interest
There is no financial support or other benefits from commercial sources for the work reported in the manuscript. The authors have declared that no competing interest exists.
Acknowledgments
This work was supported by the National Research Foundation [NRF] of Korea Grant funded by the Korean Governments by the Ministry of Education [2012R1A1A2039074] and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) [2013R1A1A2011999 and 2011-0030130]. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tchetina E.V., Squires G., Poole A.R. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J. Rheumatol. 2005;32:876–886. [PubMed] [Google Scholar]
- 2.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Loeser R.F. Molecular mechanisms of cartilage destruction: mechanics, inflammatory mediators, and aging collide. Arthritis Rheum. 2006;54:1357–1360. doi: 10.1002/art.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bau B., Gebhard P.M., Haag J., Knorr T., Bartnik E., Aigner T. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 2002;46:2648–2657. doi: 10.1002/art.10531. [DOI] [PubMed] [Google Scholar]
- 5.Neuhold L.A., Killar L., Zhao W., Sung M.L., Warner L., Kulik J., Turner J., Wu W., Billinghurst C., Meijers T. Postnatal expression in hyaline cartilage of constitutively active human collagenase-3 (MMP-13) induces osteoarthritis in mice. J. Clin. Invest. 2001;107:35–44. doi: 10.1172/JCI10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little C.B., Barai A., Burkhardt D., Smith S.M., Fosang A.J., Werb Z., Shah M., Thompson E.W. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60:3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nebbioso A., Dell'Aversana C., Bugge A., Sarno R., Valente S., Rotili D., Manzo F., Teti D., Mandrup S., Ciana P., Mol Endocrinol J. HDACs class II-selective inhibition alters nuclear receptor-dependent differentiation. J. Mol. Endocrinol. 2010;45:219–228. doi: 10.1677/JME-10-0043. [DOI] [PubMed] [Google Scholar]
- 8.Tou L., Liu Q., Shivdasani R.A. Regulation of mammalian epithelial differentiation and intestine development by class I histone deacetylases. Mol. Cell. Biol. 2004;24:3132–3139. doi: 10.1128/MCB.24.8.3132-3139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halsall J., Gupta V., O'Neill L.P., Turner B.M., Nightingale K.P. Genes are often sheltered from the global histone hyperacetylation induced by HDAC inhibitors. PLoS One. 2012;7:e33453. doi: 10.1371/journal.pone.0033453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H.S., Hu C.Y., Chan H.Y., Liew Y.Y., Huang H.P., Lepescheux L., Bastianelli E., Baron R., Rawadi G., Clément-Lacroix P. Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br. J. Pharmacol. 2007;150:862–872. doi: 10.1038/sj.bjp.0707165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villar-Garea A., Esteller M. Histone deacetylase inhibitors: understanding a new wave of anticancer agents. Int. J. Cancer. 2004;112:171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- 12.Blanchard F., Chipoy C. Histone deacetylase inhibitors: new drugs for the treatment of inflammatory diseases? Drug Discov. Today. 2005;10:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]
- 13.Nishida K., Komiyama T., Miyazawa S., Shen Z.N., Furumatsu T., Doi H., Yoshida A., Yamana J., Yamamura M., Ninomiya Y. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis Rheum. 2004;50:3365–3376. doi: 10.1002/art.20709. [DOI] [PubMed] [Google Scholar]
- 14.Flynt A.S., Lai E.C. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceman S., Saugstad J. MicroRNAs: meta-controllers of gene expression in synaptic activity emerge as genetic and diagnostic markers of human disease. Pharmacol. Ther. 2011;130:26–37. doi: 10.1016/j.pharmthera.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H., Ye C., Ramirez D., Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One. 2009;27:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oglesby I.K., McElvaney N.G., Greene C.M. MicroRNAs in inflammatory lung disease-master regulators or target practice? Respir. Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambros V. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinton A., Hunter S., Reyes G., Fogel G.B., King C.C. From pluripotency to islets: miRNAs as critical regulators of human cellular differentiation. Adv. Genet. 2012;79:1–34. doi: 10.1016/B978-0-12-394395-8.00001-3. [DOI] [PubMed] [Google Scholar]
- 20.Jovanovic M., Hengartner M.O. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 21.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Q., Seeger F.H., Castillo J., Iekushi K., Boon R.A., Farcas R., Manavski Y., Li Y.G., Assmus B., Zeiher A.M. Micro-RNA-34a contributes to the impaired function of bone marrow-derived mononuclear cells from patients with cardiovascular disease. J. Am. Coll. Cardiol. 2012;59:2107–2117. doi: 10.1016/j.jacc.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 23.Ropers H.H. X-linked mental retardation: many genes for a complex disorder. Curr. Opin. Genet. Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Liu C.J. MicroRNAs in skeletogenesis. Front. Biosci. 2009;14:2757–2764. doi: 10.2741/3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 26.Goldring M.B., Martel-Pelletier J., Abramson S.B. Osteoarthritis, an inflammatory disease: potential implications for the selection of new therapeutic targets. Arthritis Rheum. 2001;44:1237–1247. doi: 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Goldring M.B. The role of cytokines as inflammatory mediators in osteoarthritis: lessons from animal models. Connect. Tissue Res. 1999;40:1–11. doi: 10.3109/03008209909005273. [DOI] [PubMed] [Google Scholar]
- 28.Lin H.Y., Chen C.S., Lin S.P., Weng J.R., Chen C.S. Targeting histone deacetylase in cancer therapy. Med. Res. Rev. 2006;26:397–413. doi: 10.1002/med.20056. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone R.W. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 30.Kortenhorst M.S., Carducci M.A., Shabbeer S. Acetylation and histone deacetylase inhibitors in cancer. Cell. Oncol. 2006;28:191–222. doi: 10.1155/2006/760183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasu Y., Nishida K., Miyazawa S., Komiyama T., Kadota Y., Abe N., Yoshida A., Hirohata S., Ohtsuka A., Ozaki T. Trichostatin, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthr. Cartil. 2008;16:723–732. doi: 10.1016/j.joca.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Chabane N., Zayed N., Afif H., Mfuna-Endam L., Benderdour M., Boileau C., Martel-Pelletier J., Pelletier J.P., Duval N., Fahmi H. Histone deacetylase inhibitors suppress interleukin-1beta-induced nitric oxide and prostaglandin E2 production in human chondrocytes. Osteoarthritis Cartilage. 2008;16:1267–1274. doi: 10.1016/j.joca.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 34.Zemmyo M., Meharra E.J., Kühn K., Creighton-Achermann L., Lotz M. Accelerated, aging-dependent development of osteoarthritis in alpha1 integrin-deficient mice. Arthritis Rheum. 2003;48:2873–2880. doi: 10.1002/art.11246. [DOI] [PubMed] [Google Scholar]
- 35.Koskinen A., Vuolteenaho K., Nieminen R., Moilanen T., Moilanen E. Leptin enhances MMP-1, MMP-3 and MMP-13 production in human osteoarthritic cartilage and correlates with MMP-1 and MMP-3 in synovial fluid from OA patients. Clin. Exp. Rheumatol. 2011;29:57–64. [PubMed] [Google Scholar]
- 36.Young D.A., Lakey R.L., Pennington C.J., Jones D., Kevorkian L., Edwards D.R., Cawston T.E., Clark I.M. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res. Ther. 2005;7:R503–R512. doi: 10.1186/ar1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estella C., Herrer I., Atkinson S.P., Quiñonero A., Martínez S., Pellicer A., Simón C. Inhibition of histone deacetylase activity in human endometrial stromal cells promotes extracellular matrix remodelling and limits embryo invasion. PLoS One. 2012;7:e30508. doi: 10.1371/journal.pone.0030508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L.T., Chang H.C., Chiang L.C., Hung W.C. Histone deacetylase inhibitor up-regulates RECK to inhibit MMP-2 activation and cancer cell invasion. Cancer Res. 2003;63:3069–3072. [PubMed] [Google Scholar]
- 39.Venugopal B., Evans T.R. Developing histone deacetylase inhibitors as anti-cancer therapeutics. Curr. Med. Chem. 2011;18:1658–1671. doi: 10.2174/092986711795471284. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Song Y., Jacobi J.L., Tuan R.S. Inhibition of histone deacetylases antagonized FGF2 and IL-1beta effects on MMP expression in human articular chondrocytes. Growth Factors. 2009;27:40–49. doi: 10.1080/08977190802625179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W.P., Bao J.P., Tang J.L., Hu P.F., Wu L.D. Trichostatin A inhibits expression of cathepsins in experimental osteoarthritis. Rheumatol. Int. 2011;31:1325–1331. doi: 10.1007/s00296-010-1481-7. [DOI] [PubMed] [Google Scholar]
- 42.Furumatsu T., Tsuda M., Yoshida K., Taniguchi N., Ito T., Hashimoto M., Ito T., Asahara H. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J. Biol. Chem. 2005;280:35203–35208. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 43.Hong S., Derfoul A., Pereira-Mouries L., Hall D.J. A novel domain in histone deacetylase 1 and 2 mediates repression of cartilage-specific genes in human chondrocytes. FASEB J. 2009;23:3539–3552. doi: 10.1096/fj.09-133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higashiyama R., Miyaki S., Yamashita S., Yoshitaka T., Lindman G., Ito Y., Sasho T., Takahashi K., Lotz M., Asahara H. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod. Rheumatol. 2010;20:11–17. doi: 10.1007/s10165-009-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noonan E.J., Place R.F., Pookot D., Basak S., Whitson J.M., Hirata H., Giardina C., Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 48.Roccaro A.M., Sacco A., Jia X., Azab A.K., Maiso P., Ngo H.T., Azab F., Runnels J., Quang P., Ghobrial I.M. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–1514. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iliopoulos D., Malizos K.N., Oikonomou P., Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M.K., Clark I., Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 51.Shimizu E., Selvamurugan N., Westendorf J.J., Olson E.N., Partridge N.C. HDAC4 represses matrix metalloproteinase-13 transcription in osteoblastic cells, and parathyroid hormone controls this repression. J. Biol. Chem. 2010;285:9616–9626. doi: 10.1074/jbc.M109.094862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pais H., Nicolas F.E., Soond S.M., Swingler T.E., Clark I.M., Chantry A., Moulton V., Dalmay T. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA. 2010;16:489–494. doi: 10.1261/rna.1701210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdes A.M., Spector T.D., Tamm A., Kisand K., Doherty S.A., Dennison E.M., Mangino M., Tamm A., Kerna I., Hart D.J. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 2010;62:2347–2352. doi: 10.1002/art.27530. [DOI] [PubMed] [Google Scholar]
- 54.Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glasson S.S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H.L., Flannery C.R., Peluso D., Kanki K., Yang Z. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 56.Tardif G., Hum D., Pelletier J.P., Duval N., Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet. Disord. 2009;10:148. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barter M.J., Bui C., Young D.A. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage. 2012:339–349. doi: 10.1016/j.joca.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Chang J., Varghese D.S., Gillam M.C., Peyton M., Modi B., Schiltz R.L., Girard L., Martinez E.D. Differential response of cancer cells to HDAC inhibitors trichostatin A and depsipeptide. Br. J. Cancer. 2012;106:116–125. doi: 10.1038/bjc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]