Abstract
Background
HSPB1 belongs to the family of small heat shock proteins (sHSP) that have importance in protection against unfolded protein stress, in cancer cells for escaping drug toxicity stress and in neurons for suppression of protein aggregates. sHSPs have a conserved α-crystalline domain (ACD), flanked by variable N- and C-termini, whose functions are not fully understood. Dominant missense variants in HSPB1, locating mostly to the ACD, have been linked to inherited neuropathy.
Methods
Patients underwent detailed clinical and neurophysiologic characterization. Disease causing variants were identified by exome or gene panel sequencing. Primary patient fibroblasts were used to investigate the effects of the dominant defective HSPB1 proteins.
Results
Frameshift variant predicting ablation of the entire C-terminus p.(Met169Cfs2*) of HSPB1 and a missense variant p.(Arg127Leu) were identified in patients with dominantly inherited motor-predominant axonal Charcot–Marie–Tooth neuropathy. We show that the truncated protein is stable and binds wild type HSPB1. Both mutations impaired the heat stress tolerance of the fibroblasts. This effect was particularly pronounced for the cells with the truncating variant, independent of heat-induced nuclear translocation and induction of global transcriptional heat response. Furthermore, the truncated HSPB1 increased cellular sensitivity to protein misfolding.
Conclusion
Our results suggest that truncation of the non-conserved C-terminus impairs the function of HSPB1 in cellular stress response.
General significance
sHSPs have important roles in prevention of protein aggregates that induce toxicity. We showed that C-terminal part of HSPB1 is critical for tolerance of unfolded protein stress, and when lacking causes axonal neuropathy in patients.
Abbreviations: ACD, α-crystalline domain; CADD, combined annotation dependent depletion; CMT, Charcot–Marie–Tooth disease; dHMN, distal hereditary motor neuropathy; EMG, electromyography; ENMG, electroneuromyography; EVS, exome variant server; MUP, motor unit potential; QST, quantitative sensory testing; sHSP, small heat shock protein; SISu, Sequencing Initiative Suomi
Keywords: Charcot–Marie–Tooth neuropathy, heat shock protein, HSPB1, Protein misfolding
Graphical abstract
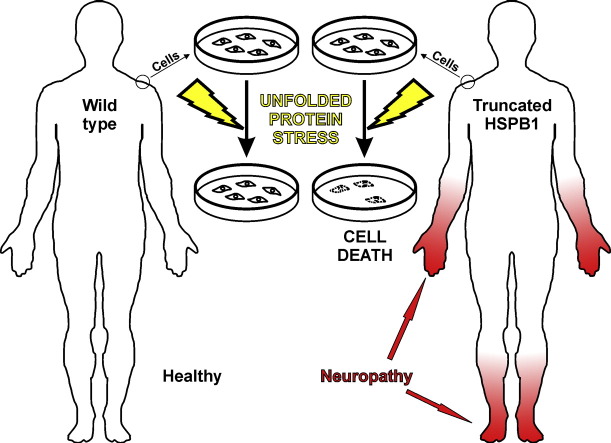
A truncating variant in the small heat shock protein HSPB1 causes neuropathy, which leads to distal weakness and sensory impairment in hands and feet. HSPB1 is important for the unfolded protein response. Cultured patient skin cells suffered and died when subjected to unfolded protein stress, whereas cells from a healthy person survived. These results demonstrate that biochemical abnormalities related to neuropathy manifest in skin cells, and suggest that neuropathy may develop as a consequence of decreased stress tolerance.
Highlights
-
•
C-terminal truncation of small heat shock protein HSPB1 causes neuropathy.
-
•
Truncated HSPB1 is stable in patient fibroblasts and binds wild type HSPB1.
-
•
C-terminus of HSPB1 is critical for tolerance to unfolded protein stress.
-
•
Neuropathy may develop as a consequence of impaired cellular stress response.
1. Introduction
Charcot–Marie–Tooth disease (CMT) is a hereditary disorder of peripheral nerves where patients suffer from chronic and progressive distal muscle weakness and sensory impairment as a result of demyelination (CMT1) or axon degeneration (CMT2) [1]. When motor symptoms predominate, the term distal hereditary motor neuropathy (dHMN) may be used, although genetic studies have shown that there is a significant overlap between CMT and dHMN [2]. The genetic heterogeneity of inherited peripheral neuropathies is very large, with more than 60 disease genes known [3].
Cellular stress responses mediated by small heat shock proteins (sHSP) are among the important pathways implicated in axon maintenance, as well as in many stress-related physiological processes and malignancies [4]. The human genome encodes genes for 10 sHSPs of which three (HSPB1, HSPB3 and HSPB8) are disease genes for CMT2 or dHMN [5], [6], [7]. In addition, HSPB5 variants cause myopathy and cardiomyopathy [8]. The common feature of the sHSPs is an approximately 85 amino acid stretch called the α-crystallin domain (ACD), which is bordered by N- and C-terminal regions [9]. Dimerization, thought to be crucial for protein function, depends on symmetrical antiparallel pairing of the β7 strands of ACD [10], [11]. The highly variable N- and C-termini have evolved independently from the ACD and may therefore be responsible for specific functional and structural effects that differentiate sHSPs from one another [12]. The C-terminus may be involved in chaperone functions through dynamic conformational changes that regulate oligomeric organization or target protein binding [13], [14]. An IxI/V domain of many sHSP C-termini binds the β4/β8 groove of an ACD, which may be important for oligomerization [15].
Ubiquitously expressed HSPB1 is a 205 amino acid protein with the ACD located at residues 86–169. The most important function of HSPB1 is thought to be the protection of the cell against stress. The ability of HSPB1 to bind and prevent the aggregation of misfolded proteins has been demonstrated in vitro [16]. Presumably, client proteins bound by HSPB1 can be transferred to ATP-dependent chaperones for active refolding, or for proteolytic degradation. A plethora of potential binding targets has been identified, including cytoskeletal components such as actin and myosin, and proteins linked to acquired neurodegenerative diseases such as synuclein, tau and β-amyloid [4], [17], [18]. Nuclear translocation of HSPB1 has been demonstrated upon heat stress in certain cell types [19], suggesting a role in chaperoning nuclear proteins or regulation of gene expression.
Nearly 20 disease causing variants in HSPB1 have been described [6], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]. They cause length-dependent, predominantly motor CMT2 or dHMN, being among the most common causes of these disorders [3]. Most of the disease-associated variants are dominantly inherited missense variants, clustering in structures that are conserved in all sHSPs, such as in the β7-strand of the ACD. Previous studies of different HSPB1 missense variants have solely utilized overexpression systems and identified pathways that may be affected in disease. Certain ACD mutations were associated with defective dimerization, increased chaperone activity, and improved heat stress tolerance [30]. Several studies have found cytoskeletal abnormalities and axonal transport defects. For instance, overexpression of HSPB1S135F or HSPB1P182L led to aggregation and altered axonal transport of neurofilament [6], [31], [32]. These mutants also displayed increased binding to tubulin, which led to stabilization and altered dynamics of microtubules [33]. Furthermore, transgenic expression of HSPB1S135F or HSPB1P182L in mouse neurons decreased the abundance of acetylated α-tubulin and induced severe axonal transport defects [34].
Two variants that predicted truncation of the C-terminus of HSPB1 have been described but the stabilities of the putative truncated proteins were not investigated [26], [27]. The disease mechanisms of the postulated C-terminal truncations have remained obscure. C-terminal missense variants produced mixed effects in vitro, including propensity to aggregate for HSPB1P182S and decreased chaperone activity for HSPB1R188W [35], whereas truncation of HSPB5 may impair oligomerization of that protein [36], [37]. Here we describe new disease-causing variants in HSPB1 in CMT2 patients, one of which leads to stable truncated HSPB1 lacking the entire C-terminus of the protein. To study the mutant proteins at their endogenous expression levels, we used primary patient fibroblasts. We demonstrate that the stable truncated HSPB1 binds wild type HSPB1, suggesting a dominant-negative effect, and strongly impairs the ability of the cells to cope with unfolded protein stress. These data have importance for understanding the molecular mechanisms behind axonal neuropathies and the chaperoning abilities of HSPB1.
2. Materials and methods
2.1. Patients and sampling
Patients and their family members gave a written informed consent to enroll in the study. Patients were clinically examined by the same neurologist at the Helsinki University Central Hospital. Approval of the study was given by the hospital's ethical committee. DNA was extracted from peripheral blood by standard methods. Skin biopsies were taken from volar antebrachium and fibroblast cultures were established. ENMG and QST were performed by established methods [38].
2.2. Exome and gene panel sequencing
We screened a panel of neuropathy-associated genes as described previously [29], with modifications in the panel including addition of disease genes for hereditary spastic paraplegia (Supplementary table). The target was designed with SureDesign software (Agilent Technologies, Santa Clara, CA, USA). Target enrichment and amplification was done with HaloPlex Target Enrichment Kit (Agilent Technologies) according to the manufacturer's instructions. For whole exome sequencing (WES) we used NimbleGen Sequence Capture method according to the manufacturer's instructions. Sequencing was done on a MiSeq sequencer. Genome alignment and variant calling were done by the pipeline developed at the Finnish Institute of Molecular Medicine [39].
2.3. NGS data filtering
Missense variant data of the exome and gene panel were filtered as follows: 1) Exclusion of non-splice site changing variants in non-coding regions, 2) exclusion of synonymous variants, 3) exclusion of variants with frequency > 0.005 in the 1000Genomes (www.100genomes.org) or Exome Variant Server (EVS, http://evs.gs.washington.edu/EVS/), and 4) exclusion of variants that scored < 10 for deleteriousness with the Combined Annotation Dependent Depletion (CADD) tool [40]. The final variants were checked for frequencies in the Sequencing Initiative Suomi (SISu, http://sisu.fimm.fi/), a database that contains exome data from > 3000 individuals of Finnish origin [41].
Insertion/deletion (indel) data were filtered by: 1) Exclusion of non-splice site changing variants in non-coding regions and 2) exclusion of variants found in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). In addition, some false positives on the gene panel data were excluded based on variant calls being present in only one restriction fragment as evaluated with Integrative Genomics Viewer (http://www.broadinstitute.org/software/igv/home).
2.4. Sanger sequencing
Findings were confirmed by Sanger sequencing. The oligonucleotide primers used for HSPB1 were ACCCGGTGTGTAATGTAACG and GCCTGAGGCTTCCTTCCAC for exon 3, and AAGTTTCTGAGAGCCCAGACC and ACAGGGAGGAGGAAACTTGG for exon 2.
2.5. Western blotting and antibodies
Whole-cell lysates were prepared by lysis in RIPA buffer (1 × PBS, 1% Nonidet-P-40, 0.5% sodium deoxycholate, 0.1% SDS). Non-reducing Western blotting conditions were essentially as previously described [30]: cells were suspended in 50 mM Tris–HCl (pH 8.0), 10% glycerol, 1% Nonidet P-40, 150 mM NaCl, 5 mM NaF, 5 μM ZnCl2, 1 mM Na3VO4, 10 mM EGTA and Complete Protease inhibitors (Roche, Basel, Switzerland); lysed on ice for 10 min and cleared by centrifugation; and then boiled for 5 min in non-reducing loading buffer (5 × solution: 250 mM Tris–HCl pH 6.8, 10% SDS, 30% glycerol, 0.02% bromophenol blue). For nuclear enrichment, cells were first homogenized by running through a 22 G syringe needle 8–12 times in buffer containing 0.3 M sucrose, 1 mM EGTA, 5 mM MOPS, 5 mM KH2PO4 and Complete Protease inhibitors at pH 7.4. Lysates were then centrifuged at 6500 rpm for 15 min at 4 °C; the supernatant was taken as the cytosolic fraction and the pellet as the nuclear fraction. The nuclear fractionations were done on non-treated cells and cells subjected to 45 °C for 1 h. SDS–PAGE was performed by standard methods.
Antibodies used were: anti-HSPB1 (18284-1-AP; Proteintech, Chicago, IL, USA), anti-GAPDH, anti-histone H3, and anti-tubulin (#2118, #4499 and #2146, respectively, Cell Signaling Technology, Danvers, MA, USA).
2.6. Stress tolerance assays
To test heat tolerance, cultured fibroblasts were subjected to 45 °C for 2 h after which the media was changed and the cells were returned to normal conditions and followed for 48 h. To induce protein misfolding stress, fibroblast media were supplemented with 20 mM canavanine (Sigma Aldrich, St Louis, MO, USA) and the cells were followed for 48 h. Cell numbers and morphology were monitored and analyzed by Cell IQ system (CM Technologies, Tampere, Finland). Each cell line was cultured in duplicate wells and 50 regions per well were imaged every second hour. Two-way ANOVA test was used for statistical analysis.
2.7. Immunocytochemistry
Immunocytochemistry to detect HSPB1 in cultured fibroblasts was done as follows: The cells were fixed with paraformaldehyde, permeabilized with Triton X and stained using the same anti-HSPB1 antibody as in Western blotting and with DAPI. Imaging was done with an Axioplan 2 fluorescence microscope (Zeiss, Jena, Germany) and the Axiovision 4.8.1.0 software. The cells were either non-treated or subjected to heat stress at 45 °C for 1 h.
2.8. Gene expression analysis
Control and HSPB1∆C-term patient fibroblasts were treated by exposure to 45 °C for 30 min. Total RNA was extracted from treated and untreated cells, using three independent replicates. Gene expression profiling of patient and control with and without treatment were carried out at Functional Genomics Unit (Biomedicum Helsinki, Finland) using an Illumina HumanHT-12 version 4 (Illumina, San Diego, CA, USA) expression array. The data analysis consisted of data pre-processing, quality analysis and detection of differentially expressed genes between the study groups. All methods used were implemented in the beadarray, limma, and BioMart packages of the Bioconductor project [42], [43], [44], [45], [46]. The result sheets from GenomeStudio were loaded to R, normalized, log2-transformed and background corrected. Additional gene information was extracted from EnsEMBL using BioMart [45]. Matching was done via EnsEMBL gene names. Pathway analysis with tests for statistical significance was done using DAVID Bioinformatics Resources 6.7 [47], [48].
3. Results
3.1. Clinical and neurophysiologic findings
Three patients from two unrelated families were studied. In family A (Fig. 1A), the index patient (II-2) was the only affected member. Her parents were both deceased, but they or her ten siblings did not have any symptoms of muscle weakness; her older sister had developmental delay but no neuropathy. In childhood the patient had difficulties in skills requiring balance and in sports, but only in her thirties she started to experience progressive symptoms and distal weakness of the feet. Clinical picture was slowly progressive requiring first peroneal support for distal muscle weakness, and after several years more aids. At the age of 65 the patient used two walking sticks, and could walk two kilometers at most. She was otherwise healthy, but due to toe deformities several operations had been performed. At neurological examination muscle atrophy concentrated at distal muscles, and there was only visible contraction without movement at ankle and distally. Muscle weakness was observed also proximally, especially in hip flexion. In hands, milder distal atrophy and weakness were noted, however proximal strength in arm abduction and elbow flexion was normal. Also sensory disturbances were observed in the feet with distally impaired touch and position senses. ENMG (electroneuromyography) showed marked sensory and active motor axonal neuropathy with distal and lower leg preponderance (Table 1). There was accentuation of needle EMG (electromyography) and sural sensory response alterations compared to ENMG carried out six years earlier. QST (quantitative sensory testing) was done to investigate small fiber involvement. It showed severe alteration of cold perception (A delta fibers) in addition to distinct alteration of warm sensory perception (C fibers) and vibration perception (A beta fibers) in the lower extremities, whereas only mild change in cold perception was recorded in the upper extremity (Table 1).
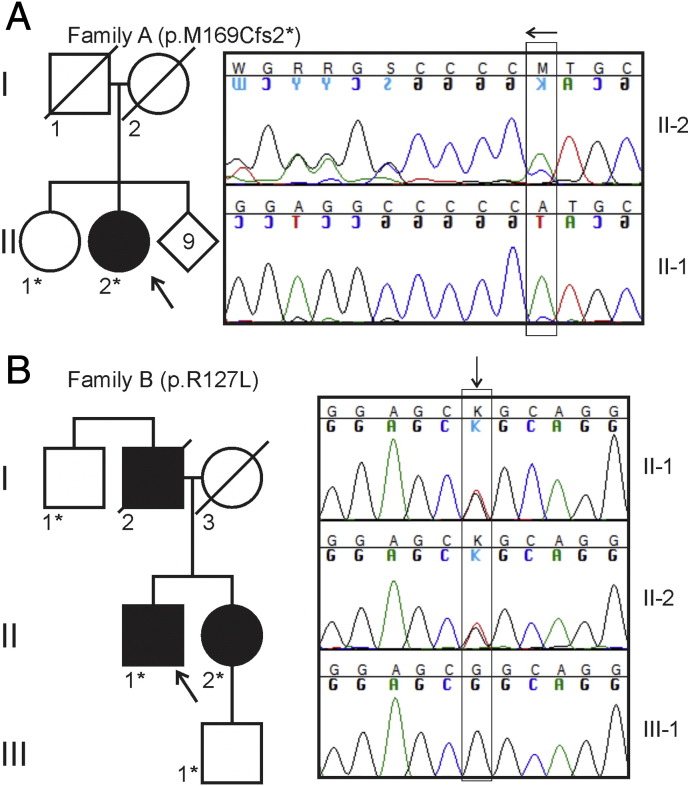
Fig. 1.
Pedigrees of the studied families. The DNA of the persons indicated by asterisk (*) was studied, persons crossed with diagonal lines were deceased. Representative capillary sequencing chromatograms are shown to the right. (A) In family A, the heterozygous deletion of a single adenine residue [c.505delA, HSPB1 (NM_001540.3)] is highlighted by the vertical box. The deletion leads to a frame shift causing superimposed curves in the sequence that follows (arrow, note that sequencing direction is right to left). (B) In family B, the heterozygous guanine to thymidine nucleotide change [c.380G > T, HSPB1 (NM_001540.3)] is indicated by the vertical box (arrow).
Table 1.
Neurophysiological investigations of the patients. APB = abductor pollicis brevis, DPN = deep peroneal nerve, EDC = extensor digitorum communis, LE = lower extremity, MN = median nerve, MUP = motor unit potential, RN = radial nerve, SN = sural nerve, SPN = superficial peroneal nerve, TN = tibial nerve, UE = upper extremity, UN = ulnar nerve.
| Patient | Age at examination | Neurography Motor fibers |
Neurography Sensory fibers |
Needle EMG | QST |
|---|---|---|---|---|---|
| A:II-2 | 65 | MN normal but DPN decreased motor conduction velocity and diminished M-response amplitude. | Normal conduction velocity but diminished amplitudes in upper and lower extremity (RN, MN, UN, SN), no response in SPN. Decreased mixed nerve amplitude (MN). | Fibrillation activity in leg muscles with distal preponderance, mild/moderate neuropathic MUP alterations in the deltoid and long finger extensor muscles, no fibrillations. Fibrillation activity and severe MUP alterations in the first interosseus dorsalis muscle | UE: Mild change A delta in the left hand. LE: Severe alteration A delta and distinct alteration in C-fibers and A beta fibers. |
| B:II-1 | 63 | Moderately decreased motor conduction velocity, increased distal motor latency, and diminished M-response amplitude (about 10% of normal, left MN). | Normal conduction velocity but diminished sensory response (left RN). Severely decreased mixed nerve conduction velocity and diminished response amplitude (MN). |
Fibrillation activity, moderate loss of motor units as well as motor unit potential alterations (right APB and left EDC). The left deltoid muscle was normal. | LE: C and A beta fiber dysfunction. |
| B:II-2 | 59 | Distal motor responses absent (DPNs) or remarkably decreased (TNs). UE: Normal |
Normal | Atrophy with no voluntarily activating MUPs (feet); fibrillation, motor unit loss and re-innervation (legs); slight motor unit loss without fibrillation (thighs). UE: Normal |
Normal |
In family B (Fig. 1B) the index male patient (II-1) developed slowly progressive polyneuropathy symptoms starting at age 35. At age 63 he moved slowly needing assistance of two walking sticks and peroneus supports. In the arms there were atrophy and weakness in distal hand muscles, but normal strength in proximal muscles. In lower limbs there was no movement at ankles and also proximal strength was reduced, more in the right. Reflexes were absent. Vibration sense was absent in the legs. The ENMG showed marked motor axonal neuropathy with distal preponderance. There was also diminished sensory response in the left radial nerve indicating axonal neuropathy of A beta fibers, in addition to decreased mixed nerve conduction velocity and decreased response amplitude in the median nerve indicating involvement of A alpha fibers. QST showed elevated warm sensation and vibration thresholds in lower extremity indicating C and A beta fiber dysfunction (Table 1).
In his younger sister (B:II-2) neuropathic symptoms began around the same age. Her main symptom was a sensation of stiffness and distal weakness in the lower extremities requiring peroneal orthoses, and occasional muscle cramps. Upon clinical examination at age 59, she was unable to walk on her heels, but could briefly rise to her toes. In the upper extremities, muscle strength, sensory testing and deep tendon reflexes were normal and no atrophy was noted. In the lower extremities, mild atrophy of intrinsic foot muscles was noted. Great toe dorsiflexion was clearly weakened, and ankle dorsi- and plantarflexion strengths were reduced. There was also proximal muscle weakness in the lower extremities, more pronounced on the right side. Lower limb deep tendon reflexes were missing. Sensation of touch and vibration were normal, but the sensation of pain was slightly pronounced bilaterally in the legs. ENMG revealed signs of moderate distal motor axonal polyneuropathy while QST was normal (Table 1). Four years earlier at the age of 55, ENMG measured in another laboratory revealed only slight signs of unspecific neuropathy as increased latencies and diminished persistencies of the lower extremity F- and H-waves as well as slight fibrillation and MUP (motor unit potential) changes in the distal leg muscles.
3.2. Genetic findings
The DNA of the index patient of family A was subjected to targeted gene panel sequencing. Filtering of missense data left no variants with frequency < 0.005 in the SISu database but filtering of indel data resulted in a single previously unknown variant, heterozygous c.505delA in HSPB1 (NM_001540.3). The variant was situated in the last exon (exon 3) of the gene and was predicted to lead to a p.(Met169Cfs2*) change, i.e. truncation of the entire C-terminus (HSPB1∆C-term) of the 205 amino acid protein. The variant was confirmed by Sanger sequencing. The patient's older sister tested negative for the mutation but the DNA of her parents or other siblings were not available for testing; they were never formally evaluated for neuropathy and had not reported symptoms. This suggested the possibility that the patient's mutation had occurred de novo (Fig. 1A).
In family B, the index patient and his sister (II-2 in Fig. 1B) underwent exome sequencing. Filtering left 159 shared heterozygous variants, out of which three were in genes previously linked to neuropathy. The first was a c.2750A > C variant in KIF1A (NM_004321.6), predicting a p.(His917Pro) amino acid change, which was found at a frequency of 0.0015 in the SISu database, i.e. being a relatively common polymorphism in Finland and unlikely to cause dominant disease.
The second was a c.684C > G variant in SCN9A (NM_002977.3), predicting a p.(Ile228Met) amino acid change. This variant was present with frequency of 0.0014 in SISu, and 0.0016 in European Americans of the EVS. Despite being relatively common, this variant has been previously suggested as a cause of small fiber neuropathy [49], [50], [51] but has not been linked to axonal neuropathy. A possibility exists that this variant could contribute to small fiber involvement in patient B:II-1. However, no small fiber involvement was documented in patient B:II-2.
The third variant, c.380G > T in HSPB1 (NM_001540.3), predicting a p.(Arg127Leu) amino acid change, was considered to be the pathogenic variant in family B, because it was not found in any of the investigated SNP databases and a change from arginine to tryptophan in the same codon has been previously described in a Belgian family with dHMN [6], and in 4 Chinese families diagnosed with CMT2 onset between ages 35 and 60 where motor symptoms dominated over sensory symptoms [28]. The variant was confirmed by Sanger sequencing and it segregated with the disease phenotype as the index patient's healthy son and paternal uncle were both negative. The father of the index patient had reported symptoms consistent with neuropathy but was already deceased and thus unavailable for the study (Fig. 1B).
3.3. Truncated HSPB1 protein is stable and binds wild-type HSPB1
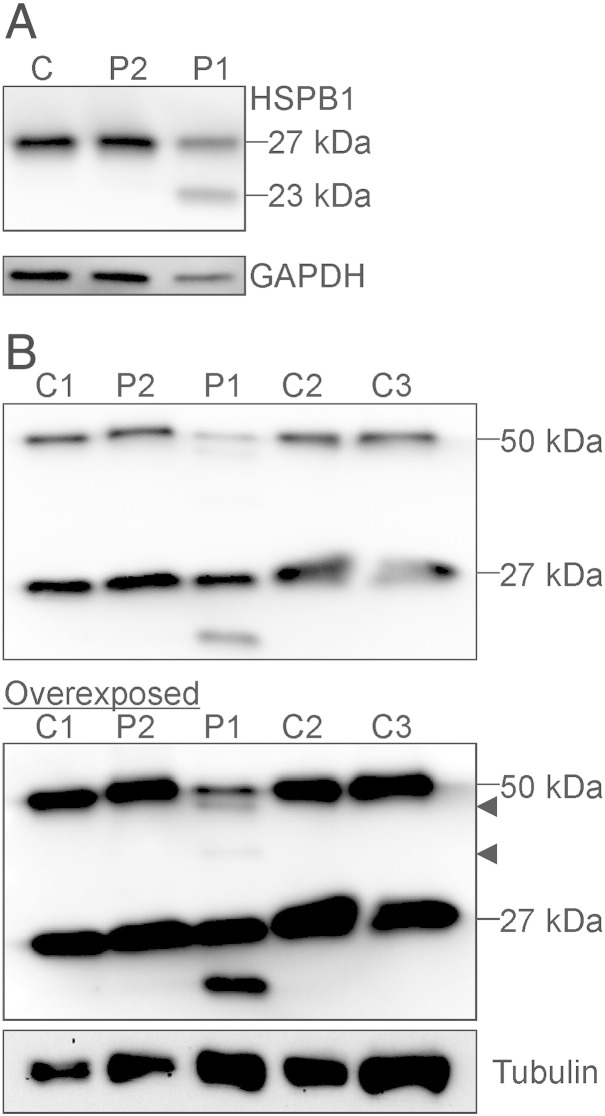
Variants leading to premature stop codons exert dominant negative effects only if the mRNAs escape nonsense-mediated decay and the stable protein is expressed. We assessed the stability of the truncated HSPB1 by SDS–PAGE on lysates of primary patient fibroblasts. While the HSPB1R127L fibroblasts had a similar amount of full-length HSPB1 protein compared to control fibroblasts, the HSPB1∆C-term fibroblasts showed two bands reacting with anti-HSPB1, corresponding to the full-length and truncated proteins (Fig. 2A).
Fig. 2.
Stability and dimerization of the truncated protein. Lysates from primary fibroblasts were analyzed by Western blotting and immunodetection with anti-HSPB1 antibody. (A) Shown is a representative blot form control (C), HSPB1R127L (P2) and HSPB1∆C-term (P1) fibroblasts. The wild type HSPB1 protein of 27 kDa size is detected in all samples. In HSPB1∆C-term fibroblasts an additional band about 23 kDa in size, corresponding to the truncated protein, is observed. (B) Western blots under non-reducing conditions were performed to detect HSPB1 dimers. At normal exposure the control fibroblasts (C1–3) show a band at ~ 27 kDa corresponding to the wild type monomer and at ~ 50 kDa corresponding to the wild type dimer. The HSPB1R127L fibroblasts (P2) are similar to wild type. In HSPB1∆C-term fibroblasts (P1), the ~ 27 kDa monomer and ~ 50 kDa dimer can be detected, but overexposure of the same blot shows two additional fainter bands (arrowheads) corresponding to dimers formed by wild type and truncated protein or two truncated proteins.
HSPB1 dimers are known to be resistant to protein denaturation but are dissociated by reducing agents [30]. We tested the abilities of the mutant proteins to form dimers in patient cells using non-reducing Western blot (Fig. 2B). The control and HSPB1R127L fibroblasts showed bands corresponding to the 27 kDa monomer and the ~ 50 kDa dimer. However, in the overexposed blot the HSPB1∆C-term fibroblasts showed two additional bands between the normal size monomer and the dimer. These bands presumably corresponded to dimers formed by a wild type and a truncated protein, and by two truncated proteins, respectively. The total abundance of wild type HSPB1 dimer was close to the expected 25% if the HSPB1∆C-term protein binds HSPB1WT protein indiscriminately. No larger bands were detected above 50 kDa, consistent with higher order multimers being susceptible to denaturation. These results suggest that the HSPB1∆C-term exerts a dominant negative effect by binding the HSPB1WT protein thus strongly decreasing the abundance of wild type dimer.
3.4. Truncated HSPB1 impairs tolerance to heat without altering nuclear translocation or gene expression
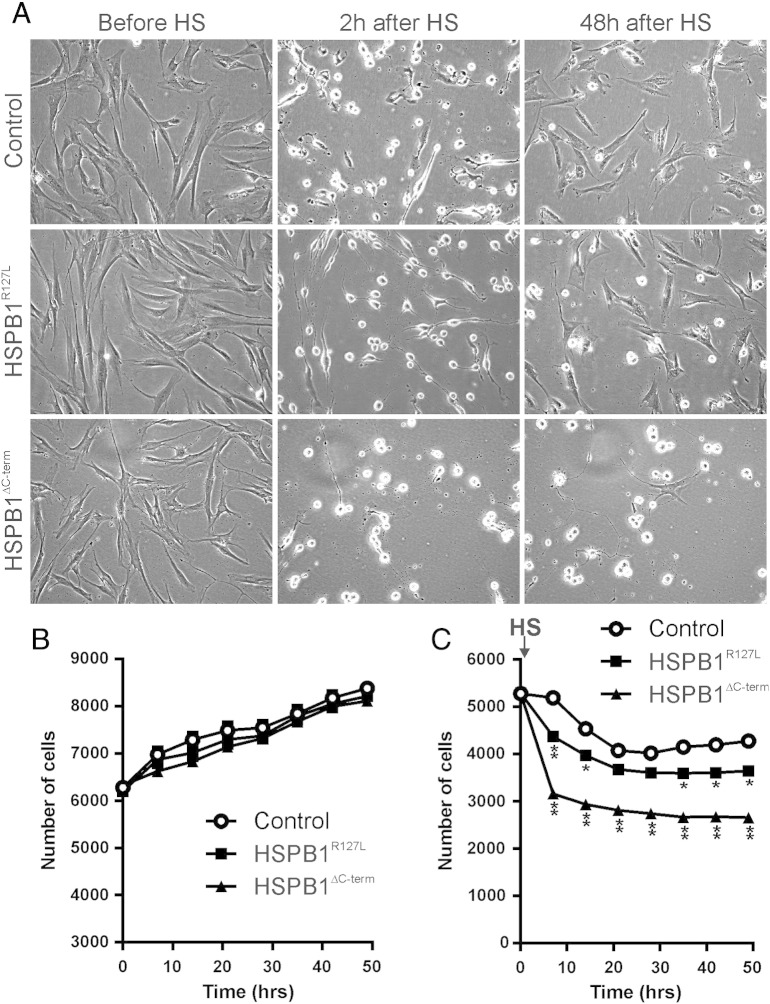
Next we assessed the effect of HSPB1 mutations on heat tolerance. Continuous cell monitoring was used to observe the growth of the fibroblasts under normal conditions and after heat stress (Fig. 3A). Quantification showed that the growth rates of patient and control fibroblasts were equal at normal temperature (Fig. 3B). Cells were then subjected to 45 °C for 2 h, washed and followed for a further 48 h. In HSPB1R127L and HSPB1∆C-term fibroblasts the proportion of cells dying from heat stress was greater, and the growth rate after heat stress significantly slower than in control fibroblasts (Fig. 4C). The impairment was more severe in HSPB1∆C-term than in HSPB1R127L fibroblasts.
Fig. 3.
Cell growth and survival after heat stress. The growth of the fibroblasts was monitored under normal culture conditions. Then the cells were subjected to 45 °C for 2 h (HS), followed by washing out of floating dead cells and automated cell monitoring for 48 h. (A) Representative images are shown for each condition in control, HSPB1R127L and HSPB1∆C-term fibroblasts. Based on cell morphology, the control and HSPB1R127L cells had mostly recovered 48 h after heat treatment. (B) Under normal conditions, cell growth of all three lines was identical. (C) After heat stress (HS, arrow) cell survival and recovery of HSPB1R127L and HSPB1∆C-term fibroblasts were worse than of control. The impairment was significantly more marked in HSPB1∆C-term fibroblasts compared to HSPB1R127L (P < 0.0001). Asterisks denotes significant differences compared to control fibroblasts *P < 0.01, **P < 0.0001 (two-way ANOVA).
Fig. 4.
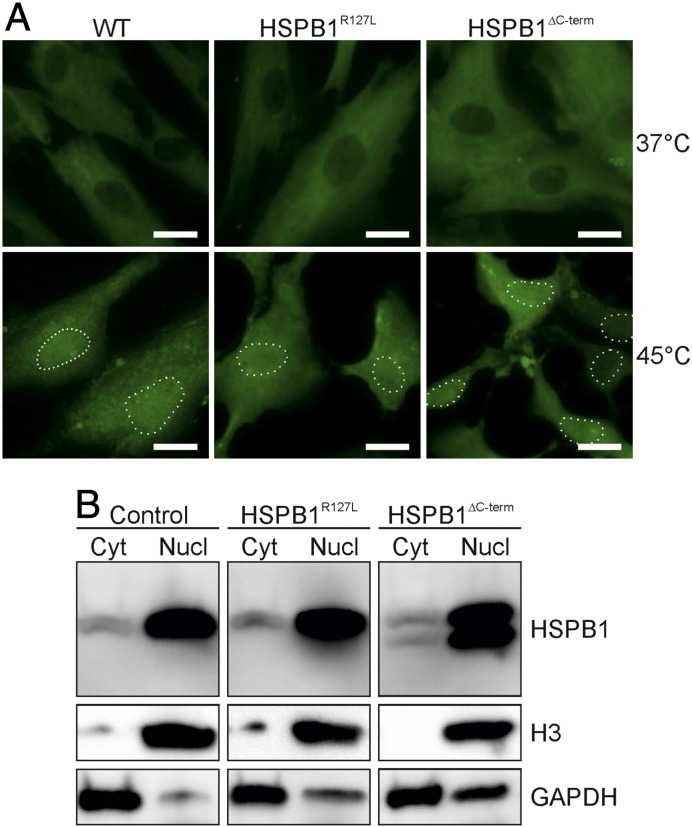
Nuclear translocation of HSPB1. (A) Immunocytochemistry of primary fibroblasts using the anti-HSPB1 antibody (green) showed that part of the cytoplasmic protein translocates to the nucleus in 45 °C in control (WT), HSPB1∆C-term and HSPB1R127L fibroblasts. Nuclei are highlighted with dotted lines. Bars = 20 μm. (B) Nuclear enrichment was performed on cells at 45 °C, followed by Western blotting to detect HSPB1. Histone H3 protein was detected as a nuclear loading control, and GAPDH as a cytoplasmic loading control. The abundance of HSPB1 was strongly increased in the nucleus (Nucl) compared to cytoplasm (Cyt) at 45 °C. Note that the HSPB1∆C-term protein is detectable and concentrated in the nucleus following heat stress, similar to the wild type protein.
HSPB1 is known to translocate to the nucleus upon heat stress [19]. We used immunocytochemistry and Western blotting to confirm the nuclear translocation of HSPB1 (Fig. 4A). Since the HSPB1∆C-term protein can be discriminated from HSPB1WT protein on Western blot, we were able to demonstrate that the truncated protein was enriched in the nuclear fraction upon heat stress (Fig. 4B).
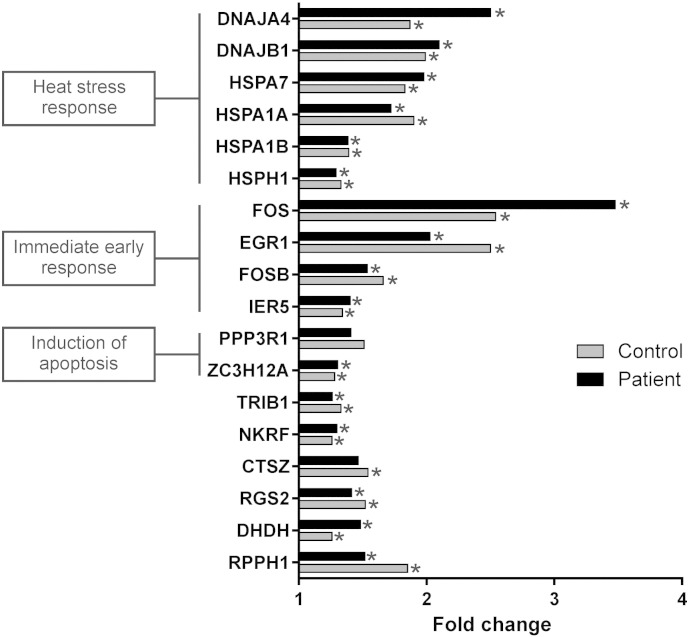
As the HSPB1∆C-term protein translocated to the nucleus, we asked whether it interfered with the global gene expression response upon heat stress by comparing the gene expression alterations in control and HSPB1∆C-term fibroblasts after a short heat treatment (30 min). Pathway analysis of genes upregulated at least 25% showed that the most significantly upregulated pathways were related to heat stress response both in patient and control cells (Fig. 5) and that the most induced genes in both cell lines were members of the heat shock response pathway. This result suggested that the impaired heat tolerance of patient cells was not caused by lack of compensatory alterations in gene expression.
Fig. 5.
Gene expression changes in response to heat stress. Control and HSPB1∆C-term (patient) fibroblasts were treated by exposure to 45 °C for 30 min and a gene expression microarray was used to measure the alterations in gene expression between treated and untreated cells. Genes that were upregulated by at least 25% in both control and patient cells are shown and categorized into biological pathways. The results show that the early transcriptional response to heat was not altered in the HSPB1∆C-term fibroblasts. Asterisk indicates p-value < 0.05.
3.5. Truncated HSPB1 impairs tolerance to unfolded protein stress
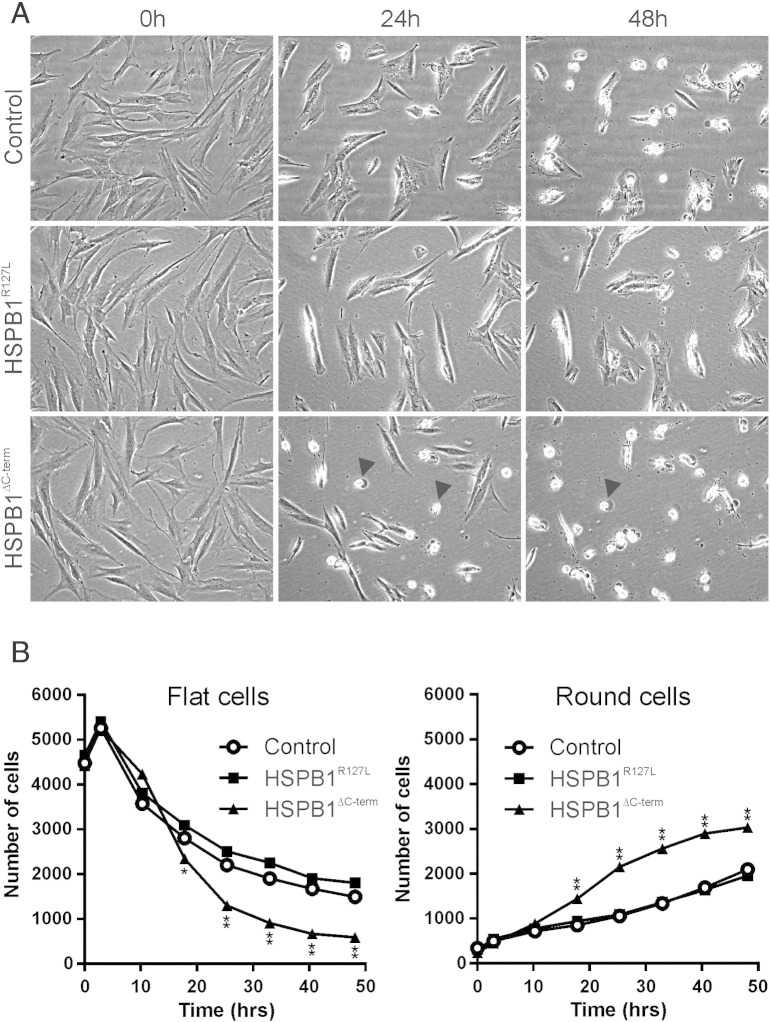
To investigate the cells' tolerance to protein misfolding, we treated the fibroblasts with canavanine, a naturally occurring analog of l-arginine that incorporates into proteins and induces misfolding [52]. With canavanine treatment, dying cells with rounded morphology were observed. In the HSPB1∆C-term fibroblasts (Fig. 6A), the number of cells with normal flat morphology decreased, and the number of dying (rounded) cells increased (Fig. 6B), significantly more compared to HSPB1R127L and control fibroblasts. This experiment demonstrated that the HSPB1∆C-term variant impaired the ability of cells to cope with unfolded protein stress.
Fig. 6.
Sensitivity to protein misfolding. The fibroblasts were exposed to canavanine and followed by continuous cell monitoring. Representative images are shown in (A) and quantifications in (B). After 24 h of canavanine treatment, HSPB1∆C-term fibroblasts started to die (rounded cells, arrowheads) and after 48 h the same was seen for control and HSPB1R127L fibroblasts. Quantification showed that the number of cells with normal flat morphology decreased in all cell lines but significantly more in HSPB1∆C-term fibroblasts. Conversely, the number of dying (round) cells increased significantly more in HSPB1∆C-term fibroblasts compared to the other cell lines. Asterisks denotes significant difference compared to control fibroblasts *P < 0.01, **P < 0.0001 (two-way ANOVA).
4. Discussion
HSPB1 has emerged as a common causative gene in neuropathy, particularly when motor symptoms predominate [3]. The mechanisms of action of HSPB1 and other sHSPs are under intense study because of their roles in protein quality control and numerous other physiologic processes [4], [53]. Understanding cell survival mechanisms in stress are important for many conditions such as neurodegeneration and cancer, and HSPB1 is an example of a protein with major significance for these processes. Cancer cells can for example escape drug-induced stress by regulating HSPB1 [54] and Parkinson's disease-related protein aggregation can be prevented by HSPB1 overexpression [55]. In this study we have characterized the molecular consequences of two new neuropathy-related HSPB1 variants in primary patient cells. The HSPB1∆C-term is of particular interest since the mutation precisely ablates the C-terminus, whose function is poorly understood, but may be critical for defining the specific functions of this particular sHSP.
Clinically, our patients displayed similar features as have been described in earlier cases of HSPB1-related neuropathy. The symptoms and signs supported a predominantly motor manifestation that was stronger in the lower extremities. However, there was also a clear sensory component, which was confirmed by ENMG and QST in two patients. In the third patient, sensory testing was normal but she was examined at an earlier age than the others and the development of sensory involvement later in her life is possible. The QST measurements also indicated that small fiber involvement may be found in HSPB1-related neuropathy. The motor-predominant phenotype with sensory involvement of our HSPB1∆C-term patient resembles the previously reported p.(Glu175*) family [27].
Molecular consequences of several HSPB1 missense mutants have previously been addressed in overexpression studies using either immortalized cell lines or primary neuronal cells from rodents [30], [31], [33], [56]. The use of primary patient fibroblasts in our study adds new understanding of the in vivo consequences of mutant proteins in cells expressing HSPB1 on physiologic endogenous levels, which is highly relevant when studying dominant defects. We showed that the truncated HSPB1 was stable in the patient cells, suggesting that the c.505delA mRNA had escaped nonsense-mediated decay, which is possible for premature stop codons occurring in the last exon of a gene.
Under normal conditions HSPB1 forms dimers and higher order oligomers up to 800 kDa in size. Oligomeric structure is regulated by post-translational modifications [4]. The dimer has been suggested to be the minimal structural unit of HSPB1 oligomers [57], [58], and dimerization is dependent on the ACD [11]. The previously discovered ACD mutant HSPB1R127W showed decreased dimerization when overexpressed in neuroblastoma cells [30]. In our HSPB1R127L fibroblasts, the amount of HSPB1 dimer under non-reducing conditions was not reduced, although the functionality of the dimer could have been affected. A possible explanation for the discrepancy is the different experimental setup regarding the cell type and expression level of the mutant. However, since HSPB1R127L and HSPB1WT are of identical size and indistinguishable on Western blot, we cannot exclude that most of the observed ~ 50 kDa dimer consists of HSPB1WT. In HSPB1∆C-term fibroblasts we detected bands corresponding to all possible combinations of HSPB1 dimers. This showed that HSPB1∆C-term bound the wild type protein, suggesting a dominant negative effect that could contribute to the neuropathy phenotype. The effect of C-terminal truncations on HSPB1 dimerization have not been reported previously, but the overexpressed C-terminal missense variant HSPB1P182L was similarly found to form dimers with both itself and HSPB1WT [30]. However, overexpressed HSPB1P182L in mouse primary cortical neurons formed insoluble intracellular aggregates [31], which were not evident upon immunofluorescence in our fibroblasts. Therefore, the pathogenic mechanisms may differ between truncations and missense variants of the C-terminus.
The functions of sHSPs converge on a theme of protection against external stress, e.g. heat, oxidative stress, heavy metals and ischemia. Heat is a generalized stressor causing protein misfolding and derangements to a host of functions, including direct effects on membranes, cytoskeleton and the cell cycle [59]. The 10 human sHSPs differ from the large heat shock proteins HSP70 and HSP90, since they do not have an ATPase domain and cannot actively refold misfolded proteins. Instead, they may operate by binding misfolded proteins to keep them in a refolding competent state [4]. HSPB1 binds denaturing proteins and prevents the formation of toxic protein aggregates [53]. We demonstrated decreased heat tolerance in the fibroblast lines from both patients, but the cell survival and recovery of cell morphology after the heat stress was significantly worse for HSPB1∆C-term cells. This suggested impairment in the ability to cope with cellular stress, particularly for the truncating variant.
Nuclear translocation of HSPB1 at high temperature has been described [19]. However, the function of the protein in the nucleus is not known. We showed that HSPB1∆C-term translocated to the nucleus in response to heat treatment and thus the decreased heat stress tolerance of the patient cells was not caused by defective nuclear translocation. Moreover, expression of the same genes of the heat shock pathway was induced in patient and control cells in response to 30 min of heat treatment, indicating that the C-terminal truncation did not interfere with the early induction of heat stress-related genes. HSPB1 may have a chaperoning function of misfolded proteins in the nucleus instead of directly regulating gene expression. The major morphological changes and cell death observed in HSPB1∆C-term fibroblasts after 2 h of heat stress were likely caused by direct denaturing effects on proteins, which the truncated HSPB1 was unable to counteract.
To more precisely pinpoint whether impaired stress tolerance in the patient cells was specifically related to defective handling of misfolded proteins, we treated cells with the arginine analog canavanine. Canavanine incorporates into polypeptide chains in place of arginine and induces protein misfolding [60]. The level of protein misfolding induced by limited doses of canavanine is small compared to high temperatures [61], and may resemble more closely what peripheral neurons encounter during normal life. As expected, the effect of canavanine on cells was milder and progressive in contrast to the immediate severe consequences of the heat treatment. The HSPB1R127L fibroblasts behaved largely as control cells during canavanine treatment. However, the HSPB1∆C-term fibroblasts suffered significantly more from canavanine than the control cells. Canavanine treatment was not associated with nuclear translocation in either control or patient cells (data not shown), which further suggested that the pathologic mechanism was cytoplasmic rather than nuclear. Collectively, these results provide evidence for the C-terminus of HSPB1 being necessary for the protein's ability to protect cells against misfolded proteins either by direct chaperoning function or through the diminished amount of HSPB1 dimer, independent of nuclear translocation.
A limitation of our study is the small sample size, as only one HSPB1∆C-term patient and two HSPB1R127L patients were identified. Collectively the known HSPB1 variants account for a significant portion of CMT2 and dHMN [3], but gathering large genetically homogeneous cohorts is difficult because individual disease variants are rare. Further studies with genetically similar patients may be needed to investigate the range of clinical variability related to truncating and missense variants in HSPB1. Our study suggested that the truncated HSPB1∆C-term was less tolerant to unfolded protein stress compared with the ACD missense variant HSPB1R127L. Interestingly, these molecular differences did not correlate with the severity of the disease in our patients, who all had rather late-onset of similar symptoms. Additional modifying factors such as the genetic background may complicate the conclusions regarding patient phenotype correlations, which is typical for the heterogeneous neuropathies.
5. Conclusion
We have shown that dominant HSPBR127L and HSPB1∆C-term cause peripheral neuropathy and impair the cell's tolerance to heat and/or misfolded proteins. Accurate disease models are needed to fully understand the pathogenesis of HSPB1-related neuropathy. Our study is the first to use primary patient fibroblasts for this aim. An interesting future possibility to investigate stress tolerance in neuronal cells is the development of differentiated neurons from patient-derived induced pluripotent stem cells. Additionally, identification of new patients with the same or similar mutations could allow more extensive testing of our conclusions. Peripheral motor and sensory neurons may be exquisitely sensitive to protein misfolding stress since they are post-mitotic and required to maintain cellular components over exceedingly large distances. When the stress response system is defective, exposure to even subtle amounts of misfolded proteins or other stressors over a life time could lead to the gradually progressive axonal degeneration that is typical for patients with HSPB1 mutations. Interventions that enhance cellular stress responses therefore offer attractive treatment opportunities.
The following is the supplementary data related to this article.
Genes were selected for our targeted sequencing panel based on association with neuropathy or related disease such as hereditary spastic paraplegia. A further set of candidate genes were included based on association with motor neuron disease, or presumed importance for maintenance of axons.
Transparency document
Transparency document.
Acknowledgments
The authors would like to thank the families for participation in the study. Riitta Lehtinen is thanked for technical help. We also acknowledge the target enrichment, sequencing and variant calling pipeline analysis performed by the Institute for Molecular Medicine Finland (FIMM). The authors wish to thank the following funding sources for support: Sigrid Jusélius Foundation (for H.T.), University of Helsinki (for H.T.), the Academy of Finland (for H.T. and E.Y.), the Finnish Neuromuscular Disorders Association (for M.A.), Svenska kulturfonden (for E.Y.), Arvid and Greta Olin's Foundation (for E.Y.) and Finska Läkaresällskapet (for E.Y.).
Footnotes
The Transparency document associated with this article can be found, in the online version.
References
- 1.Szigeti K., Lupski J.R. Charcot–Marie–Tooth disease. Eur. J. Hum. Genet. 2009;17:703–710. doi: 10.1038/ejhg.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossor A.M., Kalmar B., Greensmith L., Reilly M.M. The distal hereditary motor neuropathies. J. Neurol. Neurosurg. Psychiatry. 2012;83:6–14. doi: 10.1136/jnnp-2011-300952. [DOI] [PubMed] [Google Scholar]
- 3.Rossor A.M., Polke J.M., Houlden H., Reilly M.M. Clinical implications of genetic advances in Charcot–Marie–Tooth disease. Nat. Rev. Neurol. 2013;9:562–571. doi: 10.1038/nrneurol.2013.179. [DOI] [PubMed] [Google Scholar]
- 4.Mymrikov E.V., Seit-Nebi A.S., Gusev N.B. Large potentials of small heat shock proteins. Physiol. Rev. 2011;91:1123–1159. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- 5.Irobi J., Van Impe K., Seeman P., Jordanova A., Dierick I., Verpoorten N., Michalik A., De Vriendt E., Jacobs A., Van Gerwen V., Vennekens K., Mazanec R., Tournev I., Hilton-Jones D., Talbot K., Kremensky I., Van Den Bosch L., Robberecht W., Van Vandekerckhove J., Van Broeckhoven C., Gettemans J., De Jonghe P., Timmerman V. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat. Genet. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- 6.Evgrafov O.V., Mersiyanova I., Irobi J., Van Den Bosch L., Dierick I., Leung C.L., Schagina O., Verpoorten N., Van Impe K., Fedotov V., Dadali E., Auer-Grumbach M., Windpassinger C., Wagner K., Mitrovic Z., Hilton-Jones D., Talbot K., Martin J.J., Vasserman N., Tverskaya S., Polyakov A., Liem R.K., Gettemans J., Robberecht W., De Jonghe P., Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot–Marie–Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 7.Kolb S.J., Snyder P.J., Poi E.J., Renard E.A., Bartlett A., Gu S., Sutton S., Arnold W.D., Freimer M.L., Lawson V.H., Kissel J.T., Prior T.W. Mutant small heat shock protein B3 causes motor neuropathy: utility of a candidate gene approach. Neurology. 2010;74:502–506. doi: 10.1212/WNL.0b013e3181cef84a. [DOI] [PubMed] [Google Scholar]
- 8.Vicart P., Caron A., Guicheney P., Li Z., Prevost M.C., Faure A., Chateau D., Chapon F., Tome F., Dupret J.M., Paulin D., Fardeau M. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat. Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 9.Benndorf R., Martin J.L., Kosakovsky Pond S.L., Wertheim J.O. Neuropathy- and myopathy-associated mutations in human small heat shock proteins: characteristics and evolutionary history of the mutation sites. Mutat. Res. Rev. Mutat. Res. 2014;761:15–30. doi: 10.1016/j.mrrev.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochberg G.K., Ecroyd H., Liu C., Cox D., Cascio D., Sawaya M.R., Collier M.P., Stroud J., Carver J.A., Baldwin A.J., Robinson C.V., Eisenberg D.S., Benesch J.L., Laganowsky A. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1562–E1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baranova E.V., Weeks S.D., Beelen S., Bukach O.V., Gusev N.B., Strelkov S.V. Three-dimensional structure of alpha-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J. Mol. Biol. 2011;411:110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Kriehuber T., Rattei T., Weinmaier T., Bepperling A., Haslbeck M., Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- 13.Hilton G.R., Lioe H., Stengel F., Baldwin A.J., Benesch J.L. Small heat-shock proteins: paramedics of the cell. Top. Curr. Chem. 2013;328:69–98. doi: 10.1007/128_2012_324. [DOI] [PubMed] [Google Scholar]
- 14.Hochberg G.K., Benesch J.L. Dynamical structure of alphaB-crystallin. Prog. Biophys. Mol. Biol. 2014;115:11–20. doi: 10.1016/j.pbiomolbio.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Delbecq S.P., Jehle S., Klevit R. Binding determinants of the small heat shock protein, alphaB-crystallin: recognition of the ‘IxI’ motif. EMBO J. 2012;31:4587–4594. doi: 10.1038/emboj.2012.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stromer T., Ehrnsperger M., Gaestel M., Buchner J. Analysis of the interaction of small heat shock proteins with unfolding proteins. J. Biol. Chem. 2003;278:18015–18021. doi: 10.1074/jbc.M301640200. [DOI] [PubMed] [Google Scholar]
- 17.Markov D.I., Pivovarova A.V., Chernik I.S., Gusev N.B., Levitsky D.I. Small heat shock protein Hsp27 protects myosin S1 from heat-induced aggregation, but not from thermal denaturation and ATPase inactivation. FEBS Lett. 2008;582:1407–1412. doi: 10.1016/j.febslet.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 18.Pivovarova A.V., Chebotareva N.A., Chernik I.S., Gusev N.B., Levitsky D.I. Small heat shock protein Hsp27 prevents heat-induced aggregation of F-actin by forming soluble complexes with denatured actin. FEBS J. 2007;274:5937–5948. doi: 10.1111/j.1742-4658.2007.06117.x. [DOI] [PubMed] [Google Scholar]
- 19.Bryantsev A.L., Loktionova S.A., Ilyinskaya O.P., Tararak E.M., Kampinga H.H., Kabakov A.E. Distribution, phosphorylation, and activities of Hsp25 in heat-stressed H9c2 myoblasts: a functional link to cytoprotection. Cell Stress Chaperones. 2002;7:146–155. doi: 10.1379/1466-1268(2002)007<0146:dpaaoh>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capponi S., Geroldi A., Fossa P., Grandis M., Ciotti P., Gulli R., Schenone A., Mandich P., Bellone E. HSPB1 and HSPB8 in inherited neuropathies: study of an Italian cohort of dHMN and CMT2 patients. J. Peripher. Nerv. Syst. 2011;16:287–294. doi: 10.1111/j.1529-8027.2011.00361.x. [DOI] [PubMed] [Google Scholar]
- 21.Dierick I., Baets J., Irobi J., Jacobs A., De Vriendt E., Deconinck T., Merlini L., Van den Bergh P., Rasic V.M., Robberecht W., Fischer D., Morales R.J., Mitrovic Z., Seeman P., Mazanec R., Kochanski A., Jordanova A., Auer-Grumbach M., Helderman-van den Enden A.T., Wokke J.H., Nelis E., De Jonghe P., Timmerman V. Relative contribution of mutations in genes for autosomal dominant distal hereditary motor neuropathies: a genotype–phenotype correlation study. Brain. 2008;131:1217–1227. doi: 10.1093/brain/awn029. [DOI] [PubMed] [Google Scholar]
- 22.Houlden H., Laura M., Wavrant-De Vrieze F., Blake J., Wood N., Reilly M.M. Mutations in the HSP27 (HSPB1) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology. 2008;71:1660–1668. doi: 10.1212/01.wnl.0000319696.14225.67. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda Y., Abe A., Ishida C., Takahashi K., Hayasaka K., Yamada M. A clinical phenotype of distal hereditary motor neuronopathy type II with a novel HSPB1 mutation. J. Neurol. Sci. 2009;277:9–12. doi: 10.1016/j.jns.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 24.James P.A., Rankin J., Talbot K. Asymmetrical late onset motor neuropathy associated with a novel mutation in the small heat shock protein HSPB1 (HSP27) J. Neurol. Neurosurg. Psychiatry. 2008;79:461–463. doi: 10.1136/jnnp.2007.125179. [DOI] [PubMed] [Google Scholar]
- 25.Kijima K., Numakura C., Goto T., Takahashi T., Otagiri T., Umetsu K., Hayasaka K. Small heat shock protein 27 mutation in a Japanese patient with distal hereditary motor neuropathy. J. Hum. Genet. 2005;50:473–476. doi: 10.1007/s10038-005-0280-6. [DOI] [PubMed] [Google Scholar]
- 26.Mandich P., Grandis M., Varese A., Geroldi A., Acquaviva M., Ciotti P., Gulli R., Doria-Lamba L., Fabrizi G.M., Giribaldi G., Pizzuti A., Schenone A., Bellone E. Severe neuropathy after diphtheria–tetanus–pertussis vaccination in a child carrying a novel frame-shift mutation in the small heat-shock protein 27 gene. J. Child Neurol. 2010;25:107–109. doi: 10.1177/0883073809334387. [DOI] [PubMed] [Google Scholar]
- 27.Rossor A.M., Davidson G.L., Blake J., Polke J.M., Murphy S.M., Houlden H., Innes A., Kalmar B., Greensmith L., Reilly M.M. A novel p.Glu175X premature stop mutation in the C-terminal end of HSP27 is a cause of CMT2. J. Peripher. Nerv. Syst. 2012;17:201–205. doi: 10.1111/j.1529-8027.2012.00400.x. [DOI] [PubMed] [Google Scholar]
- 28.Tang B., Liu X., Zhao G., Luo W., Xia K., Pan Q., Cai F., Hu Z., Zhang C., Chen B., Zhang F., Shen L., Zhang R., Jiang H. Mutation analysis of the small heat shock protein 27 gene in Chinese patients with Charcot–Marie–Tooth disease. Arch. Neurol. 2005;62:1201–1207. doi: 10.1001/archneur.62.8.1201. [DOI] [PubMed] [Google Scholar]
- 29.Ylikallio E., Johari M., Konovalova S., Moilanen J.S., Kiuru-Enari S., Auranen M., Pajunen L., Tyynismaa H. Targeted next-generation sequencing reveals further genetic heterogeneity in axonal Charcot–Marie–Tooth neuropathy and a mutation in HSPB1. Eur. J. Hum. Genet. 2014;22:522–527. doi: 10.1038/ejhg.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida-Souza L., Goethals S., de Winter V., Dierick I., Gallardo R., Van Durme J., Irobi J., Gettemans J., Rousseau F., Schymkowitz J., Timmerman V., Janssens S. Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charcot–Marie–Tooth neuropathy. J. Biol. Chem. 2010;285:12778–12786. doi: 10.1074/jbc.M109.082644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackerley S., James P.A., Kalli A., French S., Davies K.E., Talbot K. A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes. Hum. Mol. Genet. 2006;15:347–354. doi: 10.1093/hmg/ddi452. [DOI] [PubMed] [Google Scholar]
- 32.Zhai J., Lin H., Julien J.P., Schlaepfer W.W. Disruption of neurofilament network with aggregation of light neurofilament protein: a common pathway leading to motor neuron degeneration due to Charcot–Marie–Tooth disease-linked mutations in NFL and HSPB1. Hum. Mol. Genet. 2007;16:3103–3116. doi: 10.1093/hmg/ddm272. [DOI] [PubMed] [Google Scholar]
- 33.Almeida-Souza L., Asselbergh B., d'Ydewalle C., Moonens K., Goethals S., de Winter V., Azmi A., Irobi J., Timmermans J.P., Gevaert K., Remaut H., Van Den Bosch L., Timmerman V., Janssens S. Small heat-shock protein HSPB1 mutants stabilize microtubules in Charcot–Marie–Tooth neuropathy. J. Neurosci. 2011;31:15320–15328. doi: 10.1523/JNEUROSCI.3266-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.d'Ydewalle C., Krishnan J., Chiheb D.M., Van Damme P., Irobi J., Kozikowski A.P., Vanden Berghe P., Timmerman V., Robberecht W., Van Den Bosch L. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot–Marie–Tooth disease. Nat. Med. 2011;17:968–974. doi: 10.1038/nm.2396. [DOI] [PubMed] [Google Scholar]
- 35.Chalova A.S., Sudnitsyna M.V., Strelkov S.V., Gusev N.B. Characterization of human small heat shock protein HspB1 that carries C-terminal domain mutations associated with hereditary motor neuron diseases. Biochim. Biophys. Acta. 2014;1844:2116–2126. doi: 10.1016/j.bbapap.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Selcen D., Engel A.G. Myofibrillar myopathy caused by novel dominant negative alpha B-crystallin mutations. Ann. Neurol. 2003;54:804–810. doi: 10.1002/ana.10767. [DOI] [PubMed] [Google Scholar]
- 37.Hayes V.H., Devlin G., Quinlan R.A. Truncation of alphaB-crystallin by the myopathy-causing Q151X mutation significantly destabilizes the protein leading to aggregate formation in transfected cells. J. Biol. Chem. 2008;283:10500–10512. doi: 10.1074/jbc.M706453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partanen J. Use of ENMG (electroneuromyography) in clinical diagnostics. Suom. Laakaril. 2014;69:2113–2120. [Google Scholar]
- 39.Sulonen A.M., Ellonen P., Almusa H., Lepisto M., Eldfors S., Hannula S., Miettinen T., Tyynismaa H., Salo P., Heckman C., Joensuu H., Raivio T., Suomalainen A., Saarela J. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim E.T., Wurtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnstrom K., Esko T., Magi R., Inouye M., Lappalainen T., Chan Y., Salem R.M., Lek M., Flannick J., Sim X., Manning A., Ladenvall C., Bumpstead S., Hamalainen E., Aalto K., Maksimow M., Salmi M., Blankenberg S., Ardissino D., Shah S., Horne B., McPherson R., Hovingh G.K., Reilly M.P., Watkins H., Goel A., Farrall M., Girelli D., Reiner A.P., Stitziel N.O., Kathiresan S., Gabriel S., Barrett J.C., Lehtimaki T., Laakso M., Groop L., Kaprio J., Perola M., McCarthy M.I., Boehnke M., Altshuler D.M., Lindgren C.M., Hirschhorn J.N., Metspalu A., Freimer N.B., Zeller T., Jalkanen S., Koskinen S., Raitakari O., Durbin R., MacArthur D.G., Salomaa V., Ripatti S., Daly M.J., Palotie A. Sequencing Initiative Suomi (SISu) Project, distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunning M.J., Smith M.L., Ritchie M.E., Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 43.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. (Article3) [DOI] [PubMed] [Google Scholar]
- 44.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 46.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Estacion M., Han C., Choi J.S., Hoeijmakers J.G., Lauria G., Drenth J.P., Gerrits M.M., Dib-Hajj S.D., Faber C.G., Merkies I.S., Waxman S.G. Intra- and interfamily phenotypic diversity in pain syndromes associated with a gain-of-function variant of NaV1.7. Mol. Pain. 2011;7 doi: 10.1186/1744-8069-7-92. (92-8069-7-92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Persson A.K., Liu S., Faber C.G., Merkies I.S., Black J.A., Waxman S.G. Neuropathy-associated Nav1.7 variant I228M impairs integrity of dorsal root ganglion neuron axons. Ann. Neurol. 2013;73:140–145. doi: 10.1002/ana.23725. [DOI] [PubMed] [Google Scholar]
- 51.Faber C.G., Hoeijmakers J.G., Ahn H.S., Cheng X., Han C., Choi J.S., Estacion M., Lauria G., Vanhoutte E.K., Gerrits M.M., Dib-Hajj S., Drenth J.P., Waxman S.G., Merkies I.S. Gain of function Nanu1.7 mutations in idiopathic small fiber neuropathy. Ann. Neurol. 2012;71:26–39. doi: 10.1002/ana.22485. [DOI] [PubMed] [Google Scholar]
- 52.Rosenthal G.A. The biological effects and mode of action of l-canavanine, a structural analogue of l-arginine. Q. Rev. Biol. 1977;52:155–178. doi: 10.1086/409853. [DOI] [PubMed] [Google Scholar]
- 53.Basha E., O'Neill H., Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katsogiannou M., Andrieu C., Rocchi P. Heat shock protein 27 phosphorylation state is associated with cancer progression. Front. Genet. 2014;5:346. doi: 10.3389/fgene.2014.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minoia M., Grit C., Kampinga H.H. HSPA1A-independent suppression of PARK2 C289G protein aggregation by human small heat shock proteins. Mol. Cell. Biol. 2014;34:3570–3578. doi: 10.1128/MCB.00698-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holmgren A., Bouhy D., De Winter V., Asselbergh B., Timmermans J.P., Irobi J., Timmerman V. Charcot–Marie–Tooth causing HSPB1 mutations increase Cdk5-mediated phosphorylation of neurofilaments. Acta Neuropathol. 2013;126:93–108. doi: 10.1007/s00401-013-1133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehrnsperger M., Lilie H., Gaestel M., Buchner J. The dynamics of Hsp25 quaternary structure. Structure and function of different oligomeric species. J. Biol. Chem. 1999;274:14867–14874. doi: 10.1074/jbc.274.21.14867. [DOI] [PubMed] [Google Scholar]
- 58.Dudich I.V., Zav'yalov V.P., Pfeil W., Gaestel M., Zav'yalova G.A., Denesyuk A.I., Korpela T. Dimer structure as a minimum cooperative subunit of small heat-shock proteins. Biochim. Biophys. Acta. 1995;1253:163–168. doi: 10.1016/0167-4838(95)00135-x. [DOI] [PubMed] [Google Scholar]
- 59.Velichko A.K., Markova E.N., Petrova N.V., Razin S.V., Kantidze O.L. Mechanisms of heat shock response in mammals. Cell. Mol. Life Sci. 2013;70:4229–4241. doi: 10.1007/s00018-013-1348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenthal G.A., Reichhart J.M., Hoffmann J.A. l-canavanine incorporation into vitellogenin and macromolecular conformation. J. Biol. Chem. 1989;264:13693–13696. [PubMed] [Google Scholar]
- 61.Trotter E.W., Kao C.M., Berenfeld L., Botstein D., Petsko G.A., Gray J.V. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes were selected for our targeted sequencing panel based on association with neuropathy or related disease such as hereditary spastic paraplegia. A further set of candidate genes were included based on association with motor neuron disease, or presumed importance for maintenance of axons.
Transparency document.