Abstract
Background
Free circulating microRNA (miRNA) in serum may be valuable biomarkers for disease diagnosis and prognosis. miR-21, the archetypal oncogenic miRNA, has been proposed as a biomarker for colorectal cancer and its benign precursor, adenomatous polyps. However, it is now becoming clear that circulating miRNA profiles may be sensitive to lifestyle and environmental influences. Dietary components involved in one-carbon metabolism are particularly well placed to modulate miRNA expression through an influence on DNA methylation pathways.
Methods
We investigated the role of methyl group donors (folate, B12, cysteine, homocysteine), polymorphisms of the enzymes of one-carbon metabolism, and serum miR-21 expression in a primary case–control cohort (colonoscopy confirmed adenomatous colon polyps vs controls; n = 253) and a secondary cross-sectional cohort (over 65s; n = 649). The relationships between these parameters and serum miR-21 levels were assessed, stratified by gender.
Conclusions
Serum miR-21 expression was related to occurrence of adenomatous polyps in females, but not males. Folate levels and MTHFR-C677T genotype was associated with miR-21 expression in both genders. Additionally, DHFR-19 del and MSR-A66G were associated with miR-21 expression in females and males, respectively. Stimulation with excess folate increased expression of miR-21 in colon cancer cell lines.
General significance
This study demonstrates that serum miR-21 expression correlates with folate status and related genetic status. This may have consequences for the proposed use of miR-21 as a colorectal cancer biomarker.
Abbreviations: AP, Adenomatous polyps; CRC, Colorectal cancer
Keywords: MiR-21, MicroRNA, Adenomatous polyps, Colorectal cancer, Folate
Highlights
-
•
Serum miR-21 is elevated in females (but not males) with adenomatous colon polyps.
-
•
Adenomatous polyps are the benign precursors to colorectal cancer.
-
•
Serum miR-21 levels correlate with blood folate levels.
-
•
Serum folate varies by genotype of enzymes involved in one carbon metabolism.
-
•
This may have implications for the proposed use of miR-21 as a biomarker.
1. Introduction
MicroRNAs (miRNAs) are an abundant class of short, non-coding RNA involved in post-transcriptional regulation of gene expression. It has been estimated that miRNAs have a post-transcriptional regulatory influence on the majority of human mRNAs, and are thought to regulate almost all cellular processes [1]. Unique signatures have been identified in a range of disease tissues [2] including circulating miRNA in plasma and serum [3]. Circulating miRNA are promising objective, non-invasive and easily accessible biomarkers for disease diagnosis and prognosis. They are remarkably stable, and can be detected with high levels of specificity and sensitivity. Therefore, significant potential exists to use miRNA as practical biomarkers for detecting or staging a wide range of diseases [4]. However, additional research is needed to unravel the complex relationships between miRNA profiles and pathophysiological conditions before this application can be fully realized.
miR-21 is the archetypal example of an oncogenic miRNA, and miR-21 expression in serum or plasma has been suggested as a biomarker for the diagnosis and prognosis in a number of cancers [5], [6], [7], [8], [9]. In the case of colorectal cancer (CRC), increasing levels of circulating miR-21 have been shown relative to healthy controls [5], [10], [11], [12]. Furthermore, serum miR-21 levels have been shown to increase with disease progression, from healthy tissue to adenomatous polyps (AP; the benign precursor to CRC) to CRC, and between TNM stages of CRC [5], [10]. Toiyama et al. have also demonstrated falling miR-21 levels in post-surgical patients [10]. Although the sources of miR-21 in circulation cannot be defined, CRC cell lines have been shown to secrete miR-21, and increased expression in serum correlates with increased expression of miR-21 in CRC tissue [10].
However, external exposures, such as diet and dietary supplements are also known to modulate miRNA profiles [13], [14], [15], [16], [17], [18], [19], [20]. miR-21 in particular has been shown to be modulated in the liver of rodents fed methyl-donor deficient [21], [22], [23] and high fat/high fructose diets [24]. High fat diets have also been shown to modulate miR-21 levels in adipose tissues in high fat induced obesity [25]. Additionally, dietary compounds such as curcumin [26] and fatty acids [27], [28] have been shown to modulate miR-21 expression. While it is not yet clear how diet modulates levels of circulating miR-21, diet is known to contribute to the modifiable determinant of risk for CRC [29].
Given the role of aberrant DNA methylation profiles in cancer, the potential of methyl-donor diets to modulate miR-21 levels is particularly interesting, especially since methyl-donor nutrients are the major source of methyl-groups in one-carbon metabolism cycles, and therefore influence the availability of methyl-groups for use in biological processes, including DNA methylation [30], [31], [32]. Methyl-donor availability may influence miR-21 expression directly, through methylation of miRNA genetic loci, or indirectly via modulation of other epigenetic marks or genetic signalling pathways. It is recognized that methyl-donor diet can influence methylation status, and therefore cancer risk [33]. Folate, a major contributor to the methyl-donor diet, has a complex and non-linear relationship with cancer risk, with both high and low folate potentially increasing risk [30], [34].
Importantly, one-carbon metabolism is regulated by multiple enzymes and co-factors. Therefore, polymorphisms in the genes that code for these enzymes and co-factor levels can influence the outcomes of this metabolic locus and potentially influence gene and miRNA expression levels. Therefore, it seems prudent to examine the relationship between levels of methyl-donors, co-factors, and polymorphisms of one-carbon metabolism related enzymes, and levels of miR-21 in circulation.
2. Methods
2.1. Subjects and sample collection
The primary cohort consisted of patients undergoing routine screening for colonic pathology at a gastroenterology clinic (n = 263 Gosford Hospital, NSW, Australia). 253 participants (57% male) who gave blood samples from which DNA and miRNA were successfully isolated, and received a definitive diagnosis as to the presence or absence of AP were included in this analysis. Participants were aged 18–89 (female mean 60.4 ± 1.0 years, male mean 64.4 ± 1.2 years).
The secondary cohort was taken from a large cross-sectional cohort of 1095 elderly participants, aged 65 years or more, living on the Central Coast of NSW, Australia. 649 participants (44.2% male), aged 65–95 years (females 78.0 ± 0.4 years, males 77.5 ± 0.4 years) who gave blood for analysis were included. Although colonoscopy was not performed in this cohort, it is an appropriate secondary cohort, as age is a known risk factor for development of AP and CRC, with the incidence rate more than 50 times higher in the 60–79 age group compared to individuals under 40 years of age [35]. Therefore, elderly patients would be an important target population for the use of miR-21 as a potential biomarker of CRC.
For both cohorts, blood was collected in heparin lined tubes (Greiner Bio-one, Frickenhausen, Germany) and plasma isolated by centrifugation. Informed consent was obtained prior to participation under the University of Newcastle Human Research Ethics Committee approval numbers H-429-0407 and H-2008-0431.
2.2. Blood biochemistry
Folate and serum B12 were measured by chemiluminescent assay [36], [37]. Plasma homocysteine was measured by a single enzyme homocysteine assay [31], [38]. Serum cysteine levels were measured by HPLC [36].
2.3. Genotyping
Eight variants in six genes involved in one-carbon metabolism [DHFR-19 del (rs70991108), TS-2R3R (rs34743033), SHMT-C1420T (rs1979277), MTHFR-C677T (rs1801133), MTHFR-A1298C (rs1801131), MTHFR-G1793A (rs2274976), MS-A2756G (rs1805087), MSR A66G (rs10380)], were examined using PCR, restriction enzyme digestions and gel electrophoresis [31].
2.4. iRNA analysis
miRNAs were isolated from plasma using Trizol LS and Glycoblue [39], [40]. Three synthetic Caenorhabditis elegans miRNA (cel-238, -54, -38) were added to plasma samples as exogenous spike-in controls to normalize data, as no appropriate endogenous house-keeping gene has been identified in plasma [40], [41]. cDNA was synthesized using universal primers and qPCR primer design was conducted as per the rules described by Balcells et al. [42]. Primer sequences used for miR-21 were (f) 5′- GCAGTAGCTTATCAGACTGAT -3′, (r) 5′- GGTCCAGTTTTTTTTTTTTTTTCAAC -3′ [42]. Data was analysed using qbase plus (Biogazelle, Technologiepark, Belgium) [43]. Data was expressed as log transformed relative expression units (REU). For reaction volumes and conditions see Supplementary methods.
2.5. Cell culture models
Three CRC cell lines (CACO2, HT29 and HCT116) were provided by Dr Jennette Sakoff (Department of Medical Oncology Calvary Mater Newcastle Hospital). Cells were adapted into folate-free RPMI (with 10% FCS) over 3 weeks. After 3 weeks cells were cultured (24 well plates; 0.2 × 106 cells/ml) and stimulated with no added folate (deficient), 0.001 mg/ml folate (control, equivalent to folate content in standard RPMI-1640; pterylmonoglutamic acid, Schircks Laboratories, Jona, Switzerland), or 0.01 mg/ml folate (excess). After 72 h media were removed for isolation of secreted miRNA and miR-21 assayed, as above. WST-1 assay (Biovision Research Products, CA, USA) was conducted as per manufacturer's instructions to assess cellular proliferation over the culture period. Results were expressed as miR-21 (REU) and normalized to relative cellular proliferation.
2.6. Statistics
Statistical analyses were performed using JMP (Version 11, SAS Institute Inc., Cary, NC, USA). T-tests, ANOVA (Tukey–Kramer post hoc test) and Walds χ2 were used to compare categories. Multivariable correlations were analysed by either least squares or nominal logistic regression analysis as appropriate. Outcomes were considered to be statistically significant at p ≤ 0.05. Descriptive statistics have been calculated as appropriate. Where more than 5 variables were to be considered in a single analysis, mixed direction stepwise regression was performed using significance levels of p ≤ 0.250 or p > 0.250 to enter (forward step) or remove (backward step) variables from the model, respectively. Mallow's Cp criterion was used for selecting the model where Cp first approaches p variables.
3. Results
3.1. Plasma miR-21 expression is elevated in female patients with adenomatous polyps
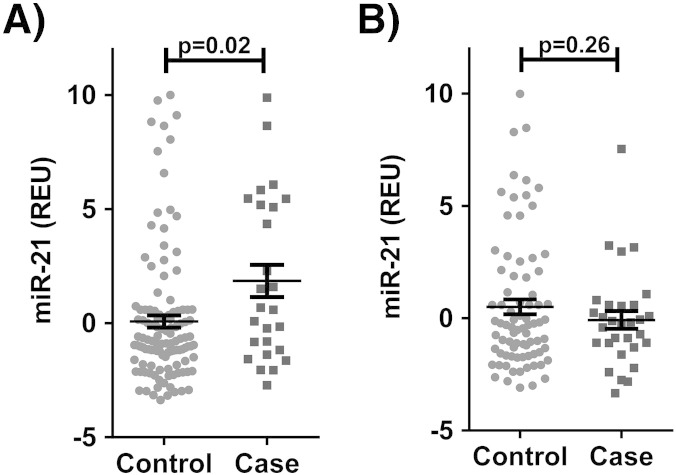
In the primary cohort cases had higher levels of plasma miR-21 than controls in females (1.85 ± 0.27 vs 0.07 ± 0.27 REU, p = 0.02; Fig. 1A), but not in males (− 0.08 ± 0.40 vs 0.50 ± 0.33, p = 0.26; Fig. 1B). Adjustment for age did not significantly alter the results (females p = 0.01, males p = 0.36). In the primary cohort females did not have significantly different levels of serum miR-21, compared to males (0.39 ± 0.26 vs 0.34 ± 0.26 REU, p = 0.89). However, in the secondary cohort females had lower serum miR-21 levels (− 0.42 ± 0.08 vs − 0.09 ± 0.09, p = 0.0065). Additional descriptive statistics for the primary and secondary cohorts are shown in Table 1, Table 2, respectively.
Fig. 1.
Serum miR-21 expression in adenomatous polyp cases versus controls in A) females (n = 144) and B) males (n = 109). REU = relative expression units.
Table 1.
Selected characteristics of the primary cohort, cases vs. controls, stratified by sex.
| Female |
Male |
Female vs male |
||||||
|---|---|---|---|---|---|---|---|---|
| Case n = 26 | Control n = 118 | p case/cntrl | Case n = 30 | Control n = 79 | p case/cntrl | p case | p cntrl | |
| Age (years)a | 65.5 ± 1.7 | 59.3 ± 1.1 | 0.004 | 67.0 ± 2.1 | 63.4 ± 1.5 | 0.16 | 0.58 | 0.03 |
| Age range | 46–82 | 18–82 | 44–89 | 28–88 | ||||
| Smoking history | ||||||||
| Currentb | 9 (6.3) | 27 (18.8) | 8 (7.3) | 18 (16.5) | ||||
| Exb | 6 (4.2) | 37 (25.7) | 9 (8.3) | 42 (38.5) | ||||
| Neverb | 11 (7.6) | 54 (37.5) | 0.42⁎ | 13 (11.9) | 19 (17.4) | 0.07⁎ | 0.76⁎ | 0.003⁎ |
| MiR-21 (REU) | 1.85 ± 0.70 | 0.07 ± 0.27 | 0.02 | − 0.07 ± 0.40 | 0.50 ± 0.33 | 0.26 | 0.02 | 0.31 |
| Red blood cell folate (μmol L− 1) | 1.29 ± 0.12 | 0.93 ± 0.04 | 0.009 | 0.89 ± 0.07 | 1.1 ± 0.05 | 0.06 | 0.006 | 0.08 |
| Serum B12 (nmol L− 1) | 0.29 ± 0.02 | 0.28 ± 0.01 | 0.80 | 0.27 ± 0.02 | 0.27 ± 13.11 | 0.96 | 0.60 | 0.53 |
| Cysteine (mmol L− 1) | 0.26 ± 0.01 | 0.26 ± 0.04 | 0.98 | 0.27 ± 0.08 | 0.27 ± 0.05 | 0.65 | 0.27 | 0.73 |
| Homocysteine (μmol L− 1) | 12.41 ± 0.79 | 12.01 ± 0.40 | 0.65 | 13.78 ± 1.21 | 13.62 ± 0.52 | 0.90 | 0.35 | 0.02 |
Mean ± SEM.
Count (%total).
p-Value χ2.
Table 2.
Selected characteristics of the secondary cohort, stratified by sex.
| Female n = 362 | Male n = 287 | p | |
|---|---|---|---|
| Age (years)a | 60.4 ± 0.96 | 64.4 ± 1.21 | 0.35 |
| Age range | 18–84 | 28–89 | |
| Smoking history | |||
| Currentb | 10 (0.02) | 11 (0.02) | |
| Exb | 122 (0.19) | 180 (0.28) | |
| Neverb | 230 (0.35) | 96 (0.15) | < 0.0001⁎ |
| MiR-21 (REU) | − 0.42 ± 0.08 | − 0.09 ± 0.09 | 0.0065 |
| Red blood cell folate (μmol L− 1) | 1.29 ± 0.02 | 1.23 ± 0.02 | 0.07 |
| Serum B12 (nmol L− 1) | 0.24 ± 0.006 | 0.22 ± 0.007 | 0.05 |
| Homocysteine (μmol L− 1) | 10.07 ± 0.28 | 11.34 ± 0.42 | 0.01 |
Mean ± SEM.
Count (%total).
p-Value χ2.
Serum miR-21 expression did not vary significantly with age in females (primary cohort r2 = 0.007, β = 0.03, p = 0.25; secondary cohort r2 = 0.0003, β = 0.004 p = 0.73; supplementary Fig. 1) or males (primary cohort r2 = 0.02, β = 0.01, p = 0.59; secondary cohort r2 = 0.008 β = − 0.02 p = 0.14; supplementary Fig. 1). Smoking status (never smoker, ex-smoker, current smoker) did not significantly influence serum miR-21 expression in females (primary cohort r2 = 0.004, F = 0.30, p = 0.74; secondary cohort: r2 = 0.0003, F = 0.06, p = 0.94; supplementary Fig. 2) or males (primary cohort r2 = 0.01, F = 0.57, p = 0.56; secondary cohort r2 = 0.006, F = 0.85, p = 0.43; supplementary Fig. 2).
3.2. Relationship between nutrients and biochemicals of one-carbon metabolism and serum miR-21 expression
Levels of folate (erythrocyte bound) and serum B12, homocysteine and cysteine were available in the primary cohort. In this cohort, cases had significantly higher folate than controls in females (1.29 ± 0.12 vs 0.93 ± 0.04 nmol L− 1, p = 0.0086; Table 1), but not males (0.89 ± 0.07 vs 1.1 ± 0.05 nmol L− 1, p = 0.06; Table 1). Serum B12, homocysteine and cysteine levels did not vary significantly between cases and controls in either sex (Table 1).
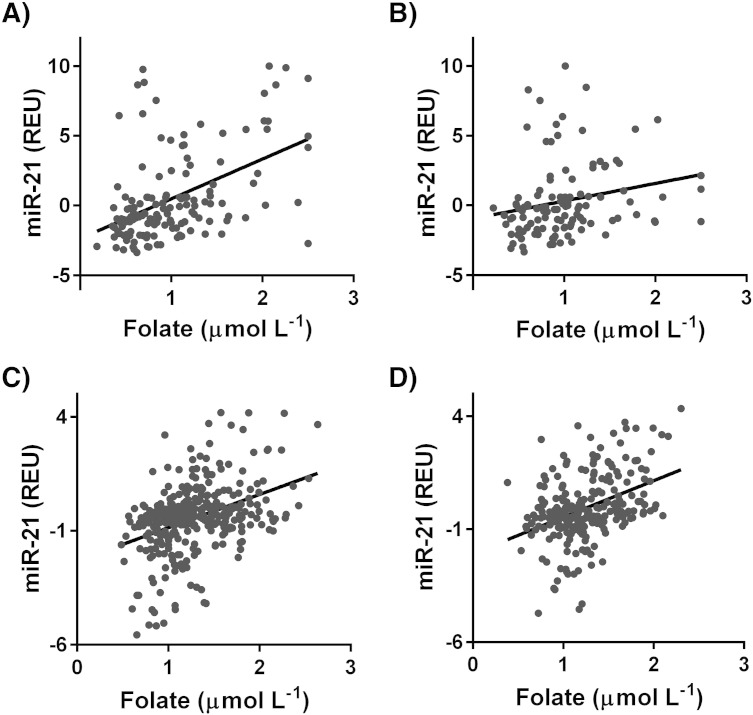
In the primary cohort miR-21 expression varied significantly with folate levels in both females (r2 = 0.248, β = 3.13 p < 0.0001; Fig. 2A) and males (r2 = 0.05, β = 1.25, p = 0.03; Fig. 2B). Serum B12, homocysteine and cysteine did not significantly influence miR-21 expression levels (supplementary Table 1). Serum miR-21 expression also varied significantly with folate levels in the secondary cohort (females: r2 = 0.140, β = 1.46 p < 0.0001, males: r2 = 0.160, β = 1.16 p < 0.0001; Fig. 2C–D) supplementary Table 2. Serum B12 and homocysteine levels did not significantly influence serum miR-21 expression in the secondary cohort (supplementary Table 2). In the secondary cohort data on blood cysteine levels were not available.
Fig. 2.
The relationship between serum miR-21 and erythrocyte folate in A) females (r2 = 0.248, β = 3.13, p = 0.0001, n = 144) and B) males (r2 = 0.05, β = 1.25, p = 0.03, n = 109) from the primary cohort and C) females (r2 = 0.140, β = 1.46, p < 0.0001, n = 362) and D) males (r2 = 0.160, β = 1.16, p < 0.0001, n = 287) from the secondary cohort. REU = relative expression units.
3.3. Relationship between polymorphisms in one-carbon metabolism enzymes and serum miR-21 expression
Multiple enzymes are involved in the production of methyl groups from methyl-donor nutrients. Polymorphisms in the genes coding for these enzymes can therefore influence methyl group production. In the primary cohort, information was available for 8 common polymorphisms of 6 genes involved in this pathway (DHFR-19 del, TS-2R3R, SHMT-C1420T, MTHFR-C677T, MTHFR-A1298C, MTHFR-G1793A, MS-A2756G, MSR A66G). Genotypic frequencies in the primary cohort are shown in supplementary Table 3.
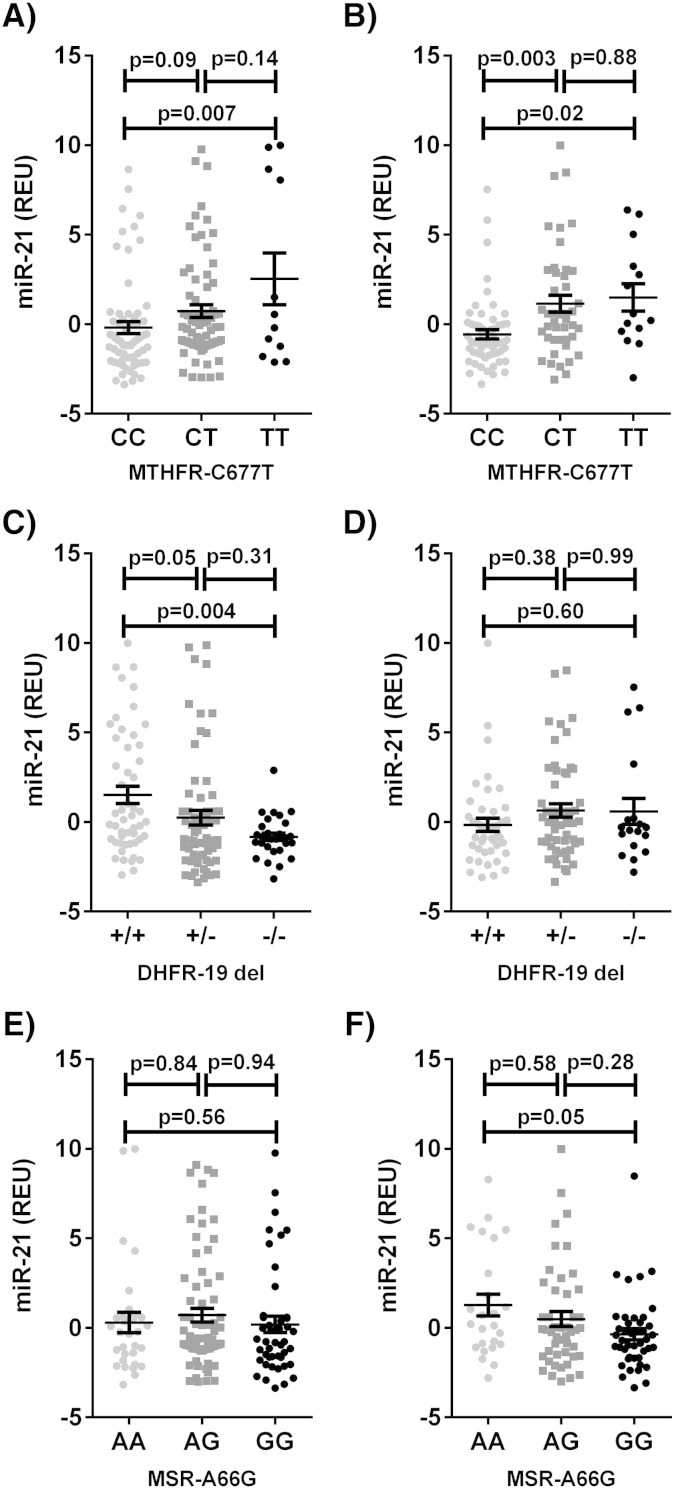
In the primary cohort stepwise regression analysis identified MTHFR-C677T and DHFR-19 del for inclusion in the models for both miR-21 expression in both females and males, and further identified MTHFR-G1793A in females and MSR-A66G and SHMT-C1420T in males. After combined modelling, DHFR-19 del (p = 0.0036) and MTHFR-C677T (p = 0.0083) were significantly related to serum miR-21 expression in females (model r2 = 0.16, p = 0.0006), and MTHFR-C677T (p = 0.0009) and MSR-A66G (p = 0.0443) were significantly related to serum miR-21 expression in males (model r2 = 0.21, p = 0.0019). In both females and males presence of the T allele of the MTHFR-C677T polymorphism corresponded to higher serum miR-21 (Fig. 3A–B). In females only, the presence of the DHFR-19 bp deletion corresponded to lower serum miR-21 (Fig. 3C–D). In males only, the presence of the G allele of the MSR-A66G variant correlated with decreased miR-21 expression in serum (Fig. 3E–F).
Fig. 3.
Serum miR-21 expression by: MTHFR-C677T genotype in A) females and B) males; DHFR-19 del genotype in C) females and D) males; and, MSR-A66G genotype in E) females and F) males of the primary cohort. REU = relative expression units; “+” = no deletion, “−” = 19 bp deletion.
In the secondary cohort MTHFR-C677T genotype data were available for all subjects, however, DHFR-19 del and MSR-A66G genotype data were only available for 200 participants. Genotypic frequencies are shown in supplementary Table 4. After combined modelling, DHFR-19 del (p = 0.0154) and MTHFR-C677T (p = 0.0011) were significantly related to serum miR-21 expression in females (model r2 = 0.137, p = 0.0017), validating the findings from the primary cohort. In males MTHFR-C677T (p = 0.0036) was significantly related to serum miR-21 expression, however the association between MSR-A66G and miR-21 expression in serum was not validated in the secondary cohort (p = 0.9851) (model r2 = 0.174, p = 0.0218).
3.4. Combined models
Combining all parameters previously identified as significant in females in the primary cohort folate, MTHFR-C677T, and DHFR-19 del produced a regression model with an adjusted r2 of 0.27 (p < 0.0001), and an adjusted r2 of 0.206 (p < 0.0001) in the secondary cohort. Similarly, combining all parameters identified as significant in males in the primary cohort (folate, MTHFR-C677T, MSR-A66G) resulted in a model with an adjusted r2 of 0.15 (p < 0.0006), and an adjusted r2 of 0.235(p = 0.0002) in the secondary cohort. When adjusted for these other variables, AP was not a significant predictor for serum miR-21 expression (females p = 0.22, males p = 0.99).
Combined regression models including all available genotypic, nutrient and biochemical variables against miR-21 expression were also conducted in the primary cohort. This resulted in an adjusted r2 of 0.32 (p < 0.0001) in females and 0.20 (p = 0.03) in males. In females, folate, MTHFR-C677T (p < 0.0001), DHFR-19 del (p = 0.04) and MTHFR-G1793A (p = 0.02) were identified as significant parameters in this model. In males MTHFR-C677T (p = 0.05), DHFR-19 del (p = 0.04) and MSR-A66G (p = 0.02) were identified as significant parameters. Exclusion of variables using a step-wise approach did not improve the predictive value of the r2. When adjusted for all variables, AP was not a significant predictor for serum miR-21 expression (females p = 0.54, males p = 0.72).
3.5. MiR-21 expression in media is increased after folic acid stimulation in colon cancer cell lines
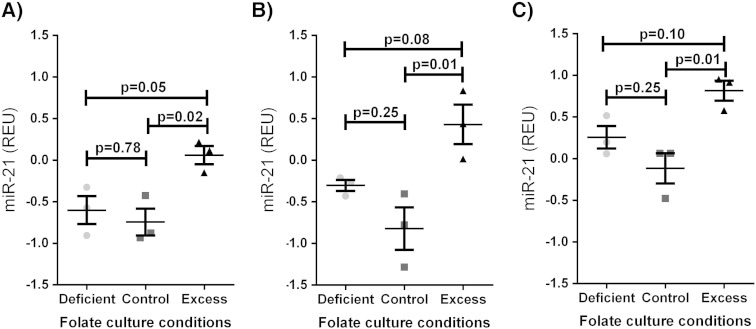
After 72 h of culture, miR-21 levels in media were significantly higher in those stimulated with excess folic acid, relative to control levels in all cell lines (CACO2: 0.06 vs − 0.74 REU, p = 0.02; HT29: 0.82 vs − 0.12, p = 0.01; HCT116: 0.43 vs − 0.82 p = 0.01; Fig. 4). miR-21 expression in folate deficient cells was not significantly different to control levels in all cell lines. miR-21 expression in media, normalized to cellular proliferation did not significantly influence these relationships (supplementary Fig. 3).
Fig. 4.
Serum miR-21 expression in colorectal cancer cell line cultures with deficient, control and excess levels of folic acid in A) CACO2, B) HT29, and C) HCT116 cells. REU = relative expression units, normalized to percentage proliferation. Data shown is mean ± standard error of mean. 3 replicates per group.
4. Discussion
A number of studies to date have suggested that miR-21 may make a practical and sensitive biomarker for diagnosis and prognosis of AP and CRC [5], [10], [11], [12]. However, study sizes and diversity of populations have been limited. The largest study to date is that of Toiyama et al. [10], which included 186 CRC patients, 43 advanced AP patients and 53 healthy controls, however, this was limited to a cohort completely of Japanese origin. As frequencies of genetic polymorphisms and sociocultural habits, including diet, can vary with ethnic diversity and location, studies such as this in additional populations are required to elucidate the relationships between genetics, environmental stimuli, disease and biomarkers.
In our Australian cohort, we have demonstrated that while serum miR-21 significantly increased in females with AP, the same trend was not seen in males. However, blood folate levels and folate related genetics were also a significant predictor of serum miR-21 expression. This may result from confounding or co-correlation, as AP and CRC, and associated surgeries and treatments, may influence diet, uptake or processing of nutrients, and CRC has known dietary risk factors [44], [45]. Importantly, when these additional factors were taken into account, serum miR-21 expression was no longer significantly associated with AP status. The observations of the correlation between folate status and serum miR-21 in human cohorts is supported by the increased release of miR-21 into media following excess folate stimulation in colon cancer cell culture models. Importantly, in cell culture, excess folate stimulation resulted in increased release of miR-21, even when results were corrected for proliferation, suggesting that the increased secretion is due to increased release, and not just an increased number of cells available for secretion.
The relationship between folate and AP and CRC risk may be a complicated one, with dose dependent biochemical and temporal considerations [30], [46]. In Australia a programme of mandatory folic acid fortification in bread flour has existed since 2009 [47]. The majority of the samples of the primary cohort were collected during the voluntary phase-in period from 2007–2009, and the samples of the secondary cohort were collected entirely post-fortification. Therefore, potential exists for modulation of these relationships in populations with different exposures from diet, including in those not exposed to folic acid fortification programmes.
All the polymorphisms that were found to be associated with serum miR-21 expression in this study are non-synonymous polymorphisms and can influence the regulation of one-carbon metabolism [31]. While MTHFR-C677T, DHFR-19 del and MSR-A66G genotypes did not differ significantly between cases and controls in the primary cohort, others have shown conflicting influences of the presence of the TT genotype on CRC risk [48], [49], [50], [51], [52], [53], [54], [55], which may be explained by complex gene–nutrient interactions [32], [49], [56], [57]. However, it is not yet clear what role differences in serum miR-21 expression play in this relationship.
The presented data demonstrates that serum miR-21 expression correlates with folate status and related genetic status. However, stratification by gender revealed that this relationship only existed in females. Although the sample size for this study was similar to previous studies, additional studies are needed to determine if this is due to a gender specific difference in diet or other gender differences.
Dietary modulation may have consequences for miR-21 and other miRNAs in their proposed uses as CRC biomarkers. While it may indeed be worthwhile pursuing miR-21 as biomarker for use in AP and CRC diagnosis and prognosis, this data suggests that dietary variables may influence the expression of miR-21, and that nutritional status may need to be considered as part of its assessment of a biomarker.
Conflict of interest statement
The authors declare that they have no conflicts of interest to disclose.
Transparency document
Transparency document
Acknowledgements
Emma Beckett receives funding from CSIRO (CSIRO OCE PhD Scholarship). This funding was not involved in decisions surrounding study design, analysis, interpretation or publication. The authors would like to thank Dr Janette Sakoff for contributing cell lines used in this work.
Footnotes
The Transparency document associated with this article can be found in the online version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbacli.2015.06.006.
Appendix A. Supplementary data
Supplementary material
References
- 1.Friedman R., Farh K., Burge C., Bartel D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calin G., Croce C. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6 doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 3.Weiland M., Gao X., Zhou L., Mi Q. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 4.Gilad S., Meiri E., Yogev Y., Benjamin S., Lebanony D., Yerushalmi N., Benjamin H., Kushnir M., Cholakh H., Melamed N., Bentwich Z., Hod M., Goren Y., Chajut A. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basati G., Emami Razavi A., Abdi S., Mirzaei A. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Med. Oncol. 2014;31:205. doi: 10.1007/s12032-014-0205-3. [DOI] [PubMed] [Google Scholar]
- 6.Li S., Yang X., Yang J., Zhen J., Zhang D. Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: a systematic review and meta-analysis. Clin. Exp. Med. 2014 doi: 10.1007/s10238-014-0332-3. [DOI] [PubMed] [Google Scholar]
- 7.Shen L., Wan Z., Ma Y., Wu L., Liu F., Zang H., Xin S. The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumour Biol. 2015;36:1993–2005. doi: 10.1007/s13277-014-2806-z. [DOI] [PubMed] [Google Scholar]
- 8.Wang G., Wang L., Sun S., Wu J., Wang Q. Quantitative measurement of serum microRNA-21 expression in relation to breast cancer metastasis in Chinese females. Ann. Lab. Med. 2015;35:226–232. doi: 10.3343/alm.2015.35.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu K., Li L., Li S. Circulating microRNA-21 as a biomarker for the detection of various carcinomas: an updated meta-analysis based on 36 studies. Tumour Biol. 2015;36:1973–1981. doi: 10.1007/s13277-014-2803-2. [DOI] [PubMed] [Google Scholar]
- 10.Toiyama Y., Takahashi M., Hur K., Nagasaka T., Tanaka K., Inoue Y., Kusunoki M., Boland C.R., Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Huang S.K., Zhao M., Yang M., Zhong J.L., Gu Y.Y., Peng H., Che Y.Q., Huang C.Z. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087451. e87451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanaan Z., Rai S.N., Eichenberger M.R., Roberts H., Keskey B., Pan J., Galandiuk S. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann. Surg. 2012;256:544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 13.Beckett E.L., Yates Z., Veysey M., Duesing K., Lucock M. The role of vitamins and minerals in modulating the expression of microRNA. Nutr. Res. Rev. 2014;27:94–106. doi: 10.1017/S0954422414000043. [DOI] [PubMed] [Google Scholar]
- 14.Tarallo S., Pardini B., Mancuso G., Rosa F., Di Gaetano C., Rosina F., Vineis P., Naccarati A. MicroRNA expression in relation to different dietary habits: a comparison in stool and plasma samples. Mutagenesis. 2014;29:385–391. doi: 10.1093/mutage/geu028. [DOI] [PubMed] [Google Scholar]
- 15.Jorde R., Svartberg J., Joakimsen R.M., Coucheron D.H. Plasma profile of microRNA after supplementation with high doses of vitamin D3 for 12 months. BMC Res. Notes. 2012;5:245. doi: 10.1186/1756-0500-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquobahrie D.A., Williams M.A., Qiu C., Siscovick D.S., Sorensen T.K. Global maternal early pregnancy peripheral blood mRNA and miRNA expression profiles according to plasma 25-hydroxyvitamin D concentrations. J. Matern. Fetal Neonatal Med. 2011;24:1002–1012. doi: 10.3109/14767058.2010.538454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimbach G., Moehring J., Huebbe P., Lodge J.K. Gene-regulatory activity of alpha-tocopherol. Molecules. 2010;15:1746–1761. doi: 10.3390/molecules15031746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu M.S., Langkamp-Henken B., Chang S.M., Shankar M.N., Cousins R.J. Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20970–20975. doi: 10.1073/pnas.1117207108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis C.D., Ross S.A. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr. Rev. 2008;66:477–482. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 20.Ross S.A., Davis C.D. MicroRNA, nutrition, and cancer prevention. Adv. Nutr. 2011;2:472–485. doi: 10.3945/an.111.001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogribny I.P., Tryndyak V.P., Ross S.A., Beland F.A. Differential expression of microRNAs during hepatocarcinogenesis induced by methyl deficiency in rats. Nutr. Rev. 2008;66(Suppl 1):S33–S35. doi: 10.1111/j.1753-4887.2008.00064.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang B., Majumder S., Nuovo G., Kutay H., Volinia S., Patel T., Schmittgen T.D., Croce C., Ghoshal K., Jacob S.T. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutay H., Bai S., Datta J., Motiwala T., Pogribny I., Frankel W., Jacob S.T., Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell. Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alisi A., Da Sacco L., Bruscalupi G., Piemonte F., Panera N., De Vito R., Leoni S., Bottazzo G.F., Masotti A., Nobili V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab. Investig. 2011;91:283–293. doi: 10.1038/labinvest.2010.166. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y.J., Hwang S.H., Cho H.H., Shin K.K., Bae Y.C., Jung J.S. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J. Cell. Physiol. 2012;227:183–193. doi: 10.1002/jcp.22716. [DOI] [PubMed] [Google Scholar]
- 26.Mudduluru G., George-William J.N., Muppala S., Asangani I.A., Kumarswamy R., Nelson L.D., Allgayer H. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci. Rep. 2011;31:185–197. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- 27.Mandal C.C., Ghosh-Choudhury T., Dey N., Choudhury G.G., Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. 2012;33:1897–1908. doi: 10.1093/carcin/bgs198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinciguerra M., Sgroi A., Veyrat-Durebex C., Rubbia-Brandt L., Buhler L.H., Foti M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology. 2009;49:1176–1184. doi: 10.1002/hep.22737. [DOI] [PubMed] [Google Scholar]
- 29.Hou N., Huo D., Dignam J.J. Prevention of colorectal cancer and dietary management. Chin. Clin. Oncol. 2013;2:13. doi: 10.3978/j.issn.2304-3865.2013.04.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucock M., Yates Z. Folic acid fortification: a double-edged sword. Curr. Opin. Clin. Nutr. Metab. Care. 2009;12:555–564. doi: 10.1097/MCO.0b013e32833192bc. [DOI] [PubMed] [Google Scholar]
- 31.Lucock M., Yates Z., Martin C., Choi J.-H., Beckett E., Boyd L., LeGras K., Ng X., Skinner V., Wai R., Kho J., Roach P., Veysey M. Methylation diet and methyl group genetics in risk for adenomatous polyp occurrence. BBA Clin. 2015;3:107–112. doi: 10.1016/j.bbacli.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niculescu M.D., Zeisel S.H. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J. Nutr. 2002;132:2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 33.Stefanska B., Karlic H., Varga F., Fabianowska-Majewska K., Haslberger A. Epigenetic mechanisms in anti-cancer actions of bioactive food components—the implications in cancer prevention. Br. J. Pharmacol. 2012;167:279–297. doi: 10.1111/j.1476-5381.2012.02002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulrich C.M. Folate and cancer prevention: a closer look at a complex picture. Am. J. Clin. Nutr. 2007;86:271–273. doi: 10.1093/ajcn/86.2.271. [DOI] [PubMed] [Google Scholar]
- 35.Haggar F.A., Boushey R.P. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucock M., Yates Z., Martin C., Choi J.H., Boyd L., Tang S., Naumovski N., Roach P., Veysey M. Hydrogen sulphide-related thiol metabolism and nutrigenetics in relation to hypertension in an elderly population. Genes Nutr. 2013;8:221–229. doi: 10.1007/s12263-012-0317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng X., Boyd L., Dufficy L., Naumovski N., Blades B., Travers C., Lewis P., Sturm J., Yates Z., Townley-Jones M., Roach P., Veysey M., Lucock M. Folate nutritional genetics and risk for hypertension in an elderly population sample. J. Nutrigenet. Nutrigenomics. 2009;2:1–8. doi: 10.1159/000160079. [DOI] [PubMed] [Google Scholar]
- 38.Tan Y., Hoffman R.M. A highly sensitive single-enzyme homocysteine assay. Nat. Protoc. 2008;3:1388–1394. doi: 10.1038/nprot.2008.117. [DOI] [PubMed] [Google Scholar]
- 39.Hastings M.L., Palma J., Duelli D.M. Sensitive PCR-based quantitation of cell-free circulating microRNAs. Methods. 2012;58:144–150. doi: 10.1016/j.ymeth.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beckett E.L., Martin C., Duesing K., Yates Z., Veysey M., Lucock M. Vitamin D receptor genotype modulates the correlation between vitamin D and circulating levels of let-7a/b and vitamin D intake in an elderly cohort. J. Nutrigenet. Nutrigenomics. 2014;7:264–273. doi: 10.1159/000381676. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O'Briant K.C., Allen A., Lin D.W., Urban N., Drescher C.W., Knudsen B.S., Stirewalt D.L., Gentleman R., Vessella R.L., Nelson P.S., Martin D.B., Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balcells I., Cirera S., Busk P.K. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol. 2011;11:70. doi: 10.1186/1472-6750-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giovannucci E. Modifiable risk factors for colon cancer. Gastroenterol. Clin. N. Am. 2002;31:925–943. doi: 10.1016/s0889-8553(02)00057-2. [DOI] [PubMed] [Google Scholar]
- 45.Neugut A.I., Garbowski G.C., Lee W.C., Murray T., Nieves J.W., Forde K.A., Treat M.R., Waye J.D., Fenoglio-Preiser C. Dietary risk factors for the incidence and recurrence of colorectal adenomatous polyps. A case–control study. Ann. Intern. Med. 1993;118:91–95. doi: 10.7326/0003-4819-118-2-199301150-00002. [DOI] [PubMed] [Google Scholar]
- 46.Lee J.E., Willett W.C., Fuchs C.S., Smith-Warner S.A., Wu K., Ma J., Giovannucci E. Folate intake and risk of colorectal cancer and adenoma: modification by time. Am. J. Clin. Nutr. 2011;93:817–825. doi: 10.3945/ajcn.110.007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A.I.o.H.a . In: Welfare, Mandatory Folic Acid and Iodine Fortification in Australia and New Zealand: Supplement to the Baseline Report for Monitoring. C.n.P. 153, editor. AIHW.; Canberra: 2011. [Google Scholar]
- 48.Chen J., Giovannucci E., Hankinson S.E., Ma J., Willett W.C., Spiegelman D., Kelsey K.T., Hunter D.J. A prospective study of methylenetetrahydrofolate reductase and methionine synthase gene polymorphisms, and risk of colorectal adenoma. Carcinogenesis. 1998;19:2129–2132. doi: 10.1093/carcin/19.12.2129. [DOI] [PubMed] [Google Scholar]
- 49.Curtin K., Bigler J., Slattery M.L., Caan B., Potter J.D., Ulrich C.M. MTHFR C677T and A1298C polymorphisms: diet, estrogen, and risk of colon cancer. Cancer Epidemiol. Biomark. Prev. 2004;13:285–292. doi: 10.1158/1055-9965.epi-03-0083. [DOI] [PubMed] [Google Scholar]
- 50.Kim J., Cho Y.A., Kim D.H., Lee B.H., Hwang D.Y., Jeong J., Lee H.J., Matsuo K., Tajima K., Ahn Y.O. Dietary intake of folate and alcohol, MTHFR C677T polymorphism, and colorectal cancer risk in Korea. Am. J. Clin. Nutr. 2012;95:405–412. doi: 10.3945/ajcn.111.020255. [DOI] [PubMed] [Google Scholar]
- 51.Le Marchand L., Wilkens L.R., Kolonel L.N., Henderson B.E. The MTHFR C677T polymorphism and colorectal cancer: the multiethnic cohort study. Cancer Epidemiol. Biomark. Prev. 2005;14:1198–1203. doi: 10.1158/1055-9965.EPI-04-0840. [DOI] [PubMed] [Google Scholar]
- 52.Levine A.J., Figueiredo J.C., Lee W., Poynter J.N., Conti D., Duggan D.J., Campbell P.T., Newcomb P., Martinez M.E., Hopper J.L., Le Marchand L., Baron J.A., Limburg P.J., Ulrich C.M., Haile R.W. Genetic variability in the MTHFR gene and colorectal cancer risk using the colorectal cancer family registry. Cancer Epidemiol. Biomark. Prev. 2010;19:89–100. doi: 10.1158/1055-9965.EPI-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma J., Stampfer M.J., Giovannucci E., Artigas C., Hunter D.J., Fuchs C., Willett W.C., Selhub J., Hennekens C.H., Rozen R. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]
- 54.Slattery M.L., Potter J.D., Samowitz W., Schaffer D., Leppert M. Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol. Biomark. Prev. 1999;8:513–518. [PubMed] [Google Scholar]
- 55.Ulrich C.M., Curtin K., Potter J.D., Bigler J., Caan B., Slattery M.L. Polymorphisms in the reduced folate carrier, thymidylate synthase, or methionine synthase and risk of colon cancer. Cancer Epidemiol. Biomark. Prev. 2005;14:2509–2516. doi: 10.1158/1055-9965.EPI-05-0261. [DOI] [PubMed] [Google Scholar]
- 56.Fallon U.B. Commentary: colon cancer, folate and genetic status. Int. J. Epidemiol. 2003;32:67–70. doi: 10.1093/ije/dyg115. [DOI] [PubMed] [Google Scholar]
- 57.Liu A.Y., Scherer D., Poole E., Potter J.D., Curtin K., Makar K., Slattery M.L., Caan B.J., Ulrich C.M. Gene–diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Mol. Nutr. Food Res. 2013;57:721–734. doi: 10.1002/mnfr.201200180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Supplementary material