Abstract
Background
Diabetes mellitus (DM), a metabolic disease, is characterized by impaired fasting glucose levels. Type 2 DM is adult onset diabetes. Long non-coding RNAs (lncRNAs) regulate gene expression and multiple studies have linked lncRNAs to human diseases.
Methods
Serum samples obtained from 96 participating veterans at JAH VA were deposited in the Research Biospecimen Repository. We used a two-stage strategy to identify an lncRNA whose levels correlated with T2DM. Initially we screened five serum samples from diabetic and non-diabetic individuals using lncRNA arrays. Next, GAS5 lncRNA levels were analyzed in 96 serum samples using quantitative PCR. Receiver operating characteristic (ROC) analysis was performed to determine the optimal cutoff GAS5 for diagnosis of DM.
Results
Our results demonstrate that decreased GAS5 levels in serum were associated with diabetes in a cohort of US military veterans. The ROC analysis revealed an optimal cutoff GAS5 value of less than or equal to 10. qPCR results indicated that individuals with absolute GAS5 < 10 ng/μl have almost twelve times higher odds of having diabetes (Exact Odds Ratio [OR] = 11.79 (95% CI: 3.97, 37.26), p < 0.001). Analysis indicated area under curve (AUC) of ROC of 0.81 with 85.1% sensitivity and 67.3% specificity in distinguishing non-diabetic from diabetic subjects. The positive predictive value is 71.4%.
Conclusion
lncRNA GAS5 levels are correlated to prevalence of T2DM.
General Significance
Assessment of GAS5 in serum along with other parameters offers greater accuracy in identifying individuals at-risk for diabetes.
Abbreviations: DM, diabetes mellitus; lncRNA, long noncoding RNA; GAS5, growth-arrest specific transcript 5; ROC, receiver operating characteristics; AUC, area under curve; BMI, body mass index; CI, confidence interval; NMD, nonsense mediated decay
Keywords: lncRNA, GAS5, Diabetes, Serum, Veterans
Highlights
-
•
Serum samples from participating veterans were analyzed.
-
•
Levels of serum lncRNAs were analyzed for correlation to diabetes.
-
•
ROC analysis was performed to determine GAS5 optimal cutoff.
-
•
Individuals with absolute GAS5 < 10 ng/μl have almost 12 times higher odds of diabetes.
-
•
We identified GAS5 lncRNA as significant indicator of diabetes.
1. Introduction
Diabetes mellitus (DM) comprises of a group of metabolic diseases characterized by impaired fasting glucose levels. Type 1 DM is juvenile onset diabetes in which the body does not produce insulin (insulin-dependent). Type 2 DM is more prevalent with 90% of adult cases being T2DM. T2DM may be insulin-sensitive or insulin-resistant. The pathogenesis of diabetes has been studied for several decades; however additional genetic and environmental factors interact to contribute towards this epidemic. Risk factors include BMI, smoking, family history, lifestyle and diet. There is overwhelming evidence linking obesity and diabetes [1], [2], [3], [4]. Within the obese population, patients show insulin-resistance though about 20% are insulin-sensitive. This is attributed to differences in oxidative stress and AMPK as well as inflammatory cytokines and SIRT1 in the adipose tissue of insulin-resistant subjects [5]. It is also known that some T2DM patients are lean. These epidemiological studies point to underlying genetic variations which may lead to onset of T2DM along with the known risk factors.
Long noncoding (lnc) RNAs have varied functions including signaling, molecular decoys, scaffolding and guiding ribonucleoprotein complexes. Multiple lines of evidence link regulatory lncRNAs to human diseases[6], [7], [8]. Sequencing of human genome determined that noncoding RNA accounts for 98% of the transcriptional output of the genome [9]. Noncoding RNAs are subdivided into transcription RNAs (rRNA and tRNA), long noncoding RNAs and short noncoding RNAs (miRNA, siRNA, snoRNA, snRNA). lncRNAs are > 200 nt in length and have distinct structural and spatial features which allow it to bind to DNA, RNA or protein partners. Most studies were done in cancer cells where genome-wide association studies (GWAS) indicated that lncRNAs are important orchestrators of essential biological networks. LncRNAs are implicated in regulation of genes in cell growth and apoptosis, epigenetic regulation, transcription and translation, and splicing. lncRNAs are transcribed by all cell types however their expression levels are specific to the cell type. This provides a high degree of specificity for its target and mode of action in the biological system.
The lncRNA growth-arrest specific transcript 5 (GAS5) is a 5′-terminal oligopyrimidine class of genes which regulates cell growth, proliferation and survival [9], [10]. The biogenesis of GAS5 is established. GAS5 gene transcribes several snoRNAs as well as four splice variants of GAS5 mRNA. However, due to the presence of STOP codon, none of the transcripts are transcribed into protein and degrade via the nonsense-mediated decay (NMD) pathway when translation is initiated. The RNA levels of GAS5 are regulated by its degradation instead of regulation at its transcriptional level [11]. GAS5 is encoded at 1q25, a locus displaying abnormalities in a number of cancers such as melanoma, prostate cancer and SLE [12], [13]. GAS5 inhibits actions of rapamycin which is an mTOR inhibitor and this affects both leukemic and untransformed human T-lymphocytes [11]. It is down-regulated in breast cancer [15]. GAS5 acts as a riborepressor by repressing transcription of glucocorticoid receptor [16]. However, the role of GAS5 lncRNA in diabetes is not known. Here, we established a correlation of expression levels of the lncRNA GAS5 detected in serum to T2DM patients.
2. Methods
2.1. Study population
Informed consent was obtained from veterans receiving care at James A Haley Veterans Hospital, Tampa, FL under IRB approved study (#Pro00012108). Serum samples are stored in the research bio-specimen repository (RBR). The de-identified and coded samples were provided to the PI along with research parameters such as age, gender, BMI (calculated as weight in kilograms/height in meter2), diabetic (determined as glucose levels > 125 mg/dL; HbA1c > 6.5%) or non-diabetic (determined as glucose levels < 100 mg/dL; HbA1c < 6.4%). To achieve statistical significance, serum samples from Caucasian male volunteers were used as they formed the predominant population volunteering their serum samples at the JAHVA at the time of initiation of this study. In addition, patients diagnosed with any form of cancer were not part of the study group. For this investigative study, 96 serum samples were analyzed (see Table 1).
Table 1.
Characteristics of patients in this study.
| Non diabetic (n = 49) | Diabetic (n = 47) | P-value | |
|---|---|---|---|
| BMI | 29.4 ± 6.6 | 34.8 ± 7.0 | < 0.001 |
| Age | 66.9 ± 9.7 | 70.3 ± 9.1 | 0.09 |
| Blood glucose | 102.8 ± 16.3 | 163.6 ± 61.3 | < 0.001 |
2.2. Quantitative Real-Time RT-PCR
Total RNA was isolated from serum samples from diabetic or non-diabetic subjects using TRIzol LS according to the manufacturer's protocol (Ambion). For the array, cDNA was prepared from 1.5 μg total RNA using lncRNA Profiler array kit (System Biosciences, cat # RA900A-1) according to manufacturer's instructions. Amplification was performed with 2X Maxima SYBR Green Masterwith ROX (Thermo Scientific) and data were analyzed using System Biosciences software to generate ΔΔCt values and calculate fold change. U6 snRNA was used as control to normalize calculations.
For GAS5 real time qPCR, 2 μg RNA was reverse-transcribed using Qiagen's RT kit. 2 μL of cDNA was amplified by real-time quantitative PCR using Syber (SYBR) Green on the ViiaA 7 system (Applied Biosystems) to quantify the absolute levels of the transcripts in the samples. GAPDH was used as the endogenous control. The primers used are: GAS5 sense primer 5ʹ- AGCTGGAAGTTGAAATGG -3ʹ and anti-sense 5′- CAAGCCGACTCTCCATACC -3ʹ; GAPDH sense primer 5ʹ- TGACGTGCCGCCTGGAGAAAC -3ʹ and anti-sense 5ʹ- CCGGCATCGAAGGTGGAAGAG -3ʹ. These primers were initially tested using cDNA and conditioned media from human preadipocytes in a RT-PCR reaction using Taq polymerase to give distinct products corresponding to the respective transcripts. Next, the optimal primer concentration was determined from a range of 50–900 nM for forward and reverse primers. The final concentration of 600/600 nM was selected to ensure efficiency and specificity for its target based on the dissociation curve that showed a single, sharp peak, indicating that the primers amplify one specific target. For absolute quantification, a standard curve was generated for each gene in every assay. To do so, 100–0.4 ng of GAS5 plasmid was used to obtain a standard curve correlating the amounts with the threshold cycle number (Ct values). A linear relationship (r2 > 0.96) was obtained for GAS5 and GAPDH. Real-time PCR was then performed on samples and standards in triplicate. The plate setup also included a standard series, no template control, no RNA control, no reverse transcriptase control, and no amplification control. The dissociation curve was analyzed for each sample. Absolute quantification of GAS5 expression levels for individual samples was calculated by normalizing the values to GAPDH. The results were analyzed as described below. A level of p < 0.05 was considered statistically significant. Significance was determined after three or more experiments.
2.3. Statistical analysis
Statistical analyses including sensitivity, specificity, positive predictive value, negative predictive value, likelihood ratios and area under the receiver operating curve analysis were performed using Stata, version 13.1 (Stata Statistical Software: Release 13. College Station, TX: StataCorp LP). The optimal cutoff point was calculated using the Cutoff Finder [12].
3. Results
3.1. Expression of GAS5 lncRNA in serum
For phase I of our study we screened serum from five diabetic and non-diabetic subjects for their long noncoding RNA (lncRNA) profiles. Total RNA was isolated and profiles of long non-coding RNAs were evaluated. To do so, human LncProfiler (SABiosciences) array comprising of 84 lncRNAs with rigorous controls was used. We observed H19, HOTAIR, GAS5, ncR-uPAR, EGOb, antiPEG11 and lincRNA-21 expression in serum from these patients. Other lncRNAs on the array were undetected for Ct analysis. Amongst the detected lncRNAs in our array, we observe a marked decrease in GAS5 expression in diabetic samples compared to non-diabetic (Fig. 1). These results showed that lncRNA GAS5 was present in circulation in serum and its levels were significantly lower (p < 0.0001 using unpaired t-test, PRISM™ software) in diabetic patients compared to non-diabetic samples. Expression of lncRNAs H19, HOTAIR, lincRNA-p21 which were reported by other studies to be associated with diabetes did not change significantly between the serum profiles of diabetic vs non-diabetic samples.
Fig. 1.
LncRNA profiler array was used to determine expression of lncRNAs in serum from diabetic (n = 5) and non-diabetic (n = 5) samples. RNA was extracted from serum samples, reverse transcribed and cDNA used in lncRNA Profiler (SBI). Average Ct was calculated for diabetic and non-diabetic samples, values normalized to hoU6 snRNA. Log graph shows fold change calculated as 2^ − (Ct(avDM) − Ct(avNDM) × ΔCt(geo mean)). Analysis was performed by software from SBI.
3.2. Analysis of serum samples
Next, we sought to evaluate circulating GAS5 levels in serum from non-diabetic and diabetic patients. Ninety six serum samples were obtained from Research Bio-specimen Repository (RBR) at the James A. Haley Veterans' Hospital. To obtain statistically significant data, male Caucasian patients were selected from the volunteer serum sample pool. Table 1 shows demographics of the patients (IRB approved protocol #Pro00012108). The prevalence of DM was 49% (95%CI: 39%, 59.4%).
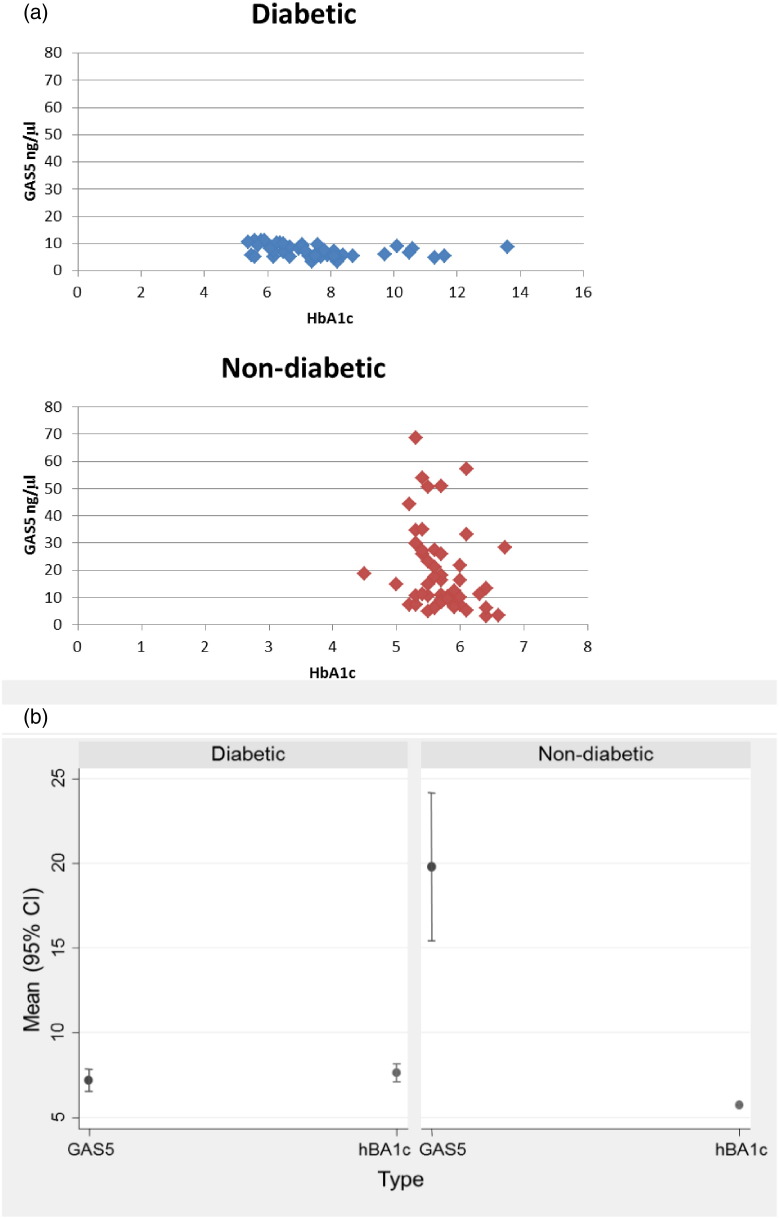
RNA was isolated and SYBR Green quantitative PCR in triplicate was used to obtain absolute GAS5 levels. Our results show that GAS5 levels were significantly decreased in diabetic samples. Markedly, GAS5 levels were decreased in patients with HbA1c > 5.9; these patients were not diagnosed as diabetic (cutoff HbA1c > 6.5) in clinic (Figs. 2a, b).
Fig. 2.
(a) Serum from non-diabetic (n = 49) and diabetic (n = 47) were analyzed for expression of GAS5 using qPCR. A standard curve was generated using 100–0.4 ng/μl GAS5 plasmid. Samples were normalized to U6snRNA. Graph shows absolute quantification of GAS5 (ng/μl) vs HbA1c. (b) Plot of mean (95% confidence interval) GAS5 and HbA1c levels by diabetic status. The confidence limits are mean ± 2 ∗ standard error.
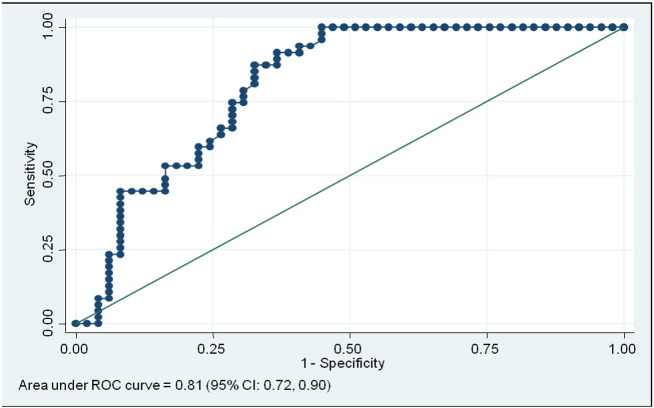
A receiver operating characteristic (ROC) curve was generated for statistical analysis of GAS5 levels in these 96 samples. ROC is widely used to investigate a reasonable cutoff level of the target. The ROC curve displays the relationship of sensitivity and specificity for GAS5. ROC analysis of GAS5 revealed the area under curve (AUC) of 0.81 (95% Confidence Interval (CI): 0.72, 0.90) (Fig. 3). The optimal cutoff GAS5 was less than or equal to 10 ng/μl measured as absolute quantification by qPCR, and this value had the sensitivity of 85.1% (95% CI: 72.3%, 92.6%), specificity of 67.3% (95% CI: 53.4%, 78.8%), positive predictive value (PPV) of 71.4% (95% CI:57.8%, 82.7%), negative predictive value of 82.5% (95% CI: 67.2%, 92.7%), positive likelihood ratio test 2.61 (95% CI: 1.71, 3.96) and negative likelihood ratio test 0.22 (95% CI: 0.11, 0.45).
Fig. 3.
Receiver operating curve (ROC) was performed on GAS5 levels in serum from diabetic patients (n = 47) and non-diabetic patients (n = 49) to determine the optimal cutoff values. Area under curve (AUC) for GAS5 is 0.81 (95% CI: 0.72, 0.90); Sensitivity of 85.1% (95% CI: 72.3%, 92.6%), specificity of 67.3% (95% CI: 53.4%, 78.8%), positive predictive value PPV = 71.4% (95% CI:57.8%, 82.7%).
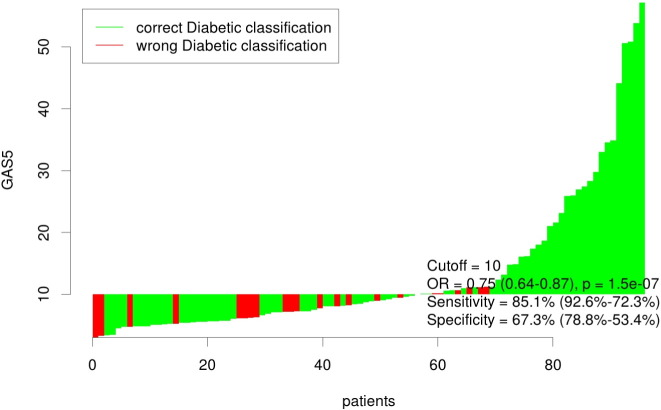
Our results indicated that individuals with absolute GAS5 < 10 ng/μl have nearly twelve times the odds of having diabetes (Odds Ratio [OR] = 11.79 (95% CI: 3.97, 37.26), p < 0.001). In clinic, individuals were classified as diabetic or non-diabetic based on their fasting glucose or HbA1c levels. The waterfall plot (Fig. 4) shows the classification accuracy of the optimal cutoff point for all 96 patients, as well as the overall odds ratio for GAS5 as a predictor of diabetes (OR = 0.75 (95% CI: 0.64, 0.87), p < 0.001). When adjusted for patient BMI, age and blood glucose, the OR for GAS5 remained significant (OR = 0.80 (95% CI: 0.65–0.97; p = 0.025)).The analysis indicates taking GAS5 levels are correlated to prevalence of diabetes.
Fig. 4.
The waterfall plot shows the classification accuracy of the optimal cutoff point for all 96 patients (diabetic (n = 47) + non-diabetic (n = 49) = 96), as well as the overall odds ratio for GAS5 as a predictor of diabetes (OR = 0.75 (95%CI: 0.64, 0.87), p < 0.001).
4. Discussion
Diabetes mellitus is a complex disease. There are several approaches and drugs available to manage the disease. However, little is known about genomic input on susceptibility and prevention. In 2012, it was estimated that diabetes costs the nation $245 billion, a 41% increase from costs incurred in 2007 (ADA study “Economic Costs of Diabetes in the US in 2012”). Further, 79 million people within USA have pre-diabetes and 1 in 4 individuals have un-diagnosed state of diabetes (CDCs 2011 Diabetes fact sheet). 25% of the veterans receiving care at VA hospitals have diabetes (Veterans' Health Administration data, 2014). In addition, the veteran population has 3.8% higher prevalence of overweight and obesity, which are predominant causes of diabetes in adults, than the general US population.
This is the first report of circulating lncRNA GAS5 as significantly correlated with diabetes. We demonstrate that GAS5 levels are decreased in serum of diabetic patients. Our results also showed decreased levels of GAS5 in certain patients with HbA1c levels > 5.9% and < 6.4%. These patients are not diagnosed as diabetic in clinic. Our study demonstrates that individuals with absolute GAS5 levels of less than 10 ng/μl have almost twelve times higher odds of having diabetes. It is of great importance to identify a biofluid molecule that indicates a change in microenvironment that may result in increased potential of developing diabetes. Presently, diabetes risk assessments include BMI measurements, family history, lifestyle and diet questionnaires. Impaired fasting glucose is an indication of advent of diabetes which may be delayed by exercise and diet. However, no valid or confirmatory test exists that predict diabetes risk or state of pre-diabetes or explains the inherent individual differences to develop diabetes in adulthood. Such a non-invasive test in serum would enable early detection of at-risk individuals. This determination could be easily integrated into a primary care setting.
Studies have identified lncRNA sets in human pancreatic islets involved in glucose-stimulated insulin secretion [13], [14]. Other studies identified ANRIL and H19 lncRNAs in the pancreas playing a role in diabetes [15], [16]. HOTAIR lncRNA is expressed in gluteal adipose tissue[17]. LncRNA NEAT1 regulates splicing duringadipogenesis [18]. Our results showed that only some of the lncRNAs (Fig. 1) were consistently detected in our samples. Amongst those detected, except for GAS5, these lncRNAs did not change significantly between serum from non-diabetic subjects compared to diabetic subjects. This may be due to the fact that lncRNAs are often specific to the cell type. Additionally, the levels of lncRNAs within cells differ from its secreted levels. Most of the lncRNAs associated with diabetes are shown in pancreatic beta cells and are specific to the cell. Interestingly, GAS5 promotes apoptosis and is decreased in breast and prostate cancer cells where survival of tumors is the underlying function attributed to GAS5 [19], [20], [21], [22]. As mentioned in our subject demographics, patients with cancer were not included in this cohort of veterans. These examples further add to our knowledge of lncRNAs in that most lncRNAs including GAS5 are multi-faceted and its function and specificity are often dictated by the cell type. Secreted lncRNAs are encapsulated within microvescicles or exosomes and are protected from degradation by RNAses in the blood [23]. This is the first study to our knowledge that identifies and correlates level of circulating lncRNA GAS5 with T2DM. Serum and plasma are a rich source for noncoding RNAs; some of which are shown as circulating biomarkers for cancer [24], [25], [26], [27].
Our mechanistic studies are currently ongoing to decipher the role of GAS5 in onset of diabetes and its target gene. This clinical study lays the foundation towards deciphering the molecular mechanisms and genetic influences underlying the pathology of diabetes mellitus. Since lncRNAs are important regulators of gene expression, it will be interesting to identify the gene targets of GAS5 that influence diabetes. Interestingly, in the breast cancer studies, inhibitors of mTOR were used to increase GAS5 levels [20]; however inhibition of mTOR also interferes with carbohydrate metabolism and increases likelihood of developing diabetes [28], [29], [30]. Additional studies are required to delineate the role of GAS5 in diabetes.
We have undertaken a longitudinal study in collaboration with the RBR at JAH Veterans Hospital. This study will address the limitations of the current study such as state of insulin resistance and metabolic syndrome, equal representation of males and females, effects of overweight and morbid obesity, age differences as well as racial and ethnic differences. Patients with diagnosed state of cancer were not part of the study as it is documented that circulating lncRNA profiles are dysregulated in cancer.
In conclusion, we identified lncRNA GAS5 as a significant indicator of diabetes which can be easily measured in serum in a cohort of US military veterans. This study can be expanded to verify the model that GAS5 levels may predict onset of diabetes in adults. The design may be easily adapted to high throughput screening of samples in a diagnostic setting and further refining of diagnosis and prediction of T2DM can be achieved by combining GAS5 levels with other molecular markers of DM.
Transparency document
Transparency document.
Acknowledgments
This work was supported by the Department of Veterans Affairs Medical Research grant (N.A.P.) (Grant # 821-MR-EN-20606). The contents do not represent the views of the Department of Veterans Affairs or the United States Government. Research Bio-specimen Repository (RBR) is supported by the James A. Haley Veterans Hospital, Tampa, FL.
Footnotes
The Transparency document associated with this article can be found in online version.
References
- 1.Colosia A.D., Palencia R., Khan S. Prevalence of hypertension and obesity in patients with type 2 diabetes mellitus in observational studies: a systematic literature review. Diabetes Metab. Syndr. Obes. Targets Ther. 2013;6:327–338. doi: 10.2147/DMSO.S51325. Epub 2013/10/02, PubMed PMID: 24082791; PubMed Central PMCID: PMC3785394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raj S.M., Halebeedu P., Kadandale J.S., Lahr M.M., Romero I.G., Yadhav J.R., Iliescu M., Rai N., Crivellaro F., Chaubey G., Villems R., Thangaraj K., Muniyappa K., Chandra H.S., Kivisild T. Variation at diabetes- and obesity-associated loci may mirror neutral patterns of human population diversity and diabetes prevalence in India. Ann. Hum. Genet. 2013 doi: 10.1111/ahg.12028. Epub 2013/07/03, PubMed PMID: 23808542. [DOI] [PubMed] [Google Scholar]
- 3.Sundborn G., Metcalf P.A., Gentles D., Scragg R., Dyall L., Black P., Jackson R. Overweight and obesity prevalence among adult Pacific peoples and Europeans in the Diabetes Heart and Health Study (DHAHS) 2002–2003, Auckland New Zealand. N. Z. Med. J. 2010;123(1311):30–42. Epub 2010/04/03, PubMed PMID: 20360794. [PubMed] [Google Scholar]
- 4.Crawford A.G., Cote C., Couto J., Daskiran M., Gunnarsson C., Haas K., Haas S., Nigam S.C., Schuette R. Prevalence of obesity, type II diabetes mellitus, hyperlipidemia, and hypertension in the United States: findings from the GE Centricity Electronic Medical Record database. Popul. Health Manag. 2010;13(3):151–161. doi: 10.1089/pop.2009.0039. Epub 2010/06/05, PubMed PMID: 20521902. [DOI] [PubMed] [Google Scholar]
- 5.Xu X.J., Pories W.J., Dohm L.G., Ruderman N.B. What distinguishes adipose tissue of severely obese humans who are insulin sensitive and resistant? Curr. Opin. Lipidol. 2013;24(1):49–56. doi: 10.1097/MOL.0b013e32835b465b. Epub 2013/01/10, PubMed PMID: 23298959; PubMed Central PMCID: PMC3575680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X., Yan G.Y. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29(20):2617–2624. doi: 10.1093/bioinformatics/btt426. PubMed PMID: 24002109. [DOI] [PubMed] [Google Scholar]
- 7.Arase M., Horiguchi K., Ehata S., Morikawa M., Tsutsumi S., Aburatani H., Miyazono K., Koinuma D. Transforming growth factor-beta-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci. 2014;105(8):974–982. doi: 10.1111/cas.12454. PubMed PMID: 24863656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q., Huang J., Zhou N., Zhang Z., Zhang A., Lu Z., Wu F., Mo Y.Y. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):4976–4987. doi: 10.1093/nar/gkt182. PubMed PMID: 23558749; PubMed Central PMCID: PMC3643595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., JD M.P., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., WR M.C., de la Bastide M., Dedhia N., Blocker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S., Chen Y.J. International human genome sequencing C. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. PubMed PMID: 11237011. [DOI] [PubMed] [Google Scholar]
- 10.Amaral P.P., Clark M.B., Gascoigne D.K., Dinger M.E., Mattick J.S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1138. (Database issue):D146-51. Epub 2010/11/30, PubMed PMID: 21112873; PubMed Central PMCID: PMC3013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams G.T., Mourtada-Maarabouni M., Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 2011;39(2):482–486. doi: 10.1042/BST0390482. Epub 2011/03/25, PubMed PMID: 21428924. [DOI] [PubMed] [Google Scholar]
- 12.Budczies J., Klauschen F., Sinn B.V., Gyorffy B., Schmitt W.D., Darb-Esfahani S., Denkert C. Cutoff finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;;7(12):e51862. doi: 10.1371/journal.pone.0051862. PubMed PMID: 23251644; PubMed Central PMCID: PMC3522617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taneera J., Fadista J., Ahlqvist E., Atac D., Ottosson-Laakso E., Wollheim C.B., Groop L. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum. Mol. Genet. 2015;24(7):1945–1955. doi: 10.1093/hmg/ddu610. PubMed PMID: 25489054. [DOI] [PubMed] [Google Scholar]
- 14.Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L., Taneera J., Storm P., Osmark P., Ladenvall C., Prasad R.B., Hansson K.B., Finotello F., Uvebrant K., Ofori J.K., Di Camillo B., Krus U., Cilio C.M., Hansson O., Eliasson L., Rosengren A.H., Renstrom E., Wollheim C.B., Groop L. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. U. S. A. 2014;111(38):13924–13929. doi: 10.1073/pnas.1402665111. PubMed PMID: 25201977; PubMed Central PMCID: PMC4183326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding G.L., Wang F.F., Shu J., Tian S., Jiang Y., Zhang D., Wang N., Luo Q., Zhang Y., Jin F., Leung P.C., Sheng J.Z., Huang H.F. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61(5):1133–1142. doi: 10.2337/db11-1314. PubMed PMID: 22447856; PubMed Central PMCID: PMC3331740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasmant E., Sabbagh A., Vidaud M., Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–448. doi: 10.1096/fj.10-172452. PubMed PMID: 20956613. [DOI] [PubMed] [Google Scholar]
- 17.Divoux A., Karastergiou K., Xie H., Guo W., Perera R.J., Fried S.K., Smith S.R. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity. 2014;22(8):1781–1785. doi: 10.1002/oby.20793. PubMed PMID: 24862299; PubMed Central PMCID: PMC4228784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper D.R., Carter G., Li P., Patel R., Watson J.E., Patel N.A. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARgamma2 splicing during adipogenesis in 3T3–L1 Cells. Genes. 2014;5(4):1050–1063. doi: 10.3390/genes5041050. PubMed PMID: 25437750; PubMed Central PMCID: PMC4276926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yacqub-Usman K., Pickard M.R., Williams G.T. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75(7):693–705. doi: 10.1002/pros.22952. PubMed PMID: 25650269. [DOI] [PubMed] [Google Scholar]
- 20.Pickard M.R., Williams G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast cancer research and treatment. 2014;145(2):359–370. doi: 10.1007/s10549-014-2974-y. PubMed PMID: 24789445. [DOI] [PubMed] [Google Scholar]
- 21.Pickard M.R., Mourtada-Maarabouni M., Williams G.T. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta. 2013;1832(10):1613–1623. doi: 10.1016/j.bbadis.2013.05.005. PubMed PMID: 23676682. [DOI] [PubMed] [Google Scholar]
- 22.Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. PubMed PMID: 18836484. [DOI] [PubMed] [Google Scholar]
- 23.Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Liang M., Kohli M., Thibodeau S.N., Boardman L., Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. PubMed PMID: 23663360; PubMed Central PMCID: PMC3653748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsui N.B., Ng E.K., Lo Y.M. Molecular analysis of circulating RNA in plasma. Methods Mol. Biol. 2006;336:123–134. doi: 10.1385/1-59745-074-X:123. Epub 2006/08/19, PubMed PMID: 16916258. [DOI] [PubMed] [Google Scholar]
- 25.Wang K., Yuan Y., Li H., Cho J.H., Huang D., Gray L., Qin S., Galas D.J. The spectrum of circulating RNA: a window into systems toxicology. Toxicol. Sci. 2013;132(2):478–492. doi: 10.1093/toxsci/kft014. Epub 2013/01/30, PubMed PMID: 23358195. [DOI] [PubMed] [Google Scholar]
- 26.Baraniskin A., Nopel-Dunnebacke S., Ahrens M., Jensen S.G., Zollner H., Maghnouj A., Wos A., Mayerle J., Munding J., Kost D., Reinacher-Schick A., Liffers S., Schroers R., Chromik A.M., Meyer H.E., Uhl W., Klein-Scory S., Weiss F.U., Stephan C., Schwarte-Waldhoff I., Lerch M.M., Tannapfel A., Schmiegel W., Andersen C.L., Hahn S.A. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int. J. Cancer. 2013;132(2):E48–E57. doi: 10.1002/ijc.27791. Epub 2012/08/22, PubMed PMID: 22907602. [DOI] [PubMed] [Google Scholar]
- 27.Payne R.E., Wang F., Su N., Krell J., Zebrowski A., Yague E., Ma X.J., Luo Y., Coombes R.C. Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br. J. Cancer. 2012;106(11):1790–1797. doi: 10.1038/bjc.2012.137. Epub 2012/04/28, PubMed PMID: 22538972; PubMed Central PMCID: PMC3364118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie J., Herbert T.P. The role of mammalian target of rapamycin (mTOR) in the regulation of pancreatic beta-cell mass: implications in the development of type-2 diabetes. Cell. Mol. Life Sci. 2012;69(8):1289–1304. doi: 10.1007/s00018-011-0874-4. PubMed PMID: 22068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ost A., Svensson K., Ruishalme I., Brannmark C., Franck N., Krook H., Sandstrom P., Kjolhede P., Stralfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol. Med. 2010;16(7–8):235–246. doi: 10.2119/molmed.2010.00023. PubMed PMID: 20386866; PubMed Central PMCID: PMC2896460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraenkel M., Ketzinel-Gilad M., Ariav Y., Pappo O., Karaca M., Castel J., Berthault M.F., Magnan C., Cerasi E., Kaiser N., Leibowitz G. mTOR inhibition by rapamycin prevents beta-cell adaptation to hyperglycemia and exacerbates the metabolic state in type 2 diabetes. Diabetes. 2008;57(4):945–957. doi: 10.2337/db07-0922. PubMed PMID: 18174523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.