Abstract
Meat source fraud and adulteration scandals have led to consumer demands for accurate meat identification methods. Nucleotide amplification assays have been proposed as an alternative method to protein-based assays for meat identification. In this study, we designed Loop-mediated isothermal amplification (LAMP) assays targeting species-specific mitochondrial DNA to identify and discriminate eight meat species; cattle, pig, horse, goat, sheep, chicken, duck, and turkey. The LAMP primer sets were designed and the target genes were discriminated according to their unique annealing temperature generated by annealing curve analysis. Their unique annealing temperatures were found to be 85.56±0.07℃ for cattle, 84.96±0.08℃ for pig, and 85.99±0.05℃ for horse in the BSE-LAMP set (Bos taurus, Sus scrofa domesticus and Equus caballus); 84.91±0.11℃ for goat and 83.90±0.11℃ for sheep in the CO-LAMP set (Capra hircus and Ovis aries); and 86.31±0.23℃ for chicken, 88.66±0.12℃ for duck, and 84.49±0.08℃ for turkey in the GAM-LAMP set (Gallus gallus, Anas platyrhynchos and Meleagris gallopavo). No cross-reactivity was observed in each set. The limits of detection (LODs) of the LAMP assays in raw and cooked meat were determined from 10 pg/μL to 100 fg/μL levels, and LODs in raw and cooked meat admixtures were determined from 0.01% to 0.0001% levels. The assays were performed within 30 min and showed greater sensitivity than that of the PCR assays. These novel LAMP assays provide a simple, rapid, accurate, and sensitive technology for discrimination of eight meat species.
Keywords: LAMP, meat identification, mitochondrial DNA, meat adulteration, food safety
Introduction
As consumption of animal resources increases and varies, excessive and unfair competition between producers has caused problems for consumers of meat-based foods. Several issues include contamination of originally highvalued meat with cheaper substitutes (beef contaminated with horse meat), choices not to intake meat or meat sources due to religious reasons, health problems (allergies) and individual preference (vegetarian) (Bottero and Dalmasso, 2011; Santos et al., 2012). These have led to the growth of consumer demands for accurate meat identification methods to combat meat source fraud and adulteration. In response to these pressures, the industry has developed a variety of species identification methods (Ahmed et al., 2010; Bottero and Dalmasso, 2011; Dooley et al., 2004).
Typical methods for meat identification rely on protein analyses, such as immunological, electrophoretic, and chromatographic assays (Bottero and Dalmasso, 2011; Hitchcock and Crimes, 1985; Proom, 1943). Enzyme-linked immunosorbent assays (ELISAs) are used for meat identification by the United States Department of Agriculture Food Safety And Inspection Service (USDA-FSIS). Several researchers have reported that ELISA provides accurate meat species identification (Macedo-Silva et al., 2000; Whittaker et al., 1983); however, protein analysis method has limitations. Protein is not favorable for discriminating processed products such as meat patties or cured, dried, and seasoned meat, because proteins are easily denatured by heat, salt, and pressure (Dincer et al., 1987). Proteins are also unsuitable tools for identifying species that are phylogenetically close, such as poultry, because of crossreactivity (Dooley et al., 2004).
Nucleotide amplification methods have been developed as an alternative to protein-based methods (Dooley et al., 2004; Koh et al., 2011; Wolf et al., 1999). Mitochondrial DNA (mtDNA) has been widely used for species identification, as it is relatively undamaged by processing and can thus be extracted intact from cooked and processed meats and meat products (Dooley et al., 2004). mtDNA sequences are frequently used instead of nuclear DNA for species- specific identification for several reasons. mtDNA exists in multiple copies (approximately 1,000 copies) per cell (Koh et al., 2011) and is relatively tolerant of environmental stresses such as heat, salt, and pressure. In addition, its rate of evolution facilitates discrimination between closely related species (Avise et al., 1987). Cytochrome b and D-loop genes are used for various methods of species identification (Dooley et al., 2004; Santos et al., 2012; Wolf et al., 1999). Meanwhile, several mtDNA genes, such as NADH dehydrogenase subunit 2 and 5 (ND2 and ND5), ATPase 6 and 8, and cytochrome oxidase subunits I and II (COI and II), have also used for species identification (Dawnay et al., 2007; Hebert et al., 2004; Kesmen et al., 2009; Kesmen et al., 2012; Verkaar et al., 2002). Several mtDNA-targeting techniques have been described for meat identification, including PCR (Koh et al., 2011), real-time PCR (Dooley et al., 2004; Santos et al., 2012), PCR-restriction fragment length polymorphism (PCR-RFLP) (Wolf et al., 1999), random amplification of polymorphic DNA (RAPD) (Martinez and Malmheden Yman, 1998), DNA hybridization (Chikuni et al., 1990), single strand conformational polymorphism (SSCP), and sequencing (Girish et al., 2004). However, some methods, such as sequencing and DNA hybridization, are expensive and others, such as PCR, RAPD, and PCR-RFLP, are generally less sensitive and laborious.
Loop-mediated isothermal amplification (LAMP) is a novel nucleotide amplification method (Francois et al., 2011; Tomita et al., 2008). It is specific, rapid, and sensitive because it employs 4 to 6 primers and specific DNA polymerases, such as Bst or Gsp DNA polymerase, which are capable of inducing auto-cycling strand displacement. The reaction occurs under isothermal conditions, eliminating the need for thermal cycling. To our knowledge, the only LAMP assay for species identification has been described with an electronic DNA sensor (Ahmed et al., 2010); none have been developed for use with intercalating dye-based analysis systems.
In this study, we developed a real-time LAMP assay for rapid, sensitive, and accurate species identification by targeting the mtDNA of eight meats. Species discrimination was performed based on annealing curve analysis of the LAMP assays. The capability of LAMP assay was assessed by determining the limit of detection (LOD) and by calculating a quantification equation for four sample types: raw meat, cooked meat, raw meat admixtures, and cooked meat admixtures. The LODs of LAMP assays were compared to those of PCR assays to evaluate the LAMP assays.
Materials and Methods
Sample preparation
Representative samples of seven meat species were purchased from grocery markets in South Korea: cattle (Bos taurus), pig (Sus scrofa domesticus), goat (Capra hircus), sheep (Ovis aries), chicken (Gallus gallus), duck (Anas platyrhynchos), and turkey (Meleagris gallopavo). Horse (Equus caballus) meat was obtained from the department of veterinary clinical pathology of Seoul National University. Samples (20 mg) were used for DNA extraction. Admixtures of horse-cattle, sheep-goat, chicken-duck, and chicken-turkey meat were prepared by combining 0.2 g of horse, sheep, and chicken meat at 0.00001 to 10% in 1.8 g of cattle, goat, duck, and turkey meat. Each sample was then homogenized by chopping with a stainless blade (Dorco, Korea) for 5 min. Heat-treated (cooked) meat samples (20 mg each) were prepared by boiling raw and admixture samples at 100℃ for 5 min. DNA was extracted with the DNeasy Tissue kit (Qiagen, USA) according to the manufacturer’s instructions. The DNA sample was then stored at 4℃ or directly used as template.
LAMP primer design
To identify each species-specific target, several mtDNA sequences of each species were collected from GenBank. Multiple sequence alignment using CLUSTALW in Meg Align program of Lasergene software (DNASTAR Inc., USA) was used to develop a consensus sequence. Three sets of primers targeting eight different regions of the consensus sequence were designed by using LAMP designer (Optigene Ltd., UK) after the in silico BLAST test provided in the program. The LAMP primers are listed in Table 1.
Table 1. Primer sequences for LAMP used and their target regions for eight meat species.
| Scientific name | Sequences (5’ to 3’) | Accession No. | Target regions | ||
|---|---|---|---|---|---|
| Bos taurus | Cattle | FIP | TCCAGCTACAATAGATGCTCCGACACATAACTGTGCTGTCAT | AY526085 | D-loop |
| BIP | GCATCTTGAGCACCAGCATAAAGTGGTGGTAGATATTTAAGGG | AY676864 | (Displacement loop) | ||
| LF | ATAGCTGAGTCCAAGCATCC | DQ124417 | |||
| LB | CAGTCAATGGTCACAGGACA | JN817306 | |||
| F3 | GCTAATCAGCCCATGCTC | NC006853 | |||
| B3 | TTGACTTTGTTTGGAGTGCT | ||||
| Sus scrofa domestica | Pig | FIP | AGGGATGGGACGGCTCATGACAATCGAGTTGTTCTACCA | KC469586 | COII |
| BIP | ACAGATGCTATCCCAGGACGATCTGAGCACTGTCCGTAA | KC469587 | (Cytochrome oxidase subunit II) | ||
| LF | GCAGTACGTCTTCAGAGGATAC | NC012095 | |||
| LB | CTCTAATATCCACACGACCTGG | AP003428 | |||
| F3 | AGACTATGAAGACCTCACCTT | ||||
| B3 | AGTGCTGACTAGCTTCTCA | ||||
| Equus caballus | Horse | FIP | CGTGGGTTTGGTGGGTCATTATTCGCAGTAGCTATAATCCAAG | AY584828 | ATP synthase F0 subunit 6 |
| BIP | CCCAGCCCATGACCACTTACACATGGCTAGTCCTGATGT | EF597513 | |||
| LF | GGCTTACCAGGAGAGTGAATAC | EU939445 | |||
| LB | CCCTATCAGCCCTCCTGA | FJ718997 | |||
| F3 | ACACCTCCTAATACACCTCAT | NC001640 | |||
| B3 | TCGGATGATGTCTCGTCA | ||||
| Capra hircus | Goat | FIP | TGTTGGCGTTTGTGTGGGTATACAGACATGCCAACAACC | GU068049 | D-loop |
| BIP | CACACAATGTTACGCGTATGCAGTCCGCGTTATATGGATGTTA | GU229280 | |||
| LF | TTGGGTTAGGATTGGGATGTTT | GU229281 | |||
| LB | AGTACATTACACCGCTCGC | GU295658 | |||
| F3 | AACACAAACTTCCCACTCC | NC005044 | |||
| B3 | GCTGGATTAGTACTGCATATGT | ||||
| Ovis aries | Sheep | FIP | GGGAGTGTTAAGTGGGTTTGCTTAGTACTATTCACGCCTGACT | HM236179 | Cyt b |
| BIP | CCATACATCAAAGCAACGGAGCAGGTCGGCTACTAGGATTC | HM236175 | (Cytochrome b) | ||
| LF | TTGTCTGGGTCTCCGAGT | HM236183 | |||
| LB | TTCCGACCAATCAGTCAATGT | NC001941 | |||
| F3 | CTACTAATCCTCATCCTCATGC | HM236185 | |||
| B3 | CCTCCAATTCATGTGAGTGT | AY858379 | |||
| Gallus gallus | Chicken | FIP | GGCTGGAAGAAGGAGTGATGGCCTGAACCTGACCATGAAC | AP003323 | ATP synthase F0 subunit 8 |
| BIP | ACTTCCATCACCAGGAAACCGATCAGAGTTGGATGGTGGA | AP003580 | ATP synthase F0 subunit 6 | ||
| LF | CTTGAGAATTGGTCGAAGAAGC | AY235570 | |||
| LB | TGGATCAACAACCGCCTC | AY235571 | |||
| F3 | CCATGATTCTCCATCATACTCC | NC001323 | |||
| B3 | AGGATAAGTGAGGTGAGTAGG | ||||
| Anas platyrhynchos | Duck | FIP | GGTTGCCTGCTAGAGGTGGACCATCATTCCTCCTTCTACT | EU009397 | COI |
| BIP | CCTCAGTGGACCTGGCTATCTGAAGTTAATGGCTCCGAGG | EU755252 | (Cytochrome oxidase subunit I) | ||
| LF | CCTCATCCACTGTAGAAGCTG | EU755253 | |||
| LB | CTCACTTCACCTGGCTGG | FJ167857 | |||
| F3 | TTCGGCAACTGATTGGTC | NC009684 | |||
| B3 | CATCCTGCTCCTCCTATCA | ||||
| Meleagris gallopavo | Turkey | FIP | AATCGTCCTGGGATTGCATCTGGCTGACGATGTATTACACTCA | EF153719 | D-loop |
| BIP | ACCAGGAGTGTTCTACGGACAGGATTCTACTACGATAGGCATG | JF275060 | |||
| LF | ACTCCGAGGGTTGGTACA | NC010195 | |||
| LB | GCGGAGCTAACCACAGTT | ||||
| F3 | AGTTGACCACCGTATAGTAGT | ||||
| B3 | AGGCTAGTGCTGATTCCA |
LAMP reaction
The LAMP assay reaction conditions were optimized using GspSSD DNA polymerase (Optigene Ltd., UK) according to the manufacturer’s instructions. Briefly, LOD tests were performed with 0.8 μM inner primers (FIP and BIP), 0.4 μM loop primers (LF and LB), 0.2 μM outer primers (F3 and B3), 1× buffer, 0.5 mM dNTPs, 4 mM MgSO2·7H2O, 1 M betaine, 0.5× Evagreen, 6 U GspSSD DNA polymerase, and 4 μL DNA template in a 25 μL reaction volume. For the cattle meat samples, 0.2 μM inner primers (FIP and BIP), 0.1 μM loop primers (LF and LB), and 0.05 μM outer primers (F3 and B3) were added. For the pig meat samples, 0.27 μM inner primers (FIP and BIP), 0.13 μM loop primers (LF and LB), 0.07 μM outer primers (F3 and B3), 3 mM MgSO2·7H2O, and 0.8 M betaine were added. For the specificity test, the LAMP sets were organized into BSE (Bos taurus, Sus scrofa domesticus and Equus caballus: cattle, pig and horse), CO (Capra hircus and Ovis aries: goat and sheep), and GAM (Gallus gallus, Anas platyrhynchos and Meleagris gallopavo: chicken, duck, and turkey) groups, with equimolar primer amounts. Nucleotide-free distilled water was used as a negative control and cross-reactivity with non-target DNA samples was tested.
Temperature optimization was performed over a range of 60 to 67℃ for 30-60 min by using the block gradient function of the Genie® II platform (Optigene Ltd., UK) before the melting curve was generated from 98 to 80℃, with a 1℃ decrease every 5 s. The best reaction temperature for each species was determined as the point of earliest inflection in the amplification ratio curve. The results were represented on the amplification graph with fluorescence (K) on the Y-axis and time (T) on the Xaxis; positivity was determined from the detection time (time taken to measure the fluorescence signal for a positive peak) and the mean species-specific annealing temperature.
Detection limit of the LAMP and PCR assays
To determine the LOD of the LAMP and PCR assays for raw and cooked samples of each species, 10-fold serially diluted template DNAs with concentrations ranging from 10 ng/μL to 10 fg/μL were tested in triplicate. In the raw and cooked meat admixtures, target species content was tested in triplicate from 10 to 0.0.00001%. Previously reported primers and conditions were used for the PCR-based comparators (Koh et al., 2011) with minor modification. Briefly, the reaction mixture contained 1× Emerald Master Mix (Takara Biotechnology, Japan), 0.5 μM each primer, and 1 μL template. The reaction was performed in a MyCycler™ Thermal Cycler (Bio-Rad Inc., Hercules, USA) with the following conditions; 1 cycle for 5 min at 95℃; followed by 30 cycles of 30 s at 95℃, 30 s at 57℃, and 35 s at 72℃; and 1 cycle for 5min at 72℃. Nucleotide-free distilled water was used as a negative control.
For each set, a standard curve based on the LAMP results was generated by plotting the log of the detection times versus the concentrations of template DNA or target species content. The quantification limits of the assay were evaluated by calculating the correlation coefficient determinants (R2) and quantification equation.
Results and Discussion
Capability of the LAMP assay
mtDNA sequences are often used to discriminate animal species. In this study, we targeted the displacement loop (D-loop), cytochrome b (cyt b), ATP synthase F0 subunits 6 and 8, and cytochrome oxidase subunits I and II (COI and II) in order to find species-specific non-variable regions and to set the annealing temperatures for successful discrimination (Table 1). The optimal reaction temperatures for each LAMP assay were identified as 64.6℃ for cattle, pig, and turkey, 64.7℃ for horse and sheep, 63.5℃ for goat, and 65.5℃ for chicken and duck. It was also demonstrated that the LAMP primers were specific to their corresponding target species. No cross-reactivity was observed within the BSE-, CO-, and GAM-LAMP groups. Indeed, no cross-reactivity was observed when the singleplex LAMP assays were performed for each of the 3 groups, indicating that the selected markers are suitable for species discrimination.
Annealing temperature differences
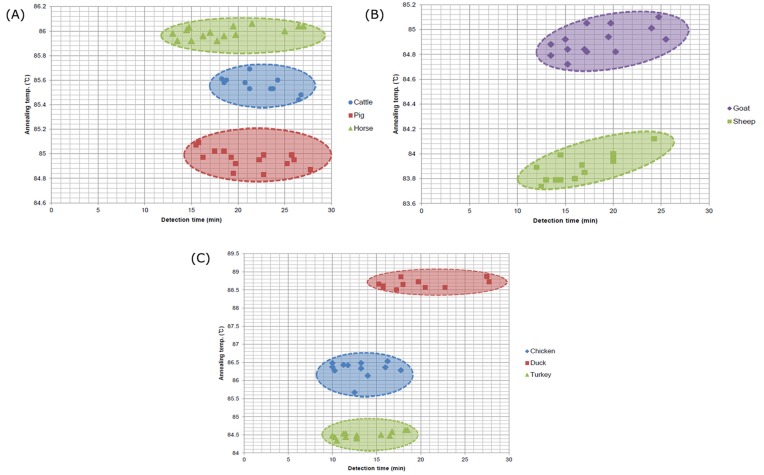
The annealing temperatures differed for each species within the margin of error: 85.56±0.07℃ for cattle, 84.96 ±0.08℃ for pig, and 85.99±0.05℃ for horse in the BSE-LAMP set; 84.91±0.11℃ for goat and 83.90±0.11℃ for sheep in the CO-LAMP set; and 86.31±0.23℃ for chicken, 88.66±0.12℃ for duck, and 84.49±0.08℃ for turkey in the GAM-LAMP set (Fig. 1). The annealing curve showed no overlaid peaks between species in each of the sets, revealing that annealing temperature analysis is an excellent tool for LAMP discrimination of these eight raw meat species. The annealing temperature is effective for species discrimination because it differed with the nucleotide composition and amplicon length of each target (Mouillesseaux et al., 2003). In contrast to probe-based real-time PCR, intercalating dye-based one has many advantages. Target detection with specific probes is more expensive than using general intercalating dyes. In addition, it is difficult to design and optimize a probe (Varga and James, 2005). For these reasons, melting curve analysis after SYBR green-based real-time PCR has been used to identify ruminant (cattle) and poultry (turkey) in foodstuffs (Şakalar and Abas yan k, 2012), discrimination of plum pox virus isolates of strain D and M (Varga and James, 2005), and discrimination of deer and six common domestic species (You et al., 2014).
Fig. 1. The annealing temperature and detection time of LAMP assay in 8 meat species. The annealing temperature and detection times of LAMP assay in (A) cattle (85.56±0.07℃), pig (84.96±0.08℃), and horse (85.99±0.05℃); (B) goat (84.91±0.11℃) and sheep (83.90±0.11℃); and (C) chicken (86.31±0.23℃), duck (88.66±0.12℃), and turkey (84.49±0.08℃) meat. The results were presented on the graph with annealing temperature (℃) on the Y-axis and detection time (min) on the X-axis.
LODs of LAMP assay in raw and cooked meat samples
The LODs of LAMP and PCR assays in raw and cooked meat samples are presented in Table 2. Amplification curves corresponding to samples at 10 ng/μL to 1 pg/μL were generated for raw pig, horse, goat, sheep, and turkey meat; samples of 10 ng/μL to 10 pg/μL were generated for cattle, chicken, and duck meat. Amplification curves corresponding to 10 ng/μL to 100 fg/μL were generated for cooked chicken, and from 10 ng/μL to 1 pg/μL for the remaining cooked samples, indicating that the LAMP assays are more sensitive than PCR by 102 to 103 times and 10 to 104 times in the raw and cooked meat samples, respectively, with strong linear relationships between detection time and template mtDNA concentration. Studies of raw horse and donkey meat samples by TaqManbased real-time PCR have yielded LODs of 1 pg DNA in water (Chisholm et al., 2005); the LOD for raw cattle meat is 35 pg DNA (Zhang et al., 2007). A study that employed a LAMP method and electrochemical DNA sensing described LODs for pork, bovine, and chicken samples as 20.33, 23.63, and 78.68 pg/μL (Ahmed et al., 2010), indicating that our LAMP assays were more sensitive in these species.
Table 2. Detection time and limits of the LAMP assays in raw and cooked meat samples for eight species.
| Detection time (min) | Cattle | Pig | Horse | Goat | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw | Cooked | Raw | Cooked | Raw | Cooked | Raw | Cooked | |||||||||
| Template concentration | 1)LAMP | 2)PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR |
| 10 ng | 18.5±0.3 | + | 16.3±0.0 | + | 15.8±0.4 | + | 16.5±0.3 | + | 13.7±0.8 | + | 11.8±0.0 | + | 13.5±0.0 | + | 14.3±0.9 | + |
| 1 ng | 21.1±0.3 | + | 19.1±0.1 | + | 18.4±0.9 | + | 18.8±0.4 | 15.3±0.8 | + | 13.0±0.3 | 15.2±0.1 | + | 16.0±1.1 | + | ||
| 100 pg | 23.8±0.4 | + | 22.0±0.3 | + | 20.5±1.5 | + | 20.7±0.1 | 17.8±0.8 | + | 15.2±0.4 | 17.2±0.1 | + | 18.1±1.4 | + | ||
| 10 pg | 26.6±0.2 | + | 24.8±0.6 | + | 23.8±1.9 | + | 22.8±1.0 | 20.3±1.1 | 16.5±0.0 | 19.8±0.4 | 21.0±1.6 | + | ||||
| 1 pg | 26.3±1.3 | + | 25.9±0.5 | 26.2±1.0 | 3)18.5 | 24.8±0.8 | 24.7±3.2 | |||||||||
| 100 fg | ||||||||||||||||
| 10 fg | ||||||||||||||||
| R2 | 0.9930 | 0.9933 | 0.9204 | 0.9717 | 0.9279 | 0.8552 | 0.9399 | 0.8407 | ||||||||
| Tm(℃) | 85.56±0.07 | 85.12±0.07 | 84.96±0.08 | 84.82±0.08 | 85.99±0.05 | 85.74±0.09 | 84.91±0.11 | 84.83±0.11 | ||||||||
| Detection time (min) | Sheep | Chicken | Duck | Turkey | ||||||||||||
| Raw | Cooked | Raw | Cooked | Raw | Cooked | Raw | Cooked | |||||||||
| Template concentration | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR |
| 10 ng | 12.5±0.5 | + | 13.9±0.3 | + | 10.1±0.1 | + | 8.9±0.3 | + | 15.6±0.3 | + | 12.3±0.1 | + | 10.3±0.3 | + | 10.2±0.3 | + |
| 1 ng | 14.3±0.3 | + | 15.8±0.1 | + | 11.8±0.6 | + | 10.2±0.3 | 17.7±0.4 | + | 14.3±0.3 | 11.4±0.1 | + | 11.2±0.1 | + | ||
| 100 pg | 16.6±0.5 | + | 18.0±0.3 | 13.5±0.4 | 11.7±0.1 | 21.0±1.6 | + | 16.0±0.0 | + | 12.8±0.0 | 12.8±0.0 | |||||
| 10 pg | 20.0±0.0 | 20.5±0.5 | 16.7±0.9 | 13.6±0.6 | 27.6±0.2 | + | 18.3±0.3 | 16.3±0.7 | 14.3±0.7 | |||||||
| 1 pg | 3)24.3 | 23.0±1.1 | 14.9±0.6 | 29.5±0.0 | 18.4±0.2 | 21.3±2.2 | ||||||||||
| 100 fg | 3)17.8 | |||||||||||||||
| 10 fg | ||||||||||||||||
| R2 | 0.9599 | 0.9768 | 0.9338 | 0.9683 | 0.9023 | 0.7814 | 0.9446 | 0.7912 | ||||||||
| Tm(℃) | 83.90±0.11 | 83.89±0.11 | 86.31±0.23 | 86.40±0.10 | 88.66±0.12 | 88.58±0.12 | 84.49±0.08 | 84.74±0.08 | ||||||||
1)LAMP results were presented with detection time (mean±standard deviation) of the triplicate. 2)PCR results were determined as positive (+) and negative (-). 3)One positive result was observed in the triplicate.
In comparison to the raw meat samples, LODs were the same or lower in cooked meat samples. Indeed, where the same LODs were found, the detection times were faster in cooked samples than in raw ones with the exception of goat, sheep, and turkey. Target regions under 150 bp are not influenced by heat treatment (Kesmen et al., 2012); thus, the amplified regions in our study should not be affected by cooking. Since the optimal size of the amplified region in LAMP is below 200 bp (Tomita et al., 2008), the method may be used for even highly degraded DNA targets.
Cattle and pig meat are the most favored meat sources in South Korea. In Europe, large quantities of horse meat are produced and imported for human consumption, as cattle meat is relatively expensive (Gill, 2005). The high cost of cattle meat could lead to intentional contamination with pig and horsemeat. In goats and sheep, the authentication of origin is important because of the close relationship between these ruminant species. In the dairy industry, inferior product substitutions may be made because sheep and goat sources have nutrient and economic advantages over cow-derived products (Cheng et al., 2006) and sheep sources are more costly than goat sources worldwide (Pappas et al., 2008). To our knowledge, no other studies have aimed to discriminate goat and sheep. Thus, our LAMP method could facilitate fast and sensitive discrimination of these small ruminant species. Poultry, chicken, duck, and turkey have also been adulterated for economic reasons. Foie gras from goose and duck have been contaminated with less expensive sources such as chicken or turkey (Rodriguez et al., 2003). Consumption of foie gras has increased along with demands for high quality and authentic foods, thus creating a need to prevent or detect adulteration. Our LAMP assays could be used for fast and sensitive monitoring for food fraud.
LODs of LAMP assay in raw and cooked meat admixtures
In the admixture samples, horse, sheep, and chicken meat were targeted because they are considered cheap substitutes. The amplification curves were generated for each target species based on detection time (Tt) and fluofluorescence for 10 to 0.0001% target in non-target species admixtures within 30 min. The LODs of LAMP and PCR assays in raw and cooked admixture samples are presented in Table 3. Amplification curves for samples of 10 to 0.01% were generated for the chicken-duck raw meat admixture; samples of 10 to 0.001% were used to generate curves for the remaining raw meat admixture samples. Samples of 10 to 0.001% were used to generate curves for sheep-goat and chicken-duck cooked meat admixtures, and samples of 10 to 0.01% and 10 to 0.0001% were used to generate curves for cooked horse-cattle and chicken-turkey admixtures, respectively. Thus, our LAMP assays were more sensitive than PCR by 102 to 104 fold. The LOD of PCR in heated ovine samples mixed with oats is 0.1% (Martin et al., 2007); in poultry and ruminant species, samples with vegetable contamination (maize) were reported at 0.002% (Dalmasso et al., 2004). However, these results cannot be directly compared because an LOD expressed as meat content (w/w) does not show the absolute capability of the assay (Ballin et al., 2009). Further study may be needed to reevaluate previous studies with a consensus unit, such as DNA/DNA equivalents (Ballin et al., 2009).
Table 3. Detection time and limits of the LAMP assays for raw and cooked meat admixture samples.
| Detection time (min) | Horse-cattle | Sheep-goat | Chicken-duck | Chicken-turkey | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw | Cooked | Raw | Cooked | Raw | Cooked | Raw | Cooked | |||||||||
| 1)Composition of target meat (%) | 2)LAMP | 3)PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR | LAMP | PCR |
| 10 | 12.3±0.4 | + | 12.8±0.0 | + | 16.7±0.7 | + | 13.3±0.0 | + | 10.2±0.5 | + | 9.3±0.0 | + | 9.0±0.4 | + | 9.1±0.1 | + |
| 1 | 13.7±0.3 | + | 14.1±0.1 | 17.8±0.8 | 15.8±0.6 | 11.1±0.1 | + | 10.0±0.5 | 10.1±0.5 | + | 10.6±0.1 | + | ||||
| 0.1 | 15.7±0.5 | 16.6±0.1 | 19.6±0.8 | 17.6±0.4 | 12.8±0.0 | 11.6±0.4 | 11.8±0.7 | + | 12.1±0.1 | |||||||
| 0.01 | 17.7±0.6 | 4)22.0 | 22.3±0.7 | 19.8±0.4 | 15.0±0.8 | 13.4±0.5 | 13.6±0.4 | 14.0±0.3 | ||||||||
| 0.001 | 4)22.3 | 29.4±0.2 | 26.0±3.2 | 18.8±1.1 | 16.2±1.9 | 17.3±1.6 | ||||||||||
| 0.0001 | 17.5±0.7 | |||||||||||||||
| 0.00001 | ||||||||||||||||
| R2 | 0.9255 | 0.8856 | 0.8430 | 0.8506 | 0.9279 | 0.8552 | 0.8939 | 0.9359 | ||||||||
| Tm(℃) | 85.75±0.16 | 85.59±0.22 | 83.96±0.12 | 83.86±0.19 | 86.35±0.15 | 86.27±0.11 | 86.34±0.11 | 86.32±0.12 | ||||||||
1)The admixtures of horse-cattle, sheep-goat, chicken-duck, and chicken-turkey meat were prepared by combining 0.2 g of horse, sheep, and chicken meat at 0.00001 to 10% in 1.8 g of cattle, goat, duck, and turkey meat.
2)LAMP results were presented with detection time (mean±standard deviation) of the triplicate.
3)PCR results were determined as positive (+) and negative (−).
4)One positive result was observed in the triplicate.
The annealing temperature of LAMP amplicons fell within the unique range for each species and the negative control was not amplified throughout the experiments. In comparison to the raw meat samples, we observed no difference in LOD for the sheep-goat raw and cooked admixture samples; the LODs for chicken-turkey and chicken- duck cooked admixtures were lower than for the raw admixtures. Detection times were faster for cooked admixtures than for raw ones (Table 3), perhaps because heating may influence the extraction efficiency of mtDNA from the sheep-goat, chicken-turkey, and chicken-duck samples. In contrast, the LODs in cooked horse-cattle admixtures were greater than in the raw admixtures; detection times and detection limits were higher for chicken-duck than for chicken-turkey admixtures. Thus, DNA from the non-target meat could inhibit target DNA amplification (Dooley et al., 2004), although the annealing temperature in raw and cooked horse-cattle admixtures was 85.75±0.16℃ and 85.59±℃, respectively, indicating successful amplification of horse mtDNA. Since the LAMP method is generally robust against pH, temperature changes, and even reagent exposure, it is more sensitive and specific than other nucleotide amplification methods (Francois et al., 2011). Our LAMP assays could be used as tools to discriminate and quantify meat species in unknown and heat-treated samples.
We developed a LAMP assay combined with annealing curve analysis to identify and discriminate eight meat species. We designed the LAMP primers for specific amplification of mtDNA regions in each target species. The annealing temperatures for each species were unique in three sets. No cross-reaction was observed in cattle-pig-horse (BSE), goat-sheep (CO), and chicken-duck-turkey (GAM) samples. The detection limits of the LAMP assays in raw meat, cooked meat, raw admixtures, and cooked admixtures were determined in 30 min and revealed greater sensitivity than PCR assays. The LAMP assays are simple, rapid, accurate, and sensitive for discrimination of eight meat species and could be used to support meat species identification to combat meat source fraud and adulteration. Further study may be needed to verify the utility of the LAMP assays for other animal products such as milk, milk powder, cheese, and smoked or frozen meats.
Acknowledgments
This work was supported by National Research Foundation of Korea Grant funded by the Korean Government (NRF-2012R1A1A1012293) and BK PLUS program for Creative Veterinary Science Research.
References
- 1.Ahmed M. U., Hasan Q., Mosharraf Hossain M., Saito M., Tamiya E. Meat species identification based on the loop mediated isothermal amplification and electrochemical DNA sensor. Food Control. (2010);21:599–605. doi: 10.1016/j.foodcont.2009.09.001. [DOI] [Google Scholar]
- 2.Avise J. C., Arnold J., Ball R. M., Bermingham E., Lamb T., Neigel J. E., Reeb C. A., Saunders N. C. Intraspecific phylogeography-the mitochondrial-DNA bridge between population-genetics and systematics. Annu. Rev. Ecol. Syst. (1987);18:489–522. [Google Scholar]
- 3.Ballin N. Z., Vogensen F. K., Karlsson A. H. Species determination-Can we detect and quantify meat adulteration? Meat Sci. (2009);83:165–174. doi: 10.1016/j.meatsci.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Bottero M. T., Dalmasso A. Animal species identification in food products: evolution of biomolecular methods. Vet. J. (2011);190:34–38. doi: 10.1016/j.tvjl.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y. H., Cheng S. D., Weng C. F. Investigation of goats' milk adulteration with cows' milk by PCR. Asian Australas. J. Anim. Sci. (2006);19:1503–1507. doi: 10.5713/ajas.2006.1503. [DOI] [Google Scholar]
- 6.Chikuni K., Ozutsumi K., Koishikawa T., Kato S. Species identification of cooked meats by DNA hybridization assay. Meat Sci. (1990);27:119–128. doi: 10.1016/0309-1740(90)90060-J. [DOI] [PubMed] [Google Scholar]
- 7.Chisholm J., Conyers C., Booth C., Lawley W., Hird H. The detection of horse and donkey using real-time PCR. Meat Sci. (2005);70:727–732. doi: 10.1016/j.meatsci.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Dalmasso A., Fontanella E., Piatti P., Civera T., Rosati S., Bottero M. T. A multiplex PCR assay for the identification of animal species in feedstuffs. Mol. Cell Probes. (2004);18:81–87. doi: 10.1016/j.mcp.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Dawnay N., Ogden R., McEwing R., Carvalho G. R., Thorpe R. S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. (2007);173:1–6. doi: 10.1016/j.forsciint.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Dincer B., Spearow J. L., Cassens R. G., Greaser M. L. The effects of curing and cooking on the detection of species origin of meat products by competitive and indirect ELISA techniques. Meat Sci. (1987);20:253–265. doi: 10.1016/0309-1740(87)90081-7. [DOI] [PubMed] [Google Scholar]
- 11.Dooley J. J., Paine K. E., Garrett S. D., Brown H. M. Detection of meat species using TaqMan real-time PCR assays. Meat Sci. (2004);68:431–438. doi: 10.1016/j.meatsci.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Francois P., Tangomo M., Hibbs J., Bonetti E. J., Boehme C. C., Notomi T., Perkins M. D., Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. (2011);62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 13.Gill C. O. Safety and storage stability of horse meat for human consumption. Meat Sci. (2005);71:506–513. doi: 10.1016/j.meatsci.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 14.Girish P. S., Anjaneyulu A. S. R., Viswas K. N., Anand M., Rajkumar N., Shivakumar B. M., Bhaskar S. Sequence analysis of mitochondrial 12S rRNA gene can identify meat species. Meat Sci. (2004);66:551–556. doi: 10.1016/S0309-1740(03)00158-X. [DOI] [PubMed] [Google Scholar]
- 15.Hebert P. D., Stoeckle M. Y., Zemlak T. S., Francis C. M. Identification of birds through DNA barcodes. PLoS Biology. (2004);2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hitchcock C. H., Crimes A. A. Methodology for meat species identification: A review. Meat Sci. (1985);15:215–224. doi: 10.1016/0309-1740(85)90077-4. [DOI] [PubMed] [Google Scholar]
- 17.Kesmen Z., Gulluce A., Sahin F., Yetim H. Identification of meat species by TaqMan-based real-time PCR assay. Meat Sci. (2009);82:444–449. doi: 10.1016/j.meatsci.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Kesmen Z., Yetiman A. E., Sahin F., Yetim H. Detection of chicken and turkey meat in meat mixtures by using real-time PCR assays. J. Food Sci. (2012);77:C167–173. doi: 10.1111/j.1750-3841.2011.02536.x. [DOI] [PubMed] [Google Scholar]
- 19.Koh B. R. D., Kim J. Y., Na H. M., Park S. D., Kim Y. H. Development of species-specific multiplex PCR assays of mitochondrial 12S rRNA and 16S rRNA for the identification of animal species. Korean J. Vet. Serv. (2011);34:417–428. doi: 10.7853/kjvs.2011.34.4.417. [DOI] [Google Scholar]
- 20.Macedo-Silva A., Barbosa S. F., Alkmin M. G., Vaz A. J., Shimokomaki M., Tenuta-Filho A. Hamburger meat identification by dot-ELISA. Meat Sci. (2000);56:189–192. doi: 10.1016/S0309-1740(00)00039-5. [DOI] [PubMed] [Google Scholar]
- 21.Martin I., Garcia T., Fajardo V., Lopez-Calleja I., Hernandez P. E., Gonzalez I., Martin R. Species-specific PCR for the identification of ruminant species in feedstuffs. Meat Sci. (2007);75:120–127. doi: 10.1016/j.meatsci.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Martinez I., Malmheden Yman I. Species identification in meat products by RAPD analysis. Food Res. Int. (1998);31:459–466. doi: 10.1016/S0963-9969(99)00013-7. [DOI] [Google Scholar]
- 23.Mouillesseaux K. P., Klimpel K. R., Dhar A. K. Improvement in the specificity and sensitivity of detection for the Taura syndrome virus and yellow head virus of penaeid shrimp by increasing the amplicon size in SYBR Green real-time RT-PCR. J. Virol. Methods. (2003);111:121–127. doi: 10.1016/S0166-0934(03)00167-8. [DOI] [PubMed] [Google Scholar]
- 24.Pappas C., Tarantilis P., Moschopoulou E., Moatsou G., Kandarakis I., Polissiou M. Identification and differentiation of goat and sheep milk based on diffuse reflectance infrared fourier transform spectroscopy (DRIFTS) using cluster analysis. Food Chem. (2008);106:1271–1277. doi: 10.1016/j.foodchem.2007.07.034. [DOI] [Google Scholar]
- 25.Proom H. The preparation of precipitating sera for the identification of animal species. J. Pathol. Bacteriol. (1943);55:419–426. doi: 10.1002/path.1700550404. [DOI] [Google Scholar]
- 26.Rodriguez M. A., Garcia T., Gonzalez I., Asensio L., Mayoral B., Lopez-Calleja I., Hernandez P. E., Martin R. Identification of goose, mule duck, chicken, turkey, and swine in foie gras by species-specific polymerase chain reaction. J. Agr. Food Chem. (2003);51:1524–1529. doi: 10.1021/jf025784+. [DOI] [PubMed] [Google Scholar]
- 27.Şakalar E., Abasıyanık M. F. The devolopment of duplex real-time PCR based on SYBR Green florescence for rapid dentification of ruminant and poultry origins in foodstuff. Food Chem. (2012);130:1050–1054. doi: 10.1016/j.foodchem.2011.07.130. [DOI] [Google Scholar]
- 28.Santos C. G., Melo V. S., Amaral J. S., Estevinho L., Oliveira M. B., Mafra I. Identification of hare meat by a species-specific marker of mitochondrial origin. Meat Sci. (2012);90:836–841. doi: 10.1016/j.meatsci.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. (2008);3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 30.Varga A., James D. Detection and differentiation of Plum pox virus using real-time multiplex PCR with SYBR Green and melting curve analysis: a rapid method for strain typing. J. Virol. Methods. (2005);123:213–220. doi: 10.1016/j.jviromet.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Verkaar E. L. C., Nijman I. J., Boutaga K., Lenstra J. A. Differentiation of cattle species in beef by PCRRFLP of mitochondrial and satellite DNA. Meat Sci. (2002);60:365–369. doi: 10.1016/S0309-1740(01)00144-9. [DOI] [PubMed] [Google Scholar]
- 32.Whittaker R. G., Spencer T. L., Copland J. An enzyme-linked immunosorbent assay for species identification of raw meat. J. Sci. Food Agr. (1983);34:1143–1148. doi: 10.1002/jsfa.2740341016. [DOI] [Google Scholar]
- 33.Wolf C., Rentsch J., Hübner P. PCR-RFLP analysis of mitochondrial DNA: a reliable method for species identification. J. Agr. Food Chem. (1999);47:1350–1355. doi: 10.1021/jf9808426. [DOI] [PubMed] [Google Scholar]
- 34.You J., Huang L., Zhuang J., Mou Z. Speciesspecific multiplex real-time PCR assay for identification of deer and common domestic animals. Food Sci. Biotechnol. (2014);23:133–139. doi: 10.1007/s10068-014-0018-3. [DOI] [Google Scholar]
- 35.Zhang C. L., Fowler M. R., Scott N. W., Lawson G., Slater A. A TaqMan real-time PCR system for the identification and quantification of bovine DNA in meats, milks and cheeses. Food Control. (2007);18:1149–1158. doi: 10.1016/j.foodcont.2006.07.018. [DOI] [Google Scholar]