Abstract
The purpose of this study was to analyze myosin heavy chain (MHC) isoforms in bovine longissimus thoracis (LT) muscle by liquid chromatography (LC) and mass spectrometry (MS). LT muscles taken from Hanwoo (Korean native cattle) steer (n=3) used to separate myosin bands by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The peptide queries were obtained from the myosin bands by LC-MS/MS analysis following in-gel digestion with trypsin. A total of 33 and 43 queries were identified as common and unique peptides, respectively, of MHC isoforms (individual ions scores >43 indicate identity or extensive homology, p<0.05). MHC-1 (IIx), -2 (IIa), -4 (IIb), and -7 (slow/I) were identified based on the Mowse score (5118, 3951, 2526, and 2541 for MHC-1, -2, -4, and -7, respectively). However, more analysis is needed to confirm the expression of MHC-4 in bovine LT muscle because any query identified as a unique peptide of MHC-4 was not found. The queries that were identified as unique peptides could be used as peptide markers to confirm MHC-1 (14 queries), -2 (8 queries), and -7 (21 queries) in bovine LT muscle; no query identified as a unique peptide of MHC-4 was found. LC-MS/MS analysis is a useful approach to study MHC isoforms at the protein level.
Keywords: myosin heavy chain, LC-MS/MS, cattle, longissimus thoracis
Introduction
The metabolic and contractile properties of muscle fiber types depend on the different myosin heavy chain (MHC) isforms, because myosin ATPase and actin-binding sites are localized at the globular head of the MHC (Rayment et al., 1993; Tokunaga et al., 1987). The MHC exists as multiple isoforms encoded for by the gene family, and nine MHC isoforms have been observed in mammalian cardiac and skeletal muscles (Berg et al., 2001; Qin et al., 2002; Weiss et al., 1999). Previous studies (Acevedo and Rivero, 2006; Chikuni et al., 2004; Kim et al., 2005; Kim et al., 2013a, b; Kjellgren et al., 2003; Lefaucheur et al., 2002) reported three or four MHC isoforms in mammals, inculding human, horse, and cattle muscles (MHC-I, -IIa, and -IIx), porcine muscle (MHC-I, -IIa, -IIx, and -IIb), and canine muscle (MHC-I, -IIa, and IIx). The relative proportions of the MHC isoforms are closely related to the differences in functional properties of muscle and in quality properties of meat both among the species and between the cuts. Efforts to analyze the expression of the MHC isoforms have been hindered be- cause of the high homology and identity among the MHC isoforms, regardless of species.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the most commonly used technique for MHC analysis, enables the separation of MHC isoforms. Until now, the method presented by Talmadge and Roy (1993) was adapted to MHC separation in various mammalian skeletal muscles. However, this method did not always clearly separate the MHC isoforms and since then many modified protocols have been introduced (Kim et al., 2013a; Kohn et al., 2007; Picard et al., 1999). Nevertheless, to confirm the specific MHC isoforms from each separated band, additional analyses, such as immunoblotting and immunohistochemistry with monoclonal antibodies, are needed.
In the present study, liquid chromatography (LC) and mass spectrometry (MS) were used for the analysis of bovine MHC isoforms. LC-MS/MS analysis allows confirmation of the unique peptides of each isoform by comparing identified peptides among the MHC isoforms. This technique does not require separation of the MHC isoforms on SDS-PAGE gel. Therefore, the purpose of this study was to develop the method to identify bovine MHC isoforms using LC-MS/MS analysis.
Materials and Methods
Sample preparation
Muscles (Longissimus Thoracis, LT) were taken from three Hanwoo steers (842±25 d of age, 653±15 kg of live weight) and promptly frozen in liquid nitrogen. Myofibrillar protein extraction and preparation were performed using the method described by Talmadge and Roy (1993). In brief, samples were mixed with sample buffer containing 4% (w/v) SDS, 1 M Tris-HCl, 20% (w/v) glycerol, 1% (v/v) mercaptoethanol, and small amount of bromophenol blue as a trace. Protein concentration was determined according to the Bradford (1976) method with bovine serum albumin as a standard.
SDS-PAGE
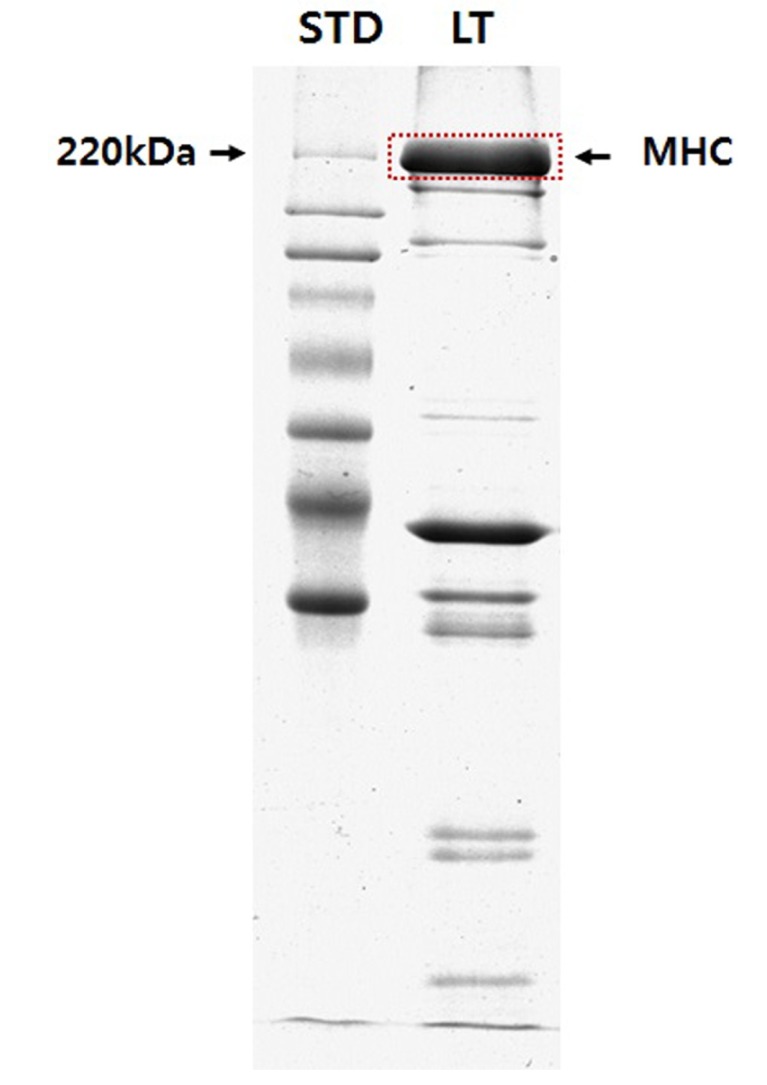
The stacking gels were composed of 4 mM EDTA, 0.4% (w/v) SDS, 4% (w/v) acrylamide/Bis (50:1), 30% (w/v) glycerol, and 70 mM Tris-HCl (pH 6.70). The separating gels were composed of 0.1 M glycine, 0.4% (w/v) SDS, 0.2 M Tris-HCl (pH 8.80), 10% (w/v) acrylamide/Bis (50 :1), and 45% (w/v) glycerol. SDS-PAGE was conducted in a mini-gel (0.75-mm-thick) system (SE-250, Amersham Biosciences, USA) at 50 mA/gel for 2 h, and the bands were visualized by Coomassie Brilliant Blue (CBB) staining. The result for SDS-PAGE was presented in Fig. 1.
Fig. 1. The result of one-dimensional gel electrophoresis for separate the myosin heavy chain (MHC) isoforms from bovine longissimus thoracis (LT) muscle. STD, standard marker.
In-gel digestion
The protein bands were excised from the CBB stained gel, destained with 50 mM NH4HCO3 buffer (pH 7.8) containing 30% (v/v) acetonitrile, and dried completely in a SpeedVac (SPD1010, Thermo Fisher Scientific Inc., USA) for 10 min. The dried gel pieces were rehydrated in 10 μL (2.5 ng/μL) of trypsin solution (Promega, UK) in 50 mM NH4HCO3 buffer (pH 7.8) at 4℃ for 2 h. After 10 μL of 50 mM NH4HCO3 was added, the gel slices were incubated at 37℃ for 12 h.
LC-MS/MS and data searching
LC-MS/MS analysis was performed with a nano-LC and LTQ mass spectrometer (Agilent 1100, Thermo Electon, USA). The capillary column was obtained from Proxeon (150 mm × 0.075 mm, Odense M, Denmark) and slurry packed in-house with 5 μm, 100 Å pore size Magic C18 stationary phase (Michrom Bioresources, USA). The mobile phase A for the LC separation was 0.1% formic acid in deionized water, and mobile phase B was 0.1% formic acid in acetonitrile. The chromatography gradient was set up to give a linear increase from 5% B to 35% B in 50 min, from 40% B to 60% B in 20 min, and from 60% B to 80% B in 5 min. The flow rate was maintained at 300 nl/min after splitting. Mass spectra were acquired using data-dependent acquisition with a full mass scan (400-1800 m/z) followed by MS/MS scans. Each MS/MS scan acquired was an average of one microscan on the LTQ. The temperature of the ion transfer tube was controlled at 200℃ and the spray was 1.5-2.0 kV. The normalized collision energy was set at 35% for the MS/MS. Sequences of the MS/MS spectra were identified by NCBInr database search using the MASCOT search engine (Matrix Science MASCOT software). Database search criteria were as described: fixed modification, no; carboxyamidomethylated at cysteine residues, variable modification, oxidized at methionine residues; maximum allowed cleavage, 1; MS tolerance, 1.2 Da; MS/MS tolerance, 0.6 D. Only peptides resulting from trypsin digests were considered.
Probability based Mowse score
Ions score is -10×log(P), where P is the probability that the observed match is a random event. Individual ions scores >43 indicate identity or extensive homology (p<0.05). Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits. Mowse scores of the MHC isoforms were 5118, 3951, 2526, and 2541 for MHC-1 (IIx), -2 (IIa), -4 (IIb), and -7 (slow/I), respectively.
Results
As shown in Fig. 1, MHC was separated from the sample mixture by SDS-PAGE analysis and LC-MS/MS analysis was performed following in-gel digestion of the MHC band with trypsin. Four MHC isoforms (three MHC-II types and one MHC-I type) including MHC-1 (2x, II type), -2 (2a, II type), -4 (2b, II type), and -7 (slow, I type) were identified (Table 1). MHC-1, which has pI of 5.57, molecular weight of 223,900 Da, and 1938 amino acids of peptide size had 524 matched queries. The sequence coverage showed 49%, and there were 53 queries with identity (the scores greater than 43 indicate identity with p<0.05). MHC-4 also has 1938 amino acids of peptide size but showed matched queries, queries with identify, and sequence coverage of 263, 27, and 33%, respectively. The other MHC-II type, MHC-2, which has pI of 5.63, molecular weight of 224,243 Da, and 1943 amino acids of peptide sequence, was identified by 397 matched queries, 39 queries with identity, and 45% sequence coverage. MHC-7, a MHC-I type, has pI of 5.58, molecular weight of 224,026 Da, and 1935 amino acids of peptide size was identified by 225 matched queries, 28 queries with identity, and 37% sequence coverage. MHC-1 had the highest sequence coverage, matched queries, and queries with identity among the MHC isoforms, whereas MHC-4 had the lowest sequence coverage and queries having identity (ion score >43).
Table 1. Proteins identified as bovine myosin heavy chain (MHC) isoforms by LC-MS/MS analysis.
| MHCa) | Accession numberb) | Species | Theoretical MW (Da)c) | pI | Size (aa)d) | Sequence coverage (%) | Queries matched | Queries with identitye) |
|---|---|---|---|---|---|---|---|---|
| MHC-1 (IIx) | gi|21743235 | Bos taurus | 223900 | 5.57 | 1938 | 49 | 524 | 53 |
| MHC-2 (IIa) | gi|261245063 | Bos taurus | 224243 | 5.63 | 1943 | 45 | 397 | 39 |
| MHC-4 (IIb) | gi|296476617 | Bos taurus | 223875 | 5.62 | 1938 | 33 | 263 | 27 |
| MHC-7 (slow/I) | gi|13560273 | Bos taurus | 224026 | 5.58 | 1935 | 37 | 225 | 28 |
a)Myosin heavy chain.
b)Accession numbers were taken from the NCBI database.
c)Theoretical molecular weight.
d)Peptide size (amino acids) recorded in NCBI database.
e)Individual ions scores greater than 43 indicate identity (p<0.05).
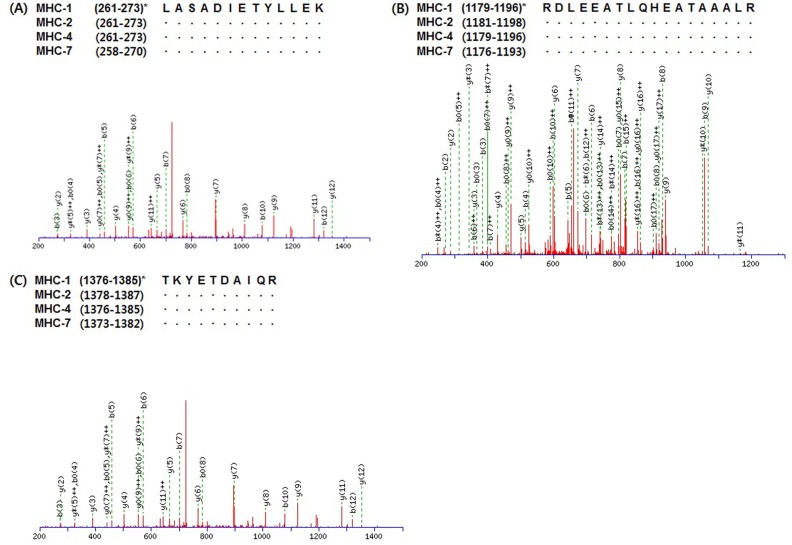
Peptides expressing in common in two or more MHC isoforms are shown in Table 2. A total of 33 queries were detected as common peptides and only three queries such as HADSVAELGEQIDNLQR, DLEEATLQHEATAAAL RKK, and VIQYFATIAVTGEK did not match the amino acid sequence of MHC-1. However, MHC-7 had just five queries (ANSEVAQWR, LASADIETYLLEK, DLEEATLQHEATAAALRKK, RDLEEATLQHEATAAALR, and TKYETDAIQR) matching its amino acid sequence. A total of 25 and 24 queries were observed as common peptides that matched MHC-2 and -4, respectively. As presented in Fig. 2, three queries (LASADIETYLLEK, RDL EEATLQHEATAAALR, and TKYETDAIQR) were found as common peptides expressing in all MHC isoforms. LASADIETYLLEK corresponds to residues 261-273 of MHC-1, -2, and -4 and to residues 258-270 of MHC-7. RDLEEATLQHEATAAALR corresponds to residues 1179-1196 of MHC-1 and -4, residues 1181-1198 of MHC-2, and residues 1176-1193 of MHC-7. TKYETDAIQR corresponds to residues 1376-1385 of MHC-1 and -4, residues 1378-1387 of MHC-2, and residues 1373-1382 of MHC-7.
Table 2. Queries expressed in common in two or more myosin heavy chain (MHC) isoforms.
| Sequence | Ion scorea) | Mr (expt) | Mr (cal) | MHC |
|---|---|---|---|---|
| ALQEAHQQTLDDLQAEEDKVNTLTK | 71 | 2839.7 | 2839.0 | MHC-1, MHC-2, MHC-4 |
| ANSEVAQWR | 57 | 1060.7 | 1060.1 | MHC-1, MHC-4, MHC-7 |
| HADSVAELGEQIDNLQR | 73 | 1895.5 | 1895.0 | MHC-2, MHC-4 |
| HADSVAELGEQIDNLQRVK | 76 | 2123.4 | 2122.3 | MHC-1, MHC-2, MHC-4 |
| IAEKDEEIDQLKR | 69 | 1587.3 | 1586.7 | MHC-1, MHC-2, MHC-4 |
| IAEQELLDASER | 67 | 1373.5 | 1373.5 | MHC-1, MHC-2 |
| IKEVTERAEDEEEINAELTAK | 71 | 2418.6 | 2417.6 | MHC-1, MHC-2 |
| KALQEAHQQTLDDLQAEEDKVNTLTK | 83 | 2967.8 | 2967.2 | MHC-1, MHC-2 |
| KHADSVAELGEQIDNLQR | 46 | 2023.2 | 2023.2 | MHC-1, MHC-2, MHC-4 |
| LAQLITR | 50 | 815.2 | 814.0 | MHC-1, MHC-4 |
| LASADIETYLLEK | 72 | 1465.7 | 1465.6 | MHC-1, MHC-2, MHC-4, MHC-7 |
| LQHELEEAEERADIAESQVNKLR | 82 | 2708.5 | 2707.9 | MHC-1, MHC-2, MHC-4 |
| LYEQHLGK | 54 | 988.1 | 987.1 | MHC-1, MHC-4 |
| SALAHALQSAR | 47 | 1125.4 | 1124.3 | MHC-1, MHC-4 |
| VGNEFVTK | 65 | 893.7 | 893.0 | MHC-1, MHC-4 |
| VLNASAIPEGQFIDSK | 50 | 1688.8 | 1688.9 | MHC-1, MHC-4 |
| DLEEATLQHEATAAALR | 93 | 1838.0 | 1839.0 | MHC-1, MHC-2, MHC-4 |
| DLEEATLQHEATAAALRKK | 46 | 2095.8 | 2095.3 | MHC-4, MHC-7 |
| ELEGEVESEQKR | 54 | 1433.5 | 1432.5 | MHC-1, MHC-2 |
| IEELEEEIEAERASR | 66 | 1803.6 | 1802.9 | MHC-1, MHC-2, MHC-4 |
| ILYADFKQR | 48 | 1154.1 | 1153.3 | MHC-1, MHC-2, MHC-4 |
| INQQLDTKQPR | 62 | 1341.4 | 1340.5 | MHC-1, MHC-2, MHC-4 |
| IQLELNQVKSEIDR | 54 | 1684.6 | 1684.9 | MHC-1, MHC-2, MHC-4 |
| KIAEQELLDASERVQLLHTQNTSLINTK | 53 | 3194.0 | 3193.6 | MHC-1, MHC-2 |
| KKHADSVAELGEQIDNLQR | 83 | 2152.1 | 2151.3 | MHC-1, MHC-2, MHC-4 |
| LQDLVDKLQAK | 63 | 1270.6 | 1270.5 | MHC-1, MHC-2 |
| LQTESGEFSR | 67 | 1153.0 | 1153.2 | MHC-1, MHC-2 |
| NVEAVKGLR | 69 | 984.6 | 985.1 | MHC-1, MHC-2 |
| RDLEEATLQHEATAAALR | 66 | 1996.1 | 1995.2 | MHC-1, MHC-2, MHC-4, MHC-7 |
| TKYETDAIQR | 58 | 1223.7 | 1224.3 | MHC-1, MHC-2, MHC-4, MHC-7 |
| VIQYFATIAVTGEK | 46 | 1540.2 | 1539.8 | MHC-2, MHC-4 |
| VRELEGEVESEQKR | 62 | 1688.8 | 1687.8 | MHC-1, MHC-2 |
| YKVLNASAIPEGQFIDSK | 50 | 1981.0 | 1980.2 | MHC-1, MHC-4 |
a)Individual ions scores greater than 43 indicate identity (p<0.05).
Fig. 2. Representative MS/MS spectra identified as common peptides of myosin heavy chain (MHC) isoforms in bovine longissimus thoracis muscle. *, residues of amino acid sequence of each MHC isoform.
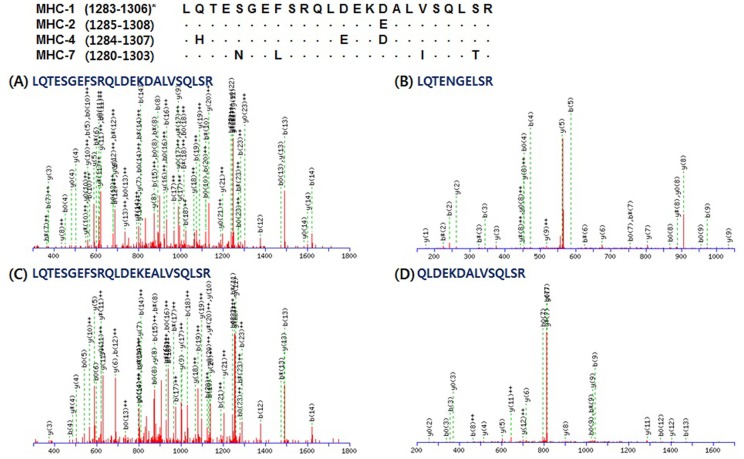
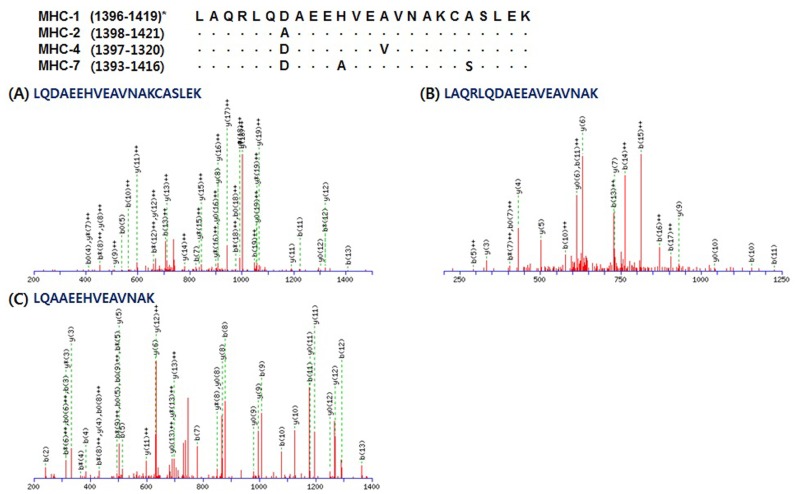
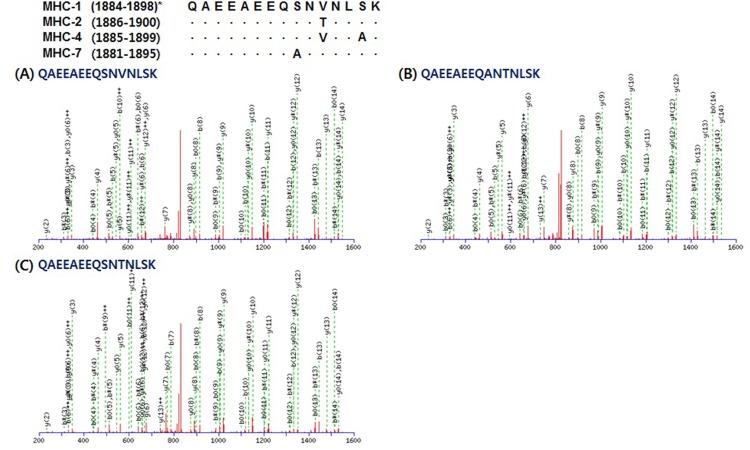
Unique peptides matched the sequence of each MHC isoform presented in Table 3. A total of 43 peptides were detected for MHC-1 (14), -2 (8), and -7 (21). LQTESGEFSRQLDEKDALVSQLSR corresponds to residues 1283-1306 of MHC-1. This is the same sequence area of residues 1285-1308 of MHC-2; however, MHC-2 has “E (glutamic acid)” instead of “D (aspartic acid)” in residue 1300 (Fig. 3). MHC-7 has different amino acids in residues 1284 (N, asparagine), 1287 (L, leucine), 1298 (I, isoleucine), and 1302 (T, threonine) instead of S (serine), F (phenylalanine), V (valine), and S of MHC-1, respectively. Unique peptides for MHC-7 were found as two queries including LQTENGELSR and QLDEKDALVSQLSR (Fig. 3C and 3D). The queries presented in Fig. 4 have different sequence length, but LQAAEEHVEAVNAK corresponds to residues 1402-1415 of MHC-2 is peptide located in the same area of the amino acid sequences in MHC-1, -2, and -7. LQDAEEHVEAVNAKCASLEK corresponds to residues 1400-1419 of MHC-1 and includes residues 1400-1413, which correspond to residues 1402-1415 of MHC -2. Only one amino acid (D) on residue 1402 is different from A (alanine) on residue 1404 of MHC-2. LAQRLQDAEEAVEAVNAK which matched to residues 1393-1410 of MHC-7 showed that D on residue 1399 and A on residue 1403 were different from A on residue 1404 of MHC-2 and H on residues 1406 of MHC-1 and 1408 of MHC-2. Lastly, QAEEAEEQSNVNLSK, which corresponds to residues 1884-1898 of MHC-1, 1886-1900 of MHC-2, 1885-1899 of MHC-4, and 1881-1895 of MHC-7, was observed as a unique peptide of MHC-1, as presented in Fig. 5A. MHC-2 has T on residue 1896 instead of V on residue 1894 of MHC-1 (Fig. 5B). QAEEAEEQ SNTNLSK, which was observed in MHC-7, showed a different amino acid (A) on residue 1889 with S on residues 1892 of MHC-1 and 1894 of MHC-2 (Fig. 5C). No unique peptides were observed for MHC-4.
Table 3. Queries identified as unique peptides of each myosin heavy chain (MHC) isoform.
| MHC | Sequence | Residue number | Ion scorea) | MW (expt) | MW (calc) |
|---|---|---|---|---|---|
| MHC-1 | TSVFVADPKESFVKATVQSR | 36-55 | 62 | 2197.6 | 2196.5 |
| MHC-1 | VGNEFVTKGQTVEQVYNAVGALAK | 409-432 | 54 | 2523.2 | 2522.8 |
| MHC-1 | GQTVEQVYNAVGALAK | 417-432 | 71 | 1648.6 | 1647.8 |
| MHC-1 | SSVKTLALLFSGPASGEAEGGPK | 615-637 | 60 | 2203.4 | 2203.4 |
| MHC-1 | ARLQTESGEFSR | 1281-1292 | 71 | 1380.1 | 1380.5 |
| MHC-1 | LQTESGEFSRQLDEKDALVSQLSR | 1283-1306 | 83 | 2737.8 | 2736.9 |
| MHC-1 | QLDEKDALVSQLSR | 1293-1306 | 88 | 1602.9 | 1601.8 |
| MHC-1 | LAQRLQDAEEHVEAVNAK | 1396-1413 | 44 | 2021.9 | 2021.2 |
| MHC-1 | LQDAEEHVEAVNAKCASLEK | 1400-1419 | 51 | 2242.1 | 2241.4 |
| MHC-1 | QKYEETHAELEASQKESR | 1461-1478 | 44 | 2163.9 | 2163.3 |
| MHC-1 | IVESMQSTLDAEIR | 1596-1609 | 49 | 1607.8 | 1607.8 |
| MHC-1 | SYKRQAEEAEEQSNVNLSK | 1880-1898 | 82 | 2210.2 | 2210.3 |
| MHC-1 | RQAEEAEEQSNVNLSK | 1883-1898 | 64 | 1832.4 | 1831.9 |
| MHC-1 | QAEEAEEQSNVNLSK | 1884-1898 | 90 | 1676.6 | 1675.7 |
| MHC-2 | SAETEKEMATMKEEFQK | 847-863 | 47 | 2050.0 | 2049.2 |
| MHC-2 | LQTESGEFSRQLDEKEALVSQLSR | 1285-1308 | 60 | 2751.5 | 2751.0 |
| MHC-2 | QLDEKEALVSQLSR | 1295-1308 | 52 | 1615.5 | 1615.8 |
| MHC-2 | ALSKANTEVAQWR | 1365-1377 | 58 | 1473.5 | 1473.6 |
| MHC-2 | ANTEVAQWR | 1369-1377 | 69 | 1074.3 | 1074.1 |
| MHC-2 | LQAAEEHVEAVNAK | 1402-1415 | 80 | 1508.4 | 1508.6 |
| MHC-2 | RQAEEAEEQSNTNLSK | 1885-1900 | 59 | 1834.5 | 1833.9 |
| MHC-2 | QAEEAEEQSNTNLSK | 1886-1900 | 73 | 1677.4 | 1677.7 |
| MHC-7 | DVFVPDDKEEFVKATILSR | 36-54 | 46 | 2209.1 | 2208.5 |
| MHC-7 | EQATGKGTLEDQIIQANPALEAFGNAK | 208-234 | 81 | 2815.4 | 2815.1 |
| MHC-7 | GTLEDQIIQANPALEAFGNAK | 214-234 | 80 | 2199.5 | 2200.4 |
| MHC-7 | VGNEYVTKGQNVQQVVYAK | 406-424 | 69 | 2124.6 | 2124.4 |
| MHC-7 | GQNVQQVVYAK | 414-424 | 58 | 1234.2 | 1233.4 |
| MHC-7 | LFDNHLGK | 552-559 | 51 | 943.8 | 943.1 |
| MHC-7 | ILNPAAIPEGQFIDSR | 724-739 | 69 | 1740.4 | 1741.0 |
| MHC-7 | LKEALEKSEAR | 859-869 | 60 | 1273.4 | 1273.4 |
| MHC-7 | ALQEAHQQALDDLQAEEDKVNTLTK | 998-1022 | 48 | 2809.0 | 2809.0 |
| MHC-7 | KKHADSVAELSEQIDNLQR | 1194-1212 | 67 | 2182.0 | 2181.4 |
| MHC-7 | SKAEETQRSVNDLTSQR | 1261-1277 | 47 | 1950.2 | 1949.0 |
| MHC-7 | SVNDLTSQRAK | 1269-1279 | 45 | 1218.2 | 1218.3 |
| MHC-7 | LQTENGELSR | 1280-1289 | 55 | 1146.1 | 1146.2 |
| MHC-7 | QLDEKEALISQLTR | 1290-1303 | 48 | 1643.8 | 1643.8 |
| MHC-7 | VLSKANSEVAQWR | 1360-1372 | 69 | 1487.5 | 1487.7 |
| MHC-7 | LAQRLQDAEEAVEAVNAK | 1393-1410 | 54 | 1955.6 | 1955.1 |
| MHC-7 | SNAAAAALDKK | 1435-1445 | 44 | 1059.3 | 1059.2 |
| MHC-7 | VVDSLQTSLDAETR | 1593-1606 | 57 | 1533.3 | 1533.6 |
| MHC-7 | VRELENELEAEQKR | 1819-1832 | 51 | 1743.4 | 1742.9 |
| MHC-7 | QAEEAEEQANTNLSK | 1881-1895 | 98 | 1662.3 | 1661.7 |
| MHC-7 | VQHELDEAEERADIAESQVNKLR | 1899-1921 | 77 | 2680.2 | 2679.9 |
a)Individual ions scores greater than 43 indicate identity (p<0.05).
Fig. 3. Representative MS/MS spectra identified unique peptides of each myosin heavy chain (MHC) isoform: LQTESGEFSRQLDEKDALVSQLSR for MHC-1, LQTESGEFSRQLDEKEALVSQLSR for MHC-2, and LQTENGELSR and QLDEKEALISQLTR for MHC-7, respectively. *, residues of amino acid sequence of each MHC isoform.
Fig. 4. Representative MS/MS spectra identified unique peptides of each myosin heavy chain (MHC) isoform: LQDAEEHVEAVNAKCASLEK for MHC-1, LQAAEEHVEAVNAK for MHC-2, and LAQRLQDAEEAVEAVNAK for MHC-7, respectively. *, residues of amino acid sequence of each MHC isoform.
Fig. 5. Representative MS/MS spectra identified unique peptides of each myosin heavy chain (MHC) isoform: QAEEAEEQSNVNLSK for MHC-1, QAEEAEEQSNTNLSK for MHC-2, and QAEEAEEQANTNLSK for MHC-7, respectively. *, residues of amino acid sequence of each MHC isoform.
Discussion
The comparative sequence analysis of MHC isoforms in bovine skeletal muscle was conducted in a previous study by Chikuni et al. (2004). They demonstrated species-specific amino acid sequences of bovine skeletal muscle compared to porcine skeletal muscle by RT-PCR analysis of MHC mRNA. Picard and Cassar-Malek (2009) also reported MHC isoform expression in bovine skeletal muscle at the gene level. Unlike these previous reports, the findings of the present study allow us to confirm the expression of MHC isoforms in bovine skeletal muscle at the protein level. A number of peptides digested with trypsin were obtained from the MHC band that separated from the bovine LT muscle. Three MHC isoforms: MHC-1 (IIx), -2 (IIa), and -7 (I/slow), were confirmed by identification of their own unique peptides after amino acid sequence matching. MHC-4 (IIB) was also found, and its Mowse score (2526), sequence coverage (33%), and the number of queries with identity (27) were high (Table 1). However, MHC-4 had no unique peptide, as shown in Table 3. Chikuni et al. (2004) also reported three MHC isoforms, including MHC-slow (-7), -2a (-2), and -2x (-1) expression in bovine skeletal muscle. MHC-4 has been found in porcine skeletal muscle (Graziotti et al., 2001; Kim et al., 2013b, 2014; Lefaucheur et al., 2002, 2004). Although Picard and Cassar-Malek (2009) demonstrated MHC-IIb (-4) expression in bovine semitendinosus and LT muscles, the authors recommended additional studies on MHC-4 gene transcription and any other mechanism.
The analysis of MHC isoforms at the protein level has been usually performed by electrophoresis, immunoblotting, and immunohistochemistry (IHC), regardless of the animal species (Abreu et al., 2006; Kim et al., 2013a; Kohn et al., 2007; Lefaucheur et al., 2004; Picard et al., 1999). Methods including immunoblotting and IHC need three or four antibodies with specificities for MHC isoforms. Moreover, electrophoresis does not clearly separate MHC isoforms because the molecular weights among MHC isoforms are similar (223.947, 223.924, 224.010, and 223.657 kDa for porcine MHC-1, -2, -4, and -7, respectively; 223.900, 224.243, 223.875, and 224.026 kDafor bovine MHC-1, -2, -4, and -7, respectively) (Kim et al., 2013a; Table 1). In the present study, 43 queries were identified as unique peptides of each MHC isoform (excluding MHC-4) by LC-MS/MS analysis, as shown in Table 3. These peptides could be markers for MHC isoforms. For example, LQTESGEFSRQLDEKDALVSQLSR can be detected in MHC-1, whereas the others have one (E on residue 1297 of MHC-2), three (H, E, and D on residues 1285, 1296, and 1299 of MHC-4), or four (N, L, I, and T on residues 1284, 1287, 1298, and 1302 of MHC-7) different amino acids compared to MHC-1 (Fig. 3). Thus, 14, 8, and 21 queries identified as unique peptides could be identification markers for MHC-1, -2, and -7, respectively. Because unique peptides of MHC-4 were not observed, it is not possible to confirm the existence of MHC-4 in bovine LT muscle. However, the high Mowse score, high sequence coverage, and the number of queries with identity for MHC-4 indicate that additional analyses, such as immunoblotting and IHC, are needed to identify this isoform.
According to a review by Pette and Staron (2000), MHC transition occurs as MHC-I (-7) MHC-2a (-2) MHC-IIx (-1) MHC-IIb (-4), and age, nutrition, and stress can change MHC expression. Picard and Cassar-Malek (2009) found MHC-IIb (-4) in a French beef breed. If we suppose that MHC-4 did not originally exist in bovine LT muscle, some variations, such as breed (Hanwoo cattle), age (842 d old), and muscle (LT), could be reasons of it.
Conclusions
The expression of bovine MHC-1, -2, and -7 was confirmed by LC-MS/MS following electrophoresis analysis and in-gel digestion with trypsin. The unique peptides identified as part of each MHC isoform could be used as peptide markers for confirmation of MHC isoforms. It is unclear if MHC-4 expresses in bovine LT muscle because no query identified as a unique peptide of MHC-4 was found.
Acknowledgments
This study was supported by National Research Foundation (NRF-2014R1A1A1008535) funded by the Ministry of Education, Republic of Korea.
References
- 1.Abreu E., Quirox-Rothe E., Mayoral A. I., Vivo J. M., Robina Á., Guillén M. T., Agüera E., Rivero J. L. Myosin heavy chain fibre types and fibre sizes in nulliparous and primiparous ovariectomized Iberian sows: Interaction with two alternative rearing systems during the fattening period. Meat Sci. (2006);74:359–372. doi: 10.1016/j.meatsci.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Acevedo L. M., Rivero J. L. New insights into skeletal muscle fibre types in the dog with particular focus towards hybrid myosin phenotypes. Cell Tissue Res. (2006);323:283–303. doi: 10.1007/s00441-005-0057-4. [DOI] [PubMed] [Google Scholar]
- 3.Berg J. S., Powell B. C., Cheney S. E. A millennium myosin census. Mol. Biol. Cell. (2001);12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. (1976);72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chikuni K., Muroya S., Nakajima I. Myosin heavy chains expressed in bovine skeletal muscles. Meat Sci. (2004);67:87–94. doi: 10.1016/j.meatsci.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Graziotti G. H., Ríos C. M., Rivero J. L. L. Evidence for three fast myosin heavy chain isoforms in type II skeletal muscle fibers in adult llama (Lama glama) J. Histochem. Cytochem. (2001);49:1033–1044. doi: 10.1177/002215540104900811. [DOI] [PubMed] [Google Scholar]
- 7.Kim G. D., Jeong J. Y., Yang H. S., Joo S. T. Identification of myosin heavy chain isoforms in porcine longissimus dorsi muscle by electrophoresis and mass spectrometry. Electrophoresis. (2013);34:1255–1261. doi: 10.1002/elps.201300049. [DOI] [PubMed] [Google Scholar]
- 8.Kim G. D., Ryu Y. C., Jeong J. Y., Yang H. S., Joo S. T. Relationship between pork quality and characteristics of muscle fibers classified by the distribution of myosin heavy chain isoforms. J. Anim. Sci. (2013);91:5525–5534. doi: 10.2527/jas.2013-6614. [DOI] [PubMed] [Google Scholar]
- 9.Kim G. D., Ryu Y. C., Jo C., Lee J. G., Yang H. S., Jeong J. Y., Joo S. T. The characteristics of myosin heavy chain-based fiber types in porcine longissimus dorsi muscle. Meat Sci. (2014);96:712–718. doi: 10.1016/j.meatsci.2013.09.028. [DOI] [PubMed] [Google Scholar]
- 10.Kim J. S., Hinchcliff K. W., Yamaguchi M., Beard L. A., Markert C. D., Devor S. T. Age-related changes in metabolic properties of equine skeletal muscle associated with muscle plasticity. Vet. J. (2005);169:397–403. doi: 10.1016/j.tvjl.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Kjellgren D., Thornell L. E., Andersen J., Pedrosa-Domellof F. Myosin heavy chain isoforms in human extraocular muscles. Invest. Ophthalmol. Vis. Sci. (2003);44:1419–1425. doi: 10.1167/iovs.02-0638. [DOI] [PubMed] [Google Scholar]
- 12.Kohn T. A., Hoffman L. C., Myburgh K. H. Identification of myosin heavy chain isoforms in skeletal muscle of four Southern African wild ruminants. Comp. Biochem. Physiol. A Physiol. Part A. (2007);148:399–407. doi: 10.1016/j.cbpa.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Lefaucheur L., Ecolan P., Plantard L., Gueguen N. New insights into muscle fiber types in the pig. J. Histochem. Cytochem. (2002);50:719–730. doi: 10.1177/002215540205000513. [DOI] [PubMed] [Google Scholar]
- 14.Lefaucheur L., Milan D., Ecolan P., Le Callennec C. Myosin heavy chain composition of different skeletal muscles in Large White and Meishan pigs. J. Anim. Sci. (2004);82:1931–1941. doi: 10.2527/2004.8271931x. [DOI] [PubMed] [Google Scholar]
- 15.Pette D., Staron R. S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. (2000);50:500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Picard B., Barboiron C., Duris M. P., Gagniére H., Jurie C., Geay Y. Electrophoretic separation of bovine muscle myosin heavy chain isoforms. Meat Sci. (1999);53:1–7. doi: 10.1016/S0309-1740(99)00021-2. [DOI] [PubMed] [Google Scholar]
- 17.Picard B., Cassar-Malek I. Evidence for expression of IIb myosin heavy chain isoform in some skeletal muscles of Blonde d’Aquitaine bulls. Meat Sci. (2009);82:30–36. doi: 10.1016/j.meatsci.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 18.Qin H., Hsu M. K. H., Morris B. J., Hoh J. F. Y. A distinct subclass of mammalian striated myosins: structure and molecular evolution of of ‘superfast’ or masticatory myosin heavy chain. J. Mol. Evol. (2002);55:544–552. doi: 10.1007/s00239-002-2349-6. [DOI] [PubMed] [Google Scholar]
- 19.Rayment I., Rypniewski W. R., Schmidt-Base K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. Three dimensional structure of myosin subfragment-1: a molecular motor. Science. (1993);261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 20.Talmadge R. J., Roy R. R. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J. Appl. Physiol. (1993);75:2337–2340. doi: 10.1152/jappl.1993.75.5.2337. [DOI] [PubMed] [Google Scholar]
- 21.Tokunaga M., Sutoh K., Toyoshima C., Wayabashi T. Location of the ATPase site of myosin determined by three-dimensional electron microscopy. Nature. (1987);329:635–638. doi: 10.1038/329635a0. [DOI] [PubMed] [Google Scholar]
- 22.Weiss A., Schiaffino S., Leinwand L. A. Comparative sequence analysis of the complete human sarcomeric myosin heavy chain family: implications for functional diversity. J. Mol. Biol. (1999);290:61–75. doi: 10.1006/jmbi.1999.2865. [DOI] [PubMed] [Google Scholar]