Abstract
The transcriptome changes hugely during development of the brain. Whole genes, alternate exons and single base pair changes related to RNA editing all show differences between embryonic and mature brain. Collectively, these changes control proteomic diversity as the brain develops. Additionally, there are many changes in non-coding RNAs (miRNA and lncRNA) that interact with mRNA to influence the overall transcriptional landscape. Here we will discuss what is known about such changes in brain development, particularly focussing on high throughput approaches and how those can be used to infer mechanisms by which gene expression is controlled in the brain as it matures.
Keywords: Adult, Editing, Embryo, Gene expression, RNA-Seq, Splicing
Introduction
The cellular and molecular complexity of the mature adult brain is influenced both by processes in development and by the experience-dependent formation of neuronal circuits (Innocenti & Price, 2005; Sur & Rubenstein, 2005). Brain development occurs throughout embryonic growth and, in most species, continues after birth with a wide variety of developmental programs between species (Borrell & Calegari, 2014). To form the mature brain, many different cell types need to differentiate from basal progenitors, migrate to their anatomical positions and, for neurons, form synapses. As such, brain development is a highly regulated process.
Part of the regulation of brain development includes the coordinated expression of many different genes in a spatially and temporally appropriate context. Virtually all levels of gene expression, from whole genes to splicing and RNA editing, show evidence of regulation during brain development. The purpose of this review is to discuss all levels of gene expression in the development of the brain with a particular emphasis on genome-wide techniques that have allowed for an overall view of the generality of expression changes in this organ.
Gene expression
A number of studies have employed microarray technology in an attempt to study the molecular changes occurring during brain development including many reports at a genome wide scale. Although these could be organized by species or brain region, here we will use the order of publication as, in general, the depth of coverage in genomewide techniques has increased over time.
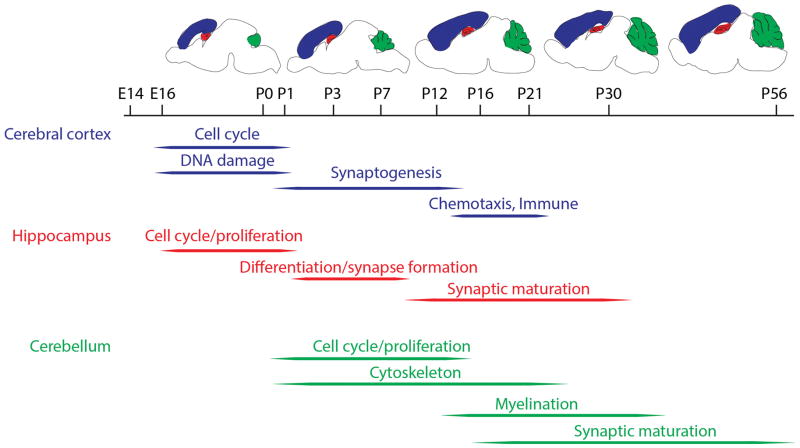
In 2001, two papers were published using arrays to look at development in the mouse hippocampus (Mody et al., 2001) and cerebellum(Kagami & Furuichi, 2001) using affymetrix arrays. These two brain regions have slightly different trajectories of development in the mouse and the two studies used slightly different choices for developmental time point. Mody et al., examined the hippocampus at embryonic day 16 (E16) and postnatal days 1 (P1), P7, P16, and P30 whereas E18, P7, P14, P21, and P56 were used in the cerebellum by Kagami and Furuichi. Despite the differences between the two regions and slight differences in the choices of time points, there are some substantial overlap in the genes and, more importantly, types of genes identified in both studies. In both studies there was a decrement in expression of gene related to neuronal proliferation or cell division from embryonic to postnatal stages(Kagami & Furuichi, 2001; Mody et al., 2001). This presumably represents the maturation from dividing neuronal precursor cells to mature, post-mitotic neurons. One example of gene expression that tended to increase with brain development noted by Mody et al was the upregulation of genes involved in glycolysis, consistent with a shift from ketone to glucose metabolism with brain maturation (Mody et al., 2001). Similarly, gene expression profiles consistent with synaptic maturation and associated signal transduction were clearly seen in both studies(Kagami & Furuichi, 2001; Mody et al., 2001). A summary of some of the key types of genes that are developmentally regulated in different brain regions are shown in Figure 1.
Figure 1.
Categories of gene expression in the developing mouse brain. The timeline from embryonic (E) to postnatal (P) gene expression is given in days and above the timeline are schematics of the brain approximately equivalent to their positions. Three brain regions where gene expression has been studied are colored in blue (cerebral cortex), red (hippocampus) and green (cerebellum). Below the time line are groups of genes that are prominently expressed at given developmental ranges in each region.
A similar study of the developing cerebellum focused particularly on granule cells which largely develop during the postnatal period in the mouse (Díaz et al., 2002). As well as examining expression patterns in the cerebellum, Diaz et al used the same technology to look at cultured granule cells developing in vitro, mutant mice where granule cells are lost in the postnatal period, as well as the pontine nucleus to which the granule cells project. Although the data series are therefore complex to interpret, they indicate that gene expression in development involves both cell autonomous and non-cell autonomous regulation.
Further studies extended these observations using whole brains using additional embryonic stages(Matsuki, Hori, & Furuichi, 2005), focusing on the prefrontal region of the cerebral cortex in postnatal development (Semeralul et al., 2006), or examining the cortex from embryonic to postnatal development (Pramparo et al., 2011). In general, these studies confirmed earlier results in that there were consistent decreases in cell division proteins and acquisition of genes that encode for synaptic proteins. Another interesting observations across several of these experiments are the sheer numbers of genes that show some evidence of regulation during brain development. For example, Matsuki et al reported that 1413 genes (~11%) showed altered expression in the prenatal period while Semeralul et al. reported 366 differentially expressed probe sets in the postnatal period. Although these two studies are not strictly comparable, they suggest that effects on gene expression are greater in the prenatal compared to postnatal period.
A similar picture of gene expression during development emerges from studies of the human prefontal cortex (Colantuoni et al., 2011). The absolute number of genes that show differential expression in prenatal development is higher than those in postnatal development, which is higher still than in aging. Furthermore, the magnitude of changes is higher in fetal development than later stages. Also consistent with mouse data were the types of genes that showed changes during development with, for example, diminishment of cell cycle genes and increases in synaptic components. One complication with human gene expression studies is the genetic diversity of humans compared to inbred mouse strains where genetic variation is minimal. Because gene expression is under genetic control, including in the brain(Gibbs et al., 2010), it is potentially important to dissect out genotypic and developmental effects on gene expression. However, single nucleotide polymorphisms (SNPs) that influence gene expression appear to influence overall expression levels rather than rates of change, that is to say that most genes that are developmentally regulated retain that regulation irrespective of genotype(Colantuoni et al., 2011).
One of the attractions of using genomewide approaches is that they might be mined to look for unexpected associations that, in turn, might be used to predict mechanisms. One useful approach is to generate self-organizing clusters of genes with similar trajectories of change with development. For example, several transcription factors (SMBP2, FE65 and Sox-M) show correlated expression with genes that increase in the post-natal time period in the hippocampus (Mody et al., 2001). As specific transcription factors are involved in neuronal specification (Thompson & Ziman, 2011), the increase in transcription factor expression in the postnatal period suggests that such proteins might also be important in the maintenance of neuronal phenotype through adulthood.
Genomewide approaches can further be used to understand how specific transcription factors work during development. For example, knockout of the transcription factor aristaless-related homeobox (Arx) results in altered migration of interneurons and abnormal neuronal differentiation. Using Affymetrix arrays in a conditional knockout where Arx was depleted in the subpallium, Fulp et al were able to reconstruct a genetic network where Arx normally represses additional transcription factors including Lmo1, Ebf3 and Shox2 (Fulp et al., 2008).
As another example of the use of mutant mice, Prampano et al., compared gene expression in several different mutations that are associated with deficits in neuronal migrations (Lis1, Dcx and Ywhae) and found alterations in classes of genes expressed (Pramparo et al., 2011). Specifically, disruption in neuronal migration genes caused alterations in cell cycle and cytoskeleton categories, but for some mutants there were also differences in genes encoding synaptic proteins. These observations suggest that there is a dependency of later gene expression events on earlier ones, i.e., that neuronal migration is a required step for synaptic maturation.
The above datasets were generated using microarrays, but there are many other ways to look at gene expression. In situ hybridization (ISH) has been applied at a genomewide level to both mouse(Liscovitch & Chechik, 2013) and human(Miller et al., 2014) brain development. The principle advantage of ISH over arrays is that gene expression can be addressed at the level of brain regions, layers and even single cells. Such analyses have generally found similar categories of genes as in the array studies but indicate that there are specific genes that are developmentally regulated in germinal zones, for example(Miller et al., 2014).
A deeper view of gene expression changes in the developing mouse cerebral cortex has been achieved using transcriptome sequencing or RNA-Seq (Dillman et al., 2013; Han et al., 2009)(Dillman et al., 2013). Such techniques have been increasingly chosen for gene expression studies as it has been reported that RNA-Seq has greater linearity and reliability compared to microarrays (’t Hoen et al., 2008). Consistent with previous array studies, genes with higher expression in the embryonic brain included many genes involved in cell division, while those that were more highly expressed in the adult brain were related to neurotransmission and ion homeostasis (Dillman et al., 2013). However, probably due to the improved dynamic range of RNA-Seq, such approaches tend to nominate many more genes as being regulated during development compared to array-based studies. In our hands, from ~24,000 genes identified, about 4,000 were differentially expressed comparing E17 mice with adult (3–4 month old) females. There was also good quantitative agreement between RNA-Seq and qRT-PCR for a subset of genes chosen for validation, again supporting the idea that RNA-Seq reliably estimates fold differences between conditions. A similar estimate (4000/16000 detected genes) of differentially expressed genes was reported by Han et al., comparing E18 and P7 mouse cortex (Han et al., 2009). RNA-Seq data is also available for the human brain at various stages of development (e.g., http://www.brainspan.org.) Analysis of this dataset again shows a distinct developmental trajectory for expression of a large number of genes including many involved in synaptic function (Parikshak et al., 2013).
An important additional utility of RNA-Seq is that as well as estimating overall gene expression, we examine more complex aspects of gene expression, which will be discussed later in the review, namely splicing and RNA editing. However, it is perhaps interesting to discuss some genetic events that may underly some of the changes in gene expression in brain development.
DNA Sequence variation and epigenetic modification in brain development
A discussion of gene expression at the RNA level should also consider the architecture of DNA itself. Within a given species, DNA is highly polymorphic and some of that variation can manifest itself as differences in expression levels rather than coding sequence of genes. Mapping the relationship between DNA variation and gene expression levels identifies expression quantitative trait loci (eQTLs). Conceptually, eQTLs are regions of the genome where polymorphic variants are statistically associated with differences in mRNA expression levels. Studies in human (Gibbs et al., 2010) and mouse (van Nas et al., 2010) brain have identified a large number of such eQTLs for many genes. Such polymorphic loci might, therefore, influence brain development. In one study overlaying genetic data with gene expression across brain development, there were very few examples of genetic polymorphisms that altered the trajectory of gene expression changes throughout development (Colantuoni et al., 2011). Nonetheless, genetic background effects do need to be considered in gene expression profiling experiments, including brain development.
As discussed above, alterations in expression levels of transcription factors may be important in the control of gene expression during brain development. However, the underlying interaction between transcription factors and DNA is dynamic as DNA is subject to a number of regulatory modifications, including methylation. DNA methylation generally occurs at cytosine bases to form 5-methylcytosine in the promoter region of genes. Cytosine methylation generally represses gene expression (Tate & Bird, 1993), although it may also be a mechanism involved in generation of alternate splicing events (Zhou, Luo, Wise, & Lou, 2014).
Methylation seems to play a particularly important role in cell differentiation in the brain. For instance, astrocyte differentiation is dependent on the transcription factor STAT3 but expression levels alone are not enough to trigger differentiation as the promoter of the astrocyte marker GFAP is methylated to prevent STAT3 binding. Once this site is demethylated the cells can respond to the presence of STAT3 and differentiation can occur (Takizawa et al., 2001). In this way, transcription factors and DNA modifications work together to control gene expression.
DNA methylation can also be involved in genomic imprinting, in which a gene is expressed in a parent-of-origin-specific manner. A substantial proportion of imprinted genes are highly expressed in the brain with unique spatial and temporal expression. For example, UBE3A has maternally based expression in specific subpopulations of neurons in the hippocampus and cerebellum but is biallelically expressed in the rest of the brain and body (Albrecht et al., 1997; Rougeulle, Glatt, & Lalande, 1997). In chimeric mice embryos, duplicated maternal genomes contributed to the development of the hypothalamsic but not to the cerebral cortex, while a duplicated paternal genome contributed to cortical but not to hypothalamic structures indicating unique differential roles for parent of origin genomes (Keverne, Fundele, Narasimha, Barton, & Surani, 1996). An example of temporal regulation is the gene Murr1, which has biallelic expression in embryonic and neonatal mice but only the maternal allele is expressed in adult brain (Wang et al., 2004).
Collectively, these examples show that the transcriptome of the brain is regulated at multiple levels in a manner that depends on epigenetic modification. As might therefore be expected, these single examples likely generalize across the genome. It has been demonstrated recently that widespread DNA methylation changes occur in development in both the mouse and human brain (Lister et al., 2013; Numata et al., 2012). As seen with expression changes, the most dramatic differences in DNA methylation occur during prenatal development with a slowing of progression after birth and even more modest changes in aging (Numata et al., 2012). In some cases, there are DNA methylation events that reverse course after initial development, ie where a sequence may undergo demethylation before birth then becoming methylated after birth. This is generally consistent with previous data using smaller sets of methylation events that showed a general increase in methylation in the human brain with age that was also confirmed using isolated neurons (Siegmund et al., 2007).
Alternative splicing
Many of the above approaches, generally considered each ‘gene’ as a single unit. However, many tissues, including the brain, show a large number of splicing events with perhaps half of all genes showing some evidence of alternate exons being incorporated into mature mRNA (Lee & Irizarry, 2003).
As might therefore be expected, there are many examples of regulated alternative splicing in neuronal development. In mice, fetal Mapt has only minor incorporation exon 10 but by postnatal day 24 all tau contains this exon (McMillan et al., 2008). Interestingly, human MAPT retains exon 10 throughout adulthood (Liu & Gong, 2008), perhaps related to the larger size of human neurons compared to neurons leading to a higher requirement for axonal stability. The glutamate receptor gene Gria2 has a pair of exons that can be spliced in or out leading to two different protein isoforms, flip and flop, that have different electrophysiological characteristics (Sommer et al., 1990). In rats, flip is expressed at stable levels throughout development while flop expression is low until postnatal day 8 (Monyer, Seeburg, & Wisden, 1991). Although not comprehensive, these examples show how alternate splicing in brain-expressed genes can be functionally important in different species.
Another level of regulation related to splicing is intron retention, where sequences that would normally be spliced out are included in the mature mRNA. In general, retention of introns is high in the brain than other tissues and is developmentally regulated, with levels of retention higher in the fetal brain than in the adult (Ameur et al., 2011). One example of intron retention during development is in the axon guidance molecule Robo3 (Colak, Ji, Porse, & Jaffrey, 2013). A Robo3 isoform containing an intronic sequence (Robo3.2) is expressed but translationally repressed and allows for neuronal attraction to the spinal cord midline. Once the axon crosses the spinal cord midline, it receives signals from the floor plate to translate Robo3.2 allowing nonsense-mediated decay to occur, causing repellence to the midline. Whether other examples of intron retention are similarly functionally important in brain development is not known, but given that intron retention is frequent in the embryonic brain(Ameur et al., 2011), it is likely that this is an important mechanism of gene regulation relevant for neuronal maturation.
The brain also has been found to have longer 3-′UTR regions (Miura, Shenker, Andreu-Agullo, Westholm, & Lai, 2013; Ramsköld, Wang, Burge, & Sandberg, 2009) than other tissues and this lengthening of UTRs occurs during development(Ji, Lee, Pan, Jiang, & Tian, 2009). This may be related to stability of mRNA transcripts, as 3′-UTR regions contain poly-adenylation signals that control the turnover of mRNA; interestingly, the brain has more alternate poly-adenylation than other tissues (Hu, Liu, & Yan, 2014). Alternate 3′-UTR signals may also be important in targeting mRNA to neuronal processes as there are signals that direct mRNAs to axons and dendrites (Mohr, 1999).
There have been several studies looking at alternative exon usage in brain development in different species including humans and other primates using genome-wide exon arrays (Johnson et al., 2009; Mazin et al., 2013). What is impressive about these studies is that, like measures of overall gene expression, they estimate that a large proportion of genes show alternative splicing. In our own work using RNA-Seq in the mouse brain, we found almost 400 exons that were differentially expressed with examples where exon inclusion were higher or lower in the adult compared to embryonic brain and these included well characterized examples such as Mapt. We also found that there were many types of alternative exon usage, including 5′ and 3′-UTR sequences (Dillman et al., 2013), showing that some of the specific examples discussed above may generalize to many genes.
An obvious mechanism for alterations in splicing during development is that splicing factors might themselves be differentially expressed. There is some support for this from large-scale experiments, which have found age-dependent changes in expression of PTBP1, PTBP2, hnRNPA1, hnRNPF, hnRNPH1, and hnRNPH3 in the developing human cerebral cortex (Mazin et al., 2013). Differential expression of RNA binding proteins also occurs in mouse development. One of the genes with the largest differences in gene expression in our own dataset (Dillman et al., 2013) was Igf2bp1, which is associated with translational repression of a subset of mRNA (Bell et al., 2013). Nova2, a neuron specific RNA binding protein, is required for the development of the spinal cord and brain stem. Using high throughput sequencing of RNA isolated by crosslinking, it was discovered that the binding of Nova2 affects alternative splicing (Licatalosi et al., 2008). These observations demonstrate that alternative splicing is therefore required for normal brain development.
It is likely that there are additional levels of complexity in transcript generation that would also be relevant to brain development. One of the limitations of that RNA-Seq we used is that sequences were limited to ~200bp, although there are technologies that allow for longer reads and hence to recover a greater depth of information about full length transcripts (Au et al., 2013). Applying a similar approach to the developing brain would be of particular interest in the future.
RNA Editing
An additional source of transcriptome diversity is generated at the single base level via RNA editing. Although there were some early claims of a huge diversity of RNA editing events in the mammalian genome(Li et al., 2011) many of the observed events were shown to be sequencing errors and other technical artifacts(Pickrell, Gilad, & Pritchard, 2012). Instead, it is generally accepted that in many species, RNA editing events are limited to Adenosine to Inosine and Cytosine to Uracil, both of which have a well defined enzymatic basis.
Adenosine to inosine substitutions in mammalian RNA are carried out by adenosine deaminases (ADARs), of which there are three isoforms. ADAR1 and ADAR2 are ubiquitously expressed, with expression levels are highest in the brain while ADAR3 is exclusively expressed in the brain(Hogg, Paro, Keegan, & O’Connell, 2011). ADARs act on double stranded RNA and may require dimerization to be enzymatically active(Cho et al., 2003; Gallo, Keegan, Ring, & O’Connell, 2003). ADARs are localized primarily in the nucleolus and are bound to ribosomal RNA(Sansam, Wells, & Emeson, 2003) but can translocate to the nucleus upon expression of specific ADAR substrates(Desterro et al., 2003).
Inosine is recognized as guanosine in translation and, as such, editing in the coding region of a gene can result in a change in the amino acid sequence(Sommer, Köhler, Sprengel, & Seeburg, 1991). Editing may be particularly important in the brain as there are multiple isoforms of neurotransmitter receptors that are targeted by ADARs(Seeburg, 2000). The majority of A-to-I editing sites are highly conserved across genetically divergent mouse strains(Danecek et al., 2012), supporting the idea that editing is biologically important. In mice, editing of Gria2 leads to a lower permeability of this glutamate channel to calcium ions. If only the unedited isoform of Gria2 is present, mice die within two weeks of birth due to seizures(Higuchi et al., 2000). In octopuses, RNA editing plays a role in the temperature adaptability of potassium channels(Garrett & Rosenthal, 2012), again showing that RNA editing may influence neuronal excitability. There are examples of genes that are both edited and that undergo alternative splicing during development (Barresi et al., 2014).
The other major editing enzyme in mammals, APOBEC1, deaminates cytidine to produce uracil (Koito & Ikeda, 2012). Genome-wide surveys suggest that C to U editing is far less numerous than A to I editing (Kleinman, Adoue, & Majewski, 2012). APOBEC1 is expressed in neurons and is thought primarily to have an antiviral role (Gee et al., 2011) and, perhaps because of this, at the time of this review any potential role in development has not been well studied.
There have been attempts to look at editing in a genome-wide manner in brain development. For example, Wahlstedt et al. described examined 28 known A to I editing sites and found that many showed an increase in editing as the brain develops (Wahlstedt, Daniel, Ensterö, & Ohman, 2009) In our own analysis of A-to-I editing, we discovered 176 sites in the mouse brain. Although some coding edits were found, the majority of sites were sin the 3′UTR of genes. We also confirmed that there was a tendency for increase in the proportion of edited transcripts with development (Dillman et al., 2013). The tendency of RNA to become more completely edited during development suggests that protein diversity is less tolerated in the mature CNS than during development.
The mechanism underlying increased completion of A-to-I editing may be partly related to increased expression of Adar enzymes through the postnatal period (Dillman et al., 2013). However, why there is variation in the level of editing between different sites, varying from less than 20% to nearly 100% edited as for Gria2, is not at all clear. One possible future experiment would be to examine RNA editing in mice lacking specific Adar isoenzymes, which might allow for estimation of the redundancy in editing between the different Adar genes.
Non-coding RNA
In recent years it has become clear that the genome contains many types of RNA distinct from protein coding mRNA species. Some small non-coding RNA (ncRNA), including micro-RNA (miRNA), have important roles in regulating stability and translation of mRNAs (Dogini et al., 2014). Others, such as long non-coding RNAs (lncRNA) influence epigenetic regulation by structural mechanisms (Peschansky & Wahlestedt, 2014) and are highly conserved across species (Chodroff et al., 2010). Human accelerated regions (HARs) are noncoding regions of the genome that are conserved throughout vertebrate evolution but have significant substitution rates in humans. There is significant enrichment adjacent to genes known to play a role in neuronal development(17082449, 16915236). One particular HAR, HAR1, had the most genomic changes in humans with 18 substitutions compared to chimpanzees, while there were only 2 base differences comparing chimpanzees to chickens. HAR1 overlaps with two ncRNAs HAR1F and HAR1R. Interestingly HAR1F is specifically expressed in the fetal brain in Cajal-Retzius neurons along with reelin a gene critical in the specification of layering in the cortex (16915236). As might be expected, there are many single examples of changes in expression of non-coding RNAs as the brain develops (Barry, 2014; Iyengar et al., 2014; Nowak & Michlewski, 2013), including instances where a neuronal specific function is impacted by miRNA expression levels during development (Schratt et al., 2006).
Several studies have attempted to use genome-wide approaches to look at miRNA in brain development. In the developing rat forebrain from E2 to P5, about 20% of mature miRNA species were shown to have altered expression patterns using a custom array (Krichevsky, King, Donahue, Khrapko, & Kosik, 2003). All of the proposed differences were validated by Northern blots, suggesting that such changes are methodologically robust. Similarly, in studies using several different microarray platforms, a large proportion of miRNA were found to show changes in expression during development of the mouse brain (Miska et al., 2004; Sempere et al., 2004), in the pig cortex and cerebellum (Podolska et al., 2011) and in many regions of the human brain (Moreau, Bruse, Jornsten, Liu, & Brzustowicz, 2013). ncRNA can also be quantified by RNA-Seq, usually by making libraries that are enriched for small RNA species. As for conventional mRNA, sequencing tends to identify a greater number of genes than arrays in brain (Juhila et al., 2011). These types of methods have been applied to the developing pig hypothalamus and pituitary and again reported a large number of differences (Zhang et al., 2013). Collectively, these results show that many miRNA are regulated during development.
Alterations in the expression levels of miRNA are an additional mechanism that might contribute to some of the changes in mRNA expression and splicing discussed above. Because miRNA generally bind multiple mature mRNA species it has been predicted that they might be important for co-ordinated control of gene expression in multiple species (Favre, Banta Lavenex, & Lavenex, 2012). It has been suggested that miRNA:mRNA interactions are particularly important in allowing for maintaining the overall stability in gene expression levels while still allowing for fine-tuning in response to developmental stimuli (Follert, Cremer, & Béclin, 2014). The lncRNA Evf2 binds to intergenic regions and influences expression of proximal genes Dlx5/6 and Gad1 (Bond et al., 2009). There is some evidence that a relationship between expression of lncRNAs and nearby protein coding genes generalizes across many examples (Mercer, Dinger, Sunkin, Mehler, & Mattick, 2008). Small RNAs may also contribute to the generation of transcript diversity. For example, a miRNA expressed selectively in the nervous system can influence splicing via the factors PTBP1 and PTBP2 (Makeyev, Zhang, Carrasco, & Maniatis, 2007).
Summary
The examples above show that the brain transcriptome undergoes a number of significant changes throughout development. Importantly, there are many levels of regulation including at the levels of whole gene, single exons and single base pairs in the case of RNA editing sites. The mechanism(s) underlying all of these changes are not always understood, but many are likely to be important in the functional specification of the brain. Future challenges include developing additional ways to look at the whole transcriptome in an unbiased manner.
References
- ’t Hoen PAC, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, de Menezes RX, Boer JM, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic acids research. 2008;36(21):e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nature genetics. 1997;17(1):75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nature structural & molecular biology. 2011;18(12):1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(50):E4821–4830. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi S, Tomaselli S, Athanasiadis A, Galeano F, Locatelli F, Bertini E, Zanni G, et al. Oligophrenin-1 (OPHN1), a gene involved in X-linked intellectual disability, undergoes RNA editing and alternative splicing during human brain development. PloS one. 2014;9(3):e91351. doi: 10.1371/journal.pone.0091351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Molecular psychiatry. 2014;19(4):410–416. doi: 10.1038/mp.2013.196. [DOI] [PubMed] [Google Scholar]
- Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cellular and molecular life sciences: CMLS. 2013;70(15):2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nature neuroscience. 2009;12(8):1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Calegari F. Mechanisms of brain evolution: Regulation of neural progenitor cell diversity and cell cycle length. Neuroscience research. 2014 doi: 10.1016/j.neures.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Cho DSC, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. The Journal of biological chemistry. 2003;278(19):17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnár Z, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome biology. 2010;11(7):R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153(6):1252–1265. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Nellåker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, Flint J, et al. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome biology. 2012;13(4):26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JMP, Keegan LP, Lafarga M, Berciano MT, O’Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. Journal of cell science. 2003;116(Pt 9):1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Díaz E, Ge Y, Yang YH, Loh KC, Serafini TA, Okazaki Y, Hayashizaki Y, et al. Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron. 2002;36(3):417–434. doi: 10.1016/s0896-6273(02)01016-4. [DOI] [PubMed] [Google Scholar]
- Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, et al. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nature neuroscience. 2013;16(4):499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogini DB, Pascoal VDB, Avansini SH, Vieira AS, Pereira TC, Lopes-Cendes I. The new world of RNAs. Genetics and molecular biology. 2014;37(1 Suppl):285–293. doi: 10.1590/s1415-47572014000200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre G, Banta Lavenex P, Lavenex P. miRNA regulation of gene expression: a predictive bioinformatics analysis in the postnatally developing monkey hippocampus. PloS one. 2012;7(8):e43435. doi: 10.1371/journal.pone.0043435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follert P, Cremer H, Béclin C. MicroRNAs in brain development and function: a matter of flexibility and stability. Frontiers in molecular neuroscience. 2014;7:5. doi: 10.3389/fnmol.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA. Identification of Arx transcriptional targets in the developing basal forebrain. Human molecular genetics. 2008;17(23):3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Keegan LP, Ring GM, O’Connell MA. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. The EMBO journal. 2003;22(13):3421–3430. doi: 10.1093/emboj/cdg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Rosenthal JJC. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science (New York, NY) 2012;335(6070):848–851. doi: 10.1126/science.1212795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee P, Ando Y, Kitayama H, Yamamoto SP, Kanemura Y, Ebina H, Kawaguchi Y, et al. APOBEC1-mediated editing and attenuation of herpes simplex virus 1 DNA indicate that neurons have an antiviral role during herpes simplex encephalitis. Journal of virology. 2011;85(19):9726–9736. doi: 10.1128/JVI.05288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS genetics. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wu X, Chung WY, Li T, Nekrutenko A, Altman NS, Chen G, et al. Transcriptome of embryonic and neonatal mouse cortex by high-throughput RNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(31):12741–12746. doi: 10.1073/pnas.0902417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406(6791):78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hogg M, Paro S, Keegan LP, O’Connell MA. RNA editing by mammalian ADARs. Advances in genetics. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- Hu W, Liu Y, Yan J. Microarray meta-analysis of RNA-binding protein functions in alternative polyadenylation. PloS one. 2014;9(3):e90774. doi: 10.1371/journal.pone.0090774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti GM, Price DJ. Exuberance in the development of cortical networks. Nature reviews. Neuroscience. 2005;6(12):955–965. doi: 10.1038/nrn1790. [DOI] [PubMed] [Google Scholar]
- Iyengar BR, Choudhary A, Sarangdhar MA, Venkatesh KV, Gadgil CJ, Pillai B. Non-coding RNA interact to regulate neuronal development and function. Frontiers in cellular neuroscience. 2014;8:47. doi: 10.3389/fncel.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, Geschwind DH, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62(4):494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhila J, Sipilä T, Icay K, Nicorici D, Ellonen P, Kallio A, Korpelainen E, et al. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PloS one. 2011;6(6):e21495. doi: 10.1371/journal.pone.0021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami Y, Furuichi T. Investigation of differentially expressed genes during the development of mouse cerebellum. Brain research. Gene expression patterns. 2001;1(1):39–59. doi: 10.1016/s1567-133x(01)00007-2. [DOI] [PubMed] [Google Scholar]
- Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain research. Developmental brain research. 1996;92(1):91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- Kleinman CL, Adoue V, Majewski J. RNA editing of protein sequences: a rare event in human transcriptomes. RNA (New York, NY) 2012;18(9):1586–1596. doi: 10.1261/rna.033233.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A, Ikeda T. Apolipoprotein B mRNA-editing, catalytic polypeptide cytidine deaminases and retroviral restriction. Wiley interdisciplinary reviews. RNA. 2012;3(4):529–541. doi: 10.1002/wrna.1117. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA (New York, NY) 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, Irizarry K. Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biological psychiatry. 2003;54(8):771–776. doi: 10.1016/s0006-3223(03)00375-5. [DOI] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science (New York, NY) 2011;333(6038):53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscovitch N, Chechik G. Specialization of gene expression during mouse brain development. PLoS computational biology. 2013;9(9):e1003185. doi: 10.1371/journal.pcbi.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, et al. Global epigenomic reconfiguration during mammalian brain development. Science (New York, NY) 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Molecular neurodegeneration. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Molecular cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Hori G, Furuichi T. Gene expression profiling during the embryonic development of mouse brain using an oligonucleotide-based microarray system. Brain research. Molecular brain research. 2005;136(1–2):231–254. doi: 10.1016/j.molbrainres.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, et al. Widespread splicing changes in human brain development and aging. Molecular systems biology. 2013;9:633. doi: 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan P, Korvatska E, Poorkaj P, Evstafjeva Z, Robinson L, Greenup L, Leverenz J, et al. Tau isoform regulation is region- and cell-specific in mouse brain. The Journal of comparative neurology. 2008;511(6):788–803. doi: 10.1002/cne.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome biology. 2004;5(9):R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome research. 2013;23(5):812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody M, Cao Y, Cui Z, Tay KY, Shyong A, Shimizu E, Pham K, et al. Genome-wide gene expression profiles of the developing mouse hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(15):8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr E. Subcellular RNA compartmentalization. Progress in neurobiology. 1999;57(5):507–525. doi: 10.1016/s0301-0082(98)00066-5. [DOI] [PubMed] [Google Scholar]
- Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6(5):799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, Jornsten R, Liu Y, Brzustowicz LM. Chronological changes in microRNA expression in the developing human brain. PloS one. 2013;8(4):e60480. doi: 10.1371/journal.pone.0060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JS, Michlewski G. miRNAs in development and pathogenesis of the nervous system. Biochemical Society transactions. 2013;41(4):815–820. doi: 10.1042/BST20130044. [DOI] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics. 2012;90(2):260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics: official journal of the DNA Methylation Society. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science (New York, NY) 2012;335(6074):1302. doi: 10.1126/science.1210484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolska A, Kaczkowski B, Kamp Busk P, Søkilde R, Litman T, Fredholm M, Cirera S. MicroRNA expression profiling of the porcine developing brain. PloS one. 2011;6(1):e14494. doi: 10.1371/journal.pone.0014494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramparo T, Libiger O, Jain S, Li H, Youn YH, Hirotsune S, Schork NJ, et al. Global developmental gene expression and pathway analysis of normal brain development and mouse models of human neuronal migration defects. PLoS genetics. 2011;7(3):e1001331. doi: 10.1371/journal.pgen.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS computational biology. 2009;5(12):e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nature genetics. 1997;17(1):14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Seeburg PH. RNA helicase participates in the editing game. Neuron. 2000;25(2):261–263. doi: 10.1016/s0896-6273(00)80891-0. [DOI] [PubMed] [Google Scholar]
- Semeralul MO, Boutros PC, Likhodi O, Okey AB, Van Tol HHM, Wong AHC. Microarray analysis of the developing cortex. Journal of neurobiology. 2006;66(14):1646–1658. doi: 10.1002/neu.20302. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome biology. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PloS one. 2007;2(9):e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, et al. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science (New York, NY) 1990;249(4976):1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Sur M, Rubenstein JLR. Patterning and plasticity of the cerebral cortex. Science (New York, NY) 2005;310(5749):805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Developmental cell. 2001;1(6):749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Current opinion in genetics & development. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- Thompson JA, Ziman M. Pax genes during neural development and their potential role in neuroregeneration. Progress in neurobiology. 2011;95(3):334–351. doi: 10.1016/j.pneurobio.2011.08.012. [DOI] [PubMed] [Google Scholar]
- van Nas A, Ingram-Drake L, Sinsheimer JS, Wang SS, Schadt EE, Drake T, Lusis AJ. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics. 2010;185(3):1059–1068. doi: 10.1534/genetics.110.116087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Ensterö M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome research. 2009;19(6):978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, Nabetani A, Beechey CV, et al. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Molecular and cellular biology. 2004;24(1):270–279. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cai Z, Wei S, Zhou H, Zhou H, Jiang X, Xu N. MicroRNA expression profiling of the porcine developing hypothalamus and pituitary tissue. International journal of molecular sciences. 2013;14(10):20326–20339. doi: 10.3390/ijms141020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic acids research. 2014;42(2):701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]