Abstract
Homotropic cooperativity of 1-alkoxy-4-nitrobenzene substrates and also their heterotropic cooperative binding interactions with the iron ligand 1,4-phenylene diisocyanide (Ph(NC)2) had been demonstrated previously with rabbit cytochrome P450 (P450) 1A2 (Miller, G. P. and Guengerich, F. P. (2001) Biochemistry 40, 7262–7272). Multiphasic kinetics were observed for the binding of Ph(NC)2 to both ferric and ferrous P450 1A2, including relatively slow steps. Ph(NC)2 induced an apparently rapid change in the circular dichroism spectrum, consistent with a structural change, but had no effect on tryptophan fluorescence. Ph(NC)2 binds the P450 iron in both the ferric and ferrous forms; ferric P450 1A2 was reduced rapidly in the absence of added ligands, and the rate was attenuated when Ph(NC)2 was bound. No oxidation products of Ph(NC)2 were detected. Docking studies with a rabbit P450 1A2 homology model based on the published structure of a human P450 1A2•α-naphthoflavone (αNF) complex indicated adequate room for a complex with either two 1-isopropoxy-4-nitrobenzene molecules or a combination of one 1-isopropoxy-4-nitrobenzene and one Ph(NC)2; in the case of αNF no space for an extra ligand was available. The patterns of homotropic cooperativity seen with 1-alkoxy-4-nitrobenzenes (biphasic plots of v vs. S) differ from those seen with polycyclic hydrocarbons (positive cooperativity), suggesting that only with the latter does the ligand interaction produce improved catalysis. Consistent with this view, Ph(NC)2 inhibited the oxidation of 1-isopropoxy-4-nitrobenzene and other substrates.
Introduction
P4503 enzymes play important roles in the metabolism of drugs, steroids, eicosanoids, fat-soluble vitamins, and carcinogens in mammals [2]. In the metabolism of drugs, ~75% of the reactions are catalyzed by P450s [3,4].
The P450s involved in drug metabolism have been studied extensively. One of the phenomena that is still not understood well is ligand cooperativity. Heterotropic cooperativity [5,6], in which one ligand stimulates catalysis involving another ligand (substrate), was first observed >35 years ago [7–9]. Homotropic cooperativity, recognized more recently [10,11], refers to several types of non-hyperbolic plots of v vs. S for a single substrate. A number of possible explanations have been proposed to explain cooperativity, including “classical” allosteric ligand site binding, multiplicity of ligand binding within a large active site, multiplicity of protein conformers, and protein subunit interactions [11–21].
Much of the research on P450 cooperativity has been directed towards several P450s now known to have large substrate binding sites, i.e. P450s 3A4, 2C8, and 2C9 [22–25]. P450 1A2 has a smaller active site, as revealed in the recently published crystal structure of a human P450 1A2•αNF complex [26]. Cooperativity has been described for rabbit P450 1A2 [27,28] and human P450 1A2 [28]. Homotropic cooperativity was seen in the O-dealkylation of 1-alkoxy-4-nitrobenzenes, with v vs. S plots being biphasic and characterized with two sets of kcat and Km values, with Km,1 < Km,2 [27]. In that study, heterotropic cooperativity for ligand binding was observed in the interaction between 1- alkoxy-4-nitrobenzenes and an isocyanide, Ph(NC)2.
Isocyanides have a long history in the study of hemoproteins, going back to work in 1929 with hemoglobin and methyl isocyanide [29]. Isocyanides were also used extensively in some of the early research on P450 [30–32]; the divalent carbon atom is liganded directly to either ferric or ferrous iron in the P450 [33] and yields a “Type II” Soret difference spectrum distinctly different than that often generated by the addition of substrates (low- to high-spin iron transition, “Type I”). Titrations of rabbit P450 1A2 with 1-alkoxy-4-nitrobenzenes and Ph(NC)2 led to the conclusion that one ligand could enhance the binding of the other, indicative of multiple ligand occupancy as opposed to competition of the two ligands for a single site [27]. More recently, we have also observed homotropic cooperativity with P450 1A2 in pyrene 1-hydroxylation (and the subsequent hydroxylation(s) of 1-hydroxypyrene), with the v vs. S behavior fitting to highly sigmoidal plots with Hill coefficients (n) of 2 to 4 [28].
In the present work, several aspects of the interaction of Ph(NC)2 with rabbit P4540 1A2 were studied, including kinetics of binding, conformational changes of the enzyme (as detected by CD spectroscopy), and effects on rates of P450 reduction and catalysis of P450 1A2 reactions. Modeling of the ligands was done using the published crystal structure of the human P450 1A2-αNF complex [26], which is now available as a template. The cooperative interactions of the 1-alkoxy-4-nitrobenzenes and Ph(NC)2 are rather different that those recently described for P450 1A2 and some polycyclic hydrocarbons [28].
Materials and methods
Chemicals
Ph(NC)2 and αNF were purchased from Aldrich Chemical Co. (Milwaukee, WI). Ph(NC)2 was recrystallized from hexanes. (CH3)2CH-O-PhNO2 was synthesized as previously described [27] and recrystallized from a mixture of C2H5OH/H2O.
Enzymes
P450 1A2 was purified from liver microsomes of rabbits treated with β-naphthoflavone using slight modification [34] of a previously described method [35]. Recombinant rat NADPH-P450 reductase was expressed in Escherichia coli and purified as described elsewhere [36].
Assays of product formation
Incubations included P450 1A2 (0.25 µM), NADPH-P450 reductase (0.50 µM), and L-α-dilauroyl-sn-glycero-3-phosphocholine (23 µM) in 0.10 M potassium phosphate buffer (pH 7.4) at 37 °C, using an NADPH-generating system [37], with either αNF (14 or 35 µM), (CH3)2CH-O-PhNO2 (50 or 700 µM), or pyrene (35 or 140 µM). In the case of aniline, slight modifications in concentrations were made (3.3 µM P450 1A2, 6.6 µM NADPH-P450 reductase, 200 µM L-α-dilauroyl-sn-glycero-3-phosphocholine, and 8 mM aniline [30]). Incubations were done for 20 min in the case of the αNF, aniline, and (CH3)2CH-O-PhNO2 experiments. αNF 5,6-epoxidation was analyzed by HPLC as described elsewhere [28,38]. (CH3)2CH-O-PhNO2 O-dealkylation was monitored spectrophotometrically immediately after adding the NADPH-regenerating system (release of 4-nitrophenol, ΔA400), as described previously, using the extinction coefficient of 4-nitrophenol, ε400 = 12 mM−1 cm−1 [27]. Rates of aniline 4-hydroxylation were estimated using a colorimetric procedure [39], utilizing an extinction coefficient for 4- aminophenol (generated with a standard curve), ε630 = 10 mM−1 cm−1 [40]. Pyrene 1- hydroxylation was analyzed by HPLC/fluorescence by slight modification of a procedure described elsewhere [28,41].
Spectroscopic measurements
Steady-state UV-visible spectroscopy was done using OLIS-Cary 14 and OLIS-Aminco DW2 instruments (On-Line Instrument Systems, Bogart, GA). Steady-state fluorescence measurements were made using an OLIS DM-45 instrument. Steady-state CD measurements were made with a JASCO 810 instrument (JASCO, Easton, MD) in the Vanderbilt Structural Biology facility. Stopped-flow UV-visible and fluorescence measurements were made using an OLIS RSM-1000 system (On-Line Instrument Systems) operating in the rapid-scanning mode. Stopped-flow CD measurements were made with a modified Applied Photophysics SX-17MV instrument (Applied Photophysics, Leatherhead, UK), kindly made available by R. N. Armstrong (Vanderbilt Univ.).
Anaerobic studies were done using custom-made glass cuvettes and tonometers and a vacuum/Ar manifold as described previously [42–44], including use of a glucose oxidase-catalase-glucose enzymatic scrubbing system and loading into the RSM-1000 stopped-flow spectrophotometer [44].
Homology model building
A crystal structure of a human P450 1A2-αNF complex [26] was used as the template to develop a homology model of rabbit P450 1A2 utilizing the automated homology modeling tool SWISS-MODEL [45]. Coordinate files for molecules of (CH3)2CH-O-PhNO2 and Ph(NC)2 were generated using the Dundee PRODRG2 server [46] and manually docked into the binding cavity (replacing αNF) using Turbo Frodo.4 Simulated annealing was then performed with the program CNS Solve 1.1 using the input file model Anneal [47,48]. The heme group was fixed during simulated annealing (energy-minimized models were not adjusted following annealing).
Results
Kinetics of ligand binding to ferric P450 1A2
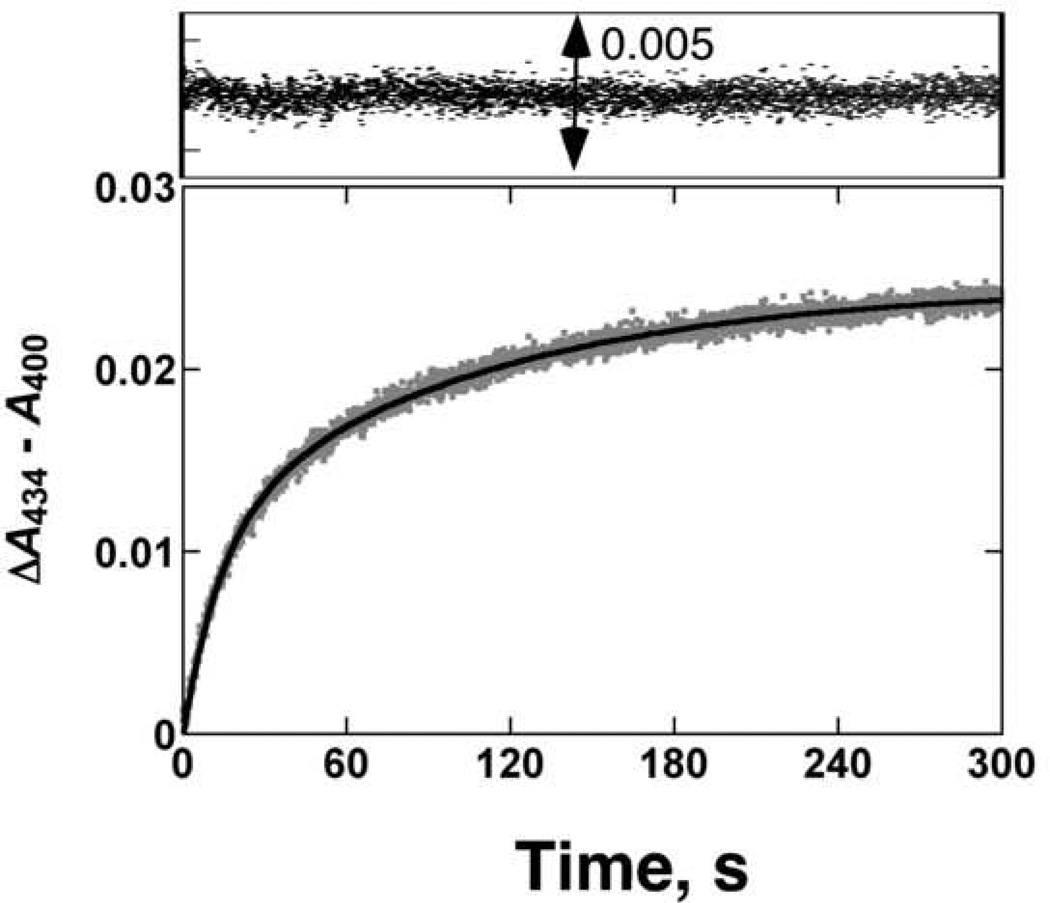
Binding of the Type II ligand Ph(NC)2 to ferric P450 1A2 was slow (Fig. 1), with estimated rates of 0.07 and 0.01 s−1 using a bi-exponential fit, at a final ligand concentration of 100 µM. Binding of the substrate (CH3)2CH-O-PhNO2 (data not presented) was also biphasic and relatively slow, with rates of 2.9 and 0.08 s−1 measured at a substrate concentration of 10 µM [28].
Fig. 1. Pre-steady-state kinetics of Ph(NC)2 binding to ferric P450 1A2.
The kinetic trace (ΔA434–A400) obtained for Ph(NC)2 (100 µM) binding to P450 1A2 (1 µM) was fit to a bi-exponential plot with rates of 0.07 and 0.01 s−1.
Kinetics of binding of Ph(NC)2 to ferrous P450 1A2
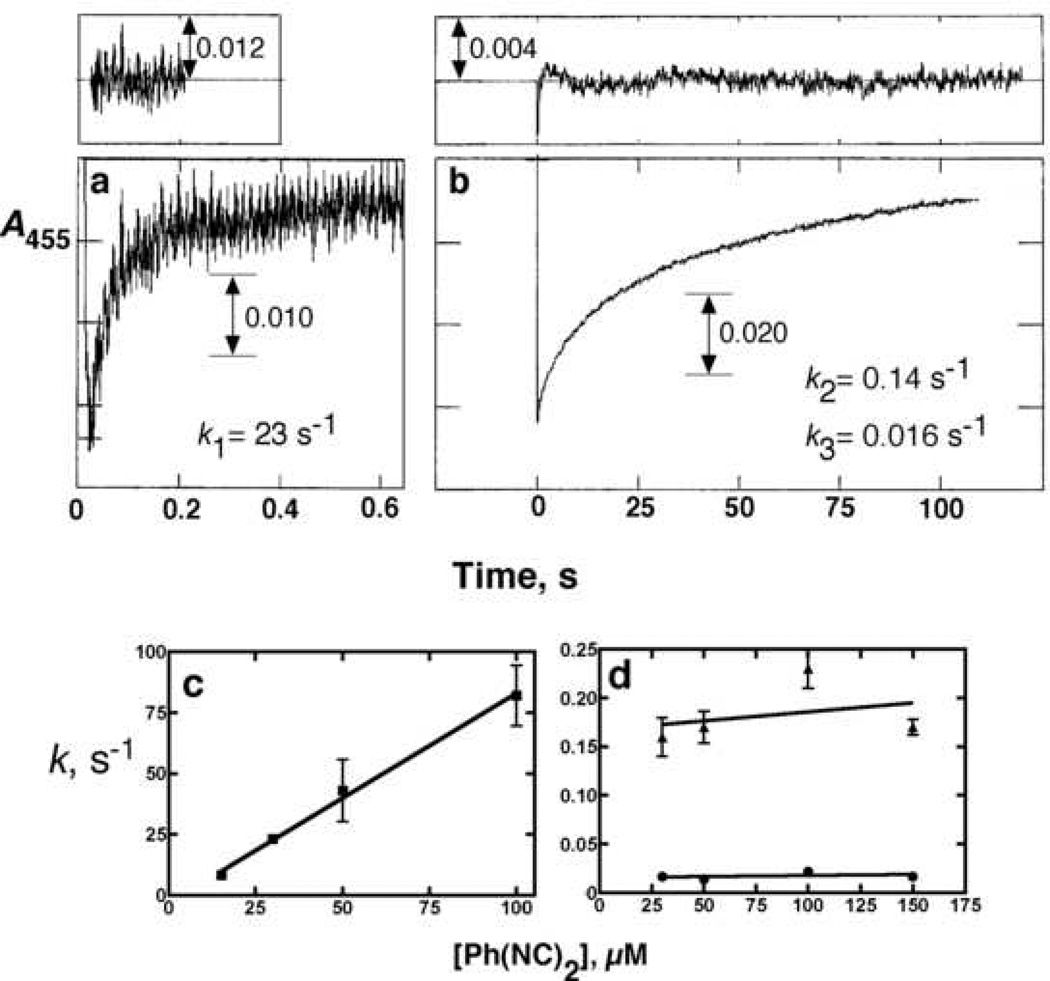
The ferrous P450-Ph(NC)2 complex had a strong absorption maximum at 455 nm, typical of isocyanide complexes [32,33], which can be used for kinetic analysis of binding. The binding of Ph(NC)2 to ferrous P450 was triphasic, with a rapid burst followed by a biphasic change that accounted for most of the absorbance increase (Figs. 2a, 2b). When the rates were measured as a function of Ph(NC)2 concentration, the fast phase yielded an apparent rate constant of 1.4 × 106 M−1 s−1 (Fig. 2c) but the rates of the latter two steps were rather invariant with the concentration (Fig. 2d).
Fig. 2. Pre-steady-state kinetics of ligand binding to ferrous P450 1A2.
P450 1A2 (1.0 µM final concentration following mixing) was reduced anaerobically in the presence of 0.25 µM NADPH-P450 reductase (and 75 µM L-α-dilauroyl-sn-glycero-3-phosphocholine and an NADPH-generating system [36]. (a) First portion of reaction with 30 µM (final concentration) of anaerobic Ph(NC)2. The initial part of the trace was fit to a single exponential of k1 = 23 ± 0.2 s−1. (b) Latter phases of the reaction were fit to a biexponential plot with k2 = 0.14 ± 0.01 s−1 and k3 = 0.016 ± 0.003 s−1. (c) Rates of k1 were determined at varying concentration of Ph(NC)2 and fit to a plot, with an intercept near 0 and a calculated second-order rate of 1.4 (± 0.1) × 106 M−1 s−1. (d) Values of k2 and k3 as a function of Ph(NC)2 concentration.
CD and fluorescence spectra
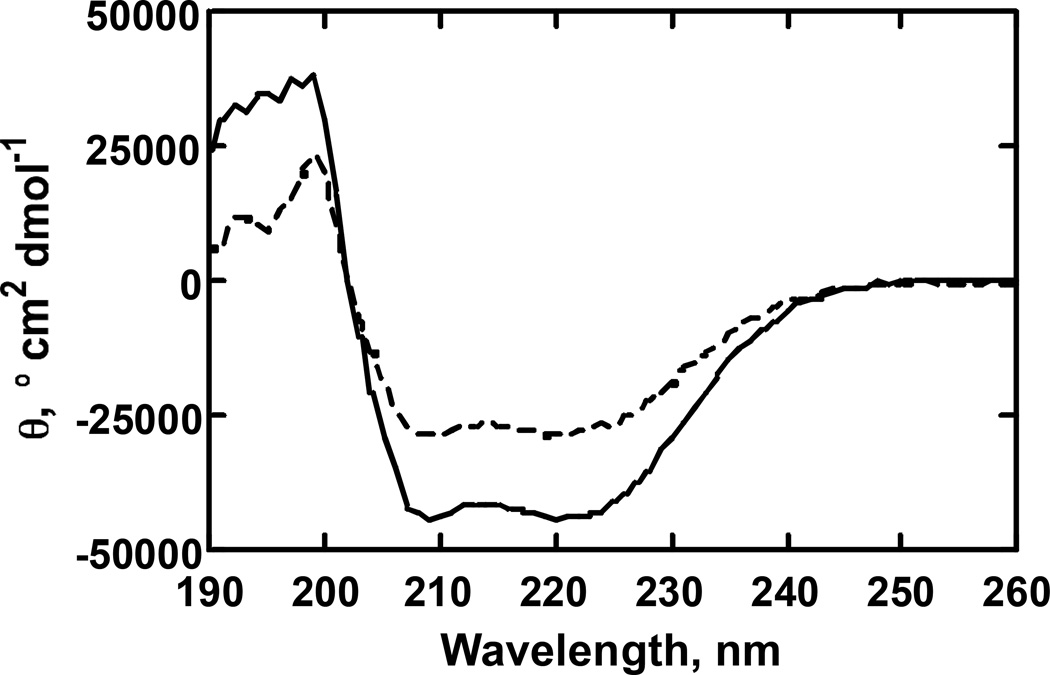
The binding of either (CH3)2CH-O-PhNO2 or Ph(NC)2 to P450 1A2 induced changes in the CD spectrum (Fig. 3). The far-UV changes are indicative of a change in the conformation of the protein and consistent with a decrease (based on [θ220) in α-helicity [49].
Fig. 3. CD spectra of Ph(NC)2 binding to rabbit P450 1A2.
The solid line (—) is a scan with 0.41 µM P450 1A2 alone and the dotted line (----) is the scan following the addition of Ph(NC)2 (40 µM), followed by a 15 min incubation. The cuvette pathlength was 1 mm and A220 was < 0.2 in both cases.
Changes in the Trp fluorescence of P450 1A2 have been observed upon binding of 1-alkoxy-4-acetanilides [50]. We also attempted to monitor the binding of Ph(NC)2 to P450 1A2 using a decrease (or increase) in P450 Trp fluorescence, but no changes were observed (for Trp or Tyr).
Effect of Ph(NC)2 on catalytic activity of P450 1A2
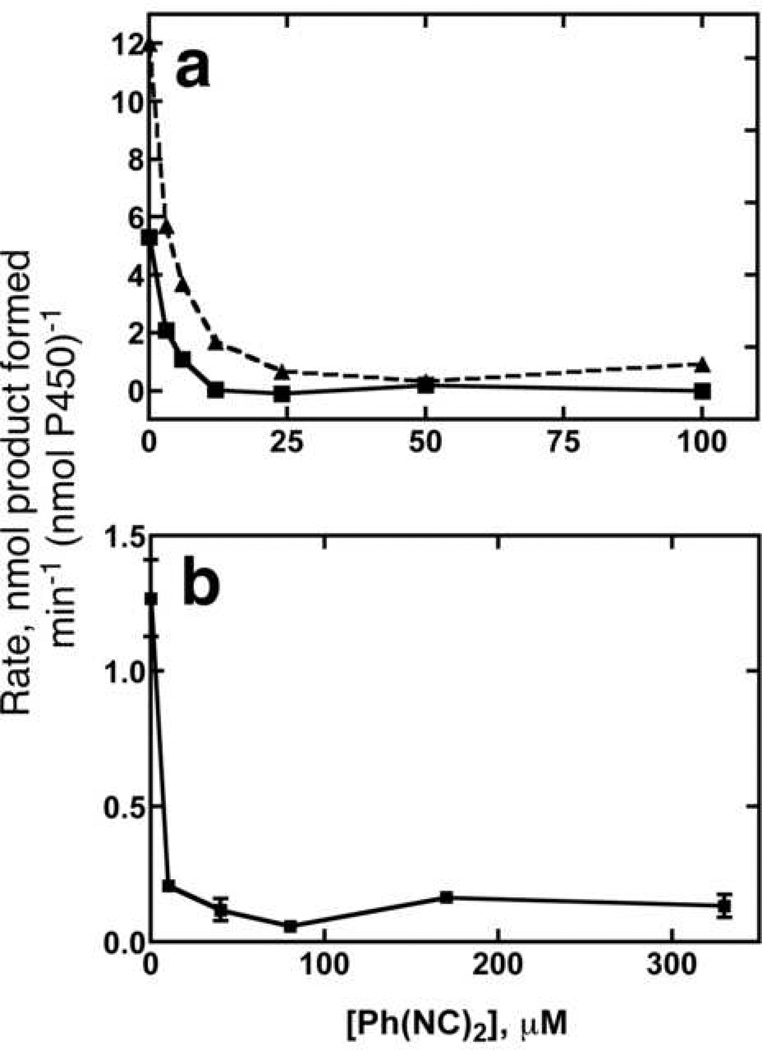
In the early literature on P450, diethylisocyanide was reported to stimulate the oxidation of aniline in rabbit liver microsomes [30,32]. The enhanced spectral changes in substrate binding with the presence of Ph(NC)2 [27] raised the possibility of stimulation of P450 1A2 activity. However, Ph(NC)2 inhibited (CH3)2CH-O-PhNO2 O-dealkylation and aniline 4-hydroxylation (Fig. 4), as well as αNF 5,6-epoxidation and pyrene 1- hydroxylation (results not shown).
Fig. 4. Inhibition of P450 1A2 catalytic activity.
(a) (CH3)2CH-O-PhNO2 O-dealkylation, as measured by absorbance at 400 nm. The data were fit to a straight line to determine the amount of 4-nitrophenol formed per min. The dotted line (----) shows the rate of O-dealkylation of 700 µM (CH3)2CH-O-P-hNO2 and the solid line (—) line shows the rates measured with 50 µM (CH3)2CH-O-PhNO2. (b) Aniline 4-hydroxylation. Formation of p-aminophenol (substrate concentration of 8 mM aniline) was monitored by measuring absorbance at 630 nm after derivatization.
Ph(NC)2 was not converted to any products detected using general HPLC-UV profiling by rabbit P450 1A2 (< 0.01 nmol of any product min−1 (nmol P450)−1, assuming a similar extinction coefficient).
Effect of Ph(NC)2 on rate of P450 1A2 reduction
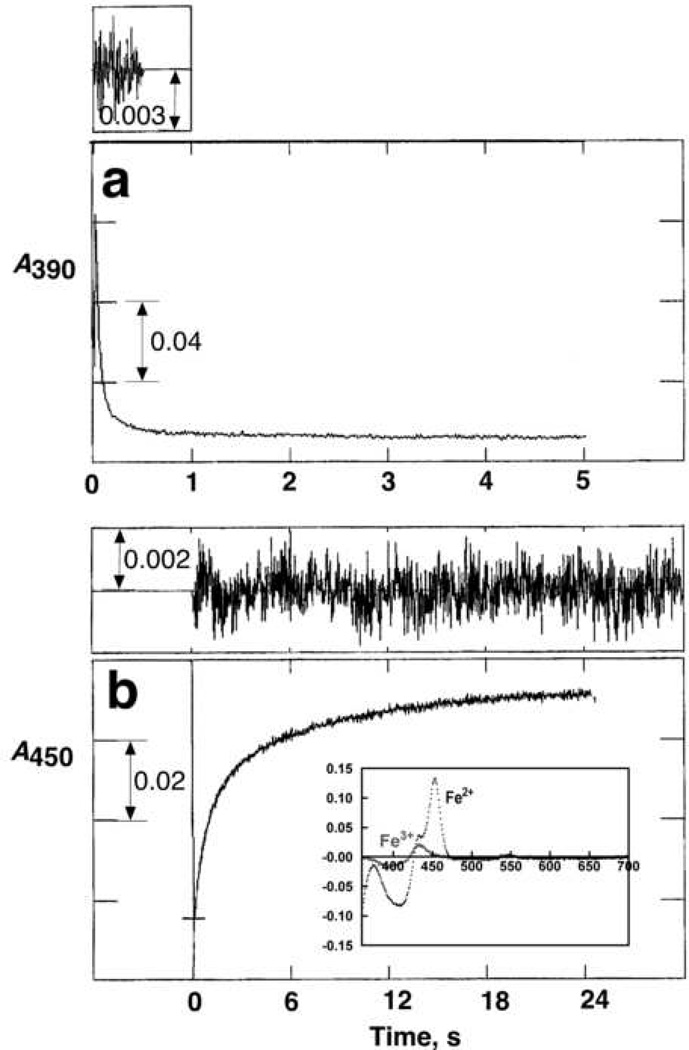
One of the interesting features of isocyanides is that, in contrast to other Type II P450 ligands, they bind to both ferric and ferrous P450 heme and the complexes can be discerned spectrally (Fig. 5). We were able to examine the effect of Ph(NC)2 on the rate of reduction of P450 1A2, which was a 10-fold attenuation (Fig. 5). However, even the attenuated rate (of heme reduction) is considerably faster than rates of substrate oxidation, and the inhibition of catalytic activity (Fig. 4) is not the result of only the decreased reduction rate. To our knowledge, a direct examination of the rates of electron transfer from NADPH-P450 reductase to a P450 with its heme iron liganded to a Type II ligand have not been determined previously, mainly because many ligands have similar spectra in the ferric and ferrous states (e.g., cyanide or azoles) and use of CO as a trap would require a dissociation step.
Fig. 5. Effect of Ph(NC)2 on rate of enzymatic reduction of P450 1A2.
All experiments included 2.0 µM P450 1A2, 4.0 µM NADPH-P450 reductase, and 75 µM L-α-dilauroylsn-glycero-3-phosphocholine, with the reaction initiated by the addition of 100 µM NADPH (23 °C, anaerobic conditions, argon atmosphere). (a) Reduction in absence of ligands, measured at 390 nm. The first part of the trace was fit to a single exponential with a rate of 25 (± 1) s−1. (b) Reduction in the presence of 100 µM Ph(NC)2, measured at 455 nm. The data were fit to a bi-exponential plot with k1 = 1.3 ± 0.02 s−1 and k2 = 0.14 ± 0.002 s−1. Residuals traces for the fits are shown in both parts a and b. The inset in Part b shows the difference spectra generated by the addition of 100 µM Ph(NC)2 to ferric (peak near 430 nm, trough at 395 nm) and ferrous (peak at 455 nm, trough at 405 nm) P450 1A2 (2.0 µM).
Fitting of ligands in a homology model of P450 1A2
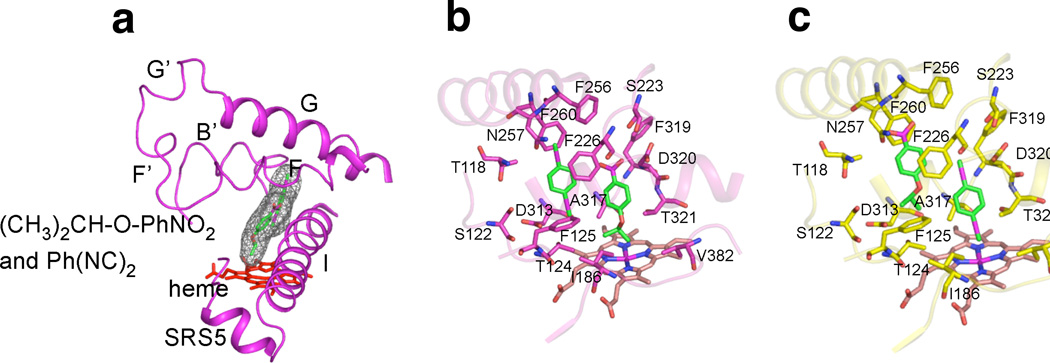
The possibility of simultaneous binding of (CH3)2CH-O-PhNO2 and Ph(NC)2 in the active site of rabbit P450 1A2, hypothesized earlier [27], was examined by docking the ligands into the active site of a homology model based on a human P450 1A2-αNF complex [26] (Fig. 6a). As with pyrene [28], both molecules can easily fit into the active site of the model, regardless of which ligand is bound closest to the heme group (Fig. 6b, 6c). Except for the disruption of the F-helix, which was observed in the human P450 1A2-αNF structure [26], none of the α-helices that comprise the active-site pocket are disrupted by the simultaneous presence of (CH3)2CH-O-PhNO2 and Ph(NC)2.
Fig. 6. Docking of ligands into homology model of rabbit P450 1A2.
(a) Rabbit P450 1A2 homology model (magenta ribbon) with heme group (red sticks) and (CH3)2CH-O-PhNO2 plus Ph(NC)2 (sticks with space-filling mesh). SRS5: Substrate recognition sequence 5 [51]. (b) Active-site of rabbit P450 1A2 homology model (magenta ribbon) with heme group (sticks) and (CH3)2CH-O-PhNO2 plus Ph(NC)2 (sticks), with (CH3)2CH-O-PhNO2 modeled close to heme Fe in the most productive mode and sidechains highlighted. (c) Active-site of rabbit P450 1A2 homology model (yellow ribbon) with heme group (sticks) and Ph(NC)2 plus (CH3)2CH-O-PhNO2 (sticks), with Ph(NC)2 liganded to heme Fe and side chains highlighted.
Discussion
The previous conclusion that both a 1-alkoxy-4-ntrobenzene and Ph(NC)2 could co-occupy the active site of P450 1A2 [27] is consistent with the protein model (Fig. 6). The model is considered reliable in that the rabbit P450 1A2 sequence has high identity (75%) with the human P450 1A2 sequence [52] and the two proteins have many similar functional characteristics [34]. The rabbit protein was used both in the previous [27] and the present studies because human P450 1A2 is isolated almost exclusively in the high-spin state [26,34] and the spectral studies with substrate binding cannot be done (at least using heme Soret perturbation approaches). The rabbit P450 1A2 used in this study was estimated to be 77% low-spin iron [28], as judged by second-derivative spectroscopy of the Soret band [53,54]. Although the protein used in these studies was isolated from rabbit liver microsomes, there was no evidence of another P450 or other impurity (i.e., sodium dodecyl sulfate polyacrylamide gel electrophoresis), and previous work with recombinant rabbit P450 1A2 showed the major cooperative features of the protein purified from liver [27].
The kinetics of binding of Ph(NC)2 were different for ferric and ferrous P450 (Figs. 1, 2). Binding to ferrous P450 1A2 showed a fast phase (Fig. 2a), followed by what appear to be two much slower phases (Fig. 2b). The rate of the fast phase increased with tc), while the two slower steps were rather invariant with the concentration (Fig. 2d). With ferric P450 (Fig. 1), only a slow biphasic change was seen, and the rates are similar to those of the second two steps with the reduced enzyme (Figs. 2b, 2d). The concentration dependence of the rates of the two steps in Fig. 1 was not examined. The similarity of the rates of the two slow steps in the ferrous and ferric reactions is not due to oxidation of the ferrous to the ferric state because the absorbance wavelength used (455 nm) is only indicative of binding to the ferrous iron (Fig. 5b). Our conclusion is that the fast rate constant with the ferrous enzyme (1.4 × 106 M−1 s−1) might represent the second-order “on” rate (although this is somewhat slow [55]) and the slower rates observed with both ferric and ferrous P450 represent conformational events in changing the protein to achieve the final spectrum. We emphasize that the multiphasic kinetics of ligand binding do not constitute any evidence for multiple ligand occupancy, in the absence of other data.
The homology model (Fig. 6) is considered reliable, at least for a ground state P450 1A2-substrate complex [26]. The active site (estimated size 375 Å3 [26]) is large enough to fit only one αNF molecule [26] or two pyrenes [28] but either two molecules of (CH3)2CH-O-PhNO2, two molecules of Ph(NC)2, or one of each ((CH3)2CH-O-PhNO2, Ph(NC)2) (Fig. 6). We propose that the co-occupancy by two molecules of (CH3)2CH-O-PhNO2 may be responsible for the two-kcat, two-Km behavior (“negative cooperativity”) observed for (CH3)2CH-O-PhNO2 O-dealkylation [27]. Fitting of one molecule each of (CH3)2CH-O-PhNO2 and Ph(NC)2 into the model of the active site (Fig. 6) is consistent with the previous conclusions based on comparisons of spectral interactions of the ligands [27]. This interaction leads to apparently enhanced binding of (CH3)2CH-O-PhNO2 in the presence of Ph(NC)2 (and vice-versa) [27] but results in inhibition of catalytic activity (Fig. 4), even when the substrate is (CH3)2CH-O-PhNO2 (Fig. 4a). We proposed that co-occupancy of the P450 1A2 active site is a common feature of numerous small substrates (and other ligands) but the nature of cooperative behavior is highly ligand dependent [27,28]. Apparently loading the active site with two molecules of (CH3)2CH-O-PhNO2 moves the substrate into a less favorable conformation for oxidation, explaining the attenuated catalytic efficiency at higher substrate concentrations [27]. The interaction of P450 1A2 with Ph(NC)2 increases the spectrally detectable binding of (CH3)2CH-O-PhNO2 to P450 1A2 [27] but apparently does so in a way that attenuates catalytic activity (Fig. 4). In our earlier work [27] with this isocyanide and substrate, the titration results provided evidence that each compound could enhance the binding of the other. We postulate that one factor in binding of Ph(NC)2 is the interaction of the phenyl ring somewhere in the active site. One option for binding is that shown in Fig. 6b, in which the substrate ((CH3)2CH-O-PhNO2) is bound near the iron atom, in juxtaposition for oxidation at the CH position. This configuration may be possible but apparently a mode such as that shown in Fig. 6c is more dominant, as reflected in the inhibition (Fig. 4). A major point of the present work is that one can get cooperativity in binding of ligands but that this does not necessarily translate to catalytic activity, e.g. see [56].
In summary, we have been able to extend earlier observations on the heterotropic cooperativity of rabbit P450 1A2 [27]. The modeling is clearly consistent with the previously presented proposal that one molecule each of Ph(NC)2 and (CH3)2CH-O-PhNO2 can co-occupy the active site. However, the heterotropic cooperativity yields enhancement of substrate binding but not an increase in catalytic activity. Apparently the juxtoposition of the 1-alkoxy-4-nitrobenzene imposed by the Ph(NC)2 ligand is unfavorable for catalysis in this case, although the nature of such an interaction was not revealed in the modeling.
Acknowledgments
This work was supported in part by U.S. Public Health Service (USPHS) Grants R37 CA090426 (F.P.G.), T32 ES007028 (C.D.S., F.P.G.), F32 CA119776 (R.L.E.), and P30 ES000267 (F.P.G.). We thank M. V. Martin for preparing some of the enzymes, W. M. Backes (Louisiana State University, New Orleans) for some of the rabbit liver samples used in enzyme purification, R. N. Armstrong for use of the stopped-flow spectrophotometer for CD studies, L. Mizoue for assistance with the JASCO CD spectrometer, and K. Trisler for assistance in preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: αNF, α-naphthoflavone (7,8-benzoflavone); (CH3)2CH-OPhNO2, (CH3)2CH-O-PhNO2; P450, cytochrome P450 (also termed “heme-thiolate protein P450 [1]; Ph(NC)2, 1,4-phenylene diisocyanide.
Cambillau, C., and Roussel, A. (1997) Turbo Frodo, Version OpenGL.1, Université Aix-Marseille II, Marseille, France.
References
- 1.Palmer G, Reedijk J. J. Biol. Chem. 1992;267:665–677. [Google Scholar]
- 2.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd ed. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 3.Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. Drug Metab. Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 4.Wienkers LC, Heath TG. Nat. Rev. Drug Discov. 2005;4:825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 5.Segel IH. Enzyme Kinetics. New York: Wiley; 1975. [Google Scholar]
- 6.Kuby SA. A Study of Enzymes, Vol. I, Enzyme Catalysis, Kinetics, and Substrate Binding. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- 7.Cinti DL. Pharmacol. Ther. 1978;2:727–749. [Google Scholar]
- 8.Kapitulnik J, Poppers PJ, Buening MK, Fortner JG, Conney AH. Clin. Pharmacol.Ther. 1977;22:475–485. doi: 10.1002/cpt1977224475. [DOI] [PubMed] [Google Scholar]
- 9.Lasker JM, Huang M-T, Conney AH. Science. 1982;216:1419–1421. doi: 10.1126/science.7089530. [DOI] [PubMed] [Google Scholar]
- 10.Guengerich FP, Kim B-R, Gillam EMJ, Shimada T. Proceedings, 8th Int. Conf. on Cytochrome P450: Biochemistry, Biophysics, and Molecular Biology. In: Lechner MC, editor. John Libbey Eurotext; Paris. 1994. pp. 97–101. [Google Scholar]
- 11.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 12.Guengerich FP. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd ed. Ortiz de Montellano PR, editor. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 377–530. [Google Scholar]
- 13.Atkins WM. Expert Opin. Drug Metab. Toxicol. 2006;2:573–579. doi: 10.1517/17425255.2.4.573. [DOI] [PubMed] [Google Scholar]
- 14.Tracy TS. Drugs Res. Devlop. 2006;7:349–363. doi: 10.2165/00126839-200607060-00004. [DOI] [PubMed] [Google Scholar]
- 15.Johnson EF, Schwab GE, Dieter HH. J. Biol. Chem. 1983;258:2785–2788. [PubMed] [Google Scholar]
- 16.Schwab GE, Raucy JL, Johnson EF. Mol. Pharmacol. 1988;33:493–499. [PubMed] [Google Scholar]
- 17.Harlow GR, Halpert JR. Proc. Natl. Acad. Sci. USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosea NA, Miller GP, Guengerich FP. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 19.Koley AP, Buters JTM, Robinson RC, Markowitz A, Friedman FK. J. Biol. Chem. 1997;272:3149–3152. doi: 10.1074/jbc.272.6.3149. [DOI] [PubMed] [Google Scholar]
- 20.Fernando H, Davydov DR, Chin CC, Halpert JR. Arch. Biochem. Biophys. 2007;460:129–140. doi: 10.1016/j.abb.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jushchyshyn MI, Hutzler JM, Schrag ML, Wienkers LC. Arch. Biochem. Biophys. 2005;438:21–28. doi: 10.1016/j.abb.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 22.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. J. Biol. Chem. 2004:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 23.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 24.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. J. Biol. Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 25.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. J. Biol. Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 26.Sansen S, Yano JK, Reynald RL, Schoch G, Griffin K, Stout CD, Johnson EF. J. Biol. Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 27.Miller GP, Guengerich FP. Biochemistry. 2001;40:7262–7272. doi: 10.1021/bi010402z. [DOI] [PubMed] [Google Scholar]
- 28.Sohl CD, Isin EM, Eoff RL, Marsch GA, Stec DF, Guengerich FP. J. Biol. Chem. 2008;283 doi: 10.1074/jbc.M709783200. in press (PMID: 18187423). [DOI] [PubMed] [Google Scholar]
- 29.Warburg O, Negelein E, Christian W. Biochem. Z. 1929;214:26–63. [Google Scholar]
- 30.Imai Y, Sato R. J. Biochem. (Tokyo) 1968;63:380–389. [PubMed] [Google Scholar]
- 31.Ichikawa Y, Yamano T. Biochim. Biophys. Acta. 1968;153:753–765. doi: 10.1016/0005-2728(68)90003-0. [DOI] [PubMed] [Google Scholar]
- 32.Imai Y, Horie S, Yamano T, Iizuka T. In: Cytochrome P-450. Sato R, Omura T, editors. New York: Academic Press; 1978. pp. 37–135. [Google Scholar]
- 33.Lee D-S, Park S-Y, Yamane K, Obayashi E, Hori H, Shiro Y. Biochemistry. 2001;40:2669–2677. doi: 10.1021/bi002225s. [DOI] [PubMed] [Google Scholar]
- 34.Sandhu P, Guo Z, Baba T, Martin MV, Tukey RH, Guengerich FP. Arch. Biochem. Biophys. 1994;309:168–177. doi: 10.1006/abbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- 35.Alterman MA, Dowgii AI. Biomed. Chromatog. 1990;4:221–222. doi: 10.1002/bmc.1130040510. [DOI] [PubMed] [Google Scholar]
- 36.Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. Arch. Biochem. Biophys. 1998;350:324–332. doi: 10.1006/abbi.1997.0534. [DOI] [PubMed] [Google Scholar]
- 37.Guengerich FP, Bartleson CJ. In: Principles and Methods of Toxicology. 5th ed. Hayes AW, editor. Boca Raton, FL: CRC Press; 2007. pp. 1981–2048. [Google Scholar]
- 38.Ueng Y-F, Kuwabara T, Chun Y-J, Guengerich FP. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 39.Imai Y, Ito A, Sato T. J. Biochem. (Tokyo) 1966;60:417–428. doi: 10.1093/oxfordjournals.jbchem.a128453. [DOI] [PubMed] [Google Scholar]
- 40.Shumyantseva VV, Bulko TV, Bachmann TT, Bilitewski U, Schmid RD, Archakov AI. Arch. Biochem. Biophys. 2000;377:43–48. doi: 10.1006/abbi.2000.1739. [DOI] [PubMed] [Google Scholar]
- 41.Chetiyanukornkul T, Toriba A, Kizu R, Makino T, Nakazawa H, Hayakaw K, Chromatog J. 2002;A 961:107–112. doi: 10.1016/s0021-9673(02)00363-1. [DOI] [PubMed] [Google Scholar]
- 42.Burleigh BD, Jr, Foust GP, Williams CH., Jr Anal. Biochem. 1969;27:536–544. doi: 10.1016/0003-2697(69)90067-0. [DOI] [PubMed] [Google Scholar]
- 43.Foust GP, Burleigh BD, Jr, Mayhew SG, Williams CH, Jr, Massey V. Anal. Biochem. 1969;27:530–535. doi: 10.1016/0003-2697(69)90066-9. [DOI] [PubMed] [Google Scholar]
- 44.Guengerich FP, Krauser JA, Johnson WW. Biochemistry. 2004;43:10775–10788. doi: 10.1021/bi0491393. [DOI] [PubMed] [Google Scholar]
- 45.Schwede T, Kopp J, Guex N, Peitsch MC. Nucl. Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuttelkopf AW, van Aalten DM. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Rice LM, Brunger AT. Proteins. 1994;19:277–290. doi: 10.1002/prot.340190403. [DOI] [PubMed] [Google Scholar]
- 49.Greenfield N, Fasman GD. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 50.Yun C-H, Miller GP, Guengerich FP. Biochemistry. 2001;40:4521–4530. doi: 10.1021/bi002906n. [DOI] [PubMed] [Google Scholar]
- 51.Gotoh O. J. Biol. Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 52.Quattrochi LC, Pendurthi UR, Okino ST, Potenza C, Tukey RH. Proc. Natl. Acad. Sci. USA. 1986;83:6731–6735. doi: 10.1073/pnas.83.18.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Haver TC, Green GL. Anal. Chem. 1976;48:312–318. [Google Scholar]
- 54.Guengerich FP. Biochemistry. 1983;22:2811–2820. doi: 10.1021/bi00281a007. [DOI] [PubMed] [Google Scholar]
- 55.Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999. pp. 158–161. [Google Scholar]
- 56.Lampe JN, Fernandez C, Nath A, Atkins WM. Biochemistry. 2008;47:509–516. doi: 10.1021/bi7013807. [DOI] [PubMed] [Google Scholar]