Abstract
The objective of this study was to identify lactic acid bacteria (LAB) isolated from kimchi and to evaluate its characteristics and functional properties for application in fermented dairy products as a probiotic or commercial starter culture. Eight stains isolated from kimchi were selected through an investigation of phenotypic characteristics. Two strains (DK211 and DK303) were identified as Lactobacillus plantarum, another two (DK207 and DK215) as Lactobacillus paracasei, and one (DK301) as Lactobacillus sakei. The remaining three strains were identified as species of Weissella. All selected Lactobacillus strains had acid and bile tolerance, even though there was wide variation in the ability of each strain. DK303 showed a remarkably higher proteolytic activity. There were no significant differences in β-galactosidase activity among the tested strains, except that DK301 showed no activity. Auto-aggregation varied between 82.1 and 90.0%, and hydrophobicity values ranged from 0.5 to 51.6%.The strongest auto-aggregation and hydrophobicity were observed in DK211. All selected strains showed better 1,1-diphenyl-2-picrylhydrzyl (DPPH) scavenging activity than commercial strains. DK211, DK215, DK301, and DK303 had effective inhibitory activity against all pathogens tested except E. coli. When selected strains were used for yogurt preparation as a single starter culture, the time required to reach target titratable acidity (0.9) was 11-12 h. The yogurt fermented with DK211 had favorable panelists ratings for most sensory attributes, which were comparable with yogurt fermented with a commercial strain. The results suggest that strains isolated from kimchi could be potential probiotic and starter cultures for use in yogurt manufacturing.
Keywords: Lactobacillus species, kimchi, acid and bile tolerance, β-galactosidase activity, yogurt
Introduction
There is an increase in consumers’ health consciousness and a growing demand for healthy foods, which leads a rapid growth of the food industries associated with probiotic bacteria and other functionalities in the marketplace (Lim and Im, 2009). Fermented foods such as yogurts, cheese, kefir, and kimchi have received attention for their health-promoting and disease preventing effects. As the fermentation process involves mixed cultures such as yeast, lactic acid bacteria, and fungi, traditional fermented foods are a plentiful source of microorganisms and some of them show probiotic characteristics (Rivera-Espinoza and Gallado-Navarro, 2010). Therefore, numerous studies have been undertaken to obtain scientific evidence of the beneficial effects of fermented foods containing probiotic bacteria (Succi et al., 2005).
Kimchi, a lactic acid-fermented vegetable product consumed raw, is considered to be a good source of potentially beneficial and useful lactic acid bacteria, and the microorganisms involved in kimchi fermentation include about 200 species of bacteria (Cho et al., 2013). The potential health benefits of lactic acid bacteria may include balancing intestinal flora, modulation of the immune system, production of bioactive substances, pathogen exclusion, and reduction of the risk of intestinal infectious diseases, cardio vascular disease, obesity, and cancer (Lee et al., 2011; Li et al., 2012; Vinderola and Reinheimer, 2003). Among lactic acid bacteria, Lactobacillus species have a great attention for their potential probiotic effects in human health, and are probably the most important bacteria in the food industry because they are widely used as starter cultures to play significant roles in the production of fermented foods (Lee et al., 2011; Zhang et al., 2011).
When selecting strains of probiotic microorganism for use in foods, a number of criteria must be considered. The most important requirements are viability and survival of probiotic bacteria in sufficient numbers in the product (Cho et al., 2013). Zubillaga et al. (2001) stated that the criteria for an ideal Lactobacillus strain for use in the dairy industry should be as follows: resistance to acid and bile, attachment to human epithelial cells, colonization of the human intestine, production of antimicrobial substances, good growth characteristics, and beneficial effects on human health. Technological requirements, including good sensory properties, cell viability during starter preparation, and survival in the manufactured products are also important, because yogurt quality, fermenting conditions, and consumer preferences may differ depending on the culture used when lactic acid bacteria is used for yogurt making as a single starter culture (Cho et al., 2013; Ko et al., 2005). Although there are a reasonable number of well-characterized probiotic strains available for commercial use around the world, the isolation and characterization of new strains is still desirable (Vinderola et al., 2008).
Therefore, this study was conducted to determine the potential use of lactic acid bacteria isolated from kimchi, a traditional Korean fermented food, as probiotics and starter culture in dairy products. We assessed the characteristics of the isolated bacteria, such as acid resistance, bile tolerance, enzymatic activity, and cell surface properties, in addition to functional properties, such as antimicrobial activity and antioxidant activity. Another goal of this study was to apply strains isolated from kimchi as starter cultures for yogurt, without reducing the product quality and sensory characteristics.
Materials and Methods
Materials
Sodium azide and MgSO4 were obtained from Junsei (Japan). Oxgall, MRS agar, MRS broth, tryptone soy agar and broth, and nutrient agar and broth were purchased from Difco (USA). HCl and Na2CO3 were obtained from Daejung (Korea). Agar powder, β-mercapto ethanol, sodium dodecyl sulfate (SDS), σ-nitrophenyl-β-D-galactopyranoside (ONPG), trichlo acetic acid, 2,2-diphenyl-1-picrylhydrazyl, 2, 4, 6-tripyridyl-s-triazine (DPPH), iron (III) chloride hexahydrate 97% A.C.S reagent, 2,4,6-tri(2-pyridyl)-1,3,5-triazine (TPTZ), FeCl3•6H2O, Pyrogallol, K2S2O8, ethylene diamine tetra acetic acid (EDTA), acetate buffer (pH 3.6), Tris-HCl buffer, xylene, sodium acetate trihydrate, and sodium taurodeoxycholate hydrate were obtained from Sigma (USA). A gram stain kit was obtained from YD Diagnostics (Korea). An API 50 CHL kit was obtained from Bio Merieux Ltd. (France). 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid was obtained from Fluka (Germany). Lactic acid was obtained from Konto (Japan). BCP agar was obtained from Aiken (Japan). Skim milk was obtained from Seoul Dairy Co-op. (Korea). Chloroform was obtained from JT Baker (USA). NaOH pellets were obtained from Mallinckrodt (USA). Finally, the sterilized 0.45 μm filters were obtained from Millipore (Ireland).
Commercial strains
Type and commercial strains were selected in this study depending on the purpose of use. Lactobacillus acidophilus (LA-5) was supplied by Christian Hansen (Denmark). Lactobacillus casei (LC-10) and Lactobacillus plantarum (LP-5) were purchased from Culture System Inc. (USA). Bacillus cereus KCTC 3624, Stapylococcus aureus KCTC 3881, Enterococcus faecalis KCTC 2011 and Escherichia coli KCTC 1682 were supplied by the Korean Culture Center of Microorganisms (Korea). Yomix-321 (mixture of Streptococcus thermophilus and Lactobacillus bulgaricus) was obtained from Danisco (Denmark).
Sample collection and isolation of lacticacid bacteria
Lactic acid bacterial strains were isolated from homemade and commercial kimchi which were collected from different regions in Korea. Each sample was mixed with PBS with the ratio of 1:1 and homogenized using a stomacher (Interscience, France). One milliliter of diluted samples was serially diluted with 0.85% saline, then 100 μL of each dilution was spread onto MRS agar containing 0.02% sodium azide and the plates were incubated at 37℃ for 48 h. Representative colonies were picked from plates and well-isolated colonies were inoculated into fresh MRS broth for stock preparation. For long term storage, stock cultures were maintained at −20℃ in MRS broth containing 15% glycerol.
Screening and identification of lactic acid bacteria
Screening of selected isolates was conducted by observation of colony shape on plates, gram staining, and observation of cell morphology using a microscope. Selected isolates were identified first by using an API kit (API 50 CHL, Bio Merieux, France) and then by 16S rDNA gene sequencing. 16S rDNA coding gene was amplified by direct PCR using universal primers: forward; 5’-GAGTTGGATCCTGGCTCAG-3’ and reverse; 5’-AAGGAGGGGATCCAGCC-3’. The reaction mixture consisted of 300 mM Tris-HCl (pH 8.8), 100 mM (NH4)2SO4, 100 mM KCl, 20 mM MgSO4, 20 pM of each primer, 20 mM of each dNTP, 2 U Taq polymerase (Genenmed, USA), and a 20 ng template. Amplification was conducted for 30 cycles of the following conditions: 1 min at 95℃, 1 min of annealing at 55℃, and 2 min of extension at 72℃ using a PCR machine (T Gradient model, Biometera, Germany). The PCR products were directly sequenced with an ABI Prism 3700 genetic analyzer at the Macrogen, Inc. (Korea). The 16S rDNA sequences were analyzed using the GenBank database, and identification was performed on the basis of 16S rDNA sequence homology.
Bile salt and acid tolerance
Selected strains were tested for their resistance to different stress factors and compared with the commercial strains, LA-5, LC-10, and LP-5. To assay acid tolerance, cultures were inoculated in the MRS broth adjusted to pH 2.0 with 1.0N HCl, and then incubated at 37℃ for 2h. Bacterial growth was observed with enumerated viable colonies on the MRS agar plates incubated at 37℃ for 48 h. For bile tolerance, cultures were inoculated in the MRS broth supplemented with 1.0% oxgall, and then incubated at 37℃ for 24 h. The survival was ascertained as described above. Survival rate (%) was calculated as follow:
Survival rate = (B / A) × 100
where A and B denote colony count (CFU/mL) of culture before treatment and colony count (CFU/mL) of culture after treatment, respectively.
Assay for β-galactosidase activity
One mL of culture was added to 50 mL of 0.1 M phosphate buffer containing 0.001 M MgSO4 and 0.05 M β-mercaptoethanol. Then, two drops of chloroform and one drop of 1% (w/v) sodium dodecyl sulfate were added to 1mL of the diluted sample. After vortexing for 10 s, the mixture was placed in a water bath at 28℃ for 5 min. Then, 0.2 mL of 4 mg/mL o-nitrophenyl-β-D-galactopyranoside (ONPG) was added to the mixture and vortexed for 10 s. The reaction catalyzed by β-galactosidase was quenched for 10 min via addition of 0.5 mL of 1 M Na2CO3. Following centrifugation at 15,000×g for 15 min, the optical density was measured at 420 nm using an ELISA reader (TECAN, Austria). The following formula was used to estimate the enzyme activity:
Activity of β-galactosidase (unit/mL) = A420/ tv × 100
where t, v, and A420 denote time of reaction (min), volume of sample (mL), and absorbance (420 nm), respectively. All chemicals used were from Merck (Germany).
Assay for proteolytic activity
Milk agar plates containing 1.6% skim milk and 1.5% agar were prepared. An aliquot of 300 μL of each strain at a final concentration of about 108 CFU/mL was inoculated onto plates and incubated 37℃ for 48 h. After incubation, each plate was examined for clear zones. The experiment was carried out in triplicate, and activity was reported as the diameter of clear zone±SD.
Auto-aggregation assay
The auto-aggregation assay was performed according to the method reported by Xu et al. (2009) with slight modifications. The strains were grown in MRS broth at 37℃ for 18 h and cells were harvested at 9,000×g for 10 min at room temperature. The cell pellet was washed twice with PBS and re-suspended in PBS to a final concentration of about 108 CFU/mL. At this point, an absorbance was measured at 600 nm (A0h). Then, 2 mL bacterial suspension were vortexed for 10 s and incubated at 37℃ for 5 h. After the incubation, 1 mL of the supernatant suspension was collected to measure the absorbance at 600 nm (A5h). Auto-aggregation (%) was calculated with the following equation:
Auto-aggregation (%) = (1 − A5h / A0h) × 100.
Cell surface hydrophobicity
The bacterial adhesion to hydrocarbons assay was performed according to the method by Xu et al. (2009) with slight modifications to determine cell surface hydrophobicity. The adhesion to xylene (an apolar solvent) demonstrates the hydrophobic surface characteristic of bacteria (Xu et al., 2009). Bacterial cells were suspended in phosphate-buffered saline (PBS; pH 7.2) to 108 CFU/mL. Equal volumes of the cell suspension in PBS buffer and xylene were mixed by vortexing and incubated at 37℃ for 10 min for temperature equilibration. The mixture was again vortexed briefly and allowed to stand for 5 min to separate into two phases. The aqueous phase was gently removed and its absorbance was measured at 600 nm. The surface hydrophobicity (H%) was calculated as follows:
H% = (1 − A / A0) × 100
Where A0 and A are the absorbance before and after extraction with xylene, respectively.
Sample preparation for the antioxidant assays
The strains were grown in MRS broth at 37℃ for 18 h, and then harvested by centrifugation (9,000×g, 4℃ for 10 min). The bacterial cells were washed twice with PBS (pH 7.2) and re-suspended in PBS to make the bacterial cell counts approximately 109 CFU/mL. Trolox was dissolved in PBS to obtain a final concentration of 0.02%.
Measurement of 1,1-diphenyl-2-picrylhydrzyl radical scavenging activity
This method is based on reduction of the free radical 1,1-diphenyl-2-picrylhydrzyl (DPPH). The DPPH free radical scavenging activity was measured by using the method reported by Li et al. (2014) with slight modification. One mL of ethanolic DPPH radical solution (0.2 mM) was added to 0.1 mL sample, and then 5 μL of ethanol was added. The mixture was shaken and incubated at 37℃ in the dark for 30 min. The absorbance was measured at 517 nm against a blank. A lower absorbance of the reaction mixture indicated higher free-radical scavenging activity. This scavenging activity (%) was expressed as a percentage using the following formula:
DPPH radical scavenging activity (%) = [(1 − (sample O.D. / blank O.D.)) × 100].
Measurement of superoxide anion scavenging activity
Superoxide anion scavenging activity was determined according to the method of Li et al. (2014) with some modifications. The reaction mixture consisted of 0.5 mL of NBT (156 μM in 0.1 M potassium phosphate buffer pH 7.4), 0.5 mL of NADH (468 μM in 0.1 M potassium phosphate buffer pH 7.4), and 0.25 mL of an appropriately diluted sample. The reaction was initiated by the addition of 50 uL of PMS (60 μM in 0.1 M potassium phosphate buffer pH 7.4) to the mixture. The tubes were incubated at ambient temperature for 20 min. A reaction mixture containing 0.25 mL of 0.1 M potassium phosphate buffer (pH 7.4) and 50 μL of PMS (60 μM in 0.1 M potassium phosphate buffer pH 7.4) served as the control. The absorbance was measured at 560 nm. A decreased absorbance of the reaction mixture indicated increased superoxide anion-scavenging activity. The percentage inhibition of superoxide anion generation was calculated using the following formula:
Scavenging activity (%) = [(1 − (sample O.D. / blank O.D.)) × 100].
Antimicrobial activity assay
Bacillus cereus KCTC 3624, Stapylococcus aureus KCTC 3881, Enterococcus faecalis KCTC 2011, and Escherichia coli KCTC 1682 were used as test strains to evaluate the antimicrobial effects of selected strains and commercial strains. These strains were incubated in nutrient broth for 18 h at 37℃, then 0.15 mL of culture broth was spread onto nutrient agar. A paper disc (diameter, 10 mm) was soaked with 100 μL of each strain at a final concentration of about 108 CFU/mL. The soaked paper discs were placed on the surface of each plate. After incubation at 37℃ for 24 h under aerobic conditions, each plate was examined for clear inhibition zones around the paper disc. A clear zone of 1 mm or more was considered positive inhibition. The experiment was carried out in triplicate and activity was reported as the diameter of the clear zone±SD.
Yogurt preparation
The yogurt was made from 92% whole milk in which the solid content was increased by the addition of 3% oligosaccharide and 5% skim milk. The milk was then homogenized, pasteurized at 95℃ for 15 min, cooled to about 40℃, and inoculated at a rate of 20 mL/L (107CFU/mL) with a probiotic isolate previously subcultured in triplicate in MRS broth. The milk was incubated at 40℃ until the desired titratable acidity of 0.9 had developed. Yogurt containing commercial strains was also prepared as above. Every 2 h, samples were taken to measure the titratable acidity. After fermentation, the viable cell counts of each yogurt product were measured.
Sensory evaluation
The samples were subjected to sensory evaluation on a 5-point Likert scale. Thirty panels were recruited. The members of this panel were university students selected for their familiarity with yogurt products but who received no training at all in order to represent the average consumer. Samples were presented to panels in identical plastic cups (50 mL) labeled with three digit random numbers and in random order. Each panelist was asked to rate the samples for the total six attributes: four attributes (overall taste, flavor, freshness, mouthfeel and aftertaste) using a 5-point hedonic scale (1 = “I extremely dislike” to 5 = “I extremely like”), and two attributes (the intensities of sweetness and sourness) using a 5-point “just-about-right” scale ranging from 1 = “much too mild” to 5 = “much too strong.”
Results and Discussion
Identification of isolates
In previous study (Cho et al., 2013), a total of 106 strains were isolated from kimchi samples. All the isolates obtained were morphologically characterized along with the observation of colony shape, Gram reaction and microscopic examination. Gram positive, non-motile, and rod-shaped bacteria showing phenotypic characters similar to the Lactobacillus species on MRS agar media and being able to grow at pH 3.0 were selected for further studies. Eight isolates of a single colony were selected and designated as DK207, DK211, DK212, DK215, DK 301, DK303, DK314, and DK318.
Table 1 gives the results of the identification of lactic acid bacteria isolated from kimchi. Identification was first tried by using an API 50 CHL kit. Four different species were identified in the present study: L. plantarum (DK 207, DK211, DK215 and DK 301), L. brevis (DK212 and DK318), L. pentosus (DK303), and W. confusa (DK314). Then, 16S rDNA gene sequencing was done to confirm the identification results from the API kit. However, only DK211 and DK314 were confirmed to be the same strain as the results from the API kit (99% similarity to Gene Bank sequences). DK207 and DK215 were identified as L. paracasei DK212 and DK318 were identified as Weissella cibaria, and DK301 and DK303 were identified as L. sakei and L. plantarum, respectively. Further studies were conducted for DK207, DK211, DK215, DK301, and DK303, the strains identified as Lactobacillus speciesand the remaining two Weissella species were excluded. The characteristics and functional properties of the selected strains were compared with those of the commercial strains, LA-5, LC-10, and LP-5.
Table 1. Identification of lactic acid bacteria isolated from kimchi by API 50 CHL and 16s-rDNA sequence.
| Strains | API 50 CHL | % ID | 16s-rDNA sequence | % ID |
|---|---|---|---|---|
| DK207 | Lactobacillus plantarum | 91.2 | Lactobacillus paracasei | 99.0 |
| DK211 | Lactobacillus plantarum | 95.5 | Lactobacillus paracasei | 99.0 |
| DK212 | Lactobacillus brevis | 90.6 | Weissella cibaria | 99.0 |
| DK215 | Lactobacillus plantarum | 99.0 | Lactobacillus paracasei | 99.0 |
| DK301 | Lactobacillus plantarum | 99.8 | Lactobacillus sakei | 99.0 |
| DK303 | Lactobacillus pentosus | 71.4 | Lactobacillus plantarum | 91.0 |
| DK314 | Weissella confusa | 99.5 | Weissella confusa | 99.0 |
| DK318 | Lactobacillus brevis | 79.7 | Weissella cibaria | 99.0 |
Acid and bile tolerance
Before reaching the gastrointestinal tract, probiotic bacteria must first survive transit through the stomach which has pH as low as 1.5 (Lim and Im, 2009). Bile secreted in the small intestine also reduces the survival of bacteria by destroying their cell membranes (Succi et al., 2005). This bile salt presence creates a more stressful condition for probiotics. Therefore, resistance to low pH and bile is an important characteristic for the screening of probiotics.
According to Lee et al. (2011), the strains that show survival rates of >10% may be expected to reach the “target” site, the small intestine, in sufficiently high numbers. As shown in Table 2, all of the strains isolated from kimchi, except L. paracasei DK207, showed a survival rate of more than 10% after a 2 h incubation in an acidic environment (pH 2.0), and had noticeably higher acid tolerances than LA-5. Two other commercial strains did not survive. DK211 was the sole strain that showed a 100% survival rate. Of the strains isolated from kimchi, the strains of L. paracasei (DK207 and DK215) had a lower acid resistance than the stains of L. plantarum (DK211 and DK303) and L. sakei (DK301). The high tolerance to pH 2.0 also makes these strains candidates for addition to fermented milk as a concentrate culture (Vinderola et al., 2008).
Table 2. Survival rate of DK strains isolated kimchi and commercial strains in acidic condition and in 1.0% Oxgall broth.
| Strains | Survivability (%) |
|
|---|---|---|
| At pH 2.0 | In 1.0% oxgall broth | |
| L. paracasei DK207 | 1.00 | 0.33 |
| L. plantarum DK211 | 100 | 25.0 |
| L. paracasei DK215 | 16.7 | 0.67 |
| L. sakei DK301 | 33.3 | 100 |
| L. plantarum DK303 | 33.3 | 25.0 |
| L. acidophilus LA-5 | 0.89 | 7.89 |
| L. casei LC-10 | 0 | 1.23 |
| L. plantarum LP-5 | 0 | 54.5 |
The mean intestinal bile concentration is 0.3%, and the staying time of food in the small intestine is suggested to be 4h (Prasad et al., 1998). The survivability of selected strains and commercial strains when exposed to 1.0% bile salt for 24 h are shown in Table 1. It was found that DK 301 had the highest tolerance against bile salt, followed by DK303. Inaccordance with the results on acid tolerance, the strains of L. paracasei (DK207 and DK 215) showed the least bile tolerance, relative to the other isolated strains. One mechanisms of bile salt resistance could be the capacity of deconjugating bile salts, which is related to the capacity to remove cholesterol from the intestinal environment (Vinderola et al., 2008). Accordingly, DK211, DK301, and DK303 could most likely survive in the stomach and the small intestine, and colonize in the large intestine.
Enzyme activities
Yogurt production begins with the breakdown of lactose into glucose and galactose, a process catalyzed by β-galactosidase (Cho et al., 2013). It is also generally accepted that most fermented milks improves lactose digestion by the action of β-galactosidase, which is one of the wellestablished probiotic effects of lactic acid bacteria (Vinderola and Reinheimer, 2003). As shown in Table 3, L. plantarum DK303 showed the highest β-galactosidase activity within the strains investigated in this study. Liang et al. (2011) reported that a β-galactosidase-producing bacterium isolated from kimchi was identified as the L. plantarum group. The least β-galactosidase activity was shown by LC-10, a commercial strain. Some research (Vinderola and Reinheimer, 2003; Vinderola et al., 2008) has indicated that, generally, no β-galactosidase activity is present in the strains of L. casei.
Table 3. Comparison of β-galactosidase activity of DK strains isolated from kimchi and commercial strains.
| Strains | β-galactosidase activity (unit/mL) |
|---|---|
| L. paracasei DK207 | 104.5±25.8 |
| L. plantarum DK211 | 143.75±42.0 |
| L. paracasei DK215 | 154.25±48.2 |
| L. sakei DK301 | 219.75±22.6 |
| L. plantarum DK303 | 390.25±24.2 |
| L. casei LC-10 | 71.75±61.2 |
| L. plantarum LP-5 | 110.00±36.9 |
All values are mean±standard deviation of triplicates.
The proteolytic activity of starter culture is also important in the preparation of various fermented milk products because it leads to the liberation of peptides and amino acids which contribute to physical structure and flavor development (Beganović et al., 2013; Gandhi, 2007).The proteolytic activity of strains isolated from kimchi was examined to assess their potential to be applied as functional starter cultures for fermented dairy products. It was found that all isolated strains produced clearing zones when tested for proteolytic activity on milk agar except L. sakei DK 301 (Table 4). Overall, the strains of L. plantarum had more efficient proteolytic activity than the other tested strains.L. paracasei DK215 was found to have the most efficient proteolytic activity on skim milk agarout of the seven strains tested. Beganović et al. (2013) also reported that L. plantarum showed higher proteolytic activity than the other Lactobacillus species, with the exception of L. helveticus.
Table 4. Comparison of proteolytic activity of DK strains isolated from kimchi and commercial strains on skim milk agar plate.
| Strains | Clear zone surrounding the discs (mm) |
|---|---|
| L. paracasei DK207 | 12.3±3.1 |
| L. plantarum DK211 | 15.7±5.0 |
| L. paracasei DK215 | 19.0±7.2 |
| L. sakei DK301 | 0.0±0.0 |
| L. plantarum DK303 | 18.3±4.0 |
| L. casei LC-10 | 11.0±2.0 |
| L. plantarum LP-5 | 15.7±4.5 |
All values are mean±standard deviation of triplicates.
Cell surface properties
Adhesion ability of probiotics to intestinal mucus and to enterocytes is an important prerequisite for colonization of the host intestinal tract, and consequently it has been proposed as an important selection criterion for potential probiotic strains (Viková et al., 2008; Xu et al., 2009). The adhesion ability of probiotic strains has been studied using in vitro model systems, which are commonly used to select and assess probiotic strains for in vivo studies (Vinderola et al., 2008). In vitro adhesion tests successfully make use of immobilized intestinal mucus and human enterocyte-like Caco-2 cell cultures, but these methods are expensive and time-consuming (Viková et al., 2008). Therefore, an auto-aggregation ability test together with cell-surface hydrophobicity have been widely used for preliminary screening in identifying potentially adherent bacteria with properties suitable for commercial purposes (Del Re et al, 2000: Viková et al., 2008).
Cell surface hydrophobicity estimated by the xylene partition has been used as an indirect test for the ability to adhere to epithelial cells (Viková et al., 2008; Vinderola et al., 2008). Strains tested in this study showed a variable hydrophobicity with values ranging from 0.5% to 51.6% (Table 5). L. plantarum DK211 and L. paracasei DK215 showed noticeably higher hydrophobicity than other selected strains and commercial strains.
Table 5. Auto-aggregation and hydrophobicity of DK strains isolated from kimchi and commercial strains.
| Strains | Auto-aggregation (%) | Hydrophobicity (%) |
|---|---|---|
| L. paracasei DK207 | 85.1±6.4 | 0.5±2.9 |
| L. plantarum DK211 | 90.0±6.2 | 51.6±2.2 |
| L. paracasei DK215 | 89.8±7.2 | 46.0±8.6 |
| L. sakei DK301 | 82.1±4.1 | 8.1±3.7 |
| L. plantarum DK303 | 87.8±8.1 | 15.1±1.2 |
| L. casei LC-10 | 88.9±0.2 | 19.1±6.4 |
| L. plantarum LP-5 | 83.0±1.6 | 5.6±3.7 |
All values are mean±standard deviation of triplicates.
The results of the auto-aggregation assay are shown in Table 5. All tested strains showed a high percentage of auto-aggregation properties, which ranged from 83.0% to 92.1%. DK301 possessed the highest potential ability to adhere to epithelial cells and mucosal surfaces among all the tested strains. Lactobacillus plantarum strains isolated from kimchi showed higher hydrophobicity and auto-aggregation abilities than a commercial L. plantarum strain, LP-10. Del Re et al. (2000) performed hydrophobicity and auto-aggregation tests with 13 strains of B. longum and demonstrated that these two properties are strongly related. Our results were in agreement with their results, as auto-aggregation and hydrophobicity showed a direct relationship in all strains tested. However, Viková et al. (2008) found no statistically significant correlations between auto-aggregation and hydrophobicity of the tested strains.
Antioxidant activity
The assays for the DPPH radical scavenging activity and superoxide anion scavenging activity were used to measure the antioxidant activity of selected Lactobacillus strains. The DPPH assay is used to evaluate the ability of antioxidant to scavenge free radicals, and is known to give reliable information concerning the antioxidant ability of the tested compounds (Li et al., 2012). Superoxides are radicals with an unpaired electron located on an oxygen. These radicals indirectly initiate lipid oxidation as a result of singlet and hydrogen peroxide serving as precursors of singlet oxygen and hydroxyl radicals (Wang et al, 2006).
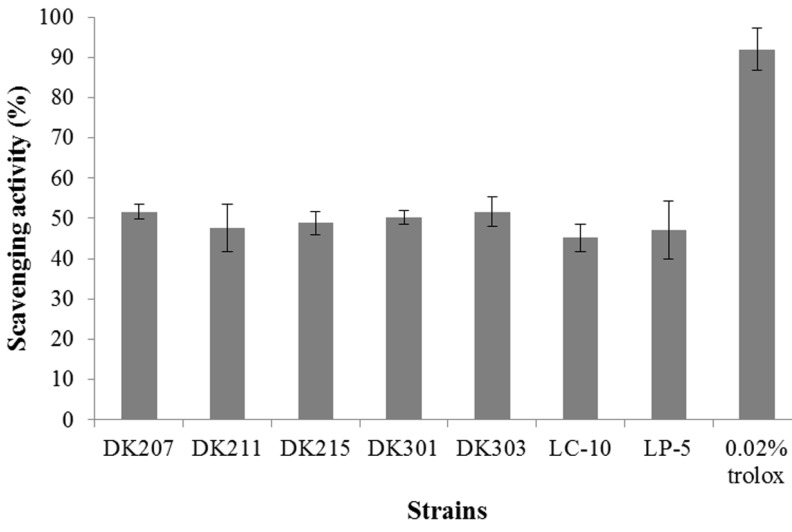
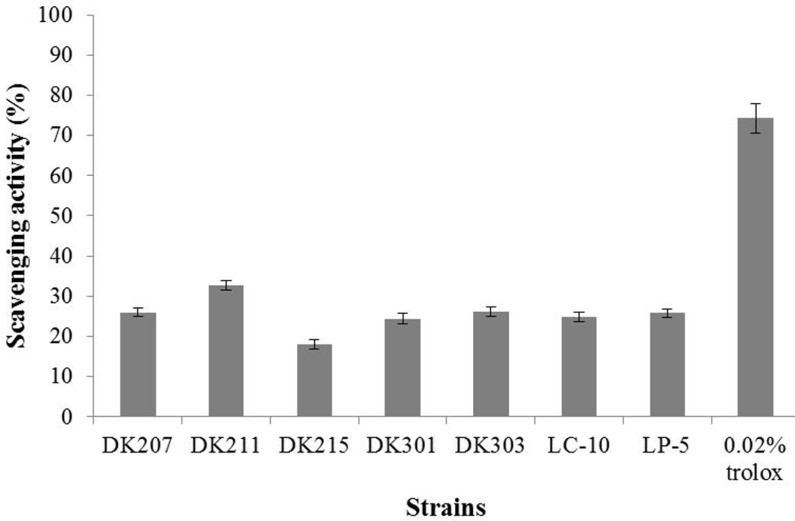
The DPPH scavenging activity of strains isolated from kimchi was around 50% when Trolox at a concentration of 0.2 mg/mL showed a DPPH scavenging activity of 90% (Fig. 1) and was higher than that of commercial strains. Of the seven strains tested, L. plantarum DK 303 and L. paracasei DK207 were found to be the most effective DPPH scavengers. Fig. 2 shows the superoxide anion scavenging activity of strains isolated from kimchi, compared against commercial strains and Trolox, the positive control. L. plantarum DK 211 had the highest superoxide anion scavenging activity (32.7%). Other strains isolated from kimchi and commercial strains showed similar scavenging activity, ranging from 24.45 to 26.1%, except L. paracasei DK215 (18.0%). It was observed that the radical scavenging activity among the different species of Lactobacillus was strain-specific. Li et al. (2012) also observed that antioxidant activity was strain-specific among 11 L. plantarum strains isolated from traditional Chinese fermented foods.
Fig. 1. DPPH radical scavenging activity of DK strains isolated from kimchi and commercial strains.
Fig. 2. Superoxide anion scavenging activity of DK strains isolated from kimchi and commercial stains.
Ramesh et al. (2012) observed a good correlation between proteolytic and antioxidant activity among the selected strains of Lactobacillus assessed. They explicated that the generation of antioxidant peptides with proteolysis contributes towards the development of antioxidant activity. However, no positive correlation between proteolytic and antioxidant activity was observed in this study. L. sakei DK301 had relatively high antioxidant activity, but showed no proteolytic activity on skim milk agar. The antioxidant activity of some lactic acid bacteria strains could also be attributed to their production of cell-surface compounds, such as extracellular polysaccharides and lipoteichoic acid (Li et al., 2012).
Antimicrobial activity
As a functional probiotic, antimicrobial activity is one of crucial properties. Lactic acid bacteria produce several metabolic compounds having antimicrobial effects, including organic acids, fatty acids, hydrogen peroxide, and diacetyl (Dunne et al., 2001). The antimicrobial ability of selected strains against some pathogen bacteria was estimated by the agar diffusion method.
The lactic acid bacteria strains showed different antimicrobial activities (Table 6). All strains effectively inhibited the growth of the Gram-positive B. cereus strain in varies degree, but had no inhibitory effect on the growth of the Gram-negative E. coli strain. Strain-specific variation was observed for E. faecalis. Lactobacillus paracasei DK215 showed the highest inhibitory effect on E. faecalis, whereas L. paracasei DK207 did not have an inhibitory effect. It has recently been reported that Lactobacillus sp. strains inhibit the growth of Gram-negative pathogenic bacteria, which has generally been attributed to the production of lactic acid during the Lactobacillus growth (Nikolova et al., 2009). Klinberg et al., (2005) also reported that L. plantarum strains isolated from fermented sausages had inhibitory activity against Gram-negative E. coli and Salmonella Typhymirium. The Lactobacillus strains selected in this study were able to inhibit the growth of Gram-positive bacteria such as E. facalis, B. cereus, and S. aureus, but were not active against the Gram-negative E. coli strain. These results are in agreement with Nieto-Lozano et al.(2002), who found that strains of L. plantarum had inhibitory activity against S. aureus but not against E. coli. Other researchers (Albano et al., 2007; Ammor et al., 2005) also observed that strains of L. plantarum could inhibit Gram-positive bacteria (S. aureus) better than Gram-negative bacteria (E. coli, P. aeuroginosa). In general, the outer membrane of Gram-negative bacteria may protect the cytoplasmic membrane from the action of antimicrobial compounds (Gao et al., 1999). It is largely accepted that the strong antimicrobial activity of lactic acid bacteria strains is attributable to the production of organic acids (Alakomi et al., 2000; Cizeikience et al., 2013; Kim et al., 2009). However some strains are able to synthesize antimicrobial substances, bacteriocins or bacteriocin-like inhibitory substances which have different inhibitory spectrum depending on the lactic acid bacteria strains (Cizeikience et al., 2013).A more specific study about antimicrobial mechanisms of strains isolated in this study is required.
Table 6. The antimicrobial activity of DK strains isolated from kimchi and commercial strains on pathogens.
| Strains | E. faecalis KCTC 2011 | B. cereus KCTC 3624 | S. aureus KCTC 3881 | E.coli KCTC 1682 |
|---|---|---|---|---|
| L. paracasei DK207 | NE | ++ | + | NE |
| L. plantarum DK211 | + | ++ | + | NE |
| L. paracasei DK215 | +++ | ++ | + | NE |
| L. sakei DK301 | + | ++ | ++ | NE |
| L. plantarum DK303 | + | ++ | ++ | NE |
| L. casei LC10 | ++ | +++ | +++ | NE |
| L. plantarum LP-5 | ++ | ++ | ++ | NE |
+ diameter of inhibition zone <1 mm, ++ diameter of inhibition zone between 2 and 4 mm, +++ diameter of inhibition zone between 5 and 8 mm, ++++ diameter of inhibition zone between 9 and 12 mm
1)NE: no effect detected.
Yogurt preparation with selected cultures
Although the most important characteristics of probiotic bacteria is their beneficial effects on host health, the evaluation of technological traits, such as growth and survival in milk-based media and during product manufacture and shelf life are important considerations for the selection of strains for dairy food applications (Vinderola et al., 2008). An ideal starter culture also should be quick and steady in acid production, produce a product with fine and clean lactic flavor, and not produce any pigments, gas, off-flavor, or bitterness in the finished product (Ghandi, 2007).
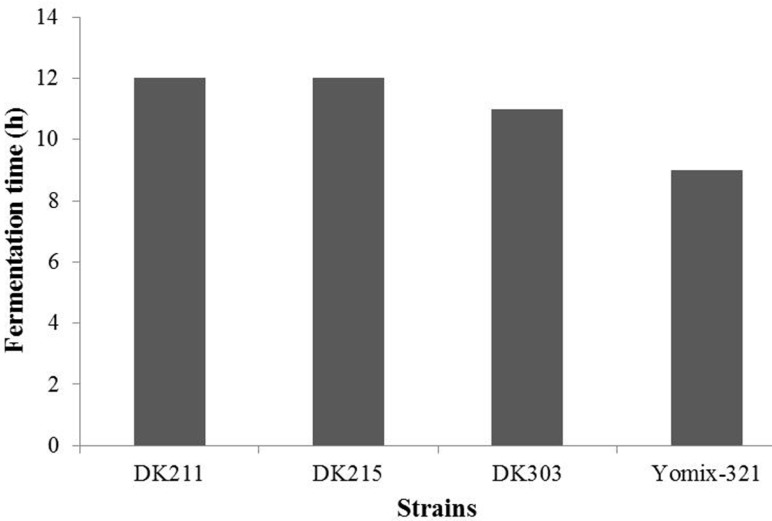
The viable cell counts and titratable acidity of yogurt fermented with selected strains were measured and compared with those of yogurt fermented with Yomix-321, a well-known commercial starter culture. As shown in Fig. 3, L. platarum DK303 developed acidity much faster than other selected strains, that is, it reached the target acidity (0.9) after 11 h. Yomix-321 reached the target acidity in 9 h, but was 12 h with L. platarum DK211 or L. paracasei DK215. DK207 and DK301 did not reach the target acidity after 16 h. The fermentation time required to reach target acidity showed relatively direct correlation with proteolytic activity, where L. sakei DK301 and L. paracasei DK207 showed no or relatively lower proteolytic activity, respectively. At the target acidity, the viable cell count of all strains tested reached around 9 log CFU/mL (data not shown). Therefore L. platarum DK211, L. paracasei DK 215, and L. platarum DK303 were selected as starter cultures for the yogurt preparation.
Fig. 3. Fermentation time required to reach the target acidity (0.9) of yogurt products fermented with L. plantarum DK211, L. paracasei DK215. L. plantarum DK303 and Yomix-321.
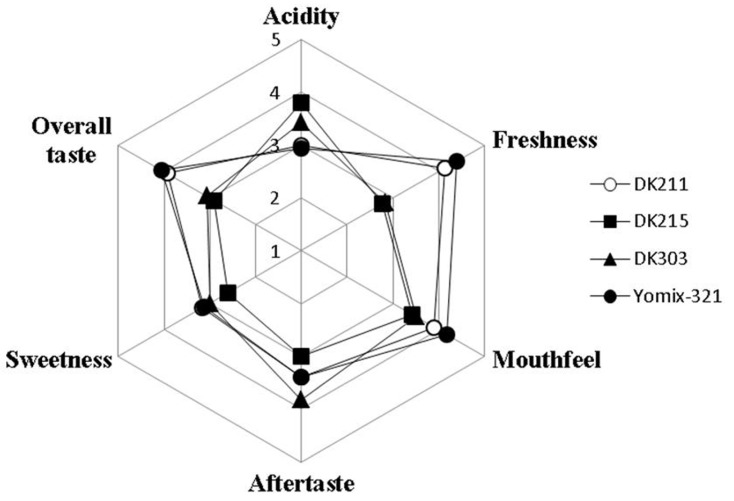
Sensory evaluation was conducted to assess the sensory characteristics of three yogurt products fermented with each selected strain in comparison to that of yogurt fermented with Yomix-321. The mean panelists rating of the sensory attributes of yogurt products fermented with each selected strain are shown in Fig. 4. Overall taste is an important sensory attribute which determines products acceptability (Emmanuel-Ikpeme et al., 2012). The overall taste rating of yogurt fermented with L. platarum DK211 was remarkably higher than that of yogurt products fermented with the other selected strains and similar to that of yogurt fermented with Yomix-321. Yogurt fermented with L. platarum DK211 also received high ratings in freshness and mouthfeel and had just right acidity and sweetness. On the other hand, yogurt fermented with L.paracasei DK215 showed the lowest ratings in all sensory attributes which might be due to its high acidity and low sweetness. According to Najgebauer-Lejko (2014), homofermentative bacteria, such as L. acidophilus and L. paracasei, yield a plain acid flavor, which is frequently perceived as too sour and lacking of real aromatic flavor. However, panelist ratings in overall taste for all the samples were above the mid-point.
Fig. 4. Sensory profile of yogurt products fermented with Lactobacillus plantarum DK211, Lactobacillus paracasei DK215, Lactobacillus plantarum DK303, and Yomix- 321. Overall taste, flavor, freshness, mouthfeel and aftertaste: 5 point hedonic scale (1=”I extremely dislike”, 5=”I extremely like”). The intensities of sweetness and sourness: 5-point “just-about-right” scale ranging from 1=“much too mild” to 5=“much too strong”.
With these results, it was found an applicability of strains isolated from kimchi for use as a starter culture in milk fermentation and probiotics. However, it was clearly observed that microorganisms belonging to the same species may develop via different mechanisms and present different characteristics.
Acknowledgments
This research was financially supported by the Ministry of Trade, Industry and Energy (MOTIE) and Korea Institute for Advancement of Technology (KIAT) through the Promoting Regional specialized Industry.
References
- 1.Alakomi H. L., Skytta E., Saarela M., Mattila-Sandholm T., Latva-Kala K., Helander I. M. Lactic acid permeabilizes Gram-negative bacteria by disrupting the outer membrane. Appl. Environ.Microbiol. (2000);66:2001–2005. doi: 10.1128/AEM.66.5.2001-2005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albano H., Oliveira M., Aroso R., Cubero N., Hogg T., Teixeira P. Antilisterial activity of lactic acid bacteria isolated from ‘‘Alherias” traditional Portuguese fermented sausages: In situ assays. Meat Sci. (2007);76:796–800. doi: 10.1016/j.meatsci.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Ammor S., Dufour E., Zagorec M., Chaillou S., Chevallier I. Characterization and selection of Lactobacillus sakei strains isolated from traditional dry sausage for their potential use as starter cultures. Food Microbiol. (2005);22:529–538. doi: 10.1016/j.fm.2004.11.016. [DOI] [Google Scholar]
- 4.Beganović J., Blaženka K., Pavunc A. L., Uroić K., Džidara P., Šušković J. Proteolytic activity of probiotic strain Lactobacillus helveticus M92. Anaeorbe. (2013);20:58–64. doi: 10.1016/j.anaerobe.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Cho Y. H., Hong S. M., Kim C. H. Isolation and characterization of lactic acid bacteria from kimchi, Korean traditional fermented food to apply into fermented dairy products. Korean J. Food Sci. An. (2013);33:75–82. doi: 10.5851/kosfa.2013.33.1.75. [DOI] [Google Scholar]
- 6.Cizeikiene D., Juodeikiene G., Paskevicius A., Bartkiene E. Antimicrobial activity of lactic acid bacteria against pathogenic andspoilage microorganism isolated from food and their control in wheatbread. Food Control. (2013);31:539–545. doi: 10.1016/j.foodcont.2012.12.004. [DOI] [Google Scholar]
- 7.Del Re B., Sgorbati M., Miglioli M., Palenzona D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifodibacteriumlongum. Lett. Appl. Microbiol. (2000);31:438–442. doi: 10.1046/j.1365-2672.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunne C., O’Mahony L., Murphy L., Thornton G., Morrissey D., O’Halloran S., Feeney M., Flynn S., Fitzgerald G., Daly C., Kiely B., O’Sullivan G. C., Shanahan R., Collins J. K. In vitro selection criteria for probiotic bacteria for human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. (2001);73:386s–392s. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 9.Emmanuel-Ikpeme C. A., Ekpeyoung I. O., Igile G. O. Nutritional and sensory characteristics of an infant food based on soybean seeds (Glycine max) and tiger nut tubers (Cyperusesculenta) British J. Appl. Sci. Technol. (2012);2:356–366. doi: 10.9734/BJAST/2012/1341. [DOI] [Google Scholar]
- 10.Gandhi D. N. Microbiology of Fermented Dairy Products. Food and Industrial Microbiology (e-book) National Science Digital Library at NISCAIR; India: (2007). pp. 1–31. [Google Scholar]
- 11.Gao Y., Van Belkum M. J., Stiles M. E. The outer membrane of Gram negative bacteria inhibits antibacterial activity of brochocin-C. Appl. Environ. Microbiol. (1999);65:4329–4333. doi: 10.1128/aem.65.10.4329-4333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M., Lee S. J., Seul K. J., Park Y. M., Ghim S. Y. Characterization of antimicrobial substance produced by Lactobacillus paraplantarum KNUC25 isolated from kimchi. Kor. J. Microbiol. Biotechnol. (2009);37:24–32. [Google Scholar]
- 13.Klinberg T. D., Axelsson L., Naterstad J., Elsser D., Budde B. B. Identification of probiotic starter cultures for Scandinavian-type fermented sausage. Int. J. Food Microbiol. (2005);105:419–431. doi: 10.1016/j.ijfoodmicro.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Ko I. H., Wang M. K., Jeon B. J., Kwak H. S. Fermentation for liquid-type yogurt with Lactobacillus casei 911C. Asian Australas J. Anim. Sci. (2005);18:102–106. doi: 10.5713/ajas.2005.102. [DOI] [Google Scholar]
- 15.Lee H., Yoon H., Ji Y., Kim H., Park H., Lee J., Shin H., Holzapfel W. Functional properties of Lactobacillus strains isolated from kimchi. Int. J. Food Microbiol. (2011);145:155–161. doi: 10.1016/j.ijfoodmicro.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Li S., Zhao Y., Zhang L., Zhang W., Huang L., Li D., Nju C., Yang Z., Wang Q. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. (2012);135:1914–1919. doi: 10.1016/j.foodchem.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 17.Li W., Ji J., Chen X., Jiang Mei., Rui X., Dong M. Structural elucidation and antioxidant activities of exopolysaccharides from Lactobacillus helveticus MB2-1. Carbohydrate Polymers. (2014);102:351–359. doi: 10.1016/j.carbpol.2013.11.053. [DOI] [PubMed] [Google Scholar]
- 18.Liang Z. Q., Srinivasan S., Kim Y. J., Kim H. B., Wang H. T., Yang D. C. Lactobacillus kimchicus sp. nov., a β-galactosidases producing bacterium isolated from kimchi. Int. J. Systematic Evolutionary Microbiol. (2011);61:894–897. doi: 10.1099/ijs.0.017418-0. [DOI] [PubMed] [Google Scholar]
- 19.Lim S. M., Im D. S. Screening and characterization of probiotic lactic acid bacteria isolated from Korean fermented foods. J. Microbiol. Biotechnol. (2009);19:178–186. doi: 10.4014/jmb.0804.269. [DOI] [PubMed] [Google Scholar]
- 20.Najgebauer-Lejko D. Effect of green tea supplementation on the microbiological, antioxidant, and sensory properties of probiotic milks. Dairy Sci. Technol. (2014);94:327–339. doi: 10.1007/s13594-014-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieto-Lozano J. C., Reguera-Useros J. I., Pelaez-Martinez M. C., de la Torre H. H. Bacteriogenic activity from starter cultures used in Spanish meat industry. Meat Sci. (2002);62:237–243. doi: 10.1016/S0309-1740(01)00252-2. [DOI] [PubMed] [Google Scholar]
- 22.Nikolova D., Petrova M., Evstatieva Y., Danova S., Atev A. Antimicrobial acitivity of Lactobacillus helveticus strain 50P1. Trakia J. Sci. (2009);7:40–44. [Google Scholar]
- 23.Prasad J., Gill H., Smart J., Gopal P. K. Selection and characterization of Lactobacillus and Bifidobacterium strains for use as probiotic. Int. Dairy J. (1998);8:993–1002. doi: 10.1016/S0958-6946(99)00024-2. [DOI] [Google Scholar]
- 24.Ramesh V., Kumar R., Singh R. R. B., Kaushik J. K., Mann B. Comparative evaluation of selected strains of lactobacilli for the development of antioxidant activity in milk. Dairy Sci. Technol. (2012);92:179–188. doi: 10.1007/s13594-011-0048-z. [DOI] [Google Scholar]
- 25.Rivera-Espinoza Y., Gallardo-Navarro Y. Nondairy probiotic products. Food Microbiol. (2010);27:1–11. doi: 10.1016/j.fm.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Succi M., Tremonte P., Reale A., Sorrentino E., Grazia L., Pacifico S., Coppola R. Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol. Lett. (2005);244:129–137. doi: 10.1016/j.femsle.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 27.Vinderola G., Capellini B., Villarreal F., Suárez V., Quiberoni A., Reinheimer J. Usefuless of a set of simple in vitro tests for the screening and identification of probiotic candidate strains for dairy use. LWT-Food Sci. Technol. (2008);41:1678–1688. doi: 10.1016/j.lwt.2007.10.008. [DOI] [Google Scholar]
- 28.Vinderola G., Reinheimer J. A. Lactic acid starter and probiotic bacteria: Comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. (2003);36:895–904. doi: 10.1016/S0963-9969(03)00098-X. [DOI] [Google Scholar]
- 29.Wang Y. C., Yu R. C., Chou C. C. Antioxidative activities of soymilk fermented with lactic acid bacteria and bifidobacteria. Food Microbiol. (2006);23:128–135. doi: 10.1016/j.fm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Xu H., Jeong H. S., Lee H. Y., Ahn J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett Appl. Microbiol. (2009);49:434–442. doi: 10.1111/j.1472-765X.2009.02684.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S., Liu S., Su Y., Li H., Sun Q., Liang X., L J. Antioxidative activity of lactic acid bacteria in yogurt. African J. Microbiol.Res. (2011);5:5194–5201. [Google Scholar]
- 32.Zubillaga M., Weill R., Postaire E., Goldman C., Caro R., Boccio J. Effect of probiotics and functional foods and their use in different diseases. Nutr. Res. (2001);21:569–579. doi: 10.1016/S0271-5317(01)00281-0. [DOI] [Google Scholar]