Abstract
Analysis of signal transduction and protein phosphorylation is fundamental to understanding physiological and pathological cell behavior and identifying novel therapeutic targets. Despite the fact that the use of physiological three-dimensional cell culture assays is increasing, 3D proteomics and phosphoproteomics remain challenging due to difficulties with easy, robust and reproducible sample preparation. Here, we present an easy-to-perform, reliable and time-efficient method for the production of 3D cell lysates that does not compromise cell adhesion before cell lysis. The samples can be used for western blotting as well as phosphoproteome array technology. This technique will be of interest for researchers working in all fields of biology and drug development.
Keywords: 3D cell culture, extracellular matrix, protein analysis, phosphoproteomics, signaling
BACKGROUND
The use of three-dimensional (3D) cell culture systems has greatly broadened the spectrum of molecular methods and also contributed to narrowing the gap between in vitro and in vivo research. Since the development of techniques like the spheroid model or the laminin-rich extracellular matrix (lrECM)-based cell cultures, several studies have shown that the results generated using these 3D assays have higher similarity with in vivo data than conventional 2D cell culture assays [1-7]. Especially in regard to signal transduction, the molecular processes in 2D and 3D cultured cells seem to differ significantly [4,5,8,9]. This could be due to multiple reasons such as altered cell-cell or cell-matrix interactions or differences in cellular morphology influencing cellular epigenetics [1,4,5,10-12].
Given that site-specific phosphorylation/dephosphorylation of proteins determines protein functionality and thus signal transduction, it is not very surprising that the phosphoproteome is under intense investigation [13]. With the invention of phosphoproteome arrays, it is now possible to efficiently analyze phosphorylation on a large scale even with limited sample volume [14]. Several studies already included results in 3D cell cultures [5,7,8,15]. However, to reduce the effects that matrix proteins or fetal bovine serum can have on array performance, some protocols include steps to dissolve the extracellular matrix or to detach cells with trypsin or accutase prior to cell lysis [8]. Importantly, this disruption of molecular connections, i.e. cell-matrix and cell-cell, may severely compromise the results.
Here we present a relatively rapid, easy and reliable method to produce 3D laminin-rich extracellular matrix (lrECM) grown cell culture lysates for phosphoproteomic analysis without cell detachment from ECM. To estimate cellular protein concentrations, detection and densitometry of housekeeping proteins like β-Actin are employed. The phosphoproteomic data have been confirmed by western blot analysis. Therefore, this new technique could be a useful tool to examine the molecular mechanisms under more physiological growth conditions.
MATERIALS
Reagents
-
✓
Laminin-rich extracellular matrix (lrECM) (BD Matrigel™, Cat. # 354248)
-
✓
Agarose type I-A (Sigma, Cat. # A0169-500G)
-
✓
Cell culture medium (e.g. DMEM; PAA, Cat. # E15-883)
-
✓
Fetal bovine serum (FBS; PAA, Cat. # A15-101)
-
✓
Trypsin-EDTA, 1× (PAA, Cat. # L11-660)
-
✓
Phosphate-buffered saline, 1× (PBS; PAA, Cat. # H15-002)
-
✓
Tubes, 50 ml, PP, sterile (greiner bio-one, Cat. # 210261)
-
✓
Safe Lock 1.5 ml reaction tubes (Eppendorf, Cat. # 0030120086)
-
✓
Serological pipette, 1 ml, sterile (greiner bio-one, Cat. # 604181)
-
✓
Serological pipette, 5 ml, sterile (greiner bio-one, Cat. # 606180)
-
✓
Serological pipette, 10 ml, sterile (greiner bio-one, Cat. # 607180)
-
✓
Serological pipette, 25 ml, sterile (greiner bio-one, Cat. # 760180)
-
✓
ART 100E Aerosol resistant tips, 100 μl (Molecular BioProducts, Cat. # 2065E)
-
✓
ART 200 Aerosol resistant tips, 100 μl (Molecular BioProducts, Cat. # 2069)
-
✓
ART 1000E Aerosol resistant tips, 1000 μl (Molecular BioProducts, Cat. # 2079E)
-
✓
24-well Microplates, Tissue culture treated, polystyrene, flat bottom, with low-evaporation lid (BD Falcon™, Cat. # 353226)
-
✓
Protein extraction buffer (Full Moon BioSystems, Cat. # EXB050)
-
✓
Complete protease inhibitor cocktail tablets (Roche, Cat. # 11697498001)
-
✓
Phosphatase inhibitor Sodium orthovanadate (AppliChem, Cat. # A2196,0005)
-
✓
Phosphatase inhibitor Sodium fluoride (AppliChem, Cat. # 7681-49-4)
-
✓
Insulin syringe with 27 G × 5/8 in. BD Micro-Fine™ IV (Orange) permanently attached needle (BD, Cat. # 309310)
-
✓
Ponceau S solution (Sigma, Cat. # P7170)
-
✓
Nonidet P-40 (Fluka, Cat. # 743858)
-
✓
EDTA (Roth, Cat. # 3619.1)
-
✓
Sodium deoxycholate (AppliChem, Cat. # A1531,0100)
-
✓
Sodium chloride (Merck, Cat. # 1.06404.1000)
-
✓
Glycerol (Roth, Cat. # 3783.1)
-
✓
DTT (AppliChem, Cat. # A1101,0025)
-
✓
Sodium dodecyl sulfate (SDS, Roth, Cat. # 2326.2)
-
✓
Tris (Roth, Cat. # 5429.3)
-
✓
Bromophenol blue (Serva, Cat. # 15375)
Recipes
-
✓
Cell culture medium: Add FBS up to 10% or 20%, according to the American Type Culture Collection (ATCC) recommendation for each specific cell line, and store at 4°C for up to 1 month.
-
✓
Agarose, 1%: Prepare 1% Agarose by weighing in 1 g Agarose type I-A and addition of double-distilled water up to 100 ml. Autoclave the solution at 121°C for 15 min, cool down and store at room temperature for up to 4 months.
-
✓
Protein extraction buffer (Full Moon): Prepare the buffer according to manufacturer’s instructions. Add Complete protease inhibitor cocktail to 1× final concentration.
-
✓
Protein extraction buffer (RIPA): Prepare a solution containing 50 mM Tris (pH = 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM sodium choride and 1 mM EDTA in double-distilled water. Add Complete protease inhibitor cocktail (1× final concentration) and phoshatase inhibitors sodium orthovanadate (final concentration 1 mM) and sodium fluoride (final concentration 2 mM) immediately before use.
-
✓
lrECM working solution: Remove the lrECM from −20 or −80°C and place in a refrigerator at 4°C 2 days before the start of the assay. After 24 h add cell culture medium (without FBS) to a final concentration of 10 mg/ml and incubate on a rotary shaker over night at 4°C. The solution must be kept on ice while pipetting since it gels rapidly. Store at 4°C for up to 2 weeks.
-
✓
6× Sample buffer: Prepare a solution containing 50% glycerol, 0.6 M DTT, 10.28% SDS, 0.35 M Tris (pH = 6.8) and 0.012% bromophenol blue in double-distilled water. Store at −20°C.
Equipment
-
✓
Cell culture incubator (humidified, 7% CO2)
-
✓
Biological hood with laminar flow
-
✓
Incubator for medium
-
✓
Inverted phase microscope with ×2.5 and ×10 objectives (Zeiss, Axiovert 25, equipped with 2.5×/0.65 A-Plan and 10×/0.25 Ph1 CP-ACHROMAT objectives)
-
✓
Centrifuge with a swing-bucket rotor, refrigerated (Eppendorf, 5804 R)
-
✓
Eppendorf Research® pipette, 10-100 μl (Eppendorf, Cat. # 3111 000.149)
-
✓
Eppendorf Research® pipette, 20-200 μl (Eppendorf, Cat. # 3111 000.157)
-
✓
Eppendorf Research® pipette, 100-1000 μl (Eppendorf, Cat. # 3111 000.165)
-
✓
Pipettor (BRAND accu-jet® pro, Cat. # 26300)
-
✓
Neubauer counting chamber (Marienfeld GmbH, Cat. # 06 401 30)
PROCEDURE
- Inserting of cells in 3D lrECM
-
1.1Heat 1% agarose in microwave until it is completely resolved. Pipette 250 μl 1% Agarose per well under sterile conditions in a 24-well plate and distribute it evenly in the well by horizontally shaking the plate.
-
1.2Trypsinize and count cells. Calculate and pipette the required amount of cells in a centrifugation tube. Centrifuge at 130 × g for 3 min at room temperature. Remove the supernatant.
-
1.3Prepare an lrECM solution with a final concentration of 0.5 mg/ml using cell culture medium (with FBS).
-
1.4Resuspend the cell pellet with lrECM solution and pipette 1 ml per well in the agarose-coated 24-well plate.
-
1.5After 2 h-incubation at 37°C, 7% CO2, carefully add 200 μl of cell culture medium (with FBS) to each well. (When cells are cultured less than 3 days, this step is optional).HINTS: Cell number is cell line- and incubation time-dependent and may range from 1 × 105 to 1 × 106 cells per well.
-
1.1
-
Treatment of 3D lrECM cell cultures
-
2.1
For treatment of cells with inhibitors/chemotherapeutics carefully add the desired amount of drug in the upper medium layer. Take the volume of the lrECM-cell layer and media (1.2 ml per well) into account. For controls, treat corresponding numbers of wells with lrECM-cell layers with the same amount of solvent.
-
2.2
If indicated, remove the drug by several (at least 5) medium changes without touching the lrECM-cell layer.
-
2.1
- Harvesting of 3D lrECM cell cultures
-
3.1For preparation of protein lysates from 24-well plates with 3D lrECM-cell cultures, place the plate on ice.
-
3.2The cells should be visible in the lower third of the lrECM-cell layer. To reduce lrECM-FBS contamination in the protein lysate, carefully remove the layer containing media and excess lrECM-media mixture without aspirating the cells.
-
3.3Immediately add 200 μl of Protein extraction buffer (supplemented with Protease inhibitor cocktail) to the wells.
-
3.4Incubate the plate on ice for 30 min. Transfer cell lysate to a 1.5 ml ice-cold reaction tube.
-
3.5Pass the cell lysate through an insulin syringe with a 27 gauge needle 5 times, while on ice, to ensure complete cell lysis.
-
3.6Incubate for an additional 30 min on ice. Centrifuge lysate for 20 min at 16,000 × g and 4°C.
-
3.7Transfer supernatant to a new 1.5 ml reaction tube and proceed with western blotting or phosphoproteome analysis.HINTS: The cell layer should be completely covered to allow cell lysis. Cell lysates should be kept on ice at all times. Make sure, that the complete lysate is passed 5 times through the needle to accomplish complete cell lysis.
-
3.1
- Western Blot and protein estimation
-
4.1Mix 15 μl of 3D lysate with 3 μl of 6× sample buffer. Heat samples at 95°C for 5 min.
-
4.2Load Western blot gels with at least two different amounts of a 2D lysate with known concentration (for example 25 μg and 20 μg per lane) mixed with sample buffer. Then load 3D samples.
-
4.3Perform western blotting and transfer protein to nitrocellulose membrane
-
4.4Analyze β-Actin expression by incubation with specific primary and secondary antibodies according to the manufacturer’s instructions.
-
4.5Perform a densitometric analysis using a suitable software (e.g. ImageJ) by measuring the mean grey value.
-
4.6Calculate the cell protein concentration of 3D lysates by comparison with densitometric values of 2D lysates.HINTS: For densitometric analysis make sure to include background correction.
-
4.1
- Phosphoproteome array
-
5.1Perform phosphoproteome array or send samples to phosphoproteome service on dry ice. Further information about available arrays from Full moon Biosystems as well as detailed user manuals can be found at the following website: http://www.fullmoonbio.com/services/antibody-array-assay-service/.
-
5.1
ANTICIPATED RESULTS
Protein expression and phosphorylation of cells cultured in a 3D matrix have been shown to better reflect results obtained in vivo compared with cells grown under 2D monolayer cell culture conditions [5,6,9,16,17].
While phosphoproteome and proteome array techniques have been customized for the widely used 2D cell culture plastic growth conditions, we established these techniques for total protein lysates harvested from 3D lrECM cell cultures to make broader examination of signal transduction feasible (Fig. 1 and Fig. 2).
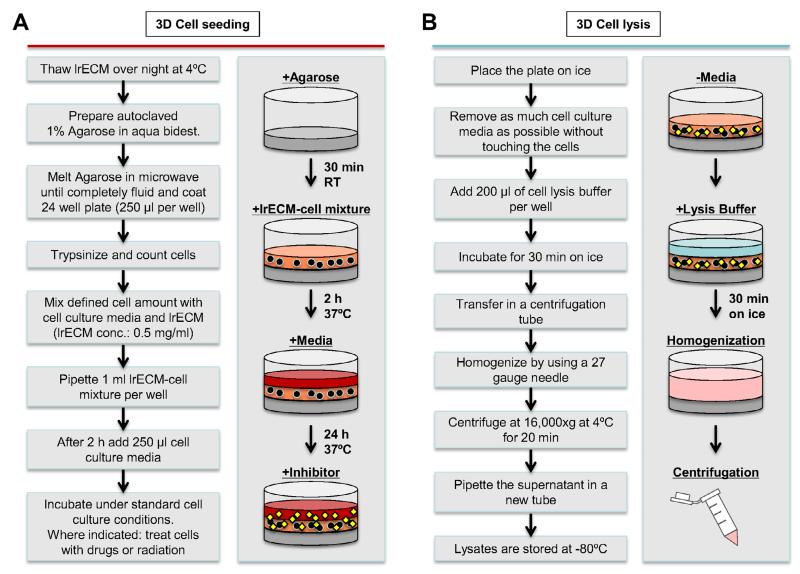
Figure 1.
A. Scheme of 24-well preparation and cell plating into 3D lrECM. Wells are first coated with agarose to prevent cell attachment. After trypsinization and counting, cell suspension is diluted with lrECM and placed into the wells. Medium is added after lrECM mixture has polymerized. B. Prior to cell lysis, redundant medium/lrECM is carefully removed without disturbing the cell layer. Cell lysates are homogenized with a 27 gauge needle and centrifuged. The supernatant is stored at −80°C.
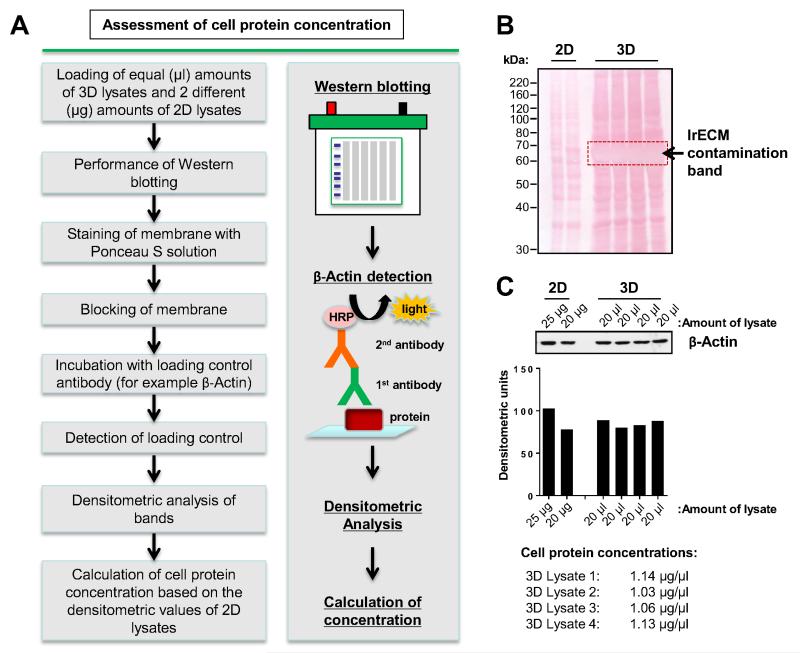
Figure 2.
A. Work flow of protein estimation using western blotting. B. Representative image of a membrane containing 2D and 3D samples stained with Ponceau S. The contamination band of the remaining laminin-rich extracellular matrix (lrECM) is around 50–70 kDa. C. Detection of β-Actin and densitometric analysis are used to estimate cell protein concentrations.
In contrast to most other techniques, cells are here lysed in 3D without detaching the cells from the surrounding ECM prior to adding cell lysis buffer. Because FBS and lrECM cannot be completely removed from the cell lysates, Ponceau S protein staining of the nitrocellulose membrane might show a pronounced band at approximately 50–70 kDa (Fig. 2B). This contamination with non-cellular proteins also interferes with bicinchoninic acid assay (BCA assay) to measure cellular protein concentration. Therefore, we used western blotting and subsequent analysis of β-Actin expression to calculate cellular protein concentration (Fig. 2C). Concentrations of 3D cell lysates might be lower than concentrations of 2D cell lysates due to the dilution with remaining lrECM and cell culture medium. Detection of proteins in the range of 40–60 kDa might run 5–10 kDa lower due to lrECM rests. In our hands, all tested antibodies for proteins with a molecular weight in this range worked perfectly. To rule out any false results, the detection of particular proteins within this kDa range requires individual attention by the experimenter.
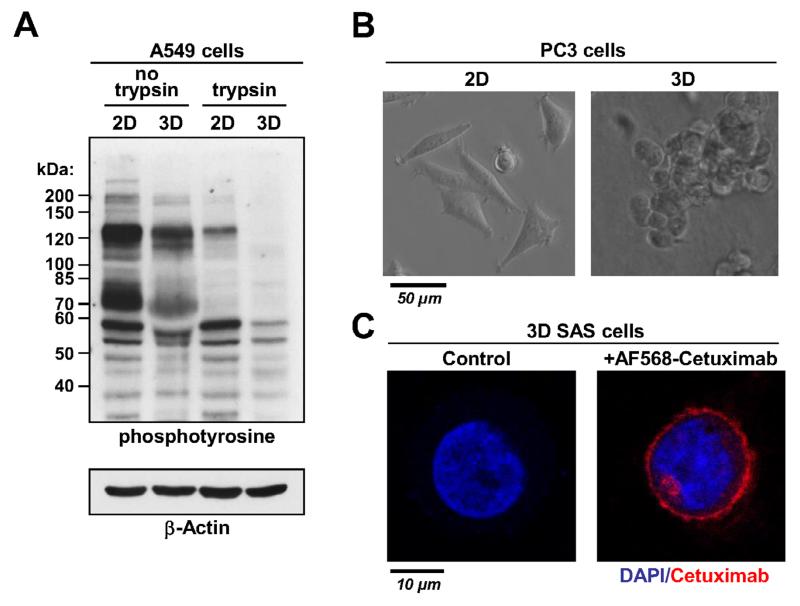
Detachment of cells has a critical and immediate effect on protein phosphorylation even when phosphatase inhibitors are used during the procedure (Fig. 3A). One advantage of the described method is the preservation of cell-ECM interactions and consequently protein phosphorylation patterns for reliable investigation of signal transduction in 3D cultured cells. As shown in Figure 3B, cell morphology is expected to be different between 2D and 3D cultured cells (Fig. 3B). While 2D cells are flat and spread out, 3D grown cells are round and show a similar cell shape as cells in vivo [5,11]. This distinct morphology is connected to differences in cytoskeletal and nuclear organization that have been shown to impact on cellular mechanisms and hereby affect protein phosphorylation [4,10,11,18-22].
Figure 3.
A. Western blot for phosphotyrosine in 2D and 3D A549 lung carcinoma cell cultures lysed either in adherent growing cells or suspension cells detached with trypsin. B. Representative images of 2D and 3D grown PC3 prostate carcinoma cells. C. Immunofluorescence staining of one SAS cell growing in 3D. Cells were incubated with fluorochrome-labeled Cetuximab for 30 min. An untreated cell is shown as control. The nucleus was stained with DAPI.
Reliable analysis of the cellular proteome and phosphoproteome after treatment with pharmacological compounds requires the unrestricted diffusion of drugs in the 3D matrix. To address this issue, we used fluorochrome-labeled Cetuximab as an example for an inhibitory antibody (Fig. 3C). As shown in Figure 3C, Cetuximab was bound to the cell membrane 30 min after treatment indicating a sufficient drug penetration through the 3D matrix.
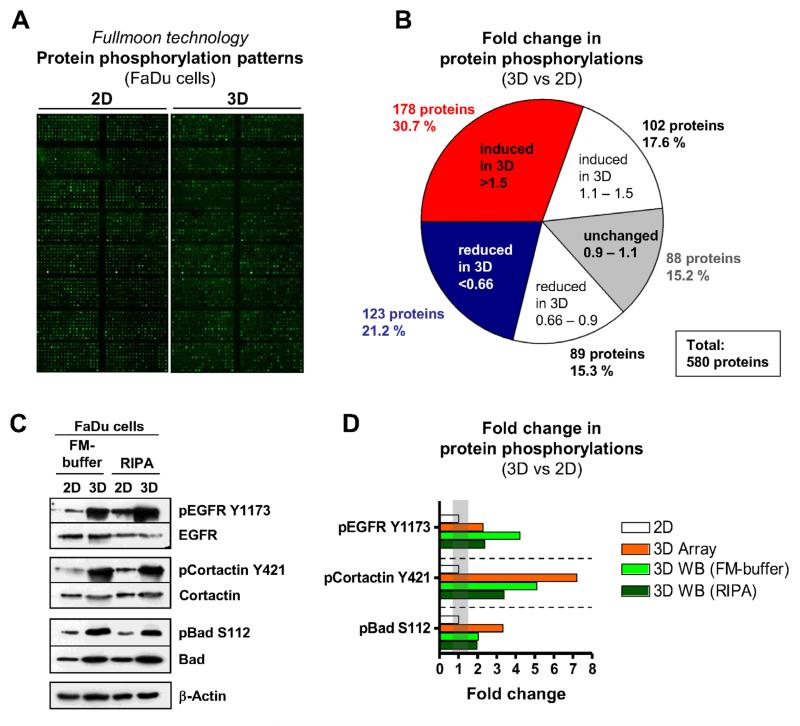
Next, we investigated the role of ECM- and growth condition-dependent signal transduction pathways more closely. Therefore, we compared protein expression and phosphorylation in 2D vs. 3D grown human FaDu cells using different buffer conditions and a high-throughput method (Fig. 4). After phosphoproteome array hybridization and analysis (Fig. 4A), changes in protein phosphorylation were calculated by normalization to total protein expression (Fig. 4B). Among 580 proteins in total, only 15.2% of proteins showed similar changes in phosphorylation in 2D and 3D cultures. Importantly, we have observed great similarity in protein phosphorylation patterns in 3D lrECM cancer cell cultures and their corresponding tumor xenografts in nude mice in contrast to 2D monolayer cell cultures [5]. In Figure 4C and D, modified RIPA and Full Moon protein extraction buffers were compared for their usefulness and reproducibility in 3D lrECM and 2D cell cultures. Exemplarily, proteins without alteration in phosphorylation pattern as well as proteins with increased or reduced phosphorylation are exhibited and matched with phosphoproteome array data (Fig. 4C and D). This comparative analysis shows growth factor receptor signaling to be more activated in 3D than 2D cultures (Fig. 4C and D) [5,6,23], which is in line with our previously published data in animal models [5].
Figure 4.
A. Full Moon Phospho Explorer Array (PEX100) (Cat. # SEV03; Σ 580 proteins, Σ 1358 phospho sites) was used to explore differences in protein phosphorylation in 2D versus 3D cell cultures. B. Protein phosphorylations examined in 3D versus 2D total protein extracts using Full Moon buffer. The numbers in the pie diagram indicate the fold change in protein phosphorylation of 3D lrECM cell cultures relative to 2D, i.e. induced (red), reduced (blue), unchanged (gray), and white (failed to reach the > 1.5- or < 0.66-fold cut-off). C. Comparison of total protein lysates extracted with Full Moon or RIPA buffer from 2D and 3D cell cultures on the basis of specific proteins and protein phosphorylations. β-Actin served as loading control. D. Densitometric protein phosphorylation analysis of protein bands shown in C in comparison to results of array analysis.
In summary, this robust method can easily be used to analyze the proteome and phosphoproteome of cells embedded in a 3D cell culture system more accurately by fast, in situ harvesting of whole cell lysates than via methods that disturb cell-ECM and cell-cell contact prior to cell lysis.
TROUBLESHOOTING
Tips for troubleshooting can be found in Table 1.
Table 1. Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1.1 | 1% Agarose layer is not evenly distributed in wells |
|
|
| 1.2 | Cells form aggregates |
|
|
| 1.2 | A higher percentage of aggregated and/or of dead cells are visible under the microscope |
|
|
| 1.3 | lrECM is solid or appears cloudy/ clumpy |
|
|
| 1.4 | lrECM-cell mixture appears clumpy or contains partial polymerized products |
|
|
| 1.4 | Wells do not contain the same amount of cells in replicate wells |
|
|
| 1.4 | Cells are not evenly distributed in wells |
|
|
| 1.4 | Cell number appears too low or too high in well |
|
|
| 1.4 | Air bubbles are visible upon the lrECM-cell layer |
|
|
| 1.5 | lrECM-cell layer appears col- lapsed/contracted |
|
|
| 3.2 | Cells are not growing in 3D |
|
|
Acknowledgments
The authors were in part supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF Contracts 03ZIK041 to N.C.), the Deutsche Forschungsgemeinschaft (CO668/4-1 to N.C.), the Deutsche Krebshilfe (108976 to N.C.), the EFRE Europäische Fonds für regionale Entwicklung, Europa fördert Sachsen (100066308) and by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (to I.E.). We are grateful to Ellen Dickreuter for excellent technical assistance and the NIH Fellows Editorial Board for editorial assistance.
Footnotes
Competing interests: Yaping Zong was employed by Full Moon BioSystems, Inc. during the time the experiments were performed and this article was written.
References
- 1.Weaver VM, Fischer AH, Peterson OW, Bissell MJ. The importance of the microenvironment in breast cancer progression: recapitulation of mammary tumorigenesis using a unique human mammary epithelial cell model and a three-dimensional culture assay. Biochem Cell Biol. 1996;74:833–851. doi: 10.1139/o96-089. PMID: 9164652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson CM, Inman JL, Bissell MJ. Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat Protoc. 2008;3:674–678. doi: 10.1038/nprot.2008.35. doi: 10.1038/nprot.2008.35. PMID: 18388950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier MV, Martin KJ. Transcriptome profiling in clinical breast cancer: from 3D culture models to prognostic signatures. J Cell Physiol. 2006;209:625–630. doi: 10.1002/jcp.20787. doi: 10.1002/jcp.20787. PMID: 17001673. [DOI] [PubMed] [Google Scholar]
- 4.Storch K, Eke I, Borgmann K, Krause M, Richter C, et al. Three-dimensional cell growth confers radioresistance by chromatin density modification. Cancer Res. 2010;70:3925–3934. doi: 10.1158/0008-5472.CAN-09-3848. doi: 10.1158/0008-5472.CAN-09-3848. PMID: 20442295. [DOI] [PubMed] [Google Scholar]
- 5.Eke I, Schneider L, Förster C, Zips D, Kunz-Schughart LA, et al. EGFR/JIP-4/JNK2 signaling attenuates cetuximab-mediated radiosensitization of squamous cell carcinoma cells. Cancer Res. 2012;73:297–306. doi: 10.1158/0008-5472.CAN-12-2021. doi: 10.1158/0008-5472.CAN-12-2021. PMID: 23074283. [DOI] [PubMed] [Google Scholar]
- 6.Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, et al. Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS One. 2013;8:e59689. doi: 10.1371/journal.pone.0059689. doi: 10.1371/journal. pone.0059689. PMID: 23555746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eke I, Zscheppang K, Dickreuter E, Hickmann L, Mazzeo E, et al. Simultaneous β1 integrin-EGFR targeting and radiosensitization of human head and neck cancer. J Natl Cancer Inst. 2015;107:dju419. doi: 10.1093/jnci/dju419. doi: 10.1093/jnci/dju419.PMID: 25663685. [DOI] [PubMed] [Google Scholar]
- 8.Levin VA, Panchabhai S, Shen L, Baggerly KA. Protein and phosphoprotein levels in glioma and adenocarcinoma cell lines grown in normoxia and hypoxia in monolayer and three-dimensional cultures. Proteome Sci. 2012;10:5. doi: 10.1186/1477-5956-10-5. doi: 10.1186/1477-5956-10-5. PMID: 22276931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo AT, Mori H, Mott J, Bissell MJ. Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J Mammary Gland Biol Neoplasia. 2012;17:103–110. doi: 10.1007/s10911-012-9251-7. doi: 10.1007/s10911-012-9251-7. PMID: 22573197. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda Y, Kawamoto Y, Teduka K, Peng W, Yamamoto T, et al. Morphological and cytoskeletal alterations of nervous system tumor cells with different culturing methods. Int J Oncol. 2011;38:1253–1258. doi: 10.3892/ijo.2011.945. doi: 10.3892/ijo.2011.945. PMID: 21331444. [DOI] [PubMed] [Google Scholar]
- 11.Eke I, Cordes N. Radiobiology goes 3D: how ECM and cell morphology impact on cell survival after irradiation. Radiother Oncol. 2011;99:271–278. doi: 10.1016/j.radonc.2011.06.007. doi: 10.1016/j.radonc.2011.06.007. PMID: 21704412. [DOI] [PubMed] [Google Scholar]
- 12.Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. doi: 10.1371/journal. pone.0002994. PMID: 18714348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. PMID: 12368087. [DOI] [PubMed] [Google Scholar]
- 14.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol. 2010;5:105–119. doi: 10.1021/cb900277e. doi: 10.1021/cb900277e. PMID: 20047291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eke I, Deuse Y, Hehlgans S, Gurtner K, Krause M, et al. β1Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J Clin Invest. 2012;122:1529–1540. doi: 10.1172/JCI61350. doi: 10.1172/JCI61350. PMID: 22378044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eke I, Leonhardt F, Storch K, Hehlgans S, Cordes N. The small molecule inhibitor QLT0267 Radiosensitizes squamous cell carcinoma cells of the head and neck. PLoS One. 2009;4:e6434. doi: 10.1371/journal.pone.0006434. doi: 10.1371/journal.pone.0006434. PMID: 19649326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Fuente G, Mollinedo P, Grande L, Vazquez-Barquero A, Fernandez-Luna JL. Culture dimensionality influences the resistance of glioblastoma stem-like cells to multikinase inhibitors. Mol Cancer Ther. 2014;13:1664–1672. doi: 10.1158/1535-7163.MCT-13-0854. doi: 10.1158/1535-7163.MCT-13-0854. PMID: 24723451. [DOI] [PubMed] [Google Scholar]
- 18.Schlaepfer DD, Hunter T. Integrin signalling and tyrosine phosphorylation: just the FAKs? Trends Cell Biol. 1998;8:151–157. doi: 10.1016/s0962-8924(97)01172-0. PMID: 9695829. [DOI] [PubMed] [Google Scholar]
- 19.Burridge K, Turner CE, Romer LH. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrix: a role in cytoskeletal assembly. J Cell Biol. 1992;119:893–903. doi: 10.1083/jcb.119.4.893. PMID: 1385444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. doi: 10.1038/nrm2957. PMID: 20729930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janmey PA. The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiol Rev. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. PMID: 9674694. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. doi: 10.1038/nature08908. PMID: 20110992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eke I, Storch K, Krause M, Cordes N. Cetuximab attenuates its cytotoxic and radiosensitizing potential by inducing fibronectin biosynthesis. Cancer Res. 2013;73:5869–5879. doi: 10.1158/0008-5472.CAN-13-0344. doi: 10.1158/0008-5472.CAN-13-0344. PMID: 23950208. [DOI] [PubMed] [Google Scholar]