Abstract
The mammalian brain is characterized by distinct classes of cells that differ in morphology, structure, signaling, and function. Dysregulation of gene expression in these cell populations leads to various neurological disorders. Neural cells often need to be acutely purified from animal brains for research, which requires complicated procedure and specific expertise. Primary culture of these cells in vitro is a viable alternative, but the differences in gene expression of cells grown in vitro and in vivo remain unclear. Here, we cultured three major neural cell classes of rat brain (ie, neurons, astrocytes, and oligodendrocyte precursor cells [OPCs]) obtained from commercial sources. We measured transcript abundance of these cell types by RNA sequencing (RNA-seq) and compared with their counterparts acutely purified from mouse brains. Cross-species RNA-seq data analysis revealed hundreds of genes that are differentially expressed between the cultured and acutely purified cells. Astrocytes have more such genes compared to neurons and OPCs, indicating that signaling pathways are greatly perturbed in cultured astrocytes. This dataset provides a powerful resource to demonstrate the similarities and differences of biological processes in mammalian neural cells grown in vitro and in vivo at the molecular level.
Keywords: transcriptome, mammalian, astrocytes, neurons, OPCs, primary culture
Introduction
The brain is one of the most complex and important organs in a mammal. A typical mammalian brain contains 108 (mouse) to 1011 (human) neurons and an even larger number of glial cells.1 Neurons possess a large cell body (soma) and projections interconnecting each other, allowing long-distance electrical communication among cells in mammals. Glial cells are nonneuronal cells, including astrocytes, oligodendrocytes (OLs), and microglia.2 Generally, glial cells are considered to be ancillary cells in the nervous system, providing physical support (eg, astrocytes), producing the myelin sheath (eg, OLs), and acting as the main form of active immune defense (eg, microglia). Oligodendrocyte precursor cells (OPCs) are cells that are able to differentiate into OLs, neurons, and astrocytes.3 Neurons and glial cells are involved in numerous neurological diseases, such as Parkinson’s disease,4 Huntington’s disease,5 Rett syndrome,6 and fragile X mental retardation.7 Understanding gene expression in these cell types may help both diagnosis and treatment of these diseases.
Technically, neural cell populations can be acutely purified from animal brains by four experimental methods: (1) laser-capture microdissection (LCM) or laser-directed microdissection (LDM),8–10 (2) fluorescence-activated cell sorting (FACS),11–15 (3) immunopanning (PAN),15–17 and (4) translating ribosome affinity purification (TRAP).18–21 Each method has its own advantages and disadvantages.22 FACS and PAN are shown to have relatively lower levels of contamination compared to LCM/LDM and TRAP.22 Since the cells are isolated from animals in a short period of time, these pooled cells are believed to reflect their physiological states in vivo.
The aforementioned experimental methods are technically complicated, which require extensive training and practice. Handling animals needs necessary facilities and personnel. A practical alternative is to use primary cultures of neural cells that are grown in vitro. Commercial availability of purified neural cells allows researchers to conduct cell-based experiments in a time- and cost-effective manner and draws meaningful conclusions about the cells’ functions in vivo. Indeed, primary cultures of astroglia have been used as in vitro proxies to astrocytes in vivo.23 Nevertheless, many argue that in vitro cultures of neural cells cannot substitute the acutely purified cells from animals. For instance, astrocytes in situ are highly polarized cells, with distinct sets of processes that project to either synapses or vascular walls.24,25 However, cultured astrocytes appear nonpolarized with an epithelioid-like shape. Moreover, several studies have found that genes that are induced in the cultured astrocytes are not necessarily expressed in vivo, suggesting that cultured astroglia do not represent the same cell type as in vivo astrocytes.15,26 However, this is not true for OLs.27 In particular, Dugas et al made a comparison of in vitro and in vivo OLs using microarrays and concluded that many aspects of the expression profile of the in vitro OLs were very similar to that of OLs acutely purified from animals.27 Overall, these microarray-based studies show that in vitro and in vivo astrocytes have different gene expression profiles, while in vitro OLs have similar profiles to their counterparts grown in vivo; similar comparison has not been done for neurons.
Recently, Zhang et al have developed a transcriptome database of various mouse neural cell types, including neurons, astrocytes, and OPCs acutely purified from cerebral cortex.28 This study identified many cell type-enriched genes using RNA sequencing (RNA-seq), a more sensitive high-throughput technique than microarray measurements.29–34 To have a full understanding of the differences in gene expression of neural cells grown in vitro and in vivo, we quantified gene expression levels in rat neurons, astrocytes and OPCs in primary cultures by RNA-seq and analyzed them with their in vivo counterparts by a cross-species RNA-seq data analysis pipeline.35 Our RNA-seq data are of high quality in which many known cell type-specific marker genes are solely expressed in the corresponding cell populations. By comparing in vitro and in vivo gene expression profiles, we identified hundreds of differentially expressed genes (DEGs), including many not found by previous studies.18,27 We found that astrocytes contain more DEGs than neurons and OPCs, suggesting that care needs to be taken when interpreting experimental results from cultured astrocytes.
Methods
Rat primary cortical astrocytes (Cat. N7745–100), neurons (Cat. A10840–01), and glial precursor cells (GPCs, N7746–100) were purchased from Life Technologies. The cells were cultured according to the protocols provided by the company (Supplementary Fig. 1). Briefly, primary rat cortical astrocytes were grown in 85% Dulbecco’s Modified Eagle’s Medium and 15% fetal bovine serum. The cells were grown in a incubator at 37 °C with 5% CO2 and a humidified atmosphere. The old medium was aspirated off and replaced with new fresh prewarmed medium every three to four days. When subculturing the cells, the medium was removed and the cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) without calcium and magnesium. The cells were then incubated with StemPro® Accutase® Cell Dissociation Reagent for 20 minutes while rocking the flasks back and forth every five minutes. The medium that was removed initially was added back to the flask, and the cells were moved to a prerinsed 15 mL centrifuge tube and spun at a centrifugation of 250 × g for five minutes. The pellet was resuspended with fresh medium and split into new flasks.
Rat neurons were recovered from a frozen vial and cultured in Neurobasal® Medium (Cat. 21103) with GlutaMAX-I™ Supplement and B-27® Supplement. Approximately 1 × 105 live cells per well were plated in a poly-d-lysine-coated (4.5 µg/cm2) 48-well plate, and then, the cells were incubated at 37 °C in an incubator with 5% CO2 and a humidified atmosphere. After six hours of incubation, half the medium was aspirated from each well and replaced with fresh medium, and then, the cells were transferred to the incubator. The cells were fed every third day by aspirating half the medium from each well and replacing with fresh medium. To eliminate the possibility of oxidative damage, the medium was left in the well while refreshing. To harvest the neuron cells for use in RNA isolation, StemPro® Accutase® was used in each well and incubated for 15 minutes. After the incubation period, a sterile cell scraper was used to gently detach the cells from the bottom of the plate. The dissociation reagent with cells were added to a prerinsed 15 mL centrifuge tube and spun at a centrifugation of 250 × g for five minutes.
Rat GPCs were raised in Complete GPC growth medium consisting of complete StemPro® NSC SFM supplemented with 2 mM GlutaMAX-I™ and 10 ng/mL PDGF-AA for their optimal growth and expansion and to keep them undifferentiated. The cells were grown in an incubator at 37 °C with 5% CO2 and a humidified atmosphere. The old medium was aspirated off and replaced with new fresh prewarmed medium every two days. GPCs were characterized with anti-A2B5 according to the description of the cell line (Cat. N7746–100), the same antibody characterizing OPCs.36,37 To avoid confusion, we named it OPCs instead of GPCs in this study.
Phenotypic characterization
Rat astrocyte cells that were to be imaged on the confocal microscope were grown on 35 mm cell culture-treated glass bottom dishes. The cells were fixed to the dish using 4% paraformaldehyde fixing solution. The cells were then incubated in blocking buffer and goat serum for 50 minutes. Following the blocking buffer, the cells were incubated at 4 °C with a 1:200 dilution of glial fibrillary acidic protein (GFAP) and DPBS, containing 5% goat serum overnight. Cells were then washed with DPBS and stained with Alexa Flour 488 fluorescent stain for 45 minutes at 37 °C. They were washed again and counterstained with a 3 ng/mL DAPI solution for five minutes. Dishes were sealed with parafilm and stored at 4 °C until imaging.
RNA extraction and sequencing
RNA samples of the rat neurons, astrocytes, and OPCs were prepared using the RNeasy Mini Kit of QIAGEN (Cat. 74104) by following the instructions of the Kit. Cells were sheared using the QIAshredder (Cat. 79654). Isolated RNA was quantified using the Thermo Scientific NanoDrop (ND-1000). A total of 250 ng RNA was sent to the UR Genomics Research Center (Rochester, NY, USA) for RNA-seq using Illumina HiSeq2500 Sequencer. Approximately 50 million single-end reads were obtained for each cell type and deposited into the NCBI Gene Expression Omnibus database (GSE72739). Each dataset has two biological replicates.
Generation of cross-species annotations
To generate cross-species annotations, we followed the method of Liu et al.32 to identify constitutive exons, ie, exons always incorporated into the final transcripts during splicing, using MISO.37 As a result, a total of 79,275 (12.1%) constitutive exons were identified in the original mm10 annotation out of a pool of 651,678 known exons. Other parts of the annotation that are not constitutive exons are discarded. Pairwise genome alignments of the chosen reference annotation to each query species are downloaded in the AXT format.38–40 Of the 79,275 constitutive exons, 68,109 (85.9%) exons are mapped perfectly to the rnor5 assembly. These exons form the cross-species annotations used in this study. A detailed protocol for the generation of cross-species annotation, differential gene expression, and pathway analysis was described in another study.35
Results and Discussion
Short reads mapping and data processing
We set out to align rat RNA-seq data to the rnor5 genome assembly using SHRiMP 41 with the default setting. The mapping statistics showed that ≥85% of the total reads was mapped successfully to the rnor5 genome, with ≥75% of them having the quality values >10 (Supplementary Table 1A), indicating that the reads are of high quality. Moreover, a small variation in the mapping statistics between biological replicates was observed, suggestive of the consistency between two replicates.
The mouse RNA-seq data (GSE52564) were originally mapped to mm9 by TopHat.42 To make a rigorous comparison with the rat RNA-seq data, we reprocessed the mouse short reads by mapping them to mm10 by also using the default setting of SHRiMP. The mapping statistics showed that ≥75% of total paired reads was mapped to the genome, with ≥70% of them having the quality values >10 and little variation observing between two biological replicates (Supplementary Table 1B).
The mapping statistics of the rat and mouse RNA-seq data to the constitutive exons in the cross-species annotation are listed in Supplementary Table 2. The percentage of the short reads properly mapped to the cross-species annotation is fairly low – between 12% and 16% because the annotation was cut down to just over 12% of its original size. Note that the percentages of mapped reads from the rat and mouse samples are similar, indicating that the quality of the annotation is not significantly changed after it has been translated across species.
Purity of rat neural cell types and data reproducibility
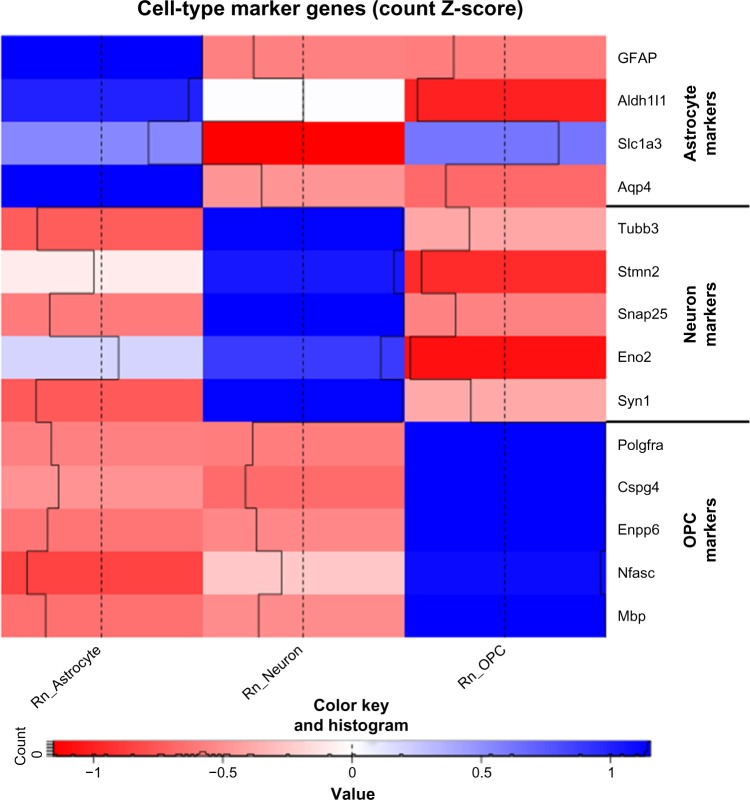
To validate the purity of the cultured brain cell types, we examined the transcriptome data for the expression of well-known cell type-specific genes15,27,43–45 for astrocytes (eg, Gfap, Aldh1l1, Slc1a3, and Aqp4), neurons (eg, Tubb3, Stmn2, Snap25, Eno2, and Syn1), and OPCs (eg, Pdgfra, Cspg4, Enpp6, Nfasc, and Mbp) (Fig. 1). All these classical cell type-specific markers exhibit high expression in the corresponding cell populations, but with no or low expression in other cell types. These data confirmed the purity and identity of the cell types cultured in vitro and established the confidence for further analysis.
Figure 1.
A heat map comparing the expression of various cell type-specific marker genes. The colors indicate the z-score of the average counts per million of each gene in each cell type.
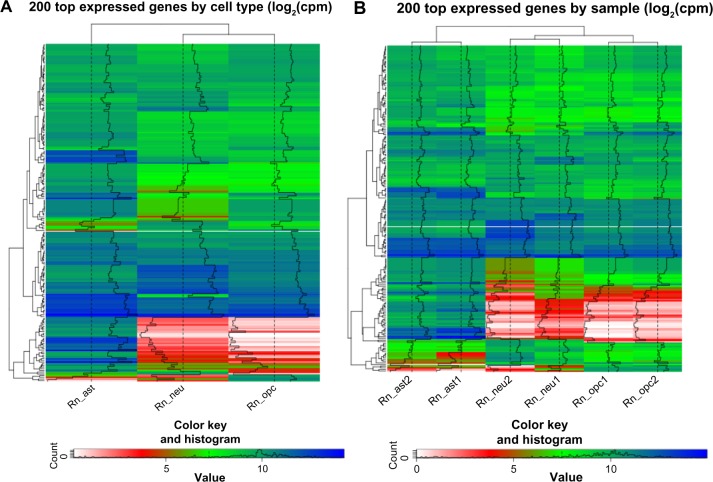
To assess the reproducibility of the rat RNA-seq data and conservation across biological replicates, we first generated the heat maps of 200 most expressed genes across all samples to visually inspect the differences within and between cell types (Fig. 2). While approximately half of the 200 genes have high expression (>10 counts per million) in all three cell types, ie, astrocytes, neurons, and OPCs, the remaining genes display differential expression across the cell types (Fig. 2A). A more detailed analysis on individual samples showed that the expression levels between two biological replicates are similar (Fig. 2B), suggesting the consistency of gene expression within each cell type.
Figure 2.
Dendrograms of 200 genes with the highest expression levels across the samples, grouped by similarity of expression patterns of cell populations (A) and individual samples (B).
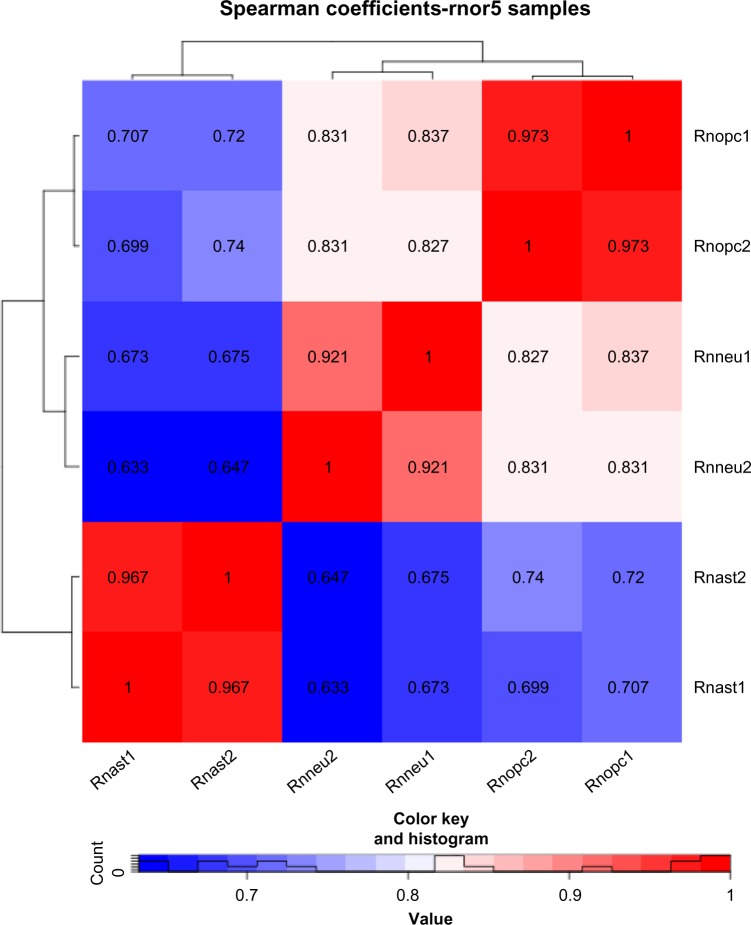
To quantify the differences within and between cell types, we calculated the correlations across all RNA-seq samples and found that the samples within each cell type are highly correlated (Spearman’s rank correlation, mean r = 0.953 ± 0.028), which illustrates a high similarity between biological replicates (Fig. 3). Note that a higher correlation is seen between astrocytes and OPCs (mean r = 0.715 ± 0.019) and between neurons and OPCs (mean r = 0.832 ± 0.004), while a lower correlation is observed between astrocytes and neurons (mean r = 0.657 ± 0.020), consistent with the fact that OPCs are able to differentiate into astrocytes and neurons.3
Figure 3.
A heat map illustrating the Spearman correlation values of the expression of each transcript in the rnor5 annotation across each individual rat sample. Rnast1 and rnast2 represent two biological replicates of rat astrocytes, while rnneu and rnopc stand for rat neuron samples and OPC samples, respectively. The colors indicate the z-score of the average counts per million of each gene in each cell type.
Differential gene expression across rat cell types
To characterize DEGs between different cell types, we first made straightforward pairwise comparisons, that is, between astrocytes and OPCs (Ast/OPC), between astrocytes and neurons (Ast/Neu), and between neurons and OPCs (Neu/OPC). The edgeR package was used to identify DEGs with P < 0.05 and FDR < 0.01.35 A total of 2102 (= 338 + 1764) DEGs that are upregulated in the Ast/OPC group are also upregulated in the Ast/Neu group (Supplementary Fig. 2A). These data indicate that 2102 genes have significantly higher expression in astrocytes than in neurons and OPCs. Similarly, we found that 1407 (= 1273 + 134) DEGs have significantly lower expression in astrocytes than in neurons and OPCs (Supplementary Fig. 2B). The heat maps of the 40 most significant DEGs in the Ast/OPC, Ast/Neu, and Neu/OPC groups clearly illustrate the differences in gene expression (Supplementary Fig. 2C–E). However, this pairwise comparison is not a good way to provide a complete picture of DEGs in three cell types. For example, if gene A is upregulated in the Ast/OPC group but downregulated in the Ast/Neu group, it is difficult to conclude if this gene is upregulated in astrocytes compared to the other two cell types.
To overcome this limitation, we made another comparison by comparing the expression level of a gene in one cell type with the average expression of this gene in the other two cell types. We found that 3107 genes have significantly higher expression in astrocytes than the average expression in neurons and OPCs (Supplementary Fig. 3A). Similarly, 2366 genes have significantly lower expression in astrocytes than the average expression in neurons and OPCs (Supplementary Fig. 3B). Differences in gene expression are ostensible for the 40 most significant DEGs (Supplementary Fig. 3C–E). A small number of genes, 129 genes, are shared between astrocytes and neurons (Supplementary Fig. 3A), which means that these genes are upregulated in both astrocytes and neurons, but are downregulated in OPCs. Therefore, these genes are DEGs for both astrocytes and neurons. Detailed examination of transcriptome data for the expression of these genes has confirmed this observation, that is, manual examination of expression levels of these 129 genes across rat cell types indeed showed that they are upregulated in astrocytes and neurons, but not in OPCs.
Pathway analysis of DEGs between rat cell populations
Although a list of DEGs is a detailed and robust way to represent differences in expression among samples, it is not a very friendly format for human beings to visualize and understand. We conducted pathway analysis based on DEGs between cell types using two different tools, GAGE46 and SPIA.47 GAGE performs a standard gene set enrichment analysis by examining all differential expressions between two samples and determining which annotated cellular pathways from KEGG48 are significantly different. SPIA performs similarly, but has the added feature of assessing the topology of the pathway. For example, if in a particular comparison, the first sample has highly expressed genes promoting a certain pathway, and the second sample has highly expressed genes repressing this pathway, SPIA rates this pathway as more significantly different than if the DEGs are randomly distributed through the pathway. However, there is a potential limitation in SPIA. If genes involved in both activation and repression of a pathway are upregulated in one sample compared to the other, it will reduce the possibility of identifying this pathway. For this reason, both GAGE and SPIA are used in this study.
To characterize the significantly enriched pathways, GAGE and SPIA were used to identify pathways statistically different (FDR-adjusted P < 0.05)35 between astrocytes and neurons (Supplementary Tables 3 and 4), between astrocytes and OPCs (Supplementary Tables 5 and 6), and between neurons and OPCs (Supplementary Tables 7 and 8). Several of these pathways identified by both GAGE and SPIA were selected for illustration to provide a coherent view at why the cells present different transcriptomes as mentioned earlier. The pathways selected for closer examination were on the basis of high significance and prior knowledge, that is, if the pathways are shown to be involved in the development of the cell type in question. The pathways were visualized by pathview49 with color-coded DEGs to represent how they are upregulated in different samples.
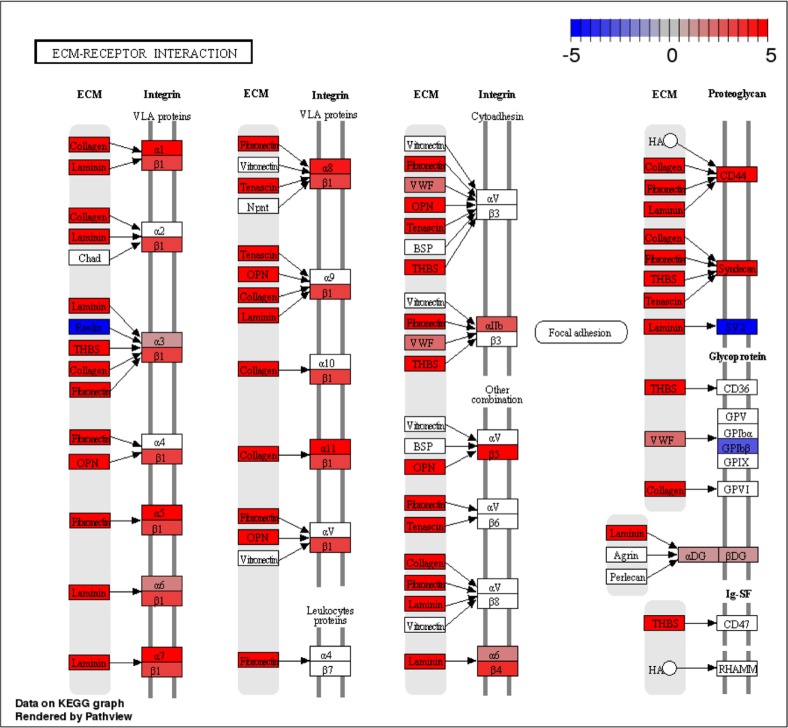
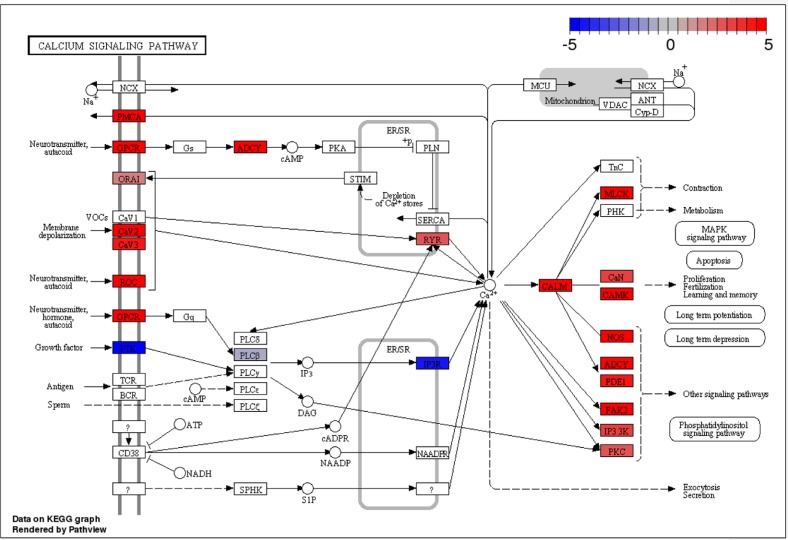
The first pathway is the ECM-receptor interaction pathway (KEGG ID: rno04512), which was ranked as one of the most significant pathways in the comparison between astrocytes and neurons (Fig. 4). It is clear that most of the genes in the ECM-receptor interaction pathway are upregulated in astrocytes, which is consistent with previous findings that astrocytes expressed ECM proteins on their surfaces in culture,50,51 and the integrin α6β4 localized on astrocytes mediates astrocyte-ECM interactions.52 The second pathway is the calcium signaling pathway (KEGG ID: rno04020) found in the comparison between neurons and OPCs (Fig. 5). Many genes that have increased expression in neurons are genes related to the production of calcium ions, which are needed for the membrane depolarization of an action potential and neuronal synaptic transmission. This pathway has previously been shown to be significantly enriched in neurons acutely purified from mouse brains.15
Figure 4.
Difference in expression of the ECM–receptor interaction pathway between rat astrocytes and neurons grown in vitro. Red indicates genes more expressed in astrocytes, while blue indicates genes more expressed in neurons.
Figure 5.
Difference in expression of the calcium signaling pathway between rat neurons and OPCs grown in vitro. Red indicates genes more expressed in neurons, while blue indicates genes more expressed in OPCs.
The basis of the cross-species comparisons between rat and mouse hinges upon the assumption that the orthology-based cross-species annotation adequately reflects changes in gene expression between rat samples and mouse samples. To illustrate the consistency of the results derived from the rnor5 annotation (aforementioned) and the cross-species annotation, we repeated the pathway analysis and analyzed significantly enriched pathways (P < 0.05) between cell types in question using the cross-species annotation. We found that the ECM-receptor interaction pathway and the calcium signaling pathway are still enriched in astrocytes and neurons, respectively, consistent with the earlier results based on the rnor5 annotation (comparing Figs. 4 and 5 and Supplementary Figs. 4 and 5). Note that the pathways from the cross-species annotation have fewer DEGs, because a large number of exons were removed from the mm10 assembly that served as the basis for the cross-species annotation.
In summary, the RNA-seq data obtained from rat astrocytes, neurons, and OPCs cultured in vitro exhibited high expression of cell type-specific genes, high reproducibility between replicates, substantial variations in gene expression between different cell types, and typical biological pathways found in the corresponding cell populations. Furthermore, the pathway analysis and DEGs were quite similar in the comparison between the rnor5 annotation and the cross-species annotation. This indicates that the process for generating the cross-species annotation works well within the rat samples. All these results suggest that the RNA-seq data generated for three rat neural cell types are of high quality, enabling meaningful cross-species comparisons with their counterparts in mice.
Differential gene expression between in vitro and in vivo
To characterize cross-species differential gene expression across neural cell types, we first visually inspected 200 most expressed genes (Supplementary Fig. 6). Gene expression based on cell type exhibited substantial changes between different cell populations (Supplementary Fig. 6A). By contrast, gene expression based on individual samples showed that expression levels between biological replicates are highly correlated, illustrating the consistency and reproducibility within each cell type for both rat and mouse data (Supplementary Fig. 6B). The top 40 genes of each cell type are listed in Supplementary Table 9.
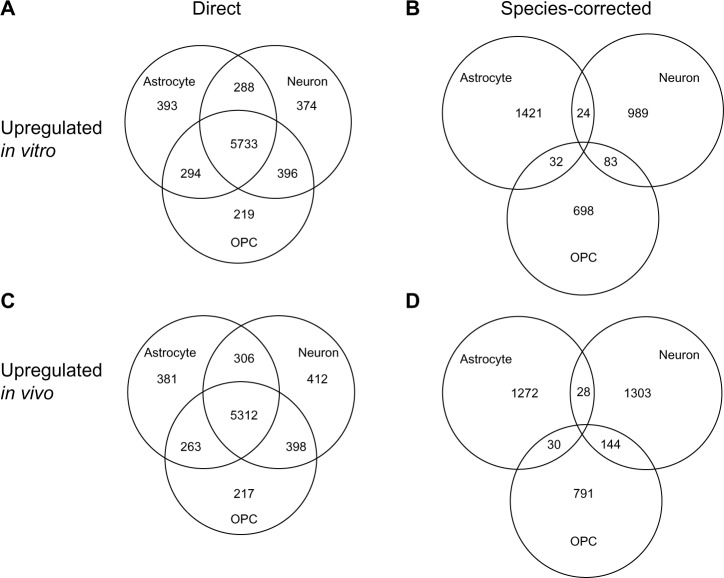
Next, we quantified the DEGs in vitro compared to in vivo (and vice versa) by making two comparisons, namely, direct comparison (Supplementary Table 10A) and species-corrected comparison (Supplementary Table 10B). For the direct comparison, we made a Venn diagram for upregulated genes in three groups: rat astrocytes vs. mouse astrocytes, rat neurons vs. mouse neurons, and rat OPCs vs. mouse OPCs (Fig. 6A). The numbers represent significantly upregulated genes in rat samples (in vitro) compared to mouse samples (in vivo). Similarly, we made a Venn diagram for upregulated genes in mouse samples (in vivo) compared to rat samples (in vitro; Fig. 6C). Note that most DEGs in the Venn diagrams are shared by all three cell types, indicating that a large number of upregulated genes (in vitro vs. in vivo and vice versa) are common among the three cell populations. This could be a combination of two factors: in vitro vs. in vivo and rat vs. mouse. To minimize the influence of the second factor, we made a species-corrected comparison. That is, we compared the expression level of a gene in rat astrocytes with the average expression of this gene in rat neurons and OPCs and identified DEGs. These DEGs were then compared with their counterparts derived from mouse cell types. As a result, for each cell type, we identified species-corrected DEGs upregulated in rat samples (in vitro; Fig. 6B) and in mouse samples (in vivo; Fig. 6D). We found that, compared to OPCs, astrocytes have a higher number of DEGs with significant changes in expression levels between in vitro and in vivo (averagely 1346 genes in astrocytes vs. 744 genes in OPCs), suggesting that the transcriptome of astrocytes is perturbed in a more drastic manner compared to that of OPCs. These data are consistent with earlier findings that gene expression profiles of cultured astrocytes are dissimilar to those of acutely purified astrocytes.15
Figure 6.
Venn diagrams compare the DEGs showing the number of DEGs in common among the different comparisons. Genes are upregulated in vitro over in vivo or vice versa with direct (A and C) or species-corrected comparison (B and D).
Comparison with published studies
To examine if DEGs identified by species-corrected expression values make sense, we assessed genes previously shown to be enriched in vitro or in vivo15 using this approach. Cahoy et al published top 80 genes from astrocytes that were enriched in vitro over in vivo, and top 80 genes enriched in vivo over in vitro.15 We calculated the fold change (FC) for these previously identified DEGs using the species-corrected expression values (Supplementary Tables 11 and 12). Note that not all the DEGs can be used for this analysis because some of these genes were not included in the cross-species annotation.
Supplementary Table 11 presents a list of 42 genes that are enriched in vitro over in vivo based on the study by Cahoy et al.15 Consistent with their observations, we found that 28 of these genes (67%) have positive log2(FC) values, meaning that these genes are more expressed in vitro, while 14 genes have negative log2(FC) values, indicating that they are more expressed in vivo. This ratio of 2:1 (28:14) is increased to 5:1 (14:3) if only the genes with FDR-adjusted P-values < 0.05 are considered. Similarly, in a list of 32 genes that are previously found to be enriched in vivo over in vitro (Supplementary Table 12),15 we found that 23 genes (72%) have negative log2(FC) values, meaning that these genes are more expressed in vivo. If only the genes with FDR-adjusted P-values < 0.05 are used, four genes have negative log(FC) values and one gene has positive log(FC) value.
In a separate study, myelin-related genes are found to be differentially expressed in OPCs in vitro vs. in vivo,27 and seven of them exist in the cross-species annotation. We found that five of these genes (71%) are indeed more expressed in vitro (Supplementary Table 13).
Overall, our data confirmed most of the genes previously shown to be expressed in vitro over in vivo, or in vivo over in vitro.15,27 The observed discrepancies between our results and the published results may be due to the following reasons. First, the experiments by Cahoy et al.15 used microarrays, while our data are based on RNA-seq. Any bias present in either experiment may have skewed the results found in these genes in either direction. It has been previously shown that microarray and RNA-seq analysis of the same samples can result in very different results.32 Second, in the study by Cahoy et al.15, in vitro astrocytes were actually astrocytes that were freshly harvested from the mouse brain, and then cultured for five days before sequencing. The astrocytes used in our experiment were ordered, frozen, and pre-isolated, from a supplier, and then were thawed and cultured for 15 days before sequencing was performed. The difference in physical treatment of the cells may be a part of the reason for this marked difference shown earlier. Finally, the study by Cahoy et al used forebrain samples, while our study used cortical samples. Since forebrain includes more than just the cortex, some of the differences in gene expression could be tissue specific.
Pathway analysis for DEGs in vitro vs. in vivo
To examine which pathways are significantly enriched in mammalian neural cells grown in vitro and in vivo, we applied GAGE and SPIA to the DEGs found in the rat and mouse data using the cross-species annotation. More pathways were found in astrocytes (Supplementary Table 14), compared to neurons (Supplementary Table 15) and OPCs (Supplementary Table 16). This result is consistent with our previous finding that, compared to the other two cell types, astrocytes have a higher number of DEGs with significant changes in expression levels between in vitro and in vivo (Fig. 6B and D). It indicates that many biological pathways are perturbed in cultured astrocytes compared to their counterparts acutely purified from animal brains.
Among the enriched pathways in astrocytes (Supplementary Table 14), the Notch signaling pathway is of interest because it was identified previously by Cahoy et al.15 These authors compared gene expression profiles from cultured astroglia and acutely purified astrocytes and found that the Notch signaling pathway is one of the most significantly enriched signaling pathway in astrocytes in vivo. In addition, they found that the Notch ligands (eg, Serrate/Jagged-1 and delta) are expressed both in vitro and in vivo. However, the downstream Notch effecter Hes5 is not expressed in vitro, and Hes1 is expressed at a much lower level in vitro. Our analysis showed that the Serrate gene is more expressed in vivo (indicated by blue in Supplementary Fig. 7). With regard to the Hes5 and Hes1 genes, they are not included in the cross-species annotation. We manually examined their expression levels using the rnor5 and mm10 annotations and found that the Hes5 gene has essentially no expression in rat samples, while the Hes1 gene has greatly reduced expression in rats compared to the homologous gene in mice, clearly in agreement with the published data.15
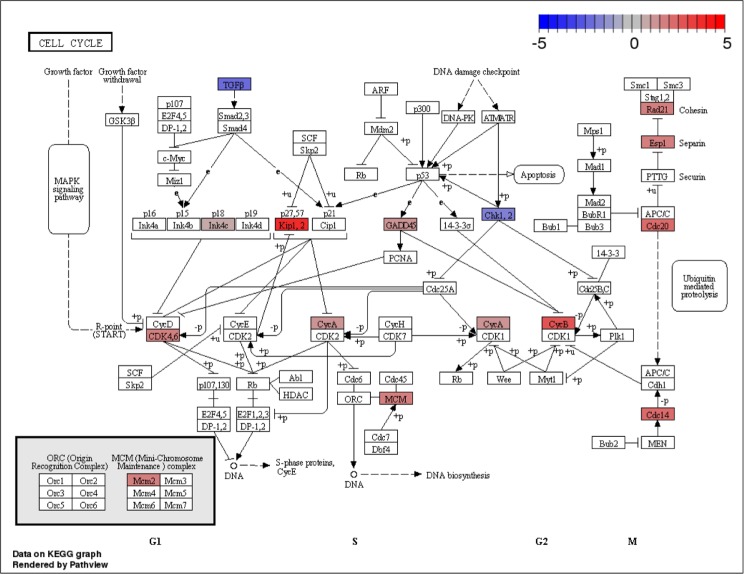
Next, we checked the pathways significantly enriched in OPCs in vitro and in vivo and found that only the cell cycle pathway was identified by both GAGE and SPIA (Supplementary Table 16). Many genes in this pathway are upregulated in rat samples (in vitro; Fig. 7), consistent with the earlier findings that cell cycle genes are often upregulated in the first few days of primary cultures of OPCs.27 Note that very few pathways were found to be significantly enriched in cultured and acutely purified neurons, with no pathway identified by both GAGE and SPIA (Supplementary Table 15). This may be due to the cross-species annotation used in this study. Future studies generating in vitro and in vivo data from the same species, namely, mice, may address this issue.
Figure 7.
Difference in expression of cell cycle-related pathways between rat and mouse OPCs correcting for species averages. Red indicates genes more expressed in rats (in vitro), while blue indicates genes more expressed in mice (in vivo).
In summary, the analysis described earlier has revealed many biological pathways that are more enriched in cells cultured in vitro vs. in cells acutely acquired from animal brains, and vice versa. These differences to some extent add to our knowledge of environment-specific gene expression. Nevertheless, other factors may also contribute to the observed differences, for example, inherent species difference (eg, rat vs. mouse) and experimental settings (eg, cultured cells vs. cells dissociated from mouse brains). Other confounding variables include, but not limited to, laboratory environment, year of experiment, models of sequencers, and orthology-based cross-species annotation. Further studies should take these factors into account.
Conclusions
We have presented the first RNA-seq analysis of mammalian neural cells grown in vitro. Three major classes of rat neural cells, ie, neurons, astrocytes, and OPCs, from commercial sources grown in primary cultures were used for sequencing. Mapping statistics of RNA short reads exhibited the consistency between two biological replicates. Known cell type-specific marker genes were only expressed in the corresponding cell types, highlighting the purity of the cell populations used in this study. Gene expression and pathway analysis have uncovered canonical signaling pathways that are well established in a specific type of cells. These data indicate that these RNA-seq datasets are of good quality. We repeated these analyses using the cross-species annotation and obtained the same results, indicating that the cross-species annotation reflects the expression of the rat cell types based on the rnor5 annotation.
We compared the rat RNA-seq data with the published data generated from mouse neural cells using the cross-species annotation. Since the mouse cells were acutely purified from neural tissues in a short period of time, these cells were believed to reflect the physiological condition in vivo. Comparison of RNA datasets between the cultured and acutely purified cells allows us to identify hundreds of DEGs in vitro and in vivo. We found more DEGs in astrocytes compared to OPCs. These data suggest that care needs to be taken when interpreting the data from primary cultures of astrocytes.
As the cross-species annotation was generated based on orthology, ie, pairwise genome alignment, it does not work well as evolutionary distance from the base species to the target species increases. Since rats are suitably closely related to mice in terms of sequence conservation, this cross-species comparison works well as expected. But it may not be the case for mice and human beings for example.
Comparisons were then presented quantifying differences in gene expression of the mouse cell populations in vivo against the corresponding rat cell populations in vitro among orthologous genes. Unfortunately, one of the major limitations of this method is its inability to control for evolutionary differences across species in cell types. This means that when reporting results regarding which genes are differentially expressed between in vitro rat cells and in vivo mouse cells, these differences could be due to the difference in cell setting (eg, in vitro and in vivo), or due to differences introduced by evolutionary divergence (eg, mouse and rat). These factors affect all the examined cell types in a similar way. Note that the bias introduced by the cross-species annotation can be controlled by correcting gene expression using the species average.
Comparisons of in vitro astrocytes against in vivo astrocytes showed that many pathways were different between the cells grown in two different settings, in vitro and in vivo. We showed that the Notch pathway, in particular, has a different expression between the settings in a way mostly consistent with previous results.15 Other pathways that are shown to be differently expressed include pathways involved in the cell’s interaction with its extracellular surroundings (eg, ECM-receptor interaction pathway) and pathways in metabolism, morphology, and intercellular connectivity. Interestingly, our preliminary study found that many DEGs from primary cultures of human neurons were found in the pathways involved in neurological diseases such as the Alzheimer’s disease, indicating that care needs to be taken when interpreting in vitro data to understand the biological processes in the Alzheimer’s disease.
Future work may involve the sequencing of mouse cells raised in vitro, which would provide data to verify the results of the comparisons aforementioned. These samples would allow for additional robustness in the in vitro/in vivo comparison as well as providing an opportunity for the quantification of the quality of the cross-species comparison performed in this study. Additionally, further downstream analysis may be applied to the existing body of data to subset it further and derive more meanings from the comparisons made in this study.
Supplementary Material
Supplementary Table 1. SHRiMP alignment statistics of each input file. Statistics are shown for (A) Rattus norvegicus cells, single-end data from neural cells grown in vitro and (B) Mus musculus cells, paired-end data from neural cells acutely purified from animals.
Supplementary Table 2. The number of input fragments that successfully matched against the generated cross-species annotations are shown here. Low percentages are to be expected, as the annotation was cut down to just over 12% of its original size.
Supplementary Table 3. GAGE pathway enrichment results between astrocytes and neurons. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 4. SPIA pathway enrichment results between astrocytes and neurons. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05. This P-value is calculated by combining the P-values of the number of DEGs with the P-value of the accumulated perturbation in the pathway.
Supplementary Table 5. GAGE pathway enrichment results between astrocytes and OPCs. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 6. SPIA pathway enrichment results between astrocytes and OPCs. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05. This P-value is calculated by combining the P-values of the number of DEGs with the P-value of the accumulated perturbation in the pathway.
Supplementary Table 7. GAGE pathway enrichment results between neurons and OPCs. This table shows the names of all pathways significantly enriched between the two samples.
Supplementary Table 8. SPIA pathway enrichment results between neurons and OPCs. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05. This P-value is calculated by combining the P-values of the numbers of DEGs with the P-value of the accumulated perturbation in the pathway.
Supplementary Table 9. The top 40 genes expressed in each cell type compared against all other cell types within its species, sorted by log FC. Genes with total expression logCPM < 1.3 have been removed.
Supplementary Table 10. The number of DEGs (q < 0.05) between in vitro and in vivo cell types in rats and mice without (A) and with (B) correction for average species differences. The total number of DEGs is shown, and then broken down by the direction in which they are differently expressed.
Supplementary Table 11. Expression of the genes listed by Cahoy et al as enriched in astrocytes in vitro. Data are shown for the rat/mouse comparison of astrocytes, corrected for species averages. A positive logFC value indicates that the gene was found to be more expressed in rats, while a negative value indicates that the gene was found to be more expressed in mice.
Supplementary Table 12. Expression of the genes listed by Cahoy et al as enriched in astrocytes in vivo. Data are shown for the rat/mouse comparison of astrocytes, corrected for species averages. A positive logFC value indicates that the gene was found to be more expressed in rats, while a negative value indicates that the gene was found to be more expressed in mice.
Supplementary Table 13. Expression information regarding the myelin-related genes examined by Dugas, corrected for differences between species. A positive logFC indicates increased expression in OPCs in rats, while a negative value indicates increased expression in OPCs in mice.
Supplementary Table 14. GAGE (A) and SPIA (B) pathway enrichment results between rat and mouse astrocytes, corrected for species averages. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 15. GAGE (A) and SPIA (B) pathway enrichment results between rats and mice in neurons. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 16. GAGE (A) and SPIA (B) pathway enrichment results between rats and mice in OPCs, corrected for species averages. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Figure 1. Cell cultures of rat astrocytes (A), neurons (B), and OPCs (C). Images were taken by a light microscope (Ziess Axiovert 40 CFL) with an INFINITY-2 camera.
Supplementary Figure 2. Venn diagrams for upregulated (A) and downregulated (B) DEGs that are in common between pairwise cell type comparisons. Heat maps are shown for the expression of the 40 most significant DEGs for astrocyte vs. neurons (C), astrocytes vs. OPCs (D), and neurons vs. OPCs (E).
Supplementary Figure 3. Venn diagrams for upregulated (A) and downregulated (B) DEGs that are in common between pairwise cell type comparisons, corrected by average expression values of two other cell populations. Heat maps are shown for the expression of the 40 most significant DEGs for astrocyte vs. neurons (C), astrocytes vs. OPCs (D), and neurons vs. OPCs (E).
Supplementary Figure 4. Difference in expression of the calcium signaling pathway between rat astrocytes and neurons grown in vitro using the cross-species annotation. Red indicates genes more expressed in astrocytes, while blue indicates genes more expressed in neurons.
Supplementary Figure 5. Difference in expression of the calcium signaling pathway between rat neurons and OPCs grown in vitro using the cross-species annotation. Red indicates genes more expressed in neurons, while blue indicates genes more expressed in OPCs.
Supplementary Figure 6. Heat maps for the 200 genes with the highest expression clustered by cell types (A) or by individual samples (B). The expression levels in each column shown by color. Both rows and columns are grouped by overall similarity, illustrated by a dendrogram. The black line on each cell is merely a histogram, providing an additional visualization of the value of each cell to complement its color.
Supplementary Figure 7. Differences in the Notch signaling pathway between rat (in vitro) and mouse (in vivo) astrocytes. The pathway is shown after species average correction. Red indicates genes more expressed in rats, while blue indicates genes more expressed in mice.
Acknowledgments
An earlier version of this research was presented by Peter R. LoVerso in fulfilment of the requirements of his M.S. in Bioinformatics at Rochester Institute of Technology.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2019 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by the RIT Undergraduate Student Summer Research Fellowship (CMW), the start-up fund, Faculty of Development (FEAD) fund and Dean’s Research Initiation Grant (D-RIG) fund from the Rochester Institute of Technology (FC), and a grant from the National Institutes of Health (R15GM116102 to FC). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: FC. Performed the experiments: CMW. Analyzed the data: PRL. Wrote the first draft of the manuscript: FC. Contributed to the writing of the manuscript: FC. Agreed with manuscript results and conclusions: PRL, CMW, FC. Jointly developed the structure and arguments for the paper: PRL, CMW, FC. Made critical revisions and approved the final version: FC. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Rowitch D, Kriegstein A. Developmental genetics of vertebrate glia-cell specification. Nature. 2010;468:214–22. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 2.Jessen K, Mirsky R. Glial cells in the enteric nervous system contain glial fibrillary acidic protein. Nature. 1980;286:736–7. doi: 10.1038/286736a0. [DOI] [PubMed] [Google Scholar]
- 3.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 4.Shulman J, De Jager P, Feany M. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 5.Walker F. Huntington’s disease. Lancet. 2007;369:218–28. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 6.Ballas N, Lioy D, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–7. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs S, Doering L. Astrocytes prevent abnormal neuronal development in the fragile X mouse. J Neurosci. 2010;30:4508–14. doi: 10.1523/JNEUROSCI.5027-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung C, Seo H, Sonntag K, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14:1709–25. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossner M, Hirrlinger J, Wichert S, et al. Global transcriptome analysis of genetically identified neurons in the adult cortex. J Neurosci. 2006;26:9956–66. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pietersen C, Lim M, Woo TU. Obtaining high quality RNA from single cell populations in human postmortem brain tissue. J Vis Exp. 2009(30):1444. doi: 10.3791/1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arlotta P, Molyneaux B, Chen J, Inoue J, Kominami R, Macklis J. Neuronal subtype-specific genes that control cortico-spinal motor neuron development in vivo. Neuron. 2005;45:207–21. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 12.Lobo M, Karsten S, Gray M, Geschwind D, Yang W. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–52. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 13.Marsh E, Minarcik J, Campbell K, Brooks-Kayal K, Golden J. FACS-array gene expression analysis during early development of mouse telencephalic interneurons. Dev Neurobiol. 2008;68:434–45. doi: 10.1002/dneu.20602. [DOI] [PubMed] [Google Scholar]
- 14.Molyneaux B, Arlotta P, Fame R, MacDonald J, MacQuarrie K, Macklis J. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–54. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahoy J, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barres B, Silverstein B, Corey D, Chun L. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988;1:791–803. doi: 10.1016/0896-6273(88)90127-4. [DOI] [PubMed] [Google Scholar]
- 17.Barres B, Hart I, Coles H, et al. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:1–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- 18.Doyle J, Dougherty J, Heiman M, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heiman M, Schaefer A, Gong S, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–48. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanz E, Yang L, Su T, Morris D, McKnight S, Amieux P. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–44. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dougherty J, Schmidt E, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–30. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okaty B, Sugino K, Nelson S. Cell type-specific transcriptomics in the brain. J Neurosci. 2011;31:6939–43. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy K, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simard M, Arcuino G, Takano T, Liu Q, Nedergaard M. Signaling at the gliovascular interface. J Neurosci. 2003;23:9254–62. doi: 10.1523/JNEUROSCI.23-27-09254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6:626–40. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- 26.Wilhelm A, Volknandt W, Langer D, Nolte C, Kettenmann H, Zimmermann H. Localization of SNARE proteins and secretory organelle proteins in astrocytes in vitro and in situ. Neurosci Res. 2004;48:249–57. doi: 10.1016/j.neures.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Dugas J, Tai Y, Speed T, Ngai J, Barres B. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–83. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Chen K, Sloan S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, Miller CJ. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics. 2010(11):282. doi: 10.1186/1471-2164-11-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal A, Koppstein D, Rozowsky J, et al. Comparison and calibration of transcriptome data from RNA-seq and tiling array. BMC Genomics. 2010(11):383. doi: 10.1186/1471-2164-11-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2010;39:578–88. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression of short read sequencing: quantitative comparison to 2-channel gene expression microarray. BMC Genomics. 2009(10):221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malone JH, Oliver B. Microarray, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011(9):34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LoVerso PR Cui F. A computational pipeline for cross-species analysis of RNA-seq data using R and Bioconductor. Bioinform Biol Insights. 2015 doi: 10.4137/BBI.S30884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietrich J, Noble M, Mayer-Proschel M. Characterization of A2B5+ glial precursor cells from cryopreserved human fetal brain progenitor cells. Glia. 2002;40:65–77. doi: 10.1002/glia.10116. [DOI] [PubMed] [Google Scholar]
- 37.Windrem MS, Nunes MC, Rashbaum WK, et al. Fetal and adult human oligodendrocyte progenitor cell isolates myelinate the congenitally dysmyelinated brain. Nat Med. 2004;10:93–7. doi: 10.1038/nm974. [DOI] [PubMed] [Google Scholar]
- 38.Chiaromonte F, Yap VB, Miller W. Scoring pairwise genomic sequence alignments. Pac Symp Biocomput. 2002:115–26. doi: 10.1142/9789812799623_0012. [DOI] [PubMed] [Google Scholar]
- 39.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 2003;100(20):11484–9. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz S, Kent WJ, Smit A, et al. Human – mouse alignments with BLASTZ. Genome Res. 2003;13(1):103–7. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5(5):e10003386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daneman R, Zhou L, Agalliu D, Cahoy JD, Kaushal A, Barres BA. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One. 2010;5:e13741. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beutner C, Linnartz-Gerlach B, Schmidt SV, et al. Unique transcriptome signature of mouse microglia. Glia. 2013;61:1429–42. doi: 10.1002/glia.22524. [DOI] [PubMed] [Google Scholar]
- 45.Chiu IM, Morimoto ET, Goodarzi H, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics. 2008;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009(10):161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M, Goto S. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liesi P, Dahl D, Vaheri A. Laminin is produced by early rat astrocytes in primary culture. J Cell Biol. 1983;96:920–4. doi: 10.1083/jcb.96.3.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asch AS, Leung LL, Shapiro J, Nachman RL. Human brain glial cells synthesize thrombospondin. Proc Natl Acad U S A. 1986;83:2904–8. doi: 10.1073/pnas.83.9.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner S, Tagaya M, Koziol JA, Qquaranta V, del Zoppo GJ. Rapid disruption of an astrocyte interaction with the extracellular matrix mediated by integrin alpha 6 beta 4 during focal cerebral ischemia/reperfusion. Stroke. 1997;28:858–65. doi: 10.1161/01.str.28.4.858. [DOI] [PubMed] [Google Scholar]
- 53.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–15. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. SHRiMP alignment statistics of each input file. Statistics are shown for (A) Rattus norvegicus cells, single-end data from neural cells grown in vitro and (B) Mus musculus cells, paired-end data from neural cells acutely purified from animals.
Supplementary Table 2. The number of input fragments that successfully matched against the generated cross-species annotations are shown here. Low percentages are to be expected, as the annotation was cut down to just over 12% of its original size.
Supplementary Table 3. GAGE pathway enrichment results between astrocytes and neurons. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 4. SPIA pathway enrichment results between astrocytes and neurons. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05. This P-value is calculated by combining the P-values of the number of DEGs with the P-value of the accumulated perturbation in the pathway.
Supplementary Table 5. GAGE pathway enrichment results between astrocytes and OPCs. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 6. SPIA pathway enrichment results between astrocytes and OPCs. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05. This P-value is calculated by combining the P-values of the number of DEGs with the P-value of the accumulated perturbation in the pathway.
Supplementary Table 7. GAGE pathway enrichment results between neurons and OPCs. This table shows the names of all pathways significantly enriched between the two samples.
Supplementary Table 8. SPIA pathway enrichment results between neurons and OPCs. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05. This P-value is calculated by combining the P-values of the numbers of DEGs with the P-value of the accumulated perturbation in the pathway.
Supplementary Table 9. The top 40 genes expressed in each cell type compared against all other cell types within its species, sorted by log FC. Genes with total expression logCPM < 1.3 have been removed.
Supplementary Table 10. The number of DEGs (q < 0.05) between in vitro and in vivo cell types in rats and mice without (A) and with (B) correction for average species differences. The total number of DEGs is shown, and then broken down by the direction in which they are differently expressed.
Supplementary Table 11. Expression of the genes listed by Cahoy et al as enriched in astrocytes in vitro. Data are shown for the rat/mouse comparison of astrocytes, corrected for species averages. A positive logFC value indicates that the gene was found to be more expressed in rats, while a negative value indicates that the gene was found to be more expressed in mice.
Supplementary Table 12. Expression of the genes listed by Cahoy et al as enriched in astrocytes in vivo. Data are shown for the rat/mouse comparison of astrocytes, corrected for species averages. A positive logFC value indicates that the gene was found to be more expressed in rats, while a negative value indicates that the gene was found to be more expressed in mice.
Supplementary Table 13. Expression information regarding the myelin-related genes examined by Dugas, corrected for differences between species. A positive logFC indicates increased expression in OPCs in rats, while a negative value indicates increased expression in OPCs in mice.
Supplementary Table 14. GAGE (A) and SPIA (B) pathway enrichment results between rat and mouse astrocytes, corrected for species averages. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 15. GAGE (A) and SPIA (B) pathway enrichment results between rats and mice in neurons. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Table 16. GAGE (A) and SPIA (B) pathway enrichment results between rats and mice in OPCs, corrected for species averages. This table shows the names of all pathways significantly enriched between the two samples to a significance level of P < 0.05.
Supplementary Figure 1. Cell cultures of rat astrocytes (A), neurons (B), and OPCs (C). Images were taken by a light microscope (Ziess Axiovert 40 CFL) with an INFINITY-2 camera.
Supplementary Figure 2. Venn diagrams for upregulated (A) and downregulated (B) DEGs that are in common between pairwise cell type comparisons. Heat maps are shown for the expression of the 40 most significant DEGs for astrocyte vs. neurons (C), astrocytes vs. OPCs (D), and neurons vs. OPCs (E).
Supplementary Figure 3. Venn diagrams for upregulated (A) and downregulated (B) DEGs that are in common between pairwise cell type comparisons, corrected by average expression values of two other cell populations. Heat maps are shown for the expression of the 40 most significant DEGs for astrocyte vs. neurons (C), astrocytes vs. OPCs (D), and neurons vs. OPCs (E).
Supplementary Figure 4. Difference in expression of the calcium signaling pathway between rat astrocytes and neurons grown in vitro using the cross-species annotation. Red indicates genes more expressed in astrocytes, while blue indicates genes more expressed in neurons.
Supplementary Figure 5. Difference in expression of the calcium signaling pathway between rat neurons and OPCs grown in vitro using the cross-species annotation. Red indicates genes more expressed in neurons, while blue indicates genes more expressed in OPCs.
Supplementary Figure 6. Heat maps for the 200 genes with the highest expression clustered by cell types (A) or by individual samples (B). The expression levels in each column shown by color. Both rows and columns are grouped by overall similarity, illustrated by a dendrogram. The black line on each cell is merely a histogram, providing an additional visualization of the value of each cell to complement its color.
Supplementary Figure 7. Differences in the Notch signaling pathway between rat (in vitro) and mouse (in vivo) astrocytes. The pathway is shown after species average correction. Red indicates genes more expressed in rats, while blue indicates genes more expressed in mice.