Abstract
The treatment paradigms for head and neck squamous cell cancer (HNSCC) are changing due to the emergence of Human Papillomavirus (HPV)-associated tumors possessing distinct molecular profiles and responses to therapy. While patients with HNSCCs are often treated with radiotherapy, preclinical models are limited by the ability to deliver precise radiation to orthotopic tumors and to monitor treatment responses accordingly. To better model this clinical scenario, we developed a novel autochthonous HPV-positive oral tumor model to track responses to small molecules and image guided radiation. We used a tamoxifen-regulated Cre recombinase system to conditionally express the HPV oncogenes E6 and E7 as well as a luciferase reporter (iHPV-Luc) in the epithelial cells of transgenic mice. In the presence of activated Cre recombinase, luciferase activity, and by proxy, HPV oncogenes were induced to 11-fold higher levels. In triple transgenic mice containing the iHPV-Luc, K14-CreERtam and LSL-Kras transgenes, tamoxifen treatment resulted in oral tumor development with increased bioluminescent activity within 6 days that reached a maximum of 74.8-fold higher bioluminescence compared to uninduced mice. Oral tumors expressed p16 and MCM7, two biomarkers associated with HPV-positive tumors. After treatment with rapamycin or image guided radiotherapy, tumors regressed and possessed decreased bioluminescence. Thus, this novel system enables us to rapidly visualize HPV-positive tumor growth in order to model existing and new interventions using clinically relevant drugs and radiotherapy techniques.
Keywords: Mice, Transgenic, Human papillomavirus type 16, Radiotherapy, Image guided, Optical Imaging, Tumorigenic Transformation
Introduction
Given their distinct oncogenic and mutational pathways (1, 2), head and neck squamous cell cancers (HNSCCs) may differentially respond to radiation and/or chemotherapy. Existing reports indicate that treatment outcomes depend on the p53 mutational status and HPV oncogene expression (3–5). To better understand HNSCC biology, several models exist to study the development of autochthonous head and neck tumors. Many groups have relied on chemical carcinogens such 4-Nitroquinolone 1-oxide (4-NQO) to induce cancers with undefined DNA lesions that mimic those caused by cigarette smoking (6, 7). To generate tumors with more homogeneous genetic profiles, other groups have genetically engineered mice to overexpress mutant cellular oncogenes such as KRAS (8) or to delete tumor suppressors such as p53 (9–12). Furthermore, groups have engineered mice to express some of these oncogenes in a spatio-temporal manner using systems such as ligand regulated Cre recombinases (8, 9, 13).
However, understanding how other oncogenes such as the HPV oncogenes E6 and E7 (E6E7) (14) impact oral tumor responses to therapy are limited by the availability preclinical models, the accurate delivery of radiotherapy and the assessment of treatment responses. While several xenotransplant models exist for HPV-associated HNSCCs, these tumors were transplanted into immunodeficient mice and may be biologically distinct from the parental cancer (15–18). Furthermore, oral tumors developed in HPV-transgenic mice treated with 4-NQO (19), but these mice constitutively expressed HPV oncogenes which may impact immune tolerance and tumor development. In addition, irradiation of oral tumors has been limited to 2 to 6 Gy due to the proximity of tumors to the central nervous system and other vital structures (17 ). Finally, monitoring treatment responses to autochthonous oral tumors has been mostly constrained to crude measurements such as survival and weight loss. Thus, understanding how the tumor genotype dictates response to therapy would benefit from novel preclinical models that monitor the response of primary HPV-positive tumors to radiation and other targeted therapies.
Here, we developed a novel head and neck tumor model to monitor the growth of HPV-positive tumors and their response to therapy using bioluminescence. We used a ligand-regulated Cre recombinase to induce the HPV oncogenes E6E7 and a luciferase reporter in vitro and in vivo. Oral tumors arose in mice when E6E7 and mutant KRASG12D oncogenes were induced in the basal epithelial layer. These oral tumors expressed HPV-associated biomarkers and grew faster than tumors arising in control mice harboring only a mutant KRAS oncogene. HPV-tumors gained bioluminescence over time which was modulated by tumoricidal agents including small molecule inhibitors and image guided radiotherapy (IGRT).
Methods
Generation of iHPV-Luc Transgenic Vector and Mice
The pB-actin E6E7 plasmid containing the HPV-16 E6E7 was a generous gift from Karl Munger (20) and was obtained from Addgene (plasmid 13712). The E6E7 gene was amplified by the 5′ primer 5′-TTGAATTCGCGGCCGCCACCATGCACCAAAAGAGAACTGC-3′ and 3′ primer 5′-TTCTCGAGTTATGGTTTCTGAGAACAGATGG-3′. The E6E7 PCR product was digested with Eco RI-Xho I and ligated to MSCV IRES Luciferase plasmid, a generous gift of Scott Lowe (Addgene plasmid 18760). An Eco RI-Sal I fragment of E6E7 IRES Luciferase construct was isolated and ligated to an Eco RI-Xho I fragment of pCAGEN, a generous gift of Connie Cepko (21) Addgene plasmid 11160 to generate the HPV-Luc vector. A LoxP EGFP polyA LoxP PCR fragment was generated by amplifying pcDNA-EGFP (a generous gift of Doug Golenbock Addgene plasmid 13031) with the forward primer 5′ TTGAATTCATAACTTCGTATAGCATACATTATACGAAGTTATTGCCACCATGGTGAGCAAGGGCGAGGAG-3′ and reverse primer 5′-TTGCGGCCGCTTATAACTTCGTATAATGTATGCTATACGAAGTTATCATAGGGAAGAAAGCGAAAGGAG-3′. This LoxP-EGFP polyA-LoxP fragment was digested with Eco RI–Not I and cloned into the HPV-Luc to generate iHPV-Luc. The resulting plasmid was linearized with SalI-BamHI and transgenic mice were made by microinjection into the nuclei of FVB/NJ (The Jackson Lab, Bar Harbor, ME) zygotes. Mice were maintained on an FVB/N background.
Mice
All mice were maintained under specific pathogen free conditions and used according to protocols approved by The University of Chicago Institutional Animal Care and Use Committee and Institutional Biosafety Committee. B6.129S4-Krastm4Tyj/J (LSL-Kras) mice were purchased from The Jackson Laboratory. Transgenic mice STOCK-Tg(KRT14-cre/ERT)20Efu/J (K14-CreERtam) have been described (22). K14-CreERtam mice were backcrossed to the FVB background using MAX BAX speed congenics Taconic (Huson, NY) for seven generations and shared 99.74% similarity to FVB mice with 2 non-FVB polymorphisms adjacent to the K14-CreERtam transgene. K14-CreERtam mice were bred with iHPV mice to generate K14-CreERtam × iHPV-Luc mice (KH mice) on an FVB background. KH mice were then bred with LSL-Kras mice to generate KHR mice or KR mice on a C57BL/6 × FVB F1 background. Mice used for experiments were between 30–40 days old.
Reagents
Rapamycin was obtained from Sigma Chemical Co (St. Louis, MO) and reconstituted in (0.2% carboxymethylcellulose and 0.25% Tween-80 and injected into mice i.p. at 4 mg/kg/d for 3d. D-Luciferin Potassium Salt was obtained from Gold Biotechnology (St. Louis, MO). Tamoxifen (Tam) was purchased from Sigma. Tam was dissolved in sunflower seed oil vehicles at 20 mg/ml and 0.1 ml was injected i.p. daily for 5 days. All oligonucleotides were synthesized by IDT (Coralville, IA).
Fluorescence cytometry
2 × 105 293 HEK cells (ATCC) were transfected with iE6E7 vector with or without PGK-Cre-bpA vector, a gift of Klaus Rajewski (Addgene plasmid 11543) using Fugene HD (Promega). Cells were trypsinized after 48h and EGFP fluorescence was assessed. Peripheral blood lymphocytes were isolated by retroorbital bleeding and red blood cells were lysed with RBC lysis buffer (eBioscience). Cells were analyzed by FACscan and data was analyzed by FlowJo software.
Western Analysis
106 293 HEK cells were transfected with iHPV-Luc vector with or without iCre vector using Fugene HD (Promega). 48h later cells were trypsinized and lysed with a modified radio-immunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 μM EDTA, 1% NP40, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 10 μM PMSF) for 30 min on ice. Samples were subjected to SDS-PAGE using a 4% stacking gel and a 10% resolving gel. The protein gel was then transferred to nitrocellulose, blocked, and probed using a 1:200 dilution of primary antibody and subsequently a 1:1000 dilution of a secondary HRP conjugated goat anti-rabbit IgG antibody (Pierce, Rockford, IL). The membrane was developed using ECL (Amersham, Arlington Heights, IL).
Luciferase Assays
2 ×105 293 HEK cells (ATCC) were transfected with iE6E7 vector with or without PGK-Cre-bpA vector, a gift of Klaus Rajewski using Fugene HD (Promega). 48h later, cells were lysed in Bright Glo lysis buffer (Promega) for 20 min luciferase activity was assessed using a Turner TD 20/20 Luminometer (Turner Designs) and luciferase activity was normalized to protein concentration of the sample.
In vivo Imaging
Luciferase activity was measured noninvasively using the IVIS-200 imaging system (Caliper LifeSciences). Mice were injected i.p. with luciferin (300 mg/kg dissolved in PBS; Caliper) and anesthetized via inhaled 3% isoflurane (Abbott Laboratories Ltd.). Exposure time for all images ranged from 1 to 5 s. All images were analyzed using Living Image software (Caliper) with a binning of 8. In vivo bioluminescent signal was quantified by taking the total photon counts for each region of interest.
Histology
Tumors were isolated and placed into 4% paraformaldyhyde solution for 24h and then dehydraded with 70% ethanol. Tissues were embedded in paraffin, sectioned and stained with hematoxyalin-eosin or the indicated antibodies at the University of Chicago Immunohistochemistry core facility.
Excision KRAS specific PCR
For detection of iE6E7 recombination, tumors were digested in lysis buffer supplemented with proteinase K and genomic DNA was isolated from tumors using ethanol precipitation. The iE6E7 gene was amplified by PCR using the primers: forward primer: 5′-CAACGTGGTTATTGTGCTG-3′ corresponding to the 3′ end of the CAG promoter and the reverse primer: 5′-GGTAACTTTCTGGGTCGCTCCT-3′ corresponding the 5′ region of the E6E7 gene with a 58 ºC annealing temperature and 72 ºC extension temperature repeated over 33 cycles. The 1629bp amplicon represents the unexcised HPV vector and the 388 bp amplicon represents the excised iE6E7 gene.
For detection of the LSL-Kras recombination, gDNA was amplified by PCR using the forward primer: GTCTTTCCCCAGCACAGTGC and the reverse primer: GGATGGCATCTTGGACCTTA that flank the floxed stop gene cassette. The resulting amplicon was digested with HindIII that cuts the unique restriction site engineered into the second exon of the mutant allele specific. The undigested 947bp amplicon represents the wild type allele and the digested 323bp and 624bp amplicons represent the mutant allele.
Image guided radiotherapy (IGRT)
Mice bearing 14–21d old tumors were irradiated using an XRAD 225Cx (Precision Xray, North Branford, CT) small animal image guided irradiator. Anesthetized mice were immobilized in the supine position and a cone beam CT was obtained using 40 kVp and 250 mA. For each mouse, the isocenter was placed in the mid-point of the tumor and anterior to the mouth in order to cover the oral tumor using half of a 1.5 cm collimator. A radiation dosimetry plan was generated using opposed lateral fields. Mice were treated with 225 kVp X-rays at 13 mA to a dose of 20 Gy.
In vivo tumor growth
Tumors were measured every three days. Given the ellipsoid growth of oral tumors, we reasoned that the oral tumor volume would approximate a solid ellipsoid minus a cylindrical volume to account for space of the oral aperture. To measure the tumor volume, we measured the intercommissural distance (a) as well as two additional orthogonal measurements (b & c). In addition, the lip thickness was measured on each side (d & e) in order to calculate the diameter of the oral aperture. The oral tumor volume (mm3) was then calculated as: (1/6*π*a*b*c) – π((1/2(a-d-e))2)*c. We illustrate this method in Supplemental Figure 1.
Statistics
Two-tailed independent Student’s t tests were done to analyze the results of in vitro luciferase assays, in vivo bioluminescence or tumor growth time points.
Results
Generation of a Cre-loxP regulated inducible HPV transgenic mouse
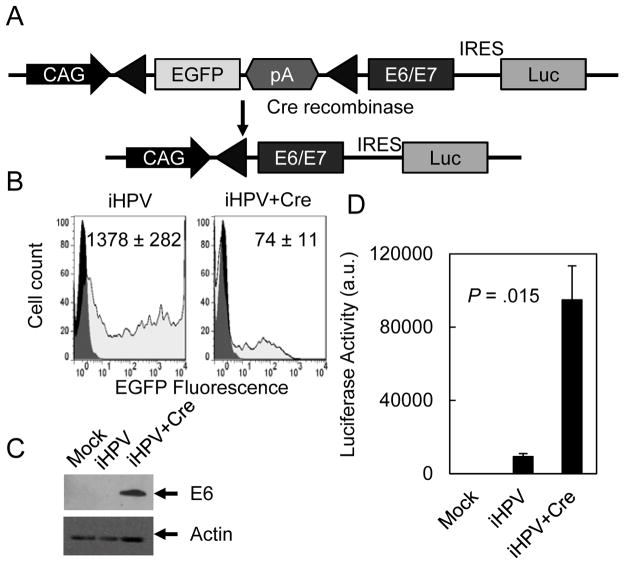
Current HPV transgenic mouse models constitutively expressed E6 and/or E7 and, therefore, do not recapitulate the HPV infections that occur in young adults who are first exposed to exogenous HPV oncogenes. To generate an inducible HPV mouse model that better reflects the clinical scenario, we generated a transgenic vector where E6E7 expression was dependent on Cre recombinase (iHPV-Luc; Fig. 1A). To monitor oncogene activation and tumor development, the E6E7 cassette was followed by an internal ribosomal entry site (IRES)-luciferase gene that enabled bicistronic expression of the E6E7 oncogenes and the luciferase reporter. iHPV-Luc contained a floxed EGFP gene that inhibited expression of the E6E7-IRES-luciferase gene cassette expression. In the absence of Cre recombinase, cells transfected with iHPV-Luc had higher levels of EGFP and lower levels of E6 expression and luciferase activity (Fig. 1B–D). In the presence of Cre recombinase, cells expressed lower levels of EGFP and 10.1-fold higher levels of luciferase activity and E6 expression consistent with the Cre mediated recombination of the floxed EGFP cassette and induction of the E6E7 genes and the luciferase reporter.
Figure 1. Development of an iHPV-Luc transgene.
(A) Scheme for iHPV-Luc transgene. A floxed EGFP reporter was placed downstream from the CAG promoter to inhibit expression of the E6E7-IRES-Luc bicistronic cassette. Excision of the floxed EGFP cassette resulted in induction of the E6E7-IRES-Luc gene cassette. (B–D) Cre recombination of iE6E7 caused decreased EGFP (grey histograms) (B), and increased E6 expression (C) and luciferase activity (D). The iHPV-Luc construct was transfected into HEK cells with or without a Cre recombinase expression vector. Transfected cells were assessed for EGFP fluorescence using flow cytometry, E6 expression by Western analysis and luciferase activity. Data represented two separate experiments performed in duplicate.
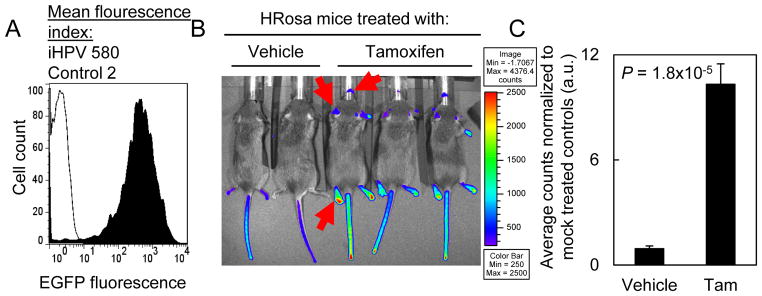
We next generated transgenic mice containing this iHPV construct. Of the 49 founder mice, 24 mice contained the iHPV-Luc transgene. In transgene positive mice, we then assessed EGFP expression in the peripheral blood leukocytes that acted as a surrogate for the potential level of oncogene induction. We selected one mouse that expressed EGFP 290-fold above background (Fig. 2A). To assess the induction of the transgene, we bred iHPV-Luc mice to Rosa-CreERtam mice to generate HRosa mice. Treatment with tamoxifen (Tam) activated the CreERtam fusion protein that was ubiquitously expressed by all tissues. After a 5d course of Tam treatment, HRosa mice had increased bioluminescence in the non-fur bearing skin that was 11-fold above background (Tam treated: 10.3±1.2-fold vs. vehicle treated: 0.9±0.2-fold; Fig. 2B and 2C).
Figure 2. Generation of an iHPV-Luc transgenic mouse line.
(A) EGFP expression in the peripheral blood leukocytes from a founder FVB/N mouse containing the iHPV-Luc transgene. Founder mice containing the iHPV-Luc transgenes were screened for EGFP expression using fluorescence cytometry. We selected the founder line with the highest level of EGFP expression. (B) Global induction of bioluminescence in iHPV-Luc × Rosa-CreERtam mice (HRosa mice). Mice were treated with vehicle or 2 mg/d Tam i.p. for 5d and bioluminescence was assessed 7 days later. Tam treated mice displayed higher levels of bioluminescence in the non-fur bearing areas. The red arrows indicate non-fur bearing skin having increased bioluminescent signal. (C) Quantitation of the bioluminescence from the paws of vehicle or Tam treated mice in (B). Results were derived six mice from the vehicle treated group and ten mice from the Tam treated group. B and C were representative from two similar experiments.
Generation of an autochthonous oral HPV-tumor model
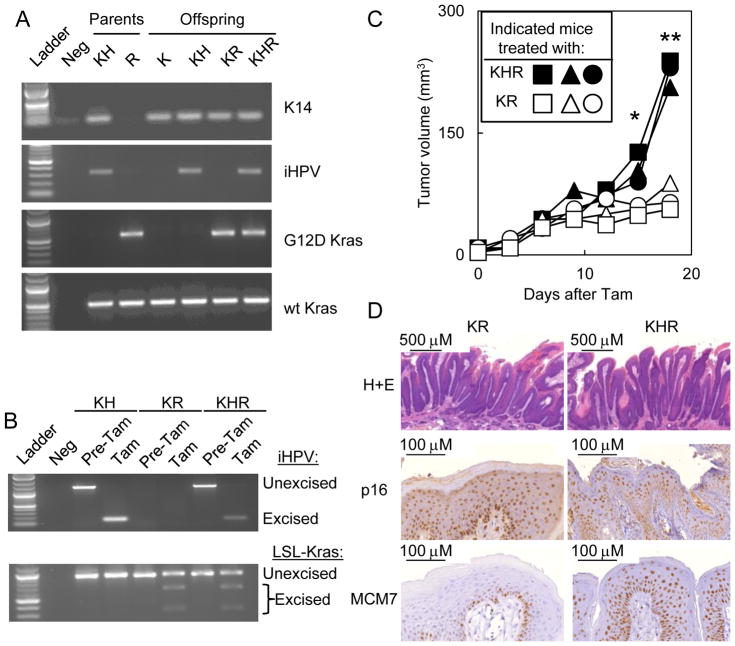
We next generated mice that developed autochthonous HPV-positive oral tumors. Based on previous reports, mice induced to express the KRASG12D mutant in the basal epithelial layer developed oral papillomas over the course of two months (8, 9). Similarly, we first bred K14-CreERtam mice to iHPV-Luc mice to generate KH mice (Fig. 3A). The K14-CreERtam transgene expresses the ligand-regulated Cre recombinase driven by the basal keratinocyte promoter, K14. KH mice were then bred to LSL-Kras mice to generate KR mice containing the K14-CreERtam and LSL-Kras transgene or KHR mice containing all three transgenes (K14-CreERtam × LSL-Kras × iHPV-Luc; Fig 3A). Tam treatment of KH, KR or KHR mice resulted in excision of the respective LSL-Kras and/or iHPV-Luc transgenes (Fig. 3B).
Figure 3. Generation of KHR mice that recombine the iHPV-Luc and LSL-Kras genes upon Tam treatment.
KH mice and LSL-Kras transgenic mice were bred to generate offspring containing all combinations of the transgenes. (A) PCR confirmed the genotype of KHR mice containing the K14-CreERtam × LSL-Kras × iHPV-Luc transgene. Transgenic mice: KH: K14-CreERtam × iHPV-Luc; R: LSL-Kras; KR: K14-CreERtam × LSL-Kras; KHR: K14-CreERtam × iHPV-Luc × LSL-Kras. (B) Treatment of these offspring with Tam resulted in excision of the iHPV-Luc and LSL-Kras transgenes. (C) After tamoxifen treatment, oral tumors grew faster in KHR mice than in KR mice. Similar results were observed in two independent experiments. Symbols indicate individual mice. Significant differences between KHR and KR mice were denoted by * for P < .05 and ** for P < .001. (D) Oral tumors arising in KHR mice were positive for p16 and MCM7, two biomarkers for HPV-positivity.
We next monitored tumor growth in KR and KHR mice. While oral tumors formed in KR and KHR mice, KHR tumors grew faster than in KR tumors (Fig. 3C). Histological analysis of KHR tumors revealed papillomas that expressed the HPV-biomarkers p16 and MCM7 (Fig. 3D). Compared to oral tumors in KR mice, oral tumors developing in KHR mice had increased MCM7 expression and similar p16 expression.
KHR tumor growth was associated with increase bioluminescence
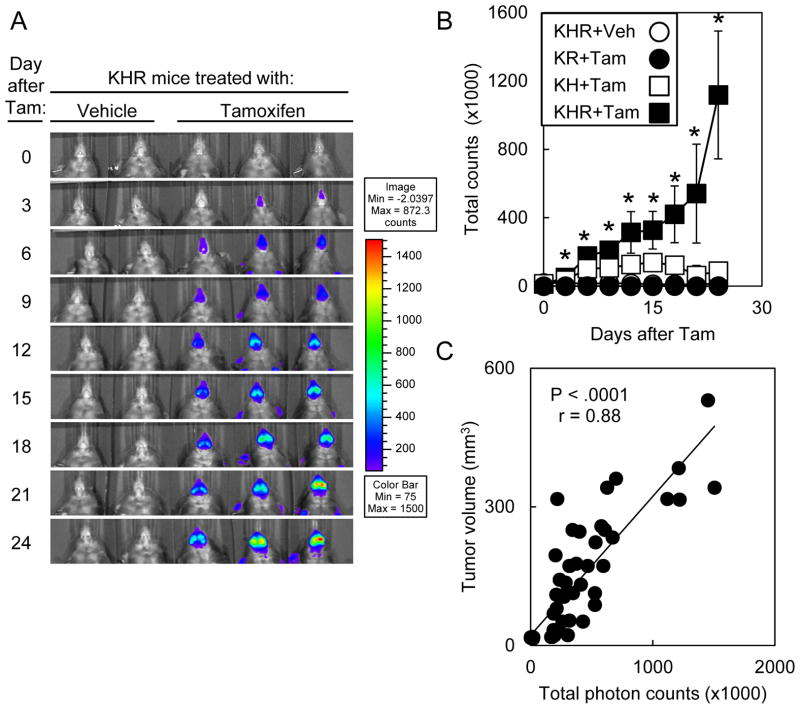
We then assessed the kinetics of bioluminescence after tamoxifen treatment (Fig 4A). Within 3d of initiating Tam treatment, mice had increased bioluminescence compared to untreated mice. By 24d after Tam treatment, KHR mice had 65.7-fold higher bioluminescent signal compared to untreated controls (Fig. 4B). KHR mice that developed oral tumors had progressively increasing oral bioluminescence that correlated with tumor growth (P < .0001, r = 0.88; Fig. 4C). By contrast, bioluminescence around the oral cavity plateaued in KH mice within 6 days at 7.4-fold above background. KR mice treated with Tam developed oral tumors that did not possess bioluminescence signal above background.
Figure 4. The increase in bioluminescence correlated with tumor growth in KHR mice.
(A) Time course for bioluminescence of KHR mice treated with vehicle or tamoxifen. Mice were imaged every 3d after the initiation of treatment. (B) Kinetics of bioluminescence in KHR (n=7), KR (n=2) and KH (n=4) mice treated with Tam or KHR mice (n=5) treated with vehicle. Significant differences between vehicle-treated or Tam-treated KHR mice were denoted by * for P < .05. Error bars represent ± 1 standard deviation. (C) Correlation of tumor volume with bioluminescence.
In vivo bioluminescent monitoring of response to tumoricidal agents
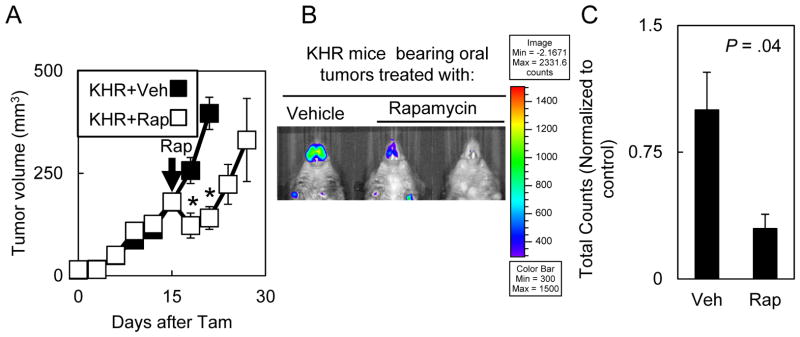
Since bioluminescent signal correlated with tumor volume, we tested the extent to which bioluminescent signal in autochthonous tumors would enable monitoring response to agents that impacted tumor growth (9). Rapamycin has been shown to impact the growth in autochthonous oral tumor models driven by a mutant KRAS, in transplanted head and neck squamous cell carcinoma models and in clinical trials. 14d after Tam treatment, we treated KHR mice with rapamycin for 3 days and monitored tumor growth. Compared to the vehicle treated controls, mice treated with rapamycin had a transient decrease in tumor growth with regrowth after rapamycin was discontinued (Fig. 5A). Compared to untreated tumors, tumors treated with rapamycin displayed 3.3-fold decreased bioluminescent signal at the completion of treatment (Fig. 5B&C).
Figure 5. Bioluminescence tracked oral tumor response to small molecule therapy.
(A) Oral tumors in Tam treated KHR mice regressed after rapamycin treatment. Mice bearing 14d old oral tumors were treated with 4 mg/kg of rapamycin for 3d (n=3) or vehicle (n=5) and tumor growth was monitored every three days. Similar results were observed in two independent experiments. Significant differences between vehicle-treated or rapamycin-treated KHR mice were denoted by * for P < .05. Error bars represent ± 1 standard deviation. Arrow indicates rapamycin (Rap) treatment. (B) 5d after rapamycin treatment, oral tumors displayed lower bioluminescence levels. (C) Quantitation of bioluminescence in KHR mice bearing oral tumors treated with rapamycin (n=3) or vehicle (n=3).
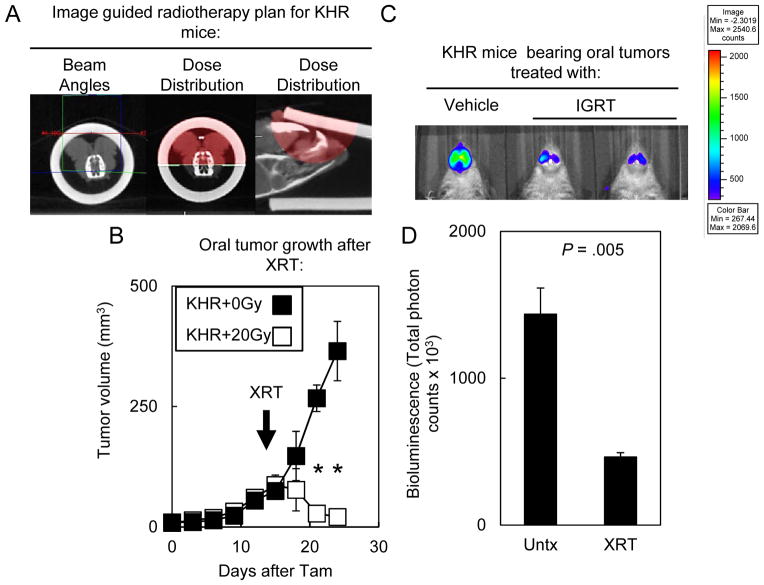
Since oral tumors in KHR mice possessed decreased bioluminescence that correlated with rapamycin treatment, we next tested the extent to which IGRT impacted KHR tumor growth and bioluminescence (Fig. 6). IGRT is rapidly being implemented in the clinic to treat HNSCC patients given the increased ability to target tumors (23). As in patients, cone beam CT images were obtained for each tumor and the isocenter was placed according to the tumor extent (Fig 6A). We developed individual radiotherapy plans using opposed lateral radiation beams to treat mice to 20 Gy. 14d after Tam treatment, mice were irradiated with 20 Gy and tumors regressed (Fig. 6B) whereas unirradiated tumors grew progressively. The final oral tumor volume was significantly smaller for irradiated KHR mice (31.63 ± 22.03 mm3; n=9) compared to unirradiated mice (414.01 ± 80.96 mm3; n=10; P = 4.96 × 10−9). 6d after radiotherapy, irradiated tumors had 3-fold less bioluminescence compared to non-irradiated controls consistent with tumor regression. Therefore, bioluminescent signal was a surrogate for response to different targeted therapies in autochthonous oral tumors.
Figure 6. IGRT caused tumor regression and decreased bioluminescence.
(A) IGRT planning for KHR mice bearing oral tumors. First, a cone beam CT scan was obtained. Then radiation beam angles were designed in order to generate a dosimetry plan. Red dose cloud represented the 100% isodose cloud. (B) 14d old primary tumors regressed after treatment with 20 Gy. Error bars represent ± 1 standard deviation. Results were representative of three similar experiments using 2–5 mice per group. Arrow indicates IGRT (XRT) treatment. Significant differences between unirradiated or irradiated KHR mice were denoted by * for P < .001. (C) 6d after irradiation, oral tumors displayed lower bioluminescence levels. (D) Quantitation of bioluminescence in KHR mice bearing oral tumors receiving 20 Gy (n=3) or non-irradiated mice (n=3). Results were representative of 2 separate experiments.
Discussion
Here, we developed a preclinical model to mimic the development and treatment of autochthonous HPV-positive oral tumors. Similar to patients who are first infected with HPV as young adults, we generated an inducible HPV oral tumor model to control HPV oncogene expression in a spatio-temporal manner. Furthermore, compared to previous models, this bioluminescent signal, linked to HPV oncogene expression, provided a non-invasive method to monitor the growth of intraoral tumors that were otherwise difficult to assess. Finally, we used this model to track responses to IGRT and small molecules, two interventions currently being used in the clinic. Therefore, we intend our novel model to study how HPV oncogenes alone or in cooperation with other genotypes impact tumor growth and response to therapy.
We recognize the limitations for our model to accurately reflect the clinical scenario of HNSCC patients. First, mice developing HPV-positive tumors also required mutant KRAS for tumor development. While mutations in the ras family account for approximately 5% of HNSCCs (1, 2), several groups have shown that the ras-MAPK pathway was aberrantly activated in HNSCCs as well as in other HPV-positive tumors (24–27). Second, HPV oncogene expression was driven by a CMV promoter, which may express oncogenes at artificial levels that do not reflect human HPV-positive tumors. Nevertheless, most, if not all, HPV-positive cancers have viral sequences that non-specifically integrated throughout the host genome likely resulting in a range of expression levels (28). In addition, HPV oncogene expression accelerated oral papilloma growth but we did not find any obvious invasive cancer. Still, given the accelerated tumor growth, our model may not have had sufficient time to develop invasive disease. Finally, HPV-positive mice developed oral tumors and not oropharyngeal cancers that are seen in HNSCC patients. Still, these oral tumors occurred at murine anatomic sites with epithelial transitional zones that are classically affected by HPV oncogenes (29).
As in previous models (8, 9), we used a tamoxifen-regulated Cre recombinase expressed in the basal epithelial layer in order to induce HPV-positive oral tumors. Previous models of autochthonous oral tumors have relied on chemical carcinogens (6) or genetically engineered transgenes (8, 9, 13). These genetically defined models expressed mutant KRAS and/or mutant p53 as well as deleted NOTCH1(12) and/or TGFβR1 (11). In addition, mouse models with inducible promoters have been used to drive oncogenes in the basal epithelial layer resulting in squamous tumors (30, 31). Finally, other groups have directly fused oncogenes to hormonal receptors that spatially sequesters oncogene activity within the cell (32). Upon treatment with estrogen or other ligands, these mice developed squamous papillomas and invasive cancers. However, these inducible promoters and oncogene fusion proteins act only transiently while the ligand is present which may impact tumorigenesis. Therefore, in our model, Cre mediated recombination enabled sustained tissue specific expression of HPV-oncogenes.
In our study, HPV oncogene expression had functional consequences as HPV-positive oral tumors grew faster than HPV-negative tumors and gained expression of MCM7, a known HPV biomarker. These results are consistent with previous studies demonstrating that the ras-MAPK pathway cooperated with HPV oncogenes in vitro and in vivo to increase tumor aggressiveness through mechanisms such as dysregulation of the cell cycle and/or cell invasiveness (29, 33). As the cooperation of HPV and Ras oncogenes have been widely studied, our model will enable us to further examine how HPV oncogenes altered the biology of oral tumors.
Our system mimics the HPV’s life cycle where HPV oncogenes expression was induced in the basal epithelial layer and continued as cells differentiated into the suprabasal layers. By contrast, previous mouse models constitutively expressed HPV oncogenes using tissue specific promoters including Keratin 10 and Keratin 14 that restricted oncogene expression based on the differentiation state of the cell (34–37). In addition, the induction of these oncogenes may minimize immune tolerance that occurs with constitutively expressed HPV antigens in order to better mimic immune responses to exogenous viral proteins (38). Thus, our inducible model may more physiologically reflect viral oncogene expression in order to enhance our understanding of HPV oncogenesis and response to therapy.
Here, we applied a bioluminescent signal to monitor the development of autochthonous HPV-positive tumors. Bioluminescent systems have overcome the difficulties in monitoring the growth of tumors transplanted in orthotopic sites such as the oral cavity which is difficult to assess (39, 40). However, orthotopic transplants were often studied in immunodeficient hosts that may not recapitulate the heterogeneous tumor biology seen in primary tumors (18). Other groups have adapted in vivo imaging technologies to monitor the development of autochthonous tumors including prostate (41, 42), pancreas (42), lymphoma (43) and others. However, in the majority of these models, the reporter transgenes were not linked to initiating oncogenic events and, therefore, it remained unclear whether regional differences existed between oncogene expression and reporter activity. In addition, others have linked reporter genes to tissue specific promoters such as prostate specific antigen and CD19 (43, 44) that are expressed in both malignant and non-malignant parenchyma. Yet, these tissue specific promoters may not be active in poorly differentiated tumors and, therefore, may not adequately track tumor growth (44). Thus, genetically linking oncogene expression to reporter activity more faithfully reflects tumor growth.
Finally, we have used our novel autochthonous HPV tumor model to monitor responses to IGRT and small molecules that are currently used in the clinic. While previous studies demonstrated that rapamycin prevented the outgrowth of autochthonous HPV-negative oral tumors (9), we observed that that rapamycin also caused regression of HPV-positive tumors. Since rapamycin inhibits mTOR, our results suggest that the mTOR/PI3-kinase pathway was active in our tumor model are are consistent with HNSCC sequencing data describing mutations in PIK3CA and PTEN leading to the activation of the mTOR pathway (1, 2, 45). As the PI3-kinase pathway is often mutated in HNSCCs, we are applying forward and reverse genetic approaches to study the mTOR/PI3-kinase pathway in our HPV-mouse model (M.T.S., unpublished observations). Paralleling the clinical setting, IGRT involves tumor localization with a cone beam CT, real time dosimetry planning and subsequent dose delivery (46, 47). Recently, several groups have treated autochthonous sarcomas and non-small cell lung cancers as well as normal tissues (8, 48–50). However, irradiation of these tumors at most resulted in growth arrest while, in our model, we observed tumor regression. While previous studies treated the head and neck region with low doses of radiation, we were able to deliver therapeutic doses of radiation using IGRT. Furthermore, IGRT will enable us to test how the tumor genotype impacts responses to biologically relevant doses delivered using different fractionation schemes (15). Thus, IGRT will more accurately target autochthonous HPV-positive tumors and enable a better understanding for how the tumor genotype impacts the radiation response.
In conclusion, we have developed a novel model to monitor the growth of HPV-positive oral tumors and their response to clinically relevant treatments. This model induced HPV oncogenes in the basal epithelial layer mimicking the natural history of this disease. Furthermore, the bioluminescent signal from these oral tumors acted as a surrogate for tumor growth and response to therapy. Finally, we have validated this model to monitor responses to small molecules and to IGRT, two modalities which are currently being used in patients. We anticipate this system to serve as a platform to better test preclinical concepts in vivo as well as to address underlying mechanisms for how the genotype of autochthonous tumors impacts responses to existing and novel therapies.
Supplementary Material
Acknowledgments
Funding: Burroughs Wellcome Career Award for Medical Scientists; Fanconi Anemia Research Fund.
M.T.S. was supported by the Burroughs Wellcome Career Award for Medical Scientists and the Fanconi Anemia Research Fund. We thank Dr. Linda Degenstein and the University of Chicago Transgenic Core Facility for expert help in producing the iHPV transgenic mouse; the University of Chicago Cytometry Core Facility; Dr. Lara Leoni and the University of Chicago Optical Imaging Facility for expert advice and assistance; Terry Li and Dr. Mark Lingen of the University of Chicago Human Tissue Resource Center for expert assistance with immunohistochemistry. Acquisition of the image-guided animal irradiator was supported by NIH Shared Instrumentation Grant 1S10RR026747-01.
Footnotes
Conflicts of Interest: None to disclose
References
- 1.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanojia D, Vaidya MM. 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol. 2006;42:655–67. doi: 10.1016/j.oraloncology.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Shin MK, Pitot HC, Lambert PF. Pocket proteins suppress head and neck cancer. Cancer Res. 72:1280–9. doi: 10.1158/0008-5472.CAN-11-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caulin C, Nguyen T, Longley MA, Zhou Z, Wang XJ, Roop DR. Inducible activation of oncogenic K-ras results in tumor formation in the oral cavity. Cancer Res. 2004;64:5054–8. doi: 10.1158/0008-5472.CAN-04-1488. [DOI] [PubMed] [Google Scholar]
- 9.Raimondi AR, Molinolo A, Gutkind JS. Rapamycin prevents early onset of tumorigenesis in an oral-specific K-ras and p53 two-hit carcinogenesis model. Cancer Res. 2009;69:4159–66. doi: 10.1158/0008-5472.CAN-08-4645. [DOI] [PubMed] [Google Scholar]
- 10.Bian Y, Hall B, Sun ZJ, Molinolo A, Chen W, Gutkind JS, et al. Loss of TGF-beta signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 31:3322–32. doi: 10.1038/onc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 13.Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, et al. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest. 2007;117:1893–901. doi: 10.1172/JCI31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 10:550–60. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 15.Kimple RJ, Harari PM, Torres AD, Yang RZ, Soriano BJ, Yu M, et al. Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clin Cancer Res. 19:855–64. doi: 10.1158/1078-0432.CCR-12-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaromina A, Kroeber T, Meinzer A, Boeke S, Thames H, Baumann M, et al. Exploratory study of the prognostic value of microenvironmental parameters during fractionated irradiation in human squamous cell carcinoma xenografts. Int J Radiat Oncol Biol Phys. 80:1205–13. doi: 10.1016/j.ijrobp.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Bray D, Yu SZ, Koprowski H, 2nd, Rhee J, Kumar S, Pericle F, et al. Combination nonviral interleukin 2 gene therapy and external-beam radiation therapy for head and neck cancer. Arch Otolaryngol Head Neck Surg. 2003;129:618–22. doi: 10.1001/archotol.129.6.618. [DOI] [PubMed] [Google Scholar]
- 18.Spiotto MT, Banh A, Papandreou I, Cao H, Galvez MG, Gurtner GC, et al. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 70:78–88. doi: 10.1158/0008-5472.CAN-09-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strati K, Pitot HC, Lambert PF. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A. 2006;103:14152–7. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–21. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–6. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu KP, Le QT. Intensity-modulated and image-guided radiation therapy for head and neck cancers. Front Radiat Ther Oncol. 43:217–54. doi: 10.1159/000322437. [DOI] [PubMed] [Google Scholar]
- 24.Albanell J, Codony-Servat J, Rojo F, Del Campo JM, Sauleda S, Anido J, et al. Activated extracellular signal-regulated kinases: association with epidermal growth factor receptor/transforming growth factor alpha expression in head and neck squamous carcinoma and inhibition by anti-epidermal growth factor receptor treatments. Cancer Res. 2001;61:6500–10. [PubMed] [Google Scholar]
- 25.Mishima K, Yamada E, Masui K, Shimokawara T, Takayama K, Sugimura M, et al. Overexpression of the ERK/MAP kinases in oral squamous cell carcinoma. Mod Pathol. 1998;11:886–91. [PubMed] [Google Scholar]
- 26.Branca M, Ciotti M, Santini D, Bonito LD, Benedetto A, Giorgi C, et al. Activation of the ERK/MAP kinase pathway in cervical intraepithelial neoplasia is related to grade of the lesion but not to high-risk human papillomavirus, virus clearance, or prognosis in cervical cancer. Am J Clin Pathol. 2004;122:902–11. doi: 10.1309/VQXF-T880-JXC7-QD2W. [DOI] [PubMed] [Google Scholar]
- 27.Landro ME, Dalbert D, Picconi MA, Cuneo N, Gonzalez J, Vornetti S, et al. Human papillomavirus and mutated H-ras oncogene in cervical carcinomas and pathological negative pelvic lymph nodes: a retrospective follow-up. J Med Virol. 2008;80:694–701. doi: 10.1002/jmv.21076. [DOI] [PubMed] [Google Scholar]
- 28.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–84. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber K, Cannon RE, Karrison T, Beck-Engeser G, Huo D, Tennant RW, et al. Strong synergy between mutant ras and HPV16 E6/E7 in the development of primary tumors. Oncogene. 2004;23:3972–9. doi: 10.1038/sj.onc.1207507. [DOI] [PubMed] [Google Scholar]
- 30.Jabbar SF, Abrams L, Glick A, Lambert PF. Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res. 2009;69:4407–14. doi: 10.1158/0008-5472.CAN-09-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–94. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 32.Tarutani M, Cai T, Dajee M, Khavari PA. Inducible activation of Ras and Raf in adult epidermis. Cancer Res. 2003;63:319–23. [PubMed] [Google Scholar]
- 33.Yoshida S, Kajitani N, Satsuka A, Nakamura H, Sakai H. Ras modifies proliferation and invasiveness of cells expressing human papillomavirus oncoproteins. J Virol. 2008;82:8820–7. doi: 10.1128/JVI.02363-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbeit JM, Munger K, Howley PM, Hanahan D. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J Virol. 1994;68:4358–68. doi: 10.1128/jvi.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carraresi L, Tripodi SA, Mulder LC, Bertini S, Nuti S, Schuerfeld K, et al. Thymic hyperplasia and lung carcinomas in a line of mice transgenic for keratin 5-driven HPV16 E6/E7 oncogenes. Oncogene. 2001;20:8148–53. doi: 10.1038/sj.onc.1205007. [DOI] [PubMed] [Google Scholar]
- 36.Auewarakul P, Gissmann L, Cid-Arregui A. Targeted expression of the E6 and E7 oncogenes of human papillomavirus type 16 in the epidermis of transgenic mice elicits generalized epidermal hyperplasia involving autocrine factors. Mol Cell Biol. 1994;14:8250–8. doi: 10.1128/mcb.14.12.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert PF, Pan H, Pitot HC, Liem A, Jackson M, Griep AE. Epidermal cancer associated with expression of human papillomavirus type 16 E6 and E7 oncogenes in the skin of transgenic mice. Proc Natl Acad Sci U S A. 1993;90:5583–7. doi: 10.1073/pnas.90.12.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doan T, Herd K, Street M, Bryson G, Fernando G, Lambert P, et al. Human papillomavirus type 16 E7 oncoprotein expressed in peripheral epithelium tolerizes E7-directed cytotoxic T-lymphocyte precursors restricted through human (and mouse) major histocompatibility complex class I alleles. J Virol. 1999;73:6166–70. doi: 10.1128/jvi.73.7.6166-6170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henson B, Li F, Coatney DD, Carey TE, Mitra RS, Kirkwood KL, et al. An orthotopic floor-of-mouth model for locoregional growth and spread of human squamous cell carcinoma. J Oral Pathol Med. 2007;36:363–70. doi: 10.1111/j.1600-0714.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim SA, Kim YC, Kim SW, Lee SH, Min JJ, Ahn SG, et al. Antitumor activity of novel indirubin derivatives in rat tumor model. Clin Cancer Res. 2007;13:253–9. doi: 10.1158/1078-0432.CCR-06-1154. [DOI] [PubMed] [Google Scholar]
- 41.Liao CP, Zhong C, Saribekyan G, Bading J, Park R, Conti PS, et al. Mouse models of prostate adenocarcinoma with the capacity to monitor spontaneous carcinogenesis by bioluminescence or fluorescence. Cancer Res. 2007;67:7525–33. doi: 10.1158/0008-5472.CAN-07-0668. [DOI] [PubMed] [Google Scholar]
- 42.Zhang N, Lyons S, Lim E, Lassota P. A spontaneous acinar cell carcinoma model for monitoring progression of pancreatic lesions and response to treatment through noninvasive bioluminescence imaging. Clin Cancer Res. 2009;15:4915–24. doi: 10.1158/1078-0432.CCR-08-2256. [DOI] [PubMed] [Google Scholar]
- 43.Scotto L, Kruithof-de Julio M, Paoluzzi L, Kalac M, Marchi E, Buitrago JB, et al. Development and characterization of a novel CD19CherryLuciferase (CD19CL) transgenic mouse for the preclinical study of B-cell lymphomas. Clin Cancer Res. 18:3803–11. doi: 10.1158/1078-0432.CCR-11-2588. [DOI] [PubMed] [Google Scholar]
- 44.Hsieh CL, Xie Z, Yu J, Martin WD, Datta MW, Wu GJ, et al. Non-invasive bioluminescent detection of prostate cancer growth and metastasis in a bigenic transgenic mouse model. Prostate. 2007;67:685–91. doi: 10.1002/pros.20510. [DOI] [PubMed] [Google Scholar]
- 45.Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW, Gutkind JS. PTEN deficiency contributes to the development and progression of head and neck cancer. Neoplasia. 2013;15:461–71. doi: 10.1593/neo.121024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhaegen F, Granton P, Tryggestad E. Small animal radiotherapy research platforms. Phys Med Biol. 56:R55–83. doi: 10.1088/0031-9155/56/12/R01. [DOI] [PubMed] [Google Scholar]
- 47.Wong J, Armour E, Kazanzides P, Iordachita I, Tryggestad E, Deng H, et al. High-resolution, small animal radiation research platform with x-ray tomographic guidance capabilities. Int J Radiat Oncol Biol Phys. 2008;71:1591–9. doi: 10.1016/j.ijrobp.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuneo KC, Mito JK, Javid MP, Ferrer JM, Kim Y, Lee WD, et al. Imaging Primary Mouse Sarcomas After Radiation Therapy Using Cathepsin-Activatable Fluorescent Imaging Agents. Int J Radiat Oncol Biol Phys. doi: 10.1016/j.ijrobp.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moding EJ, Clark DP, Qi Y, Li Y, Ma Y, Ghaghada K, et al. Dual-energy micro-computed tomography imaging of radiation-induced vascular changes in primary mouse sarcomas. Int J Radiat Oncol Biol Phys. 85:1353–9. doi: 10.1016/j.ijrobp.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez BA, Ghafoori AP, Lee CL, Johnston SM, Li Y, Moroshek JG, et al. Assessing the radiation response of lung cancer with different gene mutations using genetically engineered mice. Front Oncol. 3:72. doi: 10.3389/fonc.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.