Abstract
Chronic exposure to arsenic in drinking water causes cancer and non-cancer diseases. However, mechanisms for chronic arsenic-induced pathogeneis, especially in response to lower exposure levels, are unclear. In addition, the importance of health impacts from xeniobiotic-promoted microbiome changes is just being realized and effects of arsenic on the microbiome with relation to disease promotion are unknown. To investigate impact of arsenic exposure on both microbiome and host metabolism, the stucture and composition of colonic microbiota, their metabolic phenotype, and host tissue and plasma metabolite levels were compared in mice exposed for 2, 5, or 10 weeks to 0, 10 (low) or 250 (high) ppb arsenite (As(III)). Genotyping of colonic bacteria revealed time and arsenic concentration dependent shifts in community composition, particularly the Bacteroidetes and Firmicutes, relative to those seen in the time-matched controls. Arsenic-induced erosion of bacterial biofilms adjacent to the mucosal lining and changes in the diversity and abundance of morphologically distinct species indicated changes in microbial community structure. Bacterical spores increased in abundance and intracellular inclusions decreased with high dose arsenic. Interestingly, expression of arsenate reductase (arsA) and the As(III) exporter arsB, remained unchanged, while the dissimilatory nitrite reductase (nrfA) gene expression increased. In keeping with the change in nitrogen metabolism, colonic and liver nitrite and nitrate levels and ratios changed with time. In addition, there was a concomitant increase in pathogenic arginine metabolites in the mouse circulation. These data suggest that arsenic exposure impacts the microbiome and microbiome/host nitrogen metabolism to support disease enhancing pathogenic phenotypes.
Keywords: arsenic, colon microbiome, nitrite/nitrate, arginine, Firmicutes, Bacteroidetes
Introduction
Exposure to arsenic in drinking water is a major human health problem that causes significant disease risk in many millions of individuals worldwide. In addition to causing a number of cancers, arsenic exposure is implicated in increased risk for cardiovascular, pulmonary, liver, and metabolic disease (Parvez et al., 2010; Chen et al., 2011; Mazumder and Dasgupta, 2011; Maull et al., 2012; Moon et al., 2012; Osorio-Yanez et al., 2013). It is important to distinguish between the acute toxicities and disease promotion caused by chronic arsenic exposures when considering its pathogenic effects on metabolism. High dose arsenic and acute toxic exposure directly damage mitochondria in many cell types to impair energy metabolism and cause cell necrosis and death. In the GI, these very high exposures to arsenic cause hyperemia of the gastric mucosa, hemorrhagic injury, and gastritis. In contrast, low to moderate levels of arsenic alter cell signaling to regulate cell differentiation, phenotype and function (States et al., 2011).
Chronic exposures to environmental levels of arsenic impact vascular cell signaling and produce epigenetic inflammatory responses that promote vascular and metabolic diseases (States et al., 2009; Maull et al., 2012; Moon et al., 2012; Wu et al., 2012; Moon et al., 2013; Osorio-Yanez et al., 2013; Kuo et al., 2015). The etiologies of these diseases involve dysfunctional metabolism, altered lipid deposition, and chronic inflammation that is similar to mechanisms suggested for disease progression caused by chronic change in the microbiome (Caesar et al., 2010; Wang et al., 2011; Hazen and Smith, 2012; Lee and Hase, 2014). However, little is known of the impacts of environmental arsenic exposures on GI microbiome composition and community structure that would cause pathogenic change in the ecology of a microbiotic niche or exchange of bioactive nutrients between host and microbiome. Over the last two decades the microbial metabolism of arsenic and its biogeochemical cycle have been elucidated (Oremland and Stolz, 2003; Stolz et al., 2010). There are at least nine ars genes known to be involved in resistance including regulatory (e.g., arsR), export (e.g., arsA, arsB, acr3), and reduction (arsC) (Rosen, 2002; Stolz et al., 2006). Oxyanions of arsenic can also be used as an electron donor or acceptor in anaerobic respiration (Stolz et al., 2006). Thus one might expect the selection of arsenic resistant bacteria and the up regulation of arsenite oxidase (Aio) and resistance genes (e.g., arsA, arsB) when the gut community is exposed directly to As(III).
The gastrointestinal microbiota is essential for the health and metabolism in their host systems including humans (Leser and Molbak, 2009). Estimates of the number of commensal bacteria in the adult colon range from 1013–1014 with possibly 500 to 1,000 different species represented. Recent studies indicate humans have unique enterotypes with certain dominant species (Arumugam et al., 2011; Walter and Ley, 2011; Wu et al., 2011; Human Microbiome Project, 2012). The mucosal composition dictates the microenvironment inhabited by the microbiota and is influenced by the microbiota inhabiting the niche (Johansson et al., 2010; Ohland and Macnaughton, 2010). The mucosa of germ free animals is thin and poorly developed (Johansson et al., 2010; Ohland and Macnaughton, 2010). In response to bacteria or bacterial products, it rapidly generates a hospitable niche that harbors bacterial communities in a loose outer layer, as well as a tight layer that excludes bacteria from the colonic epithelium (Johansson et al., 2010; Ohland and Macnaughton, 2010). It is evident that the bacteria in the mucus layers (or their products) signal to regulate epithelial cell phenotype and function.
The commensal bacteria metabolize the complex structural carbohydrate glycans in the mucus to provide short chain fatty acid nutrients (e.g. butyrate) to the epithelial cells (Cherbuy et al., 1995; Cherbuy et al., 2010; Johansson et al. 2010; Lee and Hase 2014). An individual’s microbiome is fairly stable over time, but the community composition and structure can be influenced at the extremes of age, by diet, and environmental exposures (Caesar et al., 2010; Carmody and Turnbaugh, 2014; Lee and Hase, 2014; Shreiner et al., 2014). There are also direct relationships between bacterial load, bacterial products (nitrogen metabolites and patern recognition ligands) in the systemic circulation, and inflammatory state of the liver and cardiovascular system (Wang et al., 2011; Corbitt et al., 2013; Tang and Hazen, 2014). In addtion, the intestinal microbiota is also involved in detoxification and biotransformation of toxic metals, modulation of host metabolic phenotypes, metabolism of otherwise indigestible dietary compounds and metabolism of xenobiotics that can have profound effect on host health (Diaz-Bone and Van de Wiele, 2010; Lundberg and Weitzberg, 2012). Thus identifying factors that impact the indigenous microbiota is key to understanding the dynamic interactions between the microbial community and health of the host.
Bacteria metabolize oxyanions of nitrogen in both assimilatory and dissimilatory processes (Sparacino-Watkins et al., 2014). Assimilatory nitrate reduction to ammonia involves two enzymes, nitrate reductase (Nar or Nap) and nitrite reductase (Nas), the latter a siro-heme containing enzyme whose end product is ammonia. Dissimilatory nitrate reduction can produce either dinitrogen through a series of steps (e.g., dinitrification), or ammonia (DNRA). Nitrite, nitric oxide, and nitrous oxide are intermediates in denitrification, each involved unique enzymes (e.g., nitrite reducase, NirS or NirK; nitric oxide reductae, Nor; nitrous oxide reductase, Nos). DNRA metabolism involves only nitrate reductase (Nar or Nap) and a pentaheme nitrite reductase, Nrf (Stolz and Basu, 2002; Sparacino-Watkins et al., 2014). The dissimilatory microbial processes may generate reactive nitrogen species and contribute to the overall nitrogen balance in the body (Wang et al., 2009; Tang and Hazen, 2014).
Although the GI tract is one of the first organs to come in contact with ingested arsenic, the effect of this environmental contaminant on GI tract and its resident bacteria has not been studied to a great extent. Studies have shown that the gut microbiota affect metabolism of orally injested arsenic to both aid in detoxification and elimination from the body (Diaz-Bone and Van de Wiele 2010), as well as in making unique arsenic metabolites available to the host (Kubachka et al., 2009). Recent studies using high level (10 ppm), short time (4 weeks) exposure demonstrated that arsenic promotes compostional change in fecal microbes with commensurate change in bioavalable bacterial metabolites (Lu et al., 2014a). Thus, it is plausible that arsenic exposure through consumption of drinking water could affect the host microbiota but potential linkage to pathogenic change in the host was not made. These studies, as well as many other studies using metagenomic analysis of fecal contents are limited in the ability to truly capture the microbiome community and stuctural change that are relevant to host interactions, since they only measure the microbiota that is excreted and not the bacteria in biofilms or in the outer mucosal layers of the colonic epithelium where exchange of host and bacterial products occurs. Thus, we investigated fecal and gut wall microbiome changes at different segments of the colon to test the hypothesis that exposure to environmentally relevant concentrations of arsenic in drinking water impacts the microbial community of the colon to alter both microbiome and host metabolism. These studies were conducted in the context of previous published studies of arsenic-induced changes in liver and cardiovascular remodeling (Straub et al., 2007b; Straub et al., 2008) and thus both host and microbiota were monitored concurrently in the same animals. In addition, this is the first study to examine arsenic impact on the microbiome in situ, as well as to report the effects of low (10 ppb) and moderate (250 pbb) exposures over time as pathologies develop in the host relative to compositional and stuctural changes in the gut microbiota.
Materials and Methods
Animals and experimental design
Mouse exposures were performed in compliance with the institutional guidelines for animal safety and welfare at the University of Pittsburgh. C57BL/6 Tac male mice (Taconic Farms), aged 6–8 weeks, were housed and maintained as described previously (Straub et al., 2007b; Straub et al., 2008). Standard mouse chow and drinking water containing 0 (control), 10 ppb or 250 ppb of sodium arsenite (arsenic) were fed ad libitum for periods of 2, 5 and 10 weeks. Sodium meta-arsenite (Fisher Scientific) solutions were prepared triweekly using commercial bottled spring water (Straub et al., 2007b; Straub et al., 2008). At the prescribed time, five mice from each group (with the exception of the 2 week control group that had only 4 mice) were euthanized, and their colons were removed with portions flash frozen for DNA sequencing, frozen in RNAlater® (Life Technologies) and and stored at −80°C for mRNA analysis or fixed in PBS-buffered glutaraldehyde for electron microscopy.
Transmission electron microscopy (TEM)
Samples for electron microscopy were prepared as previously described (Stolz et al., 1999). Whole colons (including the gut contents) were disected separately into proximal, medial, distal, immediately fixed in PBS buffered glutaraldehyde (2.5%), and kept cold and in the dark until processed. The samples were post fixed in osmium tetroxide (1%), rinsed in PBS, and following an ethanol dehydration series (30–100%) and propylene oxide, embedded in Polybed 812 epoxy resin (Polysciences, Warrington, PA). Orientation of the blocks were determined with light microscopy of toluidene stained thick sections observed with a Nikon Mikrophot SA. Thin sections were observed on a JOEL 100CX TEM at 60 kV. Digital images were acquired using a cooled SIA digital camera.
Denaturing gradient gel electrophoresis (DGGE)
Microbial DNA was extracted from each of the 2, 5 and 10 week whole colons (including the gut contents) samples after maceration in liquid nitrogen with a mortar and pestal, using QiAmp DNA stool kit as specified by the manufacturer (Qiagen Inc., Valencia, CA, USA). The V3 region of the16S rRNA genes was amplified from the mice colon microbiota using the primer set HDA1-GC/HDA-2 (Walter et al. 2001). The reaction mixture consisted of 150 ng of genomic DNA, 10 μl of 5X buffer, 25 pmol each of forward and reverse primers, 1 μl dNTPs from a 200 μM stock, 1.25U of GoTaq DNA polymerase (Promega, Madison, WI, USA) and sterile water to a total volume of 50 μl. The touchdown program was used for amplification with annealing temperature decreasing from 60°C to 50°C at a rate of 0.5°C decrease/cycle. The products were electrophoresed on a 40–60% urea-formamide denaturing gradient gel on the D-Code system (BioRad, Hercules, CA, USA). The gel was run at 70V for 990 minutes at 60°C in 1X TAE buffer and stained in a 1: 10,000 dilution of SYBR green. To determine the degree of similarity among samples, dendrograms were constructed by the unweighted pair group method (UPGMA) using the Quantity One software (BioRad, Hercules, CA, USA).
Clone library construction
Near full length 16S rRNA genes were amplified from the microbial community DNA using the universal primers 8F (Edwards et al., 1989) and 1492R (Stackebrandt and Liesack, 1993). The PCR products obtained from each of the sample were ligated into the pCR2.1®-TOPO vector and transformed into OneShot Top10 chemically competent E. coli cells (Invitrogen, Carlsbad, CA, USA). Positive clones were picked randomly from each sample and used to inoculate 5 ml of LB broth containing ampicilllin. Plasmid extraction from the liquid cultures was performed using the Wizard SV DNA column purification kit (Promega, Madison WI), and the inserts sequenced using M13 forward and reverse primers at the Genomics and Proteomics Core Laboratory, University of Pittsburgh, PA using a ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA).
Bacterial DNA Sequence analysis
The sequences were assigned to taxonomic classification using the Ribosomal Database Project II (RDPII) classifier with a confidence threshold of 90% (Wang et al., 2007). Multiple sequence alignment of nearly full length 16S rRNA sequences obtained from the clonal libraries was done using Clustal X (Larkin et al., 2007). Sequences aligned by Clustal X were edited manually using the Seaview program (Gouy et al., 2010) to check for any ambiguity in the aligned sequences. All sequences were checked for possible chimeric artifacts using the Bellerophon (DeSantis et al., 2006), Pintail (Ashelford et al., 2006) and Chimera Slayer programs (Schloss et al., 2009). Significant differences in the composition of control and experimental clone libraries were determined using the Libshuff program of the Mothur software (Schloss et al., 2009). Comparison between libraries at the taxonomic level was also done by the Libcompare tool from RDP II database (Cole et al., 2009).
MiSeq high throughput sequencing
PCR was performed on extracted DNA using the 515F primer and 806R barcoded sequencing primers as previously described (Caporaso et al., 2012; Akyon et al., 2015). After amplification, the PCR products were purified using an Agencourt AMPure Purification Kit (Beckman Coulter, Indianapolis, IN) using a bead ratio of 0.8. PCR product was verified on a 1% (w/v) agarose gel. Purified DNA was quantified on a Qubit (Life Technologies, Carlsbad, CA). DNA was pooled at 2nM, diluted to 10 pM, and spiked with 5% 12.5 pM PhiX control (Illumina, San Diego, CA). Sequencing was performed on the Illumina MiSeq (Illumina, San Diego, CA) with a MiSeq reagent Kit V2 (300 cycles) (Illumina, San Diego, CA). Sequence reads were analyzed using QIIME 1.7.0.29 (Caporaso et al., 2010). First, reads were demultiplexed and quality filtered (split_libraries_fastq.py) to remove reads with an incorrect sample barcode or with a quality score lower than Q20. In order to save on computational requirements and to obtain an even depth of sampling for each sample, 20,000 sequences were randomly taken from each sample for further analysis. Closed reference OTUs were then predicted (pick_closed_reference_otus.py) and assigned taxonomy (assign_taxonomy.py) using the Greengenes 13_8 database (DeSantis et al., 2006). Unweighted UniFrac (Lozupone and Knight, 2005) data was obtained for beta diversity analysis (beta_diversity_through_plots.py). An Analysis of Variance model was run in Minitab statistical software Version 16 (State College, PA). The ANOVA General Linear Model was run with two factors (dose and week) consisting of three levels each (0, 10, and 250 ppb for the dose and 2, 5, and 10 for the week) along with interaction between the two factors with an individual class as the response for taxonomic diversity. Grouping information was obtained using 95% confidence level Tukey comparisons.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from the colon samples with luminal contents using TRIzol® (Life Technologies, Grand Island, NY) according to manufacturer’s instructions. After DNase I treatment, the RNA was reverse transcribed to cDNA using Superscript III first strand synthesis super mix (Invitrogen, Carlsbad, CA, USA) and random hexamers following manufacturer’s instructions. Real-time PCR reactions were performed using SYBR green PCR master mix and carried out on step one real time PCR system (Applied Biosystems, Foster City, CA, USA). Relative quantification of expression levels of the target genes (arsA, arsB and nrfA) relative to the control gene (rpoB) was done by comparative Ct method (ΔΔCt) and differences between treatment groups were compared for significance using ANOVA (p<0.05) followed by Tukey’s correction using Graphpad Prism 5.0 software (Graphpad, SanDiego, CA, USA).
Tissue nitrite and nitrate measurements
Tissue nitrite and nitrate levels were measured as previously described (Curtis et al., 2012; Cronican et al., 2013). Briefly liver and colon (flushed to remove feces) were frozen in liquid nitrogen and stored at −80°C prior to processing to prevent artifactual S-nitrosation. The tissues were thawed, homogenized, and suspended in Krebs-Henseleit buffer at a concentration of 4 mg protein/ml (measured with BCA protein assay kit; Pierce). Homogenates were deoxygenated by passing premade purchased gas mixtures (0%–21% O2, 5% CO2 balanced with N2; Matheson Gas, Pittsburgh, PA) over the homogenate in a closed chamber. Nitrite was measured by triiodide-based reduction and nitrate measured in vanadium chloride based chemiluminescene reactions in a vessel connected inline to a Nitric Oxide Analyzer (Sievers) (Curtis et al., 2012).
Plasma arginine metabolite measurements
Blood was collected by cardiac puncture into chilled tubes containing EDTA immediately after euthanizing the mice. Plasma was processed within 2 hours of blood draw, and stored at −80°C until shipped on dry ice to Dr Hazen’s laboratory for analysis. Arginine and the arginine metabolites ornithine, citrulline, MMA, ADMA, and SDMA were quantified in plasma by stable-isotope-dilution HPLC with online tandem mass spectrometry, as described (Wang et al., 2009). Briefly, [13C6]arginine (10 μmol/L final) was added to the plasma as internal standard and proteins then precipitated by addition of 4 volumes of methanol. Supernatant (20 L) was separated by HPLC and column effluent was introduced into an API 365 triple quadrupole mass spectrometer with Ionics EP 10+ upgrade (Concord). Analyses were performed using electrospray ionization in positive-ion mode with multiple reactions monitoring of parent and characteristic daughter ions specific for components monitored. The transitions monitored were mass-to-charge ratio (m/z) 133.23→70.2 for ornithine; m/z 75.13→70.0 for arginine; m/z 176.13→70.1 for citrulline; m/z 189.33→70.1 for MMA; m/z 203.2→70.3 for SDMA and ADMA; and m/z 1813→74 for [13C6]arginine. The calibration curves for quantification of ornithine, arginine, citrulline, monomethylarginine (MMA), assymetric dimethylarginine (ADMA), and symetric dimethylarginine (SDMA) were prepared by spiking different concentrations of each individual analyte to control plasma. All analytes were baseline resolved and showed unique retention times. A S/N of 3 was used as minimal for limit of detection. Assay performance characteristics included average spike and recovery of 94% (range from 84% to 110%) for all analytes monitored in plasma matrix, and assay precision of 10% across all concentration ranges monitored for all analytes under the assay conditions used. Separate analyses examined the methylation index for arginine (ArgMI), an integrated quantification of products generated from the arginine methylation pathways, and is estimated by the ratio of the known dimethylated arginine posttranslational modifications (ADMA +SDMA) to the immediate mono-methylated precursor, MMA (ie, ArgMI(ADMA+SDMA)/MMA; (Wang et al., 2009)).
Data deposition
The bacterial 16S rRNA gene sequences obtained in this study are deposited in Genbank database with accession numbers HQ681318 - HQ681412 (2 week control), JN012608 - JN012688 (2 week 10 ppb), HQ681413 - HQ681511 (2 week 250 ppb), JN012882 - JN012991 (5 week control), JN012689 - JN012791 (5 week 10 ppb), JN012792 - JN012881 (5 week 250 ppb), JN001204 - JN001304 (10 week control), JN012992 - JN013087 (10 week 10 ppb) and JN013088 - JN013183 (10 week 250 ppb).
Results
In situ community structure
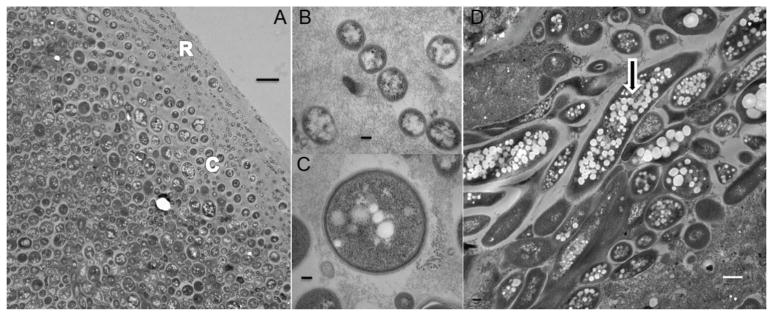
In situ light mircoscopy and TEM was used to examine microbial community structure in the proximal, medial and distal colon of control and arsenic-treated mice. The TEM of control mice revealed that the microbial community of the murine colon exhibits a structure characteristic of biofilm organization (Fig. 1). The different bacterial species appeared to be stratified into distinct populations with the small, 0.3 micron diameter coccoids nearest the epithelial wall, then a distinct layer of larger, 1 micron diameter coccoids both embedded in epithelial mucosa (Fig. 1A–C). The populations became a more heterogenous mixture of cocci, rods, and larger filamentous bacteria through the transition into the lumen interior (Fig. 1C). Many of the bacterial cells present were packed with intracellular inclusions suggesting they were actively sequestering stores of carbon and energy presumably in the form of polyhydroxyalkanoates (Fig. 1).
Figure 1.
Microbial community structure in the mouse colon as revealed by TEM. All sections shown here were taken from the medial colon from a 5 week control (unexposed) mouse. A) The transition from very small (0.3 μM) Gram positive coccoids (R) in the mucosa closest to the epithelial layer to larger (1 μM) coccoids (C). Bar 2 μM. B) the small coccus at higher magnification. Bar 100 nm. C) the larger coccus at higher magnification. Bars in both B and C are 100 nm. D) Gram negative filamentous bacteria with intracellular inclusions (arrow). Bar 500 nm.
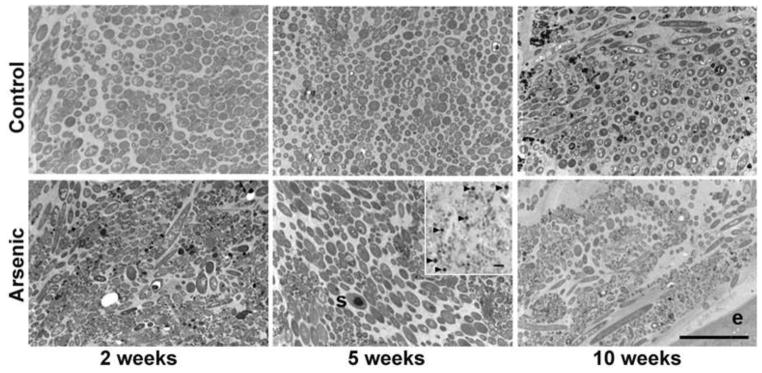
TEM of the medial colon from 2, 5, and 10 week arsenic exposed mice revealed progressive changes in microbial community structure (Fig. 2). The biofilm structure adjacent to the gut mucosal layer degraded with time and the population of small coccoids that formed the distinct microbial layer closest to the epithelium was conspicuously absent in 5 and 10 week mice. There was also evidence that the physiological state of those organisms still present had changed, as more cells were seen that lacked intracellular inclusions indicating a shift from stationary to growth phase. Bacterial spores were particularly abundant in 5 week arsenic exposed mice (Fig. 2) indicating that some of the Firmicute populations present were inactive. It also suggested that these sporulated Firmicutes would be more readily eliminated as they are no longer able to maintain their position in the gut mucosa and could present a disproportunate population in the feces. The distinct community organization (biofilm structure) was no longer evident in the 10 week arsenic-treated mice (Fig. 2). Examination by light and TEM of samples collected at the proximal, medial, and distal portions of the colon indicated that these changes in microbial community structure correlated with arsenic treatment and not location in the colon (Fig S1). The changes observed suggest a shift in microbial metabolism (change in microbial growth state), disruption of cell/cell communication (loss of biofilm organization) and changes in nutrient processing (a result of the loss of active Firmicutes).
Figure 2.
Arsenic induced changes in microbial community structure as observed by TEM at 2, 5, and 10 weeks (top row – control, bottom row - arsenic exposed), (s) spore. Insert shows abundance of spores (arrows) in 5 week arsenic exposed mice, light micrograph. The absence of the small coccoids and presence of filamentous bacteria in the mucosa closest to the epithelium is most pronounced in the 10 week arsenic exposed mice. Bars 5 μm.
Changes in microbial community composition
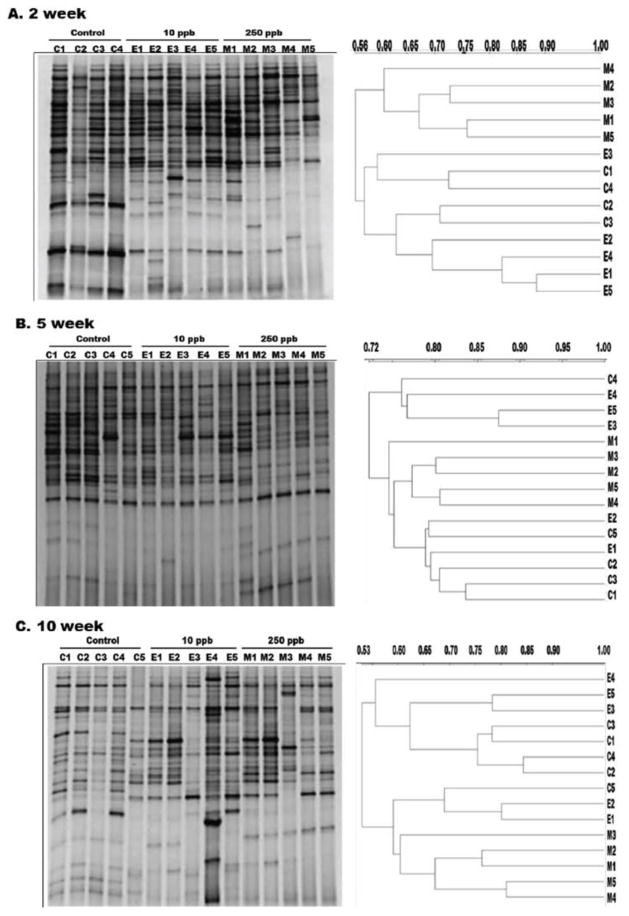
Initially, the changes in community structure were determined using DGGE (Fig. 3). A 177 bp long fragment of 16S rDNA was amplified from 2, 5 and 10 week mice colon community DNA. Alterations in DGGE profiles of arsenic treated mice were observed when compared to control mice. However these changes were more consistent and dramatic in samples exposed to 250 ppb as compared to 10 ppb exposed mice. The total number of bands did not vary significantly between control and arsenic exposed groups. This suggests that arsenic exposure did not have a marked effect on the overall species diversity of the community. Analysis of the controls also revealed a shift in the microbial profile over the course of the experiment indicating that the microbial community of the mouse was naturally changing with time. Dendrogram analysis of the DGGE gels indicated that even at 2 weeks, the 250 ppb exposed mice clustered into separate clades (Fig. 3). Although visible differences in band patterns were seen between control and 10 ppb arsenic exposed groups at 2, 5 and 10 weeks, they consistently clustered together. These results indicate that the variations observed in 10 ppb group were subtle and difficult to attribute to arsenic exposure alone.
Figure 3.
DGGE with dendrogram analysis of microbial diversity in the colon of control (C1–C5), 10ppb (E1–E5) and 250ppb (M1–M5) arsenic exposed mice at A) 2 week, B) 5 week, and C) 10 week. Scale bar on dendrograms represents similarity index values.
Dynamics of Firmicute and Bacteroidete populations
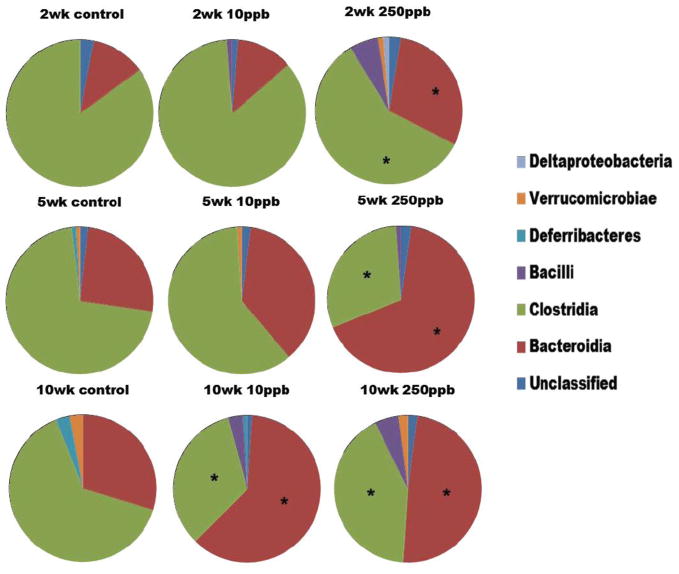
A total of 947 clones were sequenced and 123 sequences were found to be chimeras and removed from the analysis. Based on RDP II database results, the mice colon microbial community contained sequences from five bacterial phyla. For the combined data set, the majority of the sequences belonged to the Bacteroidetes (36% of the total sequenced population) and Firmicutes (60% of all sequences). Sequences from Deferribacteres, Verrucomicrobia and Proteobacteria were observed in very low number. This is not unexpected as these phyla are only a minor component of the anaerobic gut microbial community (Nava et al. 2011). Only a few sequences (< 1%) were identified as “uncultured bacterial clone” but showed identity with 16S rRNA sequences obtained from other mammalian GI tract microbes. At the Class level, the Firmicutes could be resolved to Bacilli and Clostridia (Fig. 4). At the genus level, 12 different genera were identified, representing only 34.6% of the total sequenced population. The sequences that were not classified to the genus level were classified to the next highest level in the order of family, order, class, phylum or unclassified bacteria. Bacteroidaceae and Porphyromonadaceae were the only families observed in the phylum Bacteroidetes along with some sequences that were identified as unclassified Bacteroidales. Similarly, Lachnospiraceae was the most abundant family in the Firmicutes followed by Ruminococcaceae, Lactobacillaceae, Peptostreptococcaceae and Peptococcaceae. However unclassified Clostridiales (38%) formed a major part of the Firmicute population.
Figure 4.
Diversity of the colon microbiota at the Class level with respect to time and arsenic exposure as determined by 16S rRNA gene clonal libraries. (*) indicates significantly different from control for that particular class (p<0.01).
Changes in the composition of the microbial community were observed over the course the experiment. Compared with the control group, for all time points 10 ppb and 250 ppb arsenic exposed groups had an increase in relative abundance of Bacteroidetes and proportionally fewer Firmicutes at the phylum level of taxonomy. Even at the class level, more Bacteroidia and fewer Clostridia were observed in arsenic exposed groups (Fig. 4). However based on RDPII library comparison results, this change was significant (p<0.01) only for 250 ppb groups at all time points; whereas the change was significant only by 10 weeks for the 10 ppb arsenic-exposed groups. Within the Bacteroidia, the number of Porphyromonadaceae sequences increased with time and in response to arsenic exposure whereas no such trend was observed for Bacteroidaceae (Fig. 4). Conversely, the number of Lachnospiraceae and Ruminococcaceae sequences decreased after 2 and 5 weeks of 250 ppb of arsenic exposure. In addition, Ruminococcaceae decreased significantly after 10 weeks of 10 and 250 ppb of arsenic exposure. Bacilli were generally abundant in arsenic exposed groups when compared to control groups. Paired comparison of the control and arsenic-exposed libraries using Libshuff analysis yielded a p-value of <0.001 after Bonferroni correction, indicating that the libraries are significantly different from each other.
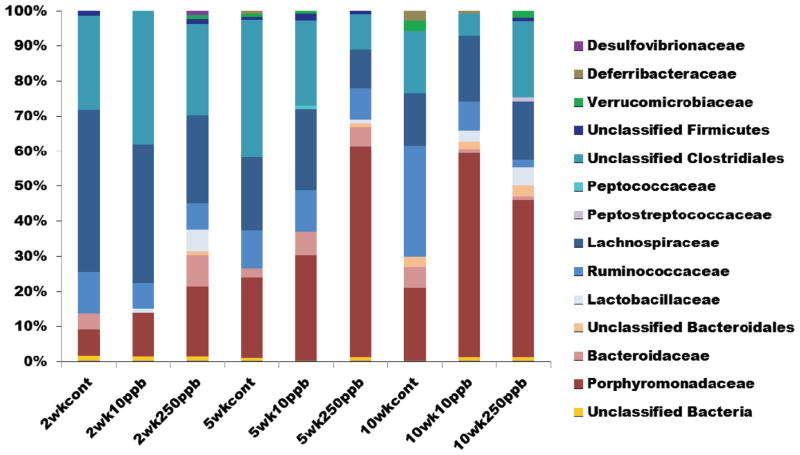
The results of the high throughput 16S rRNA sequencing analysis concurred in general, with the DGGE and clonal library results (Fig. 5). In total, over 3.6 million sequences passed quality filtering. Only one out of the 44 mouse DNA samples analyzed continually failed to produce PCR product (250 ppb, week 10, mouse 3) and thus was eliminated. The number of sequences per sample ranged from 23,000 to 137,000. To obtain an even depth of sampling for each sample, 20,000 sequences were randomly taken from each sample for further analysis. The miseq data set was posted to MGRASTAll, accession number 4654072.3. In summary, all samples were primarily made up of members of the Bacteroidetes and Firmicutes phylum. In addition, there were small percentages of Verrucomicrobia, Tenericutes, Deferribacteres, Cyanobacteria, Proteobacteria, and Actinobacteria. Similar to the DGGE and clonal library results, the Bacteroidetes increased over time while the Firmicutes abundance decreased. However, this was true for both control as well as the arsenic-exposed mice. At the phylum level, there were no significant differences for any of the dose levels at the same time point. However, when the results of dosage versus exposure are compared, only the 10 ppb population showed significant shifts at 5 weeks and 10 weeks compared to 2 weeks.
Figure 5.
Normalized plot of colon microbial diversity at the Family level with respect to time and arsenic exposure as determined by 16S rRNA gene high throughput sequencing.
Analyses at the Class level also indicated increases in bacteroidia and fewer clostridia over time, consistent with the DGGE and clonal library results. This trend, however, could be attributed to difference in arsenic dose or natural changes in the microbiome over time. Nevertheless, the 10ppb mice exposed for 10 weeks did show a significantly higher Bacteroidia population compared to the 10 week control (P=0.039). Again there was a significant community shift between the population for 5 week and 10 week 10ppb exposed mice when compared to the 2 week 10 ppb exposed mice.
At the Family level, there were decreases in Bacteroidaceae, Lachnospiraceae and Ruminococcaceae populations, but no changes in Porphyromonadaceae as had been seen in the clonal library results (Fig 5). However, all of these families appear to be present in much lower quantities in the high throughput sequencing data as compared to the clonal libraries. Instead, most of the sequences were unknown Clostridiales and Bacteroidales. Porphyromonadaceae significantly decrease (P=0.005) in the 10 week, 250 ppb samples compared to the 10 week control. However, this family is present in very low percentages throughout the study.
Unweighted UniFrac analyses was done for all the samples (Fig. S2). In general, the samples tended to group together based on time (e.g., week) rather than dosage. The control and 10 ppb samples from week 10 group together. The 250 ppb samples from week 10 also group together, although separately from the 10 ppb and control groups. All other samples grouped closely together. This suggests that the microbial community does shift but the shift may be more strongly correlated to time than to arsenic exposure.
Arsenic-induced phenotypic changes in microbiotic arsenic resistance and nitrite reductase genes
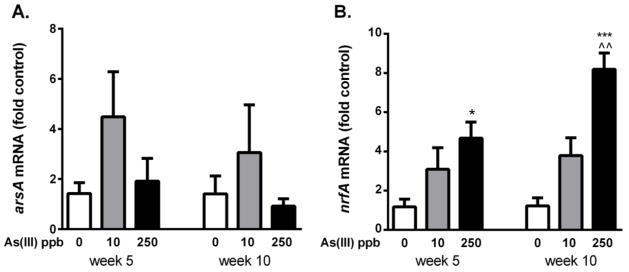
To assess the impact of arsenic exposure on the phenotype of the microbiome, mRNA expression of arsenic resistance genes, arsA and arsB, and the gene encoding the pentaheme nitrite reductase, nrfA, were measured (Fig. 6). First, to select appropriate primers for real time-qPCR quantification, a model bacteria was selected that expressed two arsenic resistance genes (arsA and arsB), a gene coding for nitrate reduction (nrfA) and the housekeeping gene rpoB. Of the abundant bacteria species observed in clone library data, B. thetaiotamicron fullfilled all criteria and, thus primers were generated to amplify the specific sequences unique to this species. Expression of all of the selected mRNA relative to rpoB was extremely low at two weeks of exposure requiring greater than 35 rounds of PCR for amplification. Expression of nrfA, but not arsA or arsB, increased in a dose dependent manner by five weeks (Fig. 6 and data not shown). The latter suggests that the structural and functional changes observed in the gastrointestinal microbiota was related to either direct or host induced effect of arsenic exposure
Figure 6.
Expression of the resistance arsenate reductase (arsA) and nitrite reductase (nrfA) in Bacteroides thetaiotamicron from the murine colon at 5 and 10 wk exposure to arsenic. * and *** designate significant difference from control at p<0.05 and <0.001 respectively. ^^ designates significantly different between 10 and 250 ppb doses at p<0.01 (n=4–5 mice per group).
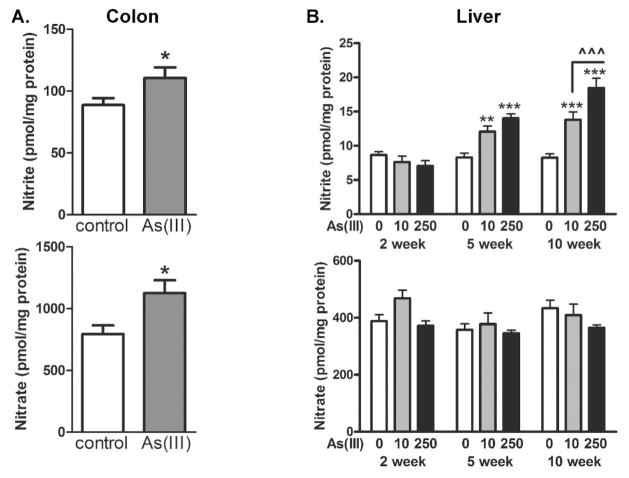
Arsenic-induced change in colonic and liver nitrite and nitrate levels
The gain of nrfA expression could functionally impact the community composition as this would favor population with bacteria that can metabolize bacteriocidal nitric oxide (NO) production. To determine whether there was a functional shift in colonic nitrogen metabolism following arsenic exposure that could pressure the community to enrich in nitrite reducing bacteia, total tissue (stripped of feces) nitrite and nitrate levels were measured. As seen in Fig. 7, both absolute nitrite and nitrate levels were increased in the colinic tissue of mice exposed to 100 ppb arsenic for five weeks. To determine whether this change was localized to the microbiome, total liver nitrite and nitrate levels were also measured. In contrast to the colon, liver, arsenic produced time and dose dependent increased in liver nitrite, but not in tissue nitrate. This may suggest that the increased microbiome nitrite reductase blunted the increased nitrite/nitrate ratio in the colon that was found in the liver.
Figure 7.
Effect of arsenic on total colon and liver nitrite and nitrate levels. A) Nitrite and nitrate levels in total proximal colon tissue (without feces) from mice exposed to 0 (control) or 100 ppb in the drinking water for 5 wk. B) liver from mice exposed to 0 (control), 10 ppb or 250 ppb As(III) in the drinking water for 2, 5, and 10 weeks. *,**, and *** designate significant difference from control at p<0.05, <0.01, and 0.001 respectively. ^^^ designates difference between doses at p<0.001 (n= colons from 8 mice or livers from 4 mice).
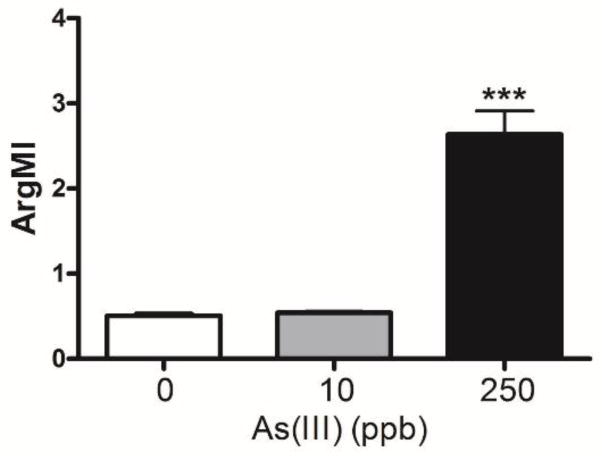
Nitrite and arginine are critical sources of NO that is essential for maintenance of both the gastrointestinal and cardiovascular system. Pathogenic shifts in nitrite and arginine metabolites have been linked to cardiovascular disease, especially coronary artery disease that is the major risk for death from cardiovascular disease in arsenic exposed individuals (Wang et al., 2009). Determination of critical arginine metabolites in control mice and mice exposed to 250 ppb arsenic for 10 weeks revealed decreased plasma arginine and monomethylarginine levels and increased levels of dimethylated arginine metabolite levels (Table 1). The shifts in levels produced an arginine methylation index (Fig. 8) that is predictive of increased risk for coronary artery disease and major adverse cardiac events (Wang et al., 2009).
Table I.
Effect of As(III) on plasma arginine and metabolite levels
| control | 10 μg/L As(III) | 250 μg/L As(III) | |
|---|---|---|---|
| Arginine metabolic index | 0.51 ± 0.03 | 0.54 ± 0.02 | 2.64 ± 0.30* |
| Plasma metabolite level (μM, mean ± SEM) | |||
| Arginine | 85.4 ± 15.7 | 67.7 ± 7.4 | 39.6 ± 12.3* |
| Citruline | 214.6 ± 21.5 | 225.1 ± 19.4 | 51.8 ± 6.1* |
| Ornithine | 368.1 ± 16.7 | 286.2 ± 16.1* | 185.8 ± 7.4*a |
| SDMA | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.65 ± 0.09* |
| ADMA | 0.38 ± 0.01 | 0.39 ± 0.02 | 0.85 ± 0.07* |
| Monomethylarginine | 1.02 ± 0.05 | 0.96 ±0.04 | 0.58 ±0.06* |
significant difference from control (p< 0.05, n=5)
significant difference from 10 μg/L As(III) (p< 0.05, n=5)
Figure 8.
Effect of arsenic on the plasma arginine methylation index (ArgMI=ADMA+SDMA/MMA). *** designates significant difference from control at p<0.001.
Discussion
Exposure to arsenic is a major global health problem that increases the risk of cancer and non-cancer diseases, such as cardiovascular and metabolic disease, in many millions of individuals worldwide. As the major route of exposure is through contaminated drinking water, the gastrointestinal tract and its large and diverse microbiome are initial targets of arsenic effects. These effects and toxicity depend on both the dose and time of exposure with acute exposure high level exposure causing GI haemorrhage, multiple organ damage and death (Armstrong et al., 1984) and chronic low to moderate level exposures contributing to chronic disease (Hughes et al., 2011). Therefore this study was designed to investigate the in vivo effect of arsenic on mouse colon microbiota using sodium arsenite added to the drinking water at concentrations that are common to human exposures. The results from this study reveal that arsenic in drinking water does affect the microbial composition of the gut. The differences between the control and arsenic exposed mouse colon microbiota were more significant than the differences that could be attributed to variability within each group.
Previous studies of arsenic impact on the gastrointestinal microbiome have relied on genomic analysis of bacteria populating eliminated feces to characterize impacts of arsenic and other environmental exposures on the microbiome composition (Lu et al., 2014a; Lu et al., 2014b). While this approach is common in the field and more convenient than investigating the microbial community in situ, it cannot capture the structural complexity and true nature of the functional impacts of arsenic on community composition, structure, and metabolism. The normal murine colon microbiota is a complex microbial community dominated primarily by Firmicutes (e.g., Clostridium, Coprococcus, Ruminococcus, Lactobacillus) and Bacteroidetes (e.g., Bacteroides, Parabacteroides), but also harbors populations of Deferribacter, Acinetobacter, Enterobacter, and Bifidobacterium (Eckburg et al., 2005; Nava et al., 2011). The in situ microscopy done in this study revealed a highly ordered microbial community structure (Fig. 1 and 2). The microbiota, especially the community populating the outer mucosa (Johansson et al., 2010), plays a pivotal role in the metabolic and protective functions of the host by providing nutrients to the epithelial lining of the gut, regulating fat storage (Backhed et al., 2004), and also by inducing host innate immunity (Sonnenburg et al., 2004). Thus the topological arrangement of the different microbial species (e.g. community structure) may be as important as the presence and abundance of specific species. The community is dynamic and naturally changes over time as was observed in the microbiome of control mice over the 10 week study by the DGGE (Figure 3), clonal libraries (Figure 4), and high throughput sequencing data (Figure 5). The dynamic and complex nature of the mammalian microbiota along with the microbial variation between individual mice can present a significant challenge to demonstrating environmental impacts on the microbiota and consequent interactions with the host. Despite these challenges, the study demonstrated that arsenic exposure produced time and dose-dependent pressure on the microbiome; eliciting change in different bacteria species populating the gastrointestinal niche, the metabolic capacity of the bacteria, and functional change in overall nitrogen metabolism of the microbiome and host.
Many studies of the murine gut microbiota have used molecular techniques that targeted the 16S rRNA gene including near full length sequencing (Eckburg et al., 2005), pyrosequencing (Nava et al., 2011)) and fingerprinting techniques such as DGGE (Zoetendal et al., 2002) and TRFLP (Osborn et al., 2000). Others have employed the variability in the spacer region between 16S and 23S rRNA gene (e.g., RISA, ARISA) (Scanlan et al., 2008; Spencer et al., 2011). Each of these techniques has their advantages and limitations (Sekirov et al., 2010). A polyphasic approach has been recently used to investigate the changes in gut microbiota of pigs fed a diet with chicory (Liu et al., 2012). Thus we initially employed DGGE and clonal libraries, then subsequently high throughput sequencing (e.g., MiSeq). The molecular approaches were complemented with in situ TEM. In situ TEM has been successfully used to identify morphologically distinct microbial species in natural systems especially biofilms, microbial mats, and stromatolites (Franks et al., 2009). Whole colon was used to address potential differences in species composition between the mucosal and luminal regions (Zoetendal et al., 2002), and between ascending (proximal), transverse (medial) and descending (distal) colon (Nava et al., 2011).
Results from the molecular approaches used in this study were consistent as they all showed shifts in host microbial populations after two weeks of 250 ppb of arsenic exposure and ten weeks of 10 ppb of arsenic exposure. Banding patterns and dendrogram analysis of the DGGE gels (Figure S1) showed that the molecular similarity between the 2, 5 and 10 week 250 ppb exposed mice is greater than the similarity between any of the 250 ppb mice to any of the control group mice from the same time point. This type of intra group similarity was observed for 10 ppb arsenic-exposed mice only after 5–10 weeks of arsenic exposure. The results obtained from both 16S rRNA clone libraries and high throughput sequencing (i.e. MiSeq) data analysed at various levels of taxonomical hierarchy showed an initial increase in Bacteroidetes and a proportionate decrease in Firmicutes was observed with increasing arsenic exposures versus control groups (Figures 3 and 4). The changes observed between control and arsenic groups were attributed to class Bacteroidia and Clostridia and families Porphyromonadaceae, Lachnospiraceae and Ruminococcaceae, results that were corroborated by the microscopy observations. At the genus level over 19% of the increase in Bacteroidetes in arsenic-exposed groups was associated with Barnesiella. In addition, an increase in Bacilli and Lactobacillus was found in the arsenic exposed group. Since many sequences remain unclassified at the order level it is not possible to conclude whether arsenic affects different genera and families within a phyla differently, causing one subgroup to increase while causing other to decrease. While the changes in community composition observed in this study were in general similar to previous studies, whether 16s rRNA gene clone libraries or high throughput formats (e.g., MiSeq)(Lu et al., 2014a), specific differences may be due to the fact that we used intact colon as opposed to fecal samples. Comparison of results obtained from microscopic and molecular studies indicate that effect of arsenic on gut community can be seen firstly in the form of changes in spatial organization followed by changes in composition.
Humans and mice show considerable similarity in the composition of their gut microbiota at higher levels of bacterial taxonomy (Croucher et al., 1983; Ley et al., 2005). Several recent studies have used murine models to show the relationship between a disease and altered gut microbial communities (Bibiloni et al., 2005; Turnbaugh et al., 2006). Some of these studies as in the case of obesity showed similar results when repeated on human samples (Ley et al., 2006; Turnbaugh et al., 2006). Thus it is plausible that the results from this work may be extended to humans and possible etiologies for arsenic-related diseases or disease modification. The recent realization that the human microbiomes have a much greater impact on environmental health (Arumugam et al., 2011; Betts, 2011; Human Microbiome Project, 2012) make it all the more imperative to have nonhuman model systems for study.
The mouse exposure studies were designed to investigate arsenic impacts on vascular remodelling in the liver and revealed that defenestration and capillarization of the liver sinusoidal endothelial layer occurred by two weeks after exposure to 10 ppb or more of arsenic (Straub et al., 2007a; Straub et al., 2007b; Straub et al., 2008). In the present studies, the changes in the microbial flora of the colons of these same mice started appearing only after 2 weeks of exposure and changes in response to 10 ppb arsenic were not significant until after 10 weeks of exposure. This suggests that arsenic-induced changes in mouse physiology strongly contributed to altering community structure and composition of the microbiota. Nevertheless, the shift towards a population dominated by species of Bacteroides possessing arsenic resistant genes may indicate direct effects to the elevated levels of arsenic in the drinking water. Furthermore, increase in Bacteroidetes and lesser proportions of Firmicutes have also been associated with type I and type II diabetes (Giongo et al., 2011) and an opposite trend has been observed in obesity (Ley et al., 2006). Higher levels of Bacilli and Lactobacillus observed in this study were also observed in type II diabetic human adults (Larsen et al., 2010). Bacteroidetes are gram negative bacteria with LPS in their outer membrane which is an important virulence factor and may cause inflammation (Allcock et al., 2001). Additionally, Bacteroidetes produce small chain fatty acids as end products of their metabolic pathways that have important neurological effects on the rat brain (MacFabe et al., 2007). Together the data suggest that arsenic induced changes in microbial community composition may cause further changes in mouse physiology, such as alterations in nutrient uptake and fat distribution, accelerated aging, weight loss, diabetes and innate immune responses.
The reciprocal impacts of arsenic exposure on nitrogen metabolites demonstrates the pathogenic potential of arsenic-induced interactions between the microbiome and host. The microbiome is both an important source of healthy dietary nitrite and is influenced by nitrogen metabolites in the GI microenvironment (Lundberg and Weitzberg, 2012). GI nitrite is a source of NO that protects the GI and vasculature, provides for vascular function, and limits metabolic disease (Lundberg and Weitzberg, 2012). However, NO is cytotoxic to many bacteria that cannot metabolize the radical species. The elevated levels of nitrite and nitrate that we found in the arsenic-exposed mouse colons (Fig 7) could provide pressure for microbial community change, as evidenced by the enrichment of the colon with nitrite reducing bacteria (Fig 6). However, since our analysis measured only tissue nitrite/nitrate and not bacterial generated nitrite/nitrate, it is not possible to determine whether elevation of tissue nitrite was due to arsenic-induced changes in bacterial nrfA expression or due to arsenic’s direct effects on the non-bacterial tissue cells.
The enrichment of nitrite relative to nitrate in the liver suggests that there is strong pressure for delivering more nitrite to the systemic system that normally could provide more beneficial NO. Unfortunately, excess NO in the setting of inflammation or oxidative stress damages tissues and signals for aberrant cell growth or differentiation (Lundberg and Weitzberg, 2012). The NO can combine with superoxide to generate damaging peroxynitrite or other nitrogen radical species and we found that the vascular remodeling in the livers of these mice was associated with peroxynitrite generation in the sinusoidal endothelium of NADPH oxidase competent mice (Straub et al., 2008). We also observed progressive increases in liver CD45/CD68 positive inflammatory cells and sinusoidal remodeling in response to Arsenic (Straub et al., 2007b; Straub et al., 2008).
In addition to nitrite, L-arginine is the main source for enzymatically generated NO and aberrant profiles of circulating arginine metabolites are linked to both microbiotic differences and cardiovascular and pulmonary disease risk (Wang et al., 2009; Holguin et al., 2012). The finding that increase of SDMA and ADMA and decreased MMA in the blood suggests that the arsenic exposure causes pathogenic shifts in both arginine total nitrogen metabolism in the microbiome and host. The resultant arginine methylation index (Fig 8) reflects an increased risk for coronary artery disease and major adverse cardiac events (Wang et al., 2009; Tang and Hazen, 2014). These observations in mice are limited in that we did not evaluate pathogenic or disease endpoints for cardiovascular disease. However, recent a recent epidemiological study associated arsenic exposure and plasma ADMA levels with increased carotid intima-media thickening (a preclinical indicator of atherosclerosis and coronary artery disease) in Mexican children with arsenic exposure through drinking water (Osorio-Yanez et al., 2013). More definitive studies are required to strengthen and integrate the linkage between the arsenic-induced host and microbial changes in nitrogen metabolism and the etiology of arsenic-promoted cardiovascular diseases.
Conclusion
This report represents a polyphasic approach to understanding the effect of chronic arsenic exposure on the murine gut microbiota and host physiology. The results show a change in microbiota over time, however, there may be a greater contribution of arsenic effects on the host rather than directy on the microbiome to produce this change. The data suggest a cascade of affects that includes changes in host physiology (e.g., cardiovascular and liver function) and nitrogen balance. Whether additional changes in the host occur in response to the shift in microbial community composition remains to be seen. Further investigation is required to elucidate the mechanisms for such selective effect on host microbiota and advance our understanding of the dynamic interplay between the host and its microbiota. Arsenic toxicological studies should include GI microbes as a possible affected organ or as an organ that may play a role in arsenic associated disease progression. Our results also emphasize that, due to inter-individual variations in microbial community structure, it is necessary to use a combination of molecular and statistical approaches to demonstrate the effect of any variable on the microbial community of the host. In addition, they underscore the importance of in situ studies (i.e., microscopy) to complement molecular investigations as they can reveal important aspects of community structure that the latter does not. Full evaluation of the complex interplay between host and microbiome structure will greatly advance the mechanistic understanding of arsenic-induced cardiovascular disease, and potentially other disease risks as well.
Supplementary Material
Oriented thick sections, toluidine blue stained, of proximal, medial, and distal colon in cross section. A–C, Control 5 wk, A) proximal, B) medial, and C) distal colon. D–F, 250 ppb As(III) 5 wk, D) proximal, E) medial, F) distal colon; bar for all figures 10 um, arrows denote spores. For this experiment each colon was dissected into proximal, medial, and distal portions, and three mice were used for each condition (e.g., 2wk, 5wk, 10wk; 0ppb,10ppb, 250ppb As(III)).
Rooted tree of unweighted UniFrac data showing that the control and 10 ppb samples from week 10 group together (indicated by red rectangle) and the 250 ppb samples from week 10 also group together (indicated by green rectangle), although separately from the 10 ppb and control groups.
Highlights.
Arsenic exposure induces changes in host and host nitrogen metabolism that cause progresive change in the microbiome.
A polyphasic approach reveals changes in microbial community structure, composition and nitrite reductase expression.
The profile of nitrogen and nitroamino acid change caused by arsenic may relect increased risk of cardiovascular pathogenesis.
Acknowledgments
This work was funded by NIEHS grant R01ES013781 (A.B., D.B.S.), R01ES013781-02S1 (A.B., D.B.S. and J.F.S), and P20HL113452 (SLH). The authors thank S. Williard for additional light and electron microscopy, P. Basu for helpful discussion, and the Center for Biologic Imaging at the University of Pittsburgh for sample preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akyon B, Stachler E, Wei N, Bibby K. Microbial mats as a biological treatment approach for saline wastewaters: the case of produced water from hydraulic fracturing. Environ Sci Technol. 2015;49:6172–6180. doi: 10.1021/es505142t. [DOI] [PubMed] [Google Scholar]
- Allcock GH, Allegra M, Flower RJ, Perretti M. Neutrophil accumulation induced by bacterial lipopolysaccharide: effects of dexamethasone and annexin 1. Clin Exp Immunol. 2001;123:62–67. doi: 10.1046/j.1365-2249.2001.01370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CW, Stroube RB, Rubio T, Siudyla EA, Miller GB., Jr Outbreak of fatal arsenic poisoning caused by contaminated drinking water. Arch Environ Health. 1984;39:276–279. doi: 10.1080/00039896.1984.10545849. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M’Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Applied and environmental microbiology. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts KS. A study in balance: how microbiomes are changing the shape of environmental health. Environ Health Perspect. 2011;119:A340–346. doi: 10.1289/ehp.119-a340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibiloni R, Simon MA, Albright C, Sartor B, Tannock GW. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Letters in applied microbiology. 2005;41:45–51. doi: 10.1111/j.1472-765X.2005.01720.x. [DOI] [PubMed] [Google Scholar]
- Caesar R, Fak F, Backhed F. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J Intern Med. 2010;268:320–328. doi: 10.1111/j.1365-2796.2010.02270.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest. 2014;124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Graziano JH, Parvez F, Liu M, Slavkovich V, Kalra T, Argos M, Islam T, Ahmed A, Rakibuz-Zaman M, Hasan R, Sarwar G, Levy D, van Geen A, Ahsan H. Arsenic exposure from drinking water and mortality from cardiovascular disease in Bangladesh: prospective cohort study. BMJ. 2011;342:d2431. doi: 10.1136/bmj.d2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbuy C, Darcy-Vrillon B, Morel MT, Pegorier JP, Duee PH. Effect of germfree state on the capacities of isolated rat colonocytes to metabolize n-butyrate, glucose, and glutamine. Gastroenterology. 1995;109:1890–1899. doi: 10.1016/0016-5085(95)90756-4. [DOI] [PubMed] [Google Scholar]
- Cherbuy C, Honvo-Houeto E, Bruneau A, Bridonneau C, Mayeur C, Duee PH, Langella P, Thomas M. Microbiota matures colonic epithelium through a coordinated induction of cell cycle-related proteins in gnotobiotic rat. Am J Physiol Gastrointest Liver Physiol. 2010;299:G348–G357. doi: 10.1152/ajpgi.00384.2009. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbitt N, Kimura S, Isse K, Specht S, Chedwick L, Rosborough BR, Lunz JG, Murase N, Yokota S, Demetris AJ. Gut Bacteria Drive Kupffer Cell Expansion via MAMP-Mediated ICAM-1 Induction on Sinusoidal Endothelium and Influence Preservation-Reperfusion Injury after Orthotopic Liver Transplantation. Am J Pathol. 2013;182:180–191. doi: 10.1016/j.ajpath.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronican AA, Fitz NF, Carter A, Saleem M, Shiva S, Barchowsky A, Koldamova R, Schug J, Lefterov I. Genome-Wide Alteration of Histone H3K9 Acetylation Pattern in Mouse Offspring Prenatally Exposed to Arsenic. PloS one. 2013;8:e53478. doi: 10.1371/journal.pone.0053478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher SC, Houston AP, Bayliss CE, Turner RJ. Bacterial populations associated with different regions of the human colon wall. Applied and environmental microbiology. 1983;45:1025–1033. doi: 10.1128/aem.45.3.1025-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis E, Hsu LL, Noguchi AC, Geary L, Shiva S. Oxygen regulates tissue nitrite metabolism. Antioxidants & redox signaling. 2012;17:951–961. doi: 10.1089/ars.2011.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Bone RA, Van de Wiele T. Biotransformation of metal(loid)s by intestinal microorganisms. Pure and Applied Chemistry. 2010;82:409–427. [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks J, Reid RP, Aspen RJ, Underwood CJC, Paterson DM, Prufert-Bebout L, Stolz JF. Ooid accreting diatom communities from the modern marine stromatolites at Highborne Cay, Bahamas, Microbial Mats. Springer; 2009. [Google Scholar]
- Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular biology and evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Hazen SL, Smith JD. An antiatherosclerotic signaling cascade involving intestinal microbiota, microRNA-10b, and ABCA1/ABCG1-mediated reverse cholesterol transport. Circ Res. 2012;111:948–950. doi: 10.1161/CIRCRESAHA.112.277277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguin F, Comhair SA, Hazen SL, Powers RJ, Khatri SS, Bleecker ER, Busse WW, Calhoun WJ, Castro M, Fitzpatrick AM, Gaston B, Israel E, Jarjour NN, Moore WC, Peters SP, Teague WG, Chung KF, Erzurum SC, Wenzel SE. An Association Between L-arginine/ADMA Balance, Obesity and the Age of Asthma Onset Phenotype. Am J Respir Crit Care Med. 2012 doi: 10.1164/rccm.201207-1270OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF, Beck BD, Chen Y, Lewis AS, Thomas DJ. Arsenic exposure and toxicology: a historical perspective. Toxicol Sci. 2011;123:305–332. doi: 10.1093/toxsci/kfr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Holmen Larsson JM, Hansson GC. Microbes and Health Sackler Colloquium: The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubachka KM, Kohan MC, Herbin-Davis K, Creed JT, Thomas DJ. Exploring the in vitro formation of trimethylarsine sulfide from dimethylthioarsinic acid in anaerobic microflora of mouse cecum using HPLC-ICP-MS and HPLC-ESI-MS. Toxicol Appl Pharmacol. 2009;239:137–143. doi: 10.1016/j.taap.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Kuo CC, Howard BV, Umans JG, Gribble MO, Best LG, Francesconi KA, Goessler W, Lee E, Guallar E, Navas-Acien A. Arsenic exposure, arsenic metabolism, and incident diabetes in the strong heart study. Diabetes Care. 2015;38:620–627. doi: 10.2337/dc14-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS one. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nature chemical biology. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- Leser TD, Molbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environmental microbiology. 2009;11:2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Liu H, Ivarsson E, Dicksved J, Lundh T, Lindberg JE. Inclusion of chicory (Cichorium intybus L.) in pigs’ diets affects the intestinal microenvironment and the gut microbiota. Applied and environmental microbiology. 2012;78:4102–4109. doi: 10.1128/AEM.07702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect. 2014a;122:284–291. doi: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Mahbub R, Cable PH, Ru H, Parry NM, Bodnar WM, Wishnok JS, Styblo M, Swenberg JA, Fox JG, Tannenbaum SR. Gut microbiome phenotypes driven by host genetics affect arsenic metabolism. Chem Res Toxicol. 2014b;27:172–174. doi: 10.1021/tx400454z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg JO, Weitzberg E. Biology of nitrogen oxides in the gastrointestinal tract. Gut. 2012 doi: 10.1136/gutjnl-2011-301649. [DOI] [PubMed] [Google Scholar]
- MacFabe DF, Cain DP, Rodriguez-Capote K, Franklin AE, Hoffman JE, Boon F, Taylor AR, Kavaliers M, Ossenkopp KP. Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behavioural brain research. 2007;176:149–169. doi: 10.1016/j.bbr.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Maull EA, Ahsan H, Edwards J, Longnecker MP, Navas-Acien A, Pi J, Silbergeld EK, Styblo M, Tseng CH, Thayer KA, Loomis D. Evaluation of the Association between Arsenic and Diabetes: A National Toxicology Program Workshop Review. Environ Health Perspect. 2012;120:1658–1670. doi: 10.1289/ehp.1104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder DG, Dasgupta UB. Chronic arsenic toxicity: Studies in West Bengal, India. Kaohsiung J Med Sci. 2011;27:360–370. doi: 10.1016/j.kjms.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Moon K, Guallar E, Navas-Acien A. Arsenic exposure and cardiovascular disease: an updated systematic review. Current atherosclerosis reports. 2012;14:542–555. doi: 10.1007/s11883-012-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KA, Guallar E, Umans JG, Devereux RB, Best LG, Francesconi KA, Goessler W, Pollak J, Silbergeld EK, Howard BV, Navas-Acien A. Association between exposure to low to moderate arsenic levels and incident cardiovascular disease. A prospective cohort study. Ann Intern Med. 2013;159:649–659. doi: 10.7326/0003-4819-159-10-201311190-00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2010;298:G807–G819. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Stolz JF. The ecology of arsenic. Science. 2003;300:939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- Osborn AM, Moore ER, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environmental microbiology. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Osorio-Yanez C, Ayllon-Vergara JC, Aguilar-Madrid G, Arreola-Mendoza L, Hernandez-Castellanos E, Barrera-Hernandez A, De Vizcaya-Ruiz A, Del Razo LM. Carotid intima-media thickness and plasma asymmetric dimethylarginine in Mexican children exposed to inorganic arsenic. Environ Health Perspect. 2013;121:1090–1096. doi: 10.1289/ehp.1205994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez F, Chen Y, Brandt-Rauf PW, Slavkovich V, Islam T, Ahmed A, Argos M, Hassan R, Yunus M, Haque SE, Balac O, Graziano JH, Ahsan H. A prospective study of respiratory symptoms associated with chronic arsenic exposure in Bangladesh: findings from the Health Effects of Arsenic Longitudinal Study (HEALS) Thorax. 2010;65:528–533. doi: 10.1136/thx.2009.119347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BP. Biochemistry of arsenic detoxification. FEBS Lett. 2002;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, Clune Y, Collins JK, O’Sullivan GC, O’Riordan M, Holmes E, Wang Y, Marchesi JR. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environmental microbiology. 2008;10:789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiological reviews. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Current opinion in gastroenterology. 2014 doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- Sparacino-Watkins C, Stolz JF, Basu P. Nitrate and periplasmic nitrate reductases. Chem Soc Rev. 2014;43:676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Liesack W. Nucleic acids and classification, Handbook of New Bacterial Systematics. Academic Press; London, England: 1993. pp. 152–189. [Google Scholar]
- States JC, Barchowsky A, Cartwright IL, Reichard JF, Futscher BW, Lantz RC. Arsenic toxicology: translating between experimental models and human pathology. Environ Health Perspect. 2011;119:1356–1363. doi: 10.1289/ehp.1103441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- States JC, Srivastava S, Chen Y, Barchowsky A. Arsenic and cardiovascular disease. Toxicol Sci. 2009;107:312–323. doi: 10.1093/toxsci/kfn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz DB, Ross MA, Salem HM, Mars WM, Michalopoulos GK, Enomoto K. Cationic colloidal silica membrane perturbation as a means of examining changes at the sinusoidal surface during liver regeneration. American Journal of Pathology. 1999;155:1487–1498. doi: 10.1016/S0002-9440(10)65464-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz JF, Basu P. Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem. 2002;3:198–206. doi: 10.1002/1439-7633(20020301)3:2/3<198::AID-CBIC198>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Stolz JF, Basu P, Oremland RS. Microbial arsenic metabolism: new twists on an old poison. Microbe. 2010;5:53–59. [Google Scholar]
- Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annual review of microbiology. 2006;60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- Straub AC, Clark KA, Ross MA, Chandra AG, Li S, Gao X, Pagano PJ, Stolz DB, Barchowsky A. Arsenic-stimulated liver sinusoidal capillarization in mice requires NADPH oxidase-generated superoxide. J Clin Invest. 2008;118:3980–3989. doi: 10.1172/JCI35092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub AC, Stolz DB, Ross MA, Hernandez-Zavala A, Soucy NV, Klei LR, Barchowsky A. Arsenic stimulates sinusoidal endothelial cell capillarization and vessel remodeling in mouse liver. Hepatology. 2007a;45:205–212. doi: 10.1002/hep.21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub AC, Stolz DB, Vin H, Ross MA, Soucy NV, Klei LR, Barchowsky A. Low level arsenic promotes progressive inflammatory angiogenesis and liver blood vessel remodeling in mice. Toxicol Appl Pharmacol. 2007b;222:327–336. doi: 10.1016/j.taap.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WHW, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. Journal of Clinical Investigation. 2014;124:4204. doi: 10.1172/JCI72331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annual review of microbiology. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tang WH, Cho L, Brennan DM, Hazen SL. Targeted metabolomic evaluation of arginine methylation and cardiovascular risks: potential mechanisms beyond nitric oxide synthase inhibition. Arterioscler Thromb Vasc Biol. 2009;29:1383–1391. doi: 10.1161/ATVBAHA.109.185645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Jasmine F, Kibriya MG, Liu M, Wojcik O, Parvez F, Rahaman R, Roy S, Paul-Brutus R, Segers S, Slavkovich V, Islam T, Levy D, Mey JL, van Geen A, Graziano JH, Ahsan H, Chen Y. Association between arsenic exposure from drinking water and plasma levels of cardiovascular markers. Am J Epidemiol. 2012;175:1252–1261. doi: 10.1093/aje/kwr464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Applied and environmental microbiology. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oriented thick sections, toluidine blue stained, of proximal, medial, and distal colon in cross section. A–C, Control 5 wk, A) proximal, B) medial, and C) distal colon. D–F, 250 ppb As(III) 5 wk, D) proximal, E) medial, F) distal colon; bar for all figures 10 um, arrows denote spores. For this experiment each colon was dissected into proximal, medial, and distal portions, and three mice were used for each condition (e.g., 2wk, 5wk, 10wk; 0ppb,10ppb, 250ppb As(III)).
Rooted tree of unweighted UniFrac data showing that the control and 10 ppb samples from week 10 group together (indicated by red rectangle) and the 250 ppb samples from week 10 also group together (indicated by green rectangle), although separately from the 10 ppb and control groups.